The promise of gene therapy and its potential to generate effective treatments for human diseases has been a subject of much debate. It is now well recognized that gene delivery technology presents the major obstacle to the success of this field, and a consensus has emerged that the development of vectors that can deliver and appropriately express relevant gene products in specific tissues in vivo is much needed. For this reason, significant effort has been placed on expanded studies in molecular virology and gene expression relevant to gene-transfer technology (reviewed in ref. 1). The challenge has been to achieve stable, regulated gene expression and to avoid immune responses. Thus, the ideal gene therapy vector would be injectable, targetable to specific sites in vivo, regulatable, able to maintain long-term gene expression, and nonimmunogenic. In this issue of the Proceedings, Burcin and colleagues (2) in Dr. Bert O’Malley’s laboratory describe a major step toward the generation of an optimized adenoviral vector. This vector could be useful for the treatment of liver-related diseases and serum-protein deficiencies that can be complemented through gene expression in hepatocytes.
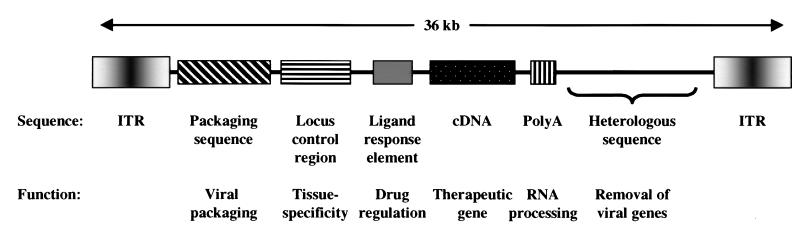
In this study, several elements have been combined into a single vector with promising features (Fig. 1). Although the individual components are not by themselves entirely novel, the fact that they work in combination represents a pragmatic advance in the development of an optimal adenoviral vector. Importantly, Burcin et al. used an adenoviral vector gutted of almost the entire adenoviral genome, replaced with a sequence from the human HPRT gene. Such vectors have been shown to sustain long-term expression in vivo (3–6). Removal of the viral genes prevents activation at low levels and hence detection by the immune system. Elimination of these sequences also reduces expression in tissues that may promote immune recognition. For example, adenoviral gene expression in minimally deleted vectors often leads to expression in antigen-presenting cells, which enhances the recognition of the products of viral genes and transgenes (7). The restriction of gene expression may therefore help to eliminate adverse consequences.
Figure 1.
Features of an optimized adenovirus gene therapy vector. Schematic diagram of a gutted adenoviral vector with an adenoviral packaging sequence and terminal repeats (ITR), containing a minimum of adenoviral genome sequences. The indicated regulatory sequences are intended to confer tissue-specific, regulatable gene expression and the inability to synthesize natural viral gene products.
The ability to regulate gene expression by using small molecules as ligands has also been explored previously. For example, O’Malley and his colleagues had described the use of progesterone-related compounds (8, 9). Other elegant studies have explored the potential of different ligands to regulate gene expression through interactions that permit DNA binding in the tetracycline repressor/operator system (10), the use of dimerizing agents to cross-link inactive domains in a chemical “two-hybrid” reaction using rapamycin-related compounds (11, 12), or the use of the insect hormone, ecdysone, which, like progesterone, modifies the binding of a transcription factor to its cognate DNA sequence (13).
The requirements for regulation of gene transfer vectors are in many ways similar to those of regulatory elements needed to maintain stable tissue-specific gene expression in transgenic animals. Despite the fact that the molecular basis of these regulatory regions has been established in vitro, it is necessary to define regulatory elements based on their activity in vivo. The use of such locus control regions to confer tissue-specific gene expression in vivo has been described previously in adenoviral vectors. A smooth muscle cell-specific promoter, SM-22, has been described by Parmacek and his colleagues to show specific, impressive restriction to smooth muscle types in vivo (14). Interestingly, this cell-specific gene expression is not always faithfully recapitulated in cell lines in vitro, in part because these cells are transformed and often contain viral transactivators that may otherwise alter tissue-specific expression. Thus, it remains necessary to demonstrate the activity of these vectors in the relevant tissues in vivo. Insulator sequences have been reported to maintain chromatin structure of actively transcribing regions, presumably isolating them from the effects of trans-acting factors that affect surrounding chromatin. For example, such an insulator sequence was described in the chicken β-globin gene (15), although the molecular basis by which it achieves this effect is not yet understood. In this report, the insulator incorporated into the vector did not exert the expected effect in maintaining high-level inducible gene expression. It thus appeared that the insulator sequence was undesirable in this vector.
Immunogenicity following gene transfer remains limiting in many vector systems and is more problematic with adenoviral vectors. The problems of the immune response relate not only to the vector but also to the transgene expressed by the vector (16–19). In the work of O’Malley and colleagues, the transactivation domain of an immunogenic viral transactivator, VP16, was replaced using the transactivation domain of a cellular transcription factor, the RelA subunit of NF-κB. This transactivation domain has been used in other ligand-regulated gene expression vectors and is well defined (12). Although replacement of this transactivation domain reduces immunogenicity, this fusion protein retains the yeast-derived GAL4 DNA-binding domain foreign to its mammalian host. Because it localizes to the nucleus, it is not yet clear whether this protein will be presented to the immune system and be immunogenic, an issue that remains to be addressed. There are other immunologic hurdles for gutted adenoviral vectors. Adenoviral proteins injected into patients could serve as targets for antibody neutralization because there is widespread immunity to a variety of adenovirus serotypes in humans. This issue will require careful evaluation in humans as clinical studies proceed. Should it prove problematic, adenoviruses from other species may need to be adapted as vectors to evade immune inactivation.
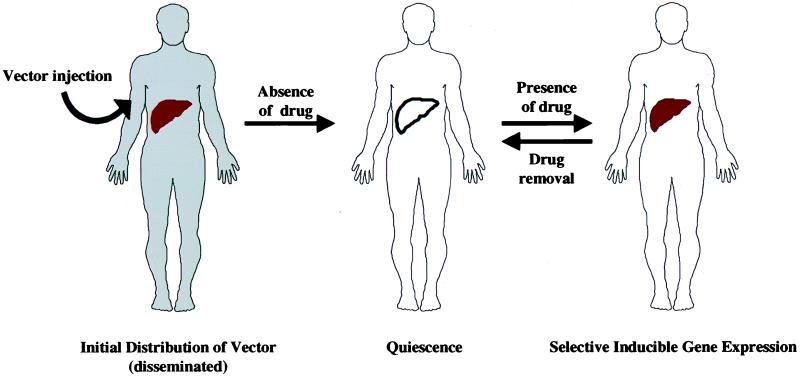
Several additional questions remain for the promising vector described by O’Malley and colleagues. Although the majority of the vector is probably delivered to the liver, the efficacy of the liver-specific regulatory element has not yet been established in vivo. Immunity to the GAL4 regulatory proteins also will require evaluation. Nonetheless, the ability to develop an injectable vector that shows targeted and reversible ligand-regulated gene expression (Fig. 2) represents a major advance. A recent report by Blau and colleagues (20) exemplifies the need for such regulation to avoid adverse consequences related to overexpression of potent biological effectors. In this case, abnormal blood vessel formation after high-level expression of an angiogenic factor was found to be deleterious. Similarly, unregulated expression of erythropoietin can lead to polycythemia (18, 21). Regulated expression of this hematopoietic growth factor would provide a better way to stimulate the production of red blood cells in anemia (22), which is currently responsive to repetitive administration of recombinant erythropoietin. The vector described by Burcin et al. provides a means to regulate expression of such potent gene products.
Figure 2.
Model of selective and inducible gene expression with ligand-regulated adenoviral vector. Gray area (Left) shows initial distribution of vector after intravenous injection. In the absence of drug treatment, vector gene expression remains minimal (Center). It is markedly increased, primarily in the liver, in a reversible fashion after drug administration (Right).
Another encouraging aspect of this work is that the long-term and regulated gene expression established in this adenoviral vector can presumably be extended to other vectors. Previous studies have shown regulated gene expression by using retroviral vectors (23), although tissue specificity was not achieved. By analogy, such regulation should be achievable using other viral vectors, such as adeno-associated, lentiviral and herpesvirus vectors, and nonviral vectors. It is thus encouraging that gene-transfer technology is progressing logically, productively, and expeditiously. The present study exemplifies the type of gene delivery refinement that will substantively contribute to more effective gene therapy for human diseases.
Footnotes
A commentary on this article begins on page 355.
References
- 1.Verma I M, Somia N. Nature (London) 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 2.Burcin M M, Schiedner G, Kochanek S, Tsai S Y, O’Malley B W. Proc Natl Acad Sci USA. 1999;96:355–360. doi: 10.1073/pnas.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemens P R, Kochanek S, Sunada Y, Chan S, Chen H H, Campbell K P, Caskey C T. Gene Ther. 1996;3:965–972. [PubMed] [Google Scholar]
- 4.Haecker S E, Stedman H H, Balice-Gordon R J, Smith D B, Greelish J P, Mitchell M A, Wells A, Sweeney H L, Wilson J M. Hum Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- 5.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 7.Jooss K, Yang Y, Fisher K J, Wilson J M. J Virol. 1998;72:4212–4223. doi: 10.1128/jvi.72.5.4212-4223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, O’Malley B W, Jr, Tsai S Y, O’Malley B W. Proc Natl Acad Sci USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, DeMayo F J, Tsai S Y, O’Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 10.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 11.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 12.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, et al. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 13.No D, Yao T P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Lin H, Barr E, Chu L, Leiden J M, Parmacek M S. J Clin Invest. 1997;100:1006–1014. doi: 10.1172/JCI119611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 16.Chirmule N, Hughes J V, Gao G P, Raper S E, Wilson J M. J Virol. 1998;72:6138–6145. doi: 10.1128/jvi.72.7.6138-6145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai Y, Schwarz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Su Q, Wilson J M. J Virol. 1996;70:7209–7212. doi: 10.1128/jvi.70.10.7209-7212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Springer M L, Chen A S, Kraft P E, Bednarski M, Blau H M. Mol Cell. 1998;2:549–558. doi: 10.1016/s1097-2765(00)80154-9. [DOI] [PubMed] [Google Scholar]
- 21.Svensson E C, Black H B, Dugger D L, Tripathy S K, Goldwasser E, Hao Z, Chu L, Leiden J M. Hum Gene Ther. 1997;8:1797–1806. doi: 10.1089/hum.1997.8.15-1797. [DOI] [PubMed] [Google Scholar]
- 22.Tripathy S K, Svensson E C, Black H B, Goldwasser E, Margalith M, Hobart P M, Leiden J M. Proc Natl Acad Sci USA. 1996;93:10876–10880. doi: 10.1073/pnas.93.20.10876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann A, Nolan G P, Blau H M. Proc Natl Acad Sci USA. 1996;93:5185–5190. doi: 10.1073/pnas.93.11.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]