Abstract
Neural crest cells are a population of multipotent stem cell-like progenitors that arise at the neural plate border in vertebrates, migrate extensively, and give rise to diverse derivatives such as melanocytes, craniofacial cartilage and bone, smooth muscle, peripheral and enteric neurons and glia. The neural crest gene regulatory network (NC-GRN) includes a number of key factors that are used reiteratively to control multiple steps in the development of neural crest cells, including the acquisition of stem cell attributes. It is therefore essential to understand the mechanisms that control the distinct functions of such reiteratively used factors in different cellular contexts. The context-dependent control of neural crest specification is achieved through combinatorial interaction with other factors, post-transcriptional and post-translational modifications, and the epigenetic status and chromatin state of target genes. Here we review the current understanding of the NC-GRN, including the role of the neural crest specifiers, their links to the control of “stemness,” and their dynamic context-dependent regulation during the formation of neural crest progenitors.
Keywords: Neural crest, NC-GRN, Epithelial-Mesenchymal Transition, epigenetics, Snail, SoxE, Foxd3, c-Myc
I. Neural Crest Progenitors as a Stem Cell Population
Understanding the processes that govern the establishment and maintenance of multipotency at the molecular level is of great interest and importance to both developmental biology and regenerative medicine. The embryonic neural crest is an excellent system in which to ask questions about these mechanisms. During vertebrate development, the neural crest arises in the ectodermal germ layer as a consequence of instructive cues generated at the border between the presumptive neural plate and epidermis (Moury and Jacobson, 1990; Selleck and Bronner-Fraser, 1995; LaBonne and Bronner-Fraser, 1998). These cells are a developmental and evolutionary novelty. Whereas development can be generally viewed as a process of progressive restriction in potential, neural crest progenitors represent one of the few examples during embryonic development where, as a consequence of an inductive event, cells arise that possesses greater developmental potential than the cells from which they were derived. Despite their ectodermal origin at the neural plate border, neural crest cells acquire the potential to give rise to cell types that are both ectodermal and mesodermal in nature. Indeed, because it gives rise to cell types characteristic of more than one of the “classic” germ layers, neural crest stem cells can, from an evolutionary perspective, be viewed as a fourth germ layer (Hall, 1999).
Stem cells have been classically defined as progenitor cells that possess at least some capacity for self-renewal, and that are capable of giving rise to one or more differentiated cell types (Morrison et al., 1997). This suggests that stem cells must express regulatory factors tasked with maintaining their multipotency and stem cell characteristics, including the repression of genes linked to cell commitment/differentiation and the maintenance of developmental potency, via genetic or epigenetic mechanisms. It will be important to learn how these characteristics are governed in neural crest precursor cells. Both in vitro clonal analyses and in vivo cell labeling/transplantation experiments have established that neural crest cells are both multipotent and self-renewing (Baroffio et al., 1991; Bronner-Fraser and Fraser, 1988; Bronner-Fraser et al., 1980; Ito and Sieber-Blum, 1991; Sieber-Blum and Cohen, 1980; Trentin et al., 2004). Multipotentecy of individual neural crest progenitors was elegantly demonstrated in experiments in which a cell–autonomous dye, lysinated rhodamine dextran (LRD), was injected into single dorsal neural tube cells in chick embryos. It was found that the labeled individual cells could give rise to daughter cells that contributed to multiple neural crest derivatives (Bronner-Fraser and Fraser, 1988). The ability of neural crest progenitors to self renew was demonstrated using neural crest cells isolated from rat neural tubes, serially diluted, and cultured at clonal density (Stemple and Anderson, 1992). These cells could give rise to multipotent neural crest cells, neurons and glia. The self-renewal property of the neural crest was further demonstrated by additional rounds of clonal dilution and subculture, and self-renewal ability was found to be maintained up to 10 days in culture (Morrison et al., 1997; Stemple and Anderson, 1992; Le Douarin and Dupin, 1993). Understanding the mechanisms that contribute to the stem cell-like characteristics of neural crests cells is of profound importance, both because these mechanisms may prove relevant to the development and maintenance of other stem cell populations, and because the formation of neural crest cells represents such a fundamental milestone in vertebrate evolution.
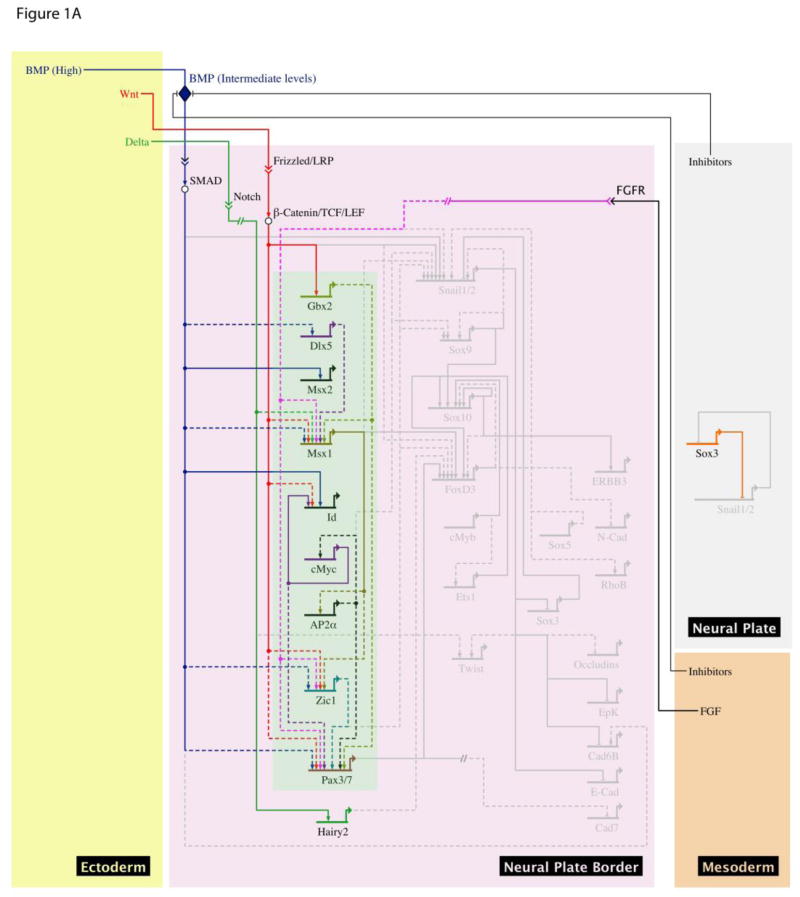
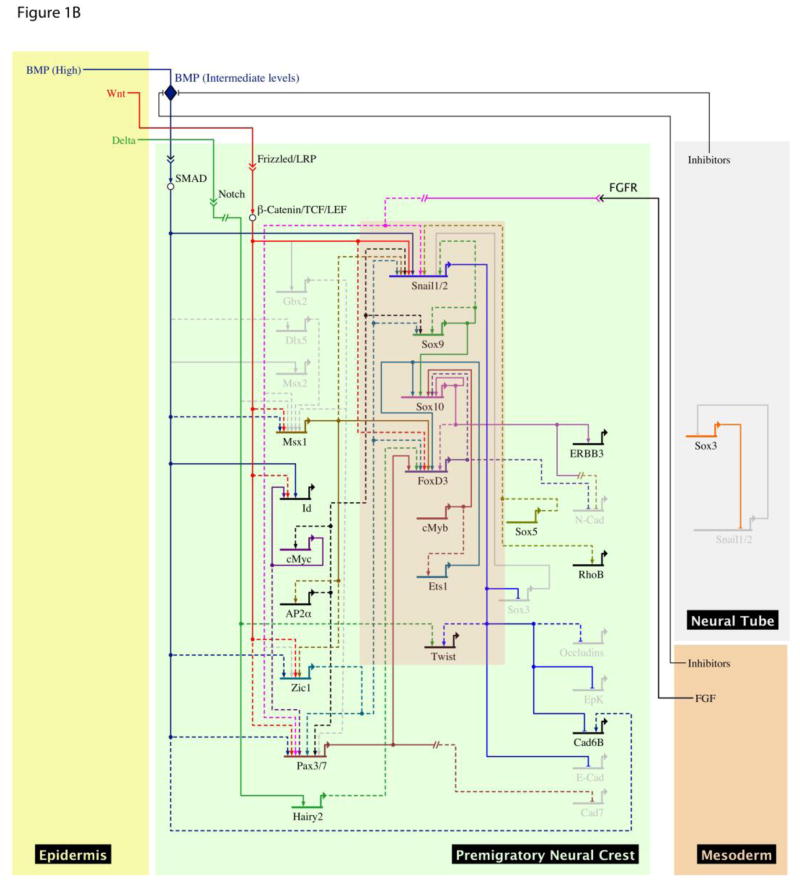
Neural crest progenitors are induced at the neural plate border, and subsequently in the dorsal neural tube, as a consequence of complex signaling events involving the BMP, Wnt and FGF pathways. Neural crest cells will ultimately differentiate into a diverse array of cell types distributed throughout the vertebrate body plan, including neurons and glia, of the peripheral nervous system, myofibroblasts, chondrocytes, and melanocytes (Le Douarin and Kalcheim, 1999). Experiments in chick embryos point to an induction process that commences during early gastrulation (Basch et al., 2006) and in anamniotes such as Xenopus, the expression of early neural crest markers at the neural plate border is apparent by mid gastrulation (Selleck and Bronner-Fraser, 1996; Mancilla & Mayor, 1996; Labosky & Kaestner, 1998; Spokony et al., 2002). Evidence from grafting experiments implicates both ectodermally and mesodermally- derived signals in neural crest induction (reviewed in LaBonne and Bronner-Fraser, 1998). Specifically, paraxial mesoderm from chick or Xenopus (dorsolateral marginal zone, DLMZ) can induce neural crest when combined with the neural plate of chick or animal caps of Xenopus embryos (Selleck and Bronner-Fraser, 1996; Monsoro-Burq, 2003). A dynamic interplay of BMP, Wnt and FGF signals, along with inhibitors of BMP signaling, are involved in inducing the neural plate border (See review by Milet and Monsoro-Burq in this issue) (Figure 1a). They subsequently contribute to the induction of early neural crest specifiers, including the transcription factors Snail2 (Slug), Snail1, and Sox9 (LaBonne and Bronner-Fraser, 1999; Sauka-Spengler and Bronner-Fraser, 2008) (Figure 1b). Indeed, Snail2 can cooperate with canonical Wnt signals to convert animal cap tissue to neural crest, bypassing the need for BMP inhibition (LaBonne and Bronner Fraser, 1998)
Figure 1.
A, B Gene regulatory network (GRN) view of regulatory networks involved in neural crest induction using data from multiple vertebrate models. GRNs show active genes and interactions (white) inactive genes and interactions (grey) in neural plate border (A) and premigratory neural crest (B) stages, and include neural plate border specifiers (green) and neural crest specifiers (red). The GRN summarizes both perturbation data (dashed lines) and cis-regulatory data (solid lines) from different model systems. Proteins denoted by white circles, intracellular interactions by double arrows extracellular ligands by diamond shape. Indirect or presumed interactions depicted by dashed line. The model was build using BioTapestry software (Longabaugh et al., 2009).
Interestingly, some transcriptional regulatory factors expressed in newly formed neural crest cells have strong links to the attributes of stemness and multipotency. Notable among these is c-myc, which is first expressed in a broad domain at the neural plate border that includes neural crest and placodal precursors and then subsequently becomes more restricted to neural crest cells (Bellmeyer et al., 2003). Myc family proteins are required for both Embryonic Stem Cell (ESC) and Induced Pluripotent Stem Cell (iPSC) self-renewal (Smith and Dalton, 2010). These factors also control potency in a number of other contexts. For example N-myc is essential for maintenance of neural and lung progenitor cells, and c-myc has been found to regulate interactions between epidermal stem cells and their niche (Smith and Dalton, 2010). It has been suggested that c-myc’s role in pluripotency is at least partially related to regulation of the chromatin remodeling machinery, and a number of histone-modifying and Swi/Snf chromatin remodeling factors are c-myc targets (Kidder et al., 2008; Kim et al., 2008). Indeed, evidence suggests that factors important for pluripotency are also involved in the epigenetic status of iPSCs. Id (inhibitor of DNA binding) proteins have also been shown to be critical effectors of Myc-family proteins in a variety of cell types, including the neural crest (Light et al., 2005; Kee and Bronner-Fraser, 2005; Lasorella et al., 2002). Additionally, forced expression of Id3 in the neural crest results in persistent expression of markers characteristic of multipotent neural crest progenitors, and blocks differentiation into neural crest derivatives, suggesting that Id3 is an important effector of c-myc’s ability to impart stem cell properties (Light et al., 2005).
Downstream of c-myc/Id3 a number of NC-GRN factors have links to the regulation of multipotency, including Snail proteins, Sox10 and FoxD3. Sox10, for example, can inhibit the differentiation of neural crest stem cells into neural cell types, thus maintaining their potential to form glia (Kim et al., 2003b). FoxD3 has been implicated in maintaining the neural crest multipotent progenitor state by inhibiting non-neural differentiation (Mundell and Labosky, 2011). Snail transcription factors, have recently been linked to the formation of cancer stem cells, in addition to their more broadly characterized role in regulating tumor invasion and metastasis. Additional insights into the control of neural crest cell multipotency may derive from recent studies in which neural crest stem cells were generated from human embryonic stem cells and human induced pluripotent stem cells by mimicking endogenous induction events and exposing them to a combination of Wnt and low-level Smad activity (Menendez et al., 2011). These induced neural crest cells were found to be multipotent, and could differentiate into an array of neural crest derivatives including peripheral neurons, and mesenchymal cell-derived osteocytes, chondrocytes, and adipocytes (Menendez et al., 2011). It will be essential to identify the downstream factors that maintain neural crest multipotency in response to these factors, and to dissect their function.
II. Neural Crest Inducing Signals
The signaling inputs and transcription factors involved in neural crest specification, migration and differentiation can be described as a gene regulatory network that defines their individual and combinatorial roles in transcriptional regulation (Meulemans and Bronner-Fraser, 2004; Sauka-Spengler and Bronner-Fraser, 2008; Betancur et al., 2010) The signaling pathways involved in neural crest precursor formation, BMP, Wnt, FGF, and Notch, act in concert to activate distinct sets of transcription factors during different stages of neural crest development. These transcription factors can be grouped into neural plate border specifiers, neural crest specifiers, neural crest EMT/migration factors, and groups of factors that direct the differentiation of neural crest cells into specific derivative cell types. Importantly, a number of key neural crest factors are used reiteratively to control more that one of these processes, and thus are required during multiple stages of neural crest development (reviewed in Taylor and LaBonne, 2007).
Neural crest precursors are believed to arise in regions of intermediate BMP signaling at the neural plate border. In Xenopus, while high levels of BMP signaling induces epidermal fate and inhibition of BMPs leads to neural induction, partial inhibition of BMP signaling in cells fated to give rise to epidermis leads to an expansion of the neural crest progenitor domain (LaBonne and Bronner-Fraser, 1998). Endogenously, the intermediate levels of BMP signaling at the neural plate border are generated by antagonistic interactions between the high intrinsic levels of BMP signals in the ectoderm and the BMP antagonists (Cerberus, noggin, chordin, and follistatins) produced by the organizer and neural plate forming regions (Sauka-Spengler and Bronner-Fraser, 2008; Tribulo et al., 2003). Low level BMP signaling is permissive but not sufficient for neural crest formation, indicating that additional signals are required (García-Castro et al., 2002; LaBonne and Bronner-Fraser, 1999).
FGF signaling has also been implicated in the induction of the neural crest in Xenopus, in concert with attenuated BMP signaling (Monsoro-Burq et al., 2003). However, mouse embryos lacking FGF receptor and zebrafish embryos without mesoderm undergo normal neural crest development (Trokovic et al., 2003; Ragland and Raible, 2004). Wnt signaling is involved in neural crest development from induction to migration. Various Wnt ligands, Wnt1, Wnt3a, Wnt6, Wnt7b, and Wnt8, are expressed in different tissues that are involved in neural crest induction (Ikeya et al., 1997; Knecht and Bronner-Fraser, 2002; Jones and Trainor, 2005). Wnts are secreted from the paraxial mesoderm in Xenopus and from non-neural ectoderm adjacent to the neural folds in chick (Saint-Jeannet et al., 1997; Garcia-Castro et al., 2002). The essential role of Wnt signaling during neural crest induction in chick and Xenopus has been demonstrated using gain and loss of function studies (Garcia-Castro et al., 2002; LaBonne and Bronner-Fraser, 1998; Monsoro-Burq et al., 2003). Notch/Delta signaling has also been implicated in early neural crest development in both frog and chick embryos (Endo et al., 2002). In zebrafish, Notch signaling appears to regulate trunk but not cranial neural crest cells (Cornell and Eisen, 2005). While the distinct contributions that each of these signaling pathways makes to neural crest precursor formation remains to be determined, together they control the expression of downstream effector transcription factors broadly classified as neural plate border and neural crest specifiers.
III. The Neural Plate Border – a zone of competence
During neural crest development, a subset of neural plate border cells begin to express factors classified as neural crest specifiers. The neural plate border itself is defined by collective expression of border specifier genes, including Msx1/2, Pax3/7, Dlx3/5, Gbx2, and Zic1 (See review by Milet and Monsoro-Burq in this issue). The presumption is that these factors act to restrict the adoption of neural plate and epidermal fates, while maintaining competence to form cell types derived from the border zone, including the neural crest. In Xenopus, BMP, Wnt and FGF are required for inducing the expression of Msx1, Zic1, and Pax3 (Monsoro-Burq et al., 2005; Sato et al., 2005). Gain of function experiments in Xenopus have shown that BMP and FGF can induce Zic1 and Pax3 expression, and that both transcription factors might be required for endogenous specification of neural crest (Hong and Saint-Jeannet, 2007). FGF8 and Wnt signals can cooperate to activate Pax3 expression (Monsoro-Burq et al., 2005), while perturbation experiments in Xenopus have implicated FGF, Wnt, retinoids and Pax3 as regulators of Pax7 expression (Maczkowiak et al., 2010). While in most vertebrates Pax3 and Pax7 share similar expression domains, these factors show distinct functional specialization in both chick and Xenopus. Interestingly, their roles appear to be reversed between the two species: in Xenopus Pax3 is essential for ectoderm and neural crest induction and Pax7 localized to paraxial mesoderm, whereas in chick, Pax7 plays a role in neural crest induction (Sato et al., 2005; Basch et al., 2006). Intermediate levels of BMP signaling can directly activate Msx2, (Brugger 2004). Wnt signaling directly activates Gbx2, which in turn induces expression of neural plate border specifier genes, Msx1 and Pax3 (Li et al., 2009). Thus, current understanding suggests that interaction of signals from neural plate, epidermis (BMP, Wnt, and Notch/Delta) and the underlying paraxial mesoderm (Wnt and FGF) induces neural crest at the neural plate border (Huang and Saint-Jeannet, 2004). While much has been learned about the transcription factors induced by these signaling pathways, little is known about the direct transcriptional targets of these neural plate border and neural crest specifiers. Improving the resolution of the current neural crest GRN will require a better understanding of the distinct contributions that each of the border specificers makes to the subsequent formation of neural crest precursors.
IV. The Neural Crest Specifiers
The onset of expression of neural crest specifier genes occurs during mid to late gastrulation in Xenopus and at mid-gastrula stage (HH4+) in the chick (Basch et al., 2006). The earliest expressed neural crest specifier genes include Snail1, Snail2 (Slug), Sox8, Sox9, FoxD3, Twist, Ets1, AP2α, cMyc and Id genes. The temporal expression of these genes can vary among vertebrates, particularly with respect to paralogs that arose as a consequence of genome duplications. It is also worth noting that c-myc, which is expressed at the neural plate border as early as stage 11 (mid gastrula) in Xenopus, is also expressed in the anterior border region/transverse neural fold that does not give rise to neural crest (Bellmeyer et al., 2003). Its expression commences earlier that that of other neural crest specifiers such as Snail2 and Sox9, and is broader in neural crest forming regions. This suggests that c-myc, and its downstream target Id3 (Light et al., 2005), might function to bridge the “neural plate border” and “neural crest precursor” state, and this will be an important area of future study. It is also essential to understand the contributions that each of the neural crest specifier factors make to the neural crest precursor state. This is complicated by the reiterative use of many of these factors for the regulation of multiple steps in neural crest development. We discuss below our current understanding of the role and regulation of key neural crest specifiers.
Snail family of transcription factors
Snail2 (Slug) and Snail1 are paralogous factors that arose as a consequence of genomic duplication. While one of these zinc finger transcriptional repressors is always expressed in the premigratory neural crest (Locascio et al., 2002) they have each subfunctionalized differently in various model organisms. For example, in chick and Xenopus Snail2 plays the predominant role in premigratory neural crest whereas in mouse Snail1 is expressed in these cells (Sefton et al., 1998). This “swapping” suggests a high degree of functional conservation.
The induction and regulation of Snail2 expression in neural crest forming regions is the focus of much study. Snail2 can be induced by canonical Wnt and intermediate levels of BMP signaling in Xenopus animal pole explants (LaBonne and Bronner-Fraser, 1998). Consistent with this neural crest regulatory regions of the mouse Snail2 promoter contain Smad1 and Tcf/Lef1 sites (Conacci-Sorrell et al., 2003), and the Xenopus Snail2 promoter contains a required LEF-1 binding site (Sakai et al., 2005; Vallin et al., 2001). Notch signaling, and its downstream target Hairy2, have also been implicated in regulation of Snail2 in Xenopus (Glavic et al., 2004). Neural plate border specifiers regulate Snail2 expression; Zic1 and Pax3/7 can both induce Snail2 expression in Xenopus whereas Msx1 has been shown to do so in chick (Meulemans and Bronner-Fraser, 2004; Sato et al., 2005; Tribulo et al., 2003), although in none of these cases has the regulation been shown to be direct. Work in Xenopus has suggested that co-activation of both Zic1 and Pax3/7 is a decisive event in induction of Snail2 at the neural plate border, although here Wnt signals are required as well (Sato et al., 2005).
Once induced, Snail family proteins play multiple essential roles in neural crest development. In Xenopus and in chick, Snail2 is required for both specification of neural crest precursors and for the subsequent migration of these cells (Mancilla and Mayor, 1996; LaBonne and Bronner-Fraser, 1998; Sefton et al., 1998). Ectopic expression of Snail2 in the chick neural tube leads to increased production of migratory neural crest in cranial regions (Locascio and Manzanares, 2002). However, in chick neural tube the delamination of trunk neural crest cells can be blocked by overexpressing the BMP antagonist Noggin, in chick neural tube without altering Snail2 expression (Sela-Donenfeld and Kalcheim, 1999). This suggests both that Snail2 is not sufficient for delamination and that there may be at least partially distinct mechanisms for controlling neural crest delamination at different axial levels. Importantly, despite its widely conserved role as a core EMT regulatory factor, Snail2 does not appear to be required for neural crest migration in mice (Jiang et al., 1998), again suggesting the possibility of distinct regulatory mechanisms. Little is known about the direct targets of Snail-mediated repression in premigratory neural crest cells although a recent study presented evidence that Snail2 and the neural plate factor Sox3 reciprocally inhibit each others expression at the neural plate border (Acloque et al., 2011).
As discussed above, Snail family proteins play additional roles in neural crest development beyond their role as neural crest specifiers, most prominently in the triggering of EMT/migration. Their role as core EMT regulatory factors is conserved in other developmental and pathological contexts, including gastrulation (Carver et al., 2001; Mayor et al., 2000), formation of the cardiac cushions (Romano and Runyan, 2000), closure of the palate (Martínez-Alvarez et al., 2004) as well as tumor metastasis (Alves et al., 2009; Hemavathy et al., 2000). In contrast to the paucity of known Snail targets in the premigratory neural crest, there are a number of well characterized regulatory targets related to EMT in other cellular systems including E-cadherin, tight junction molecules such as claudins and occludins, and cell polarity molecules including Crumbs3 and Discs large (Peinado and Olmeda, 2007; Moreno-Bueno and Portillo, 2008; Ikenouchi et al., 2003). Moreover, Snail2 represses the expression of Cadherin6B in the premigratory neural crest cells (Taneyhill et al., 2007). The involvement of Snail family proteins in both the formation of the stem cell like neural crest precursors and in the profound behavioral changes associated with EMT/migration suggests that there must be tight context dependent control over the activity of these proteins. Mechanisms for accomplishing this are beginning to be uncovered and will be discussed below. Intriguingly, Snail and Snail2 have recently been linked to the generation and maintenance of cancer stem cells. (Inoue et al., 2002; Kurrey et al., 2009 Mani et al., 2008 Guo et al., 2012). This suggests that a role in imparting stem cell-like characteristics may be a fundamental function of these proteins, and that “stemness” may be in some way coupled to the potential for EMT and invasive behavior.
SoxE family of transcription factors
In addition to Snail proteins, the SoxE family of transcription factors, Sox8, Sox9 and Sox10 are among the central players regulating the development of neural crest cells. In every vertebrate examined to date, one or more of these factors is required for specifying neural crest precursor cells, maintaining their multipotency, and promoting their survival (Haldin and LaBonne, 2010). Subsequently, SoxE proteins play instructive roles in the formation of multiple neural crest lineages including chondrocytes, melanocytes, and peripheral nervous system components such as Schwann cells (peripheral glia). Interestingly, SoxE factors play multiple context-dependent roles in the neural crest. In Xenopus, all three SoxE genes, Sox8, Sox9 and Sox10, are coexpressed in neural crest progenitors at the neural plate border (Aoki et al., 2003; Spokony et al., 2002). In chick and mouse, Sox9 and Sox10 are both expressed in neural crest progenitors prior to Sox8 (Southard-Smith et al., 1998; Cheung and Briscoe, 2003).
As with many vertebrate factors that arose via duplication, SoxE factors, expressed differentially at later stages, have subfunctionalized. Sox9 becomes restricted to ectomesenchymal crest in the border of cranial regions (Spokony et al., 2002; Cheung and Briscoe, 2003; Zhao et al., 1997) whereas, following transient expression in migratory neural crest, Sox10 expression persists in cells that will give rise to the cranial glia as well as in melanocyte precursors (Kim et al., 2003; Carney et al., 2006; Bondurand et al., 2001). Sox8 expression overlaps with both Sox9 and Sox10 in several neural crest domains (Aoki et al., 2003; Montero et al., 2002). In zebrafish, while Sox8 is undetectable until after hatching, one of the two more recent, teleost-specific Sox9 paralogs, Sox9b, is expressed in early neural crest progenitors (Yan et al., 2005; Chiang et al., 2001) and Sox10 expression commences subsequently. By contrast, Sox9a is not expressed in the neural crest at these stages (Dutton et al., 2001).
The role and regulation of Sox9 and Sox10 during neural crest development has been the focus of considerable study. With respect to how expression of these factors is established in neural crest forming regions, enhancers driving their expression have been analyzed in a number of systems. A detailed study on mouse Sox10 gene regulation identified multiple functional enhancers with binding sites for Sox9, Sox10, Pax3, AP2α, Lef1, FoxD3 and Slug (Werner et al., 2007). In zebrafish, a cis-regulatory element has been characterized in the first intron of Sox10 that includes essential Tcf/LEF sites, suggesting regulation by Wnt signals, as well as binding sites for SoxE proteins and FoxD3 (Dutton et al., 2008). A Sox10 regulatory region identified in the chick is directly controlled by Ets1, cMyb and Sox9 transcription factors, confirming studies suggesting that Sox10 is a direct SoxE target (Betancur et al., 2010). Multiple tissue specific Sox9 enhancers have also been identified in the mouse, with binding sites for AP2α, Lef1, Ets, Dlx, Otx, and Pbx (Bagheri-Fam et al., 2006).
SoxE function is essential for the formation of neural crest precursor cells. Morpholino-mediated depletion of Sox9 in Xenopus results in loss of expression of other neural crest specifiers including Snail2, FoxD3, and Sox10 (Aoki et al., 2003; Lee et al., 2004; Spokony et al., 2002). This loss of neural crest precursors led to subsequent defects in the craniofacial skeleton, similar to what is seen in Sox9 knockout mice (Bi et al., 1999). Gain and loss of functional experiments in Xenopus embryos have indicated that both Sox9 and Snail2 act as upstream regulators of Sox10 expression in the neural crest (Aoki et al., 2003), however given that Snail2 functions as a repressor, its regulatory contributions promoting Sox10 are likely to be indirect. In the chick it has also been shown that Sox9 functions in the formation of neural crest progenitors, as well as by instructing the formation of specific neural crest derivatives, and it may also influence neural crest delamination (Cheung and Briscoe, 2003; Cheung et al., 2005).
It is intriguing that SoxE factors act first to instruct the formation of neural crest stem cells, and then subsequently to direct a loss of potency and the adoption of specific derivative fates. Sox10, for example, directs the formation of neural crest derived melanocytes, in part by activating the major melanocyte differentiation factor, Mitf (Aoki et al., 2003). Consistent with this, one of the main defects in Sox10 mutant embryos, including the zebrafish colorless (cls) mutant and the Dominantmegacolon (Dom) mouse, is in the melanocyte lineage. Sox10 also regulates genes important for the formation of glial cells in the peripheral nervous system (Stolt and Wegner, 2010). Sox9, by contrast, directs the formation of ectomesenchynal neural crest, where it has regulatory targets that include the chondrocyte-specific enhancer of the collagen gene Col2a1 (Lefebvre et al., 1997).
Several studies link SoxE function with maintenance of stem cell state. In the developing peripheral nervous system, Sox10 maintains multipotency by preserving both neuronal and glial potential. In a dose dependent manner Sox10 also functions to inhibit neuronal differentiation and promote gliogenesis (Kim et al., 2003). Of particular interest with respect to neural crest progenitor formation is a recent report that Sox9 can function together with Snail2 to determine the mammary stem cell state (Guo et al., 2012). Snail1 can substitute for Snail2 in mediating the formation of these stem cells, but neither Foxd3 nor Twist, nor surprisingly c-myc, could replace Sox9 (although other SoxE family factors were not assayed). Together with some recent data from cancer stem cells (Mani et al., 2008; Morel et al., 2008) these findings suggest a fundamental link between the neural crest regulators Sox9 and Snail2, and the stem cell state. Moreover, while this and other studies suggested that Snail2 may contribute to stemness by virtue of its ability to promote EMT, Sox9 activates a distinct gene regulatory program that cooperates with the EMT program to promote stemness (Guo et al., 2012).
Foxd3
Another transcription factor that plays a key regulatory role in the maintenance of neural crest cell multipotency is the winged helix transcription factor FoxD3 (Teng et al., 2008). In mouse, Foxd3 is expressed in both pre-migratory and early migrating neural crest cells, and in most lineages its expression is down regulated as cells differentiate (Labosky and Kaestner, 1998). Thus, Foxd3 expression suggests a link to multipotency, and an elegant study using lineage mapping and clonal analysis in mouse has recently provided a direct link between FoxD3 function and neural crest stemness and self-renewal (Mundell and Labosky, 2011). Prior loss-of-function studies in Xenopus, zebrafish, and mouse had suggested a central role for Foxd3 in early neural crest development (Teng and Labosky, 2006; Lister et al., 2006; Montero-Balaguer et al., 2006; Sasai et al., 2001; Stewart et al., 2006). The more recent work demonstrates a cell-autonomous requirement for Foxd3 in maintaining both self-renewal and multipotency of neural crest cells (Mundell and Labosky, 2011). Moreover, this study further demonstrates that FoxD3 subsequently functions to repress ectomesenchymal cell fates and preserve neuronal/glial potential. Interestingly, FoxD3 is also linked to the maintenance of multipotency in other progenitor cells (Hanna et al., 2002; Liu and Labosky, 2008; Tompers et al., 2005). It will be important to determine the key Foxd3 regulatory targets in these cell populations. In chick and mouse ectopic expression of FoxD3 leads to upregulation of Sox10, cadherin-7 and β1-integrin, although it is not known if these are direct targets (Dottori et al., 2001; McKeown et al., 2005; Cheung et al., 2005). FoxD3 mediated control of multipotency is context dependent, however, as this factor can also repress melanogenesis and promotes neural/glial fates (Kos et al., 2001).
Other neural crest specifier transcription factors
While Snail, SoxE and Foxd3 families of transcription factors are clearly among the central neural crest specifier factors, and all have links to the control of multipotency in multiple systems, a large number of other, less well-studied, factors are included in this category. The key roles of c-myc and the Id genes (Bellmeyer et al., 2003; Light et al., 2005; Kee and Bronner-Fraser, 2005) have already been discussed, and these may act upstream of the other “specifier” factors. Another example is AP2, reiteratively used during neural crest formation; first at the neural plate border, as a mediator of Wnt signaling in induction of Pax3 and later in neural crest specification (De Crozé et al., 2011). Importantly, this hierarchical relationship seems also to be present in Lamprey, the extant proxy for the basal vertebrate (Nikitina et al., 2011).
Like the Snail family, the bHLH protein Twist is both a neural crest specifier and a core EMT regulatory factor that is linked to turmor cell metastasis (Yang et al., 2006). Twist proteins contain a basic domain that interacts with Ebox DNA recognition sequences (‘CANNTG’) and a helix-loop-helix domain that mediates dimerization with another Twist protein or with E12/E47 (Connerney et al., 2006). Twist is distinguished from other neural crest specifier factors by the restriction of its expression to cranial regions of the embryo, suggesting that this protein could play a role in endowing cranial neural crest precursors with the ability to give rise to mesectodermal derivatives such as cartilage and bone. Like Snail, Twist has recently been linked to the formation of cancer stem cells (Fang et al., 2010; Vesuna et al., 2009; Yang et al., 2010), although it cannot substitute for Slug/Snail in cooperating with Sox9 to promote the mammary stem cell state (Guo et al., 2012). A better understanding of the function and regulation of Twist is thus essential to understanding neural crest stem cell formation and migration, as well as the related states in tumor formation and metastasis.
cMyb and Ets1 are additional, recently identified neural crest specifier genes. In trunk neural crest, knockdown of cMyb causes reduction in Snail2 expression (Karafiat et al., 2005). Ets factors are common downstream effectors of Ras/Map kinase signaling (Nelson et al., 2010) which makes Ets1 a good candidate for mediating FGF signals during neural crest formation. However, current evidence seems to implicate Ets1 in cell cycle regulation as well as in the regulation of integrins, cadherins and MMPs (Fafeur et al., 1997; Rosen et al., 1994; Wasylyk et al., 1998). The presence of Ets1 expression in the cranial neural crest that delaminate in a sheet-like fashion may obviate the need of those cells to arrest in G1 phase prior to emigration, as trunk neural crest do (Théveneau et al., 2007). Ectopic expression of Ets1 in trunk region of chick embryos causes cell cycle independent migration of neural crest similar to cranial neural crest (Sauka-Spengler and Bronner-Fraser, 2008; Betancur et al., 2010). Also in chick, ectopic expression of Ets1 in cranial neural crest leads to progressive migration at the basal side of neural tube, but these cells do not express neural crest markers and undergo apoptosis (Théveneau et al., 2007). The differential expression of Ets1, and its role in neural crest emigration, suggests variable control of neural crest GRN function at different axial levels.
An additional family of Sox transcription factors important for neural crest development is the SoxD factors, including Sox5 (L-Sox5) and Sox6. Consistent with an essential role for these factors, mice mutant for Sox5 and Sox6 show a virtual absence of all cartilage (Smits et al., 2001). Interestingly, expression of both SoxD factors appears to be under the control of Sox9 (Akiyama et al., 2002; Perez-Alcala et al., 2004). L-Sox5 is expressed in premigratory and early migrating neural crest cells in the chick (Perez-Alcala et al., 2004) and in Xenopus (Nordin and LaBonne, unpublished) it co-localizes with Sox10 and Mitf in the melanocyte lineage (Stolt et al., 2008). Sox5 is also expressed in the Peripheral Nervous Sysem (PNS), in the NC-derived trigeminal ganglion, and differentiating neurons of the cranial ganglia. It is co-expressed with Sox10 in the satellite glial cells of the cranial ganglia (Morales et al., 2007) and in Schwann cells (Perez-Alcala et al., 2004).
SoxD family proteins appear to function, at least in part, by modulating the activity of SoxE proteins such as Sox9 and Sox10. L-Sox5 and Sox6 bind HMG-like consensus sites in the Col2A1 enhancer as homodimers, and cooperatively enhance the activation of Col2A1 by Sox9 (Lefebvre et al., 1998). SoxD proteins are also likely to modulate SoxE function during other aspects of neural crest development. For example, L-Sox5 can inhibit Sox10 mediated activation of the Mitf and Dct promoters. This may be mediated, in part, by the ability of L-Sox5 to recruit co-repressors such as HDAC1 and CtBP2 (Stolt et al., 2008). These effects contrast greatly with what occurs on the Col2a1 promoter, where L-Sox5 and Sox9 cooperatively recruit co-activators (Hattori et al., 2008), highlighting the importance of context in determining the functional output of these factors.
V. Post-Transcriptional Regulation of Neural Crest Specifiers
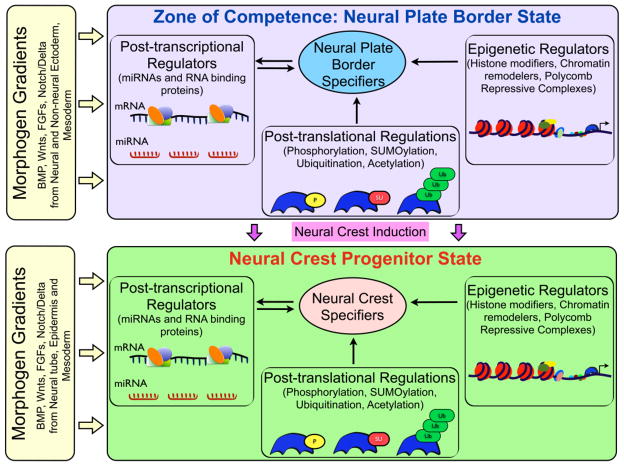
Much work has been done toward understanding the upstream signals and transcriptional response factors that direct the development of neural crest cells. These components have also been incorporated into a systems level model in the NC-GRN (Figure 1). It is important to keep in mind, however, that most of these regulatory proteins are used reiteratively during neural crest development, and therefore mechanisms must exist to control their function in a context dependent manner. While combinatorial transcriptional control is clearly one way that context can be imposed, recent work suggests that post-translational regulatory mechanisms make key contributions (Taylor and LaBonne, 2007).
Post-translational modifications (PTMs) can play essential roles in regulating the functional output of a protein. For example, in the neural crest, ubiquitination of Snail proteins is an important mechanism of context-dependent control. As discussed above, Snail proteins are used reiteratively for the formation of neural crest stem cells and the subsequent EMT/migration of these cells. Recent work has indicated that the cellular levels of Snail proteins are an important determinant of the outcome of their expression on neural crest cell development (Vernon and LaBonne, 2006). Snail1/Snail2 protein levels are regulated by the ubiquitin-proteasome system (UPS), and they can be targeted for proteasomal degradation by the F-box protein, Partner of paired (Ppa, also known as FBXL14), which serves as the substrate recognition component of an SCF (Skp-Cullin-F-box) E3 ligase. Ppa expression is dynamically expressed during neural crest development, and stabilized Snail2 protein that cannot be targeted by Ppa induces premature neural crest migration, demonstrating the necessity of tight regulation (Vernon and LaBonne, 2006). It is likely that many additional mechanisms also contribute to controlling Snail protein function in a context-dependent manner. Mammalian Snail, for example, is regulated by GSK3β phosphorylation, which regulates both its sub-cellular localization and beta-Trcp-mediated ubiquitination (Zhou et al., 2004). Interestingly, however, this regulation is not conserved in Snail2 or in anamniote Snail proteins (Vernon and LaBonne, 2006).
Like Snail1 and Snail2, Twist is both a neural crest specifier and a core EMT regulatory factor. It was recently demonstrated that despite their structural diversity, Twist, a bHLH factor, and the zinc finger transcriptional repressors Snail1/Snail2, share a common regulatory mechanism. These factors, together with another core EMT factor Sip1 (also called Zeb2), are all targeted for proteasomal degradation by the same F-box protein Ppa (Lander et al., 2011). The functions of numerous developmental regulatory proteins are regulated, at least in part, by the threshold concentrations of that protein and the net accumulation of protein product as determined by expression and protein turnover. What is remarkable here is that a common targeting mechanism has evolved to control the activity of a core group of functionally linked but structurally diverse factors. Studies in tumor cells have also identified Protein kinase B (PKB/Akt) mediated phosphorylation of Twist-1 at serine 42 as an important means of controlling its activity (Vichalkovski et al., 2010). It will be of interest to examine possible roles for PKB in neural crest development.
SoxE factors can be regulated post-translationally by both phosphorylation and SUMOylation. PKA (cAMP-dependent protein kinase A)-mediated phosphorylation of Sox9 regulates its transcriptional activity as well as its nuclear localization (Huang et al., 2000). Interestingly, PKA activity has been reported to be high in the murine dorsal neural tube (Chen et al., 2005). Mutation of the Sox9 PKA sites to alanine, preventing its phosphorylation, impaired the ability of Sox9 to mediate EMT, but did not affect its ability to induce ectopic Snail2 expression (Sakai et al 2005). PKA-mediated phosphorylation can thus contribute to context-dependent control of Sox9 function.
SUMOylation of transcription factors can affect their sub-cellular localization, DNA binding, protein–protein interactions and transcriptional activity (Gill, 2004). SUMO modification of SoxE transcription factors profoundly affects their function during early ectodermal patterning (Taylor and LaBonne, 2005). SUMO modification of Sox9 or Sox10 was found to inhibit the ability of these factors to induce neural crest progenitor cells; instead they promoted inner ear formation. SoxE factors with mutations in the SUMO acceptor sites displayed enhanced neural crest inducing activity and antagonized ear formation. SUMOylation of Sox10 has also been shown to inhibit activation of MITF (Girard and Goossens, 2006). SUMOylation converts SoxE factors to transcriptional repressors by mediating the recruitment of Grg4 (P.C. Lee and C. LaBonne, unpublished). These findings highlight the importance of SUMOylation as a versatile post-translational modification that can contribute to the context dependent control of reiteratively used regulatory factors.
SUMOylation of Ets1 has also been reported, although not in neural crest cells. SUMO modification here leads to reduced transactivation capacity (Ji et al., 2007). Ets1 is a downstream target of RAS/MapKinase signaling (Nelson et al., 2010), and thus Map kinase dependent Ets1 phosphorylation is likely to occur in response to FGF signaling in the neural crest. Studies using human fibroblasts have also demonstrated that Ets1 can be acetylated in response to TGF-β, and the acetylated form of Ets1 preferentially associates with p300/CBP complexes (Czuwara-Ladykowska et al., 2002). Acetylation of non-histone proteins has been implicated in a growing number of transcriptional regulatory processes (Spange et al., 2009). Reversible acetylation can influence subcellular localization, protein-protein interactions, degradation, and many other aspects of protein function, and it will be important to determine if Ets1 and other neural crest specifiers are modified in this manner.
MicroRNAs
MicroRNAs can control protein levels by repressing mRNA translation (Carthew, 2006) or by mRNA cleavage. It is intriguing to speculate that miRNA families implicated in the regulation of cancer stem cells and EMT/metastasis might also play a role in neural crest development. These include the miR-200 family, miR-10b, miR-373, and miR-520c (Huang et al., 2008). The miR-200 family is known to downregulate EMT factor Zeb1 (Park et al., 2008), while miR-141 and miR-200c expression can be suppressed by Zeb1 and Snail1 to maintain the mesenchymal phenotype in colon carcinoma cells (Burk et al., 2008). Given the central role that other EMT regulatory factors play in the neural crest, it will be important to determine if these miRNAs also play essential roles.
In support of this possibility, it has been shown in Xenopus, that loss of Dicer, or of miR-200b, miR-96 and miR-196a, leads to severe neural crest migration defects, and may also be involved in neural crest induction (Gessert et al., 2010). Conditional Dicer knockout in murine neural crest led to failure of neural crest differentiation (Liu et al., 2011). A comprehensive study has identified a range of miRNA expressed in developing neural tube and their gene targets in mouse embryos (Mukhopadhyay et al., 2011). Two of the miRNAs identified, miR-19a and miR-19b, could be of significant interest during neural crest development. Both are expressed in the neural tube between gestational days 8.5–9.5, and in silico analysis predicts targets that include TGFβ signaling ligands, Wnt ligands (Wnt3a and Wnt7a), and Id2, all of which are involved in neural crest development. Thus, studies are starting to uncover essential post-transcriptional regulation by miRNAs in neural crest development, and it will be important to build a comprehensive view of miRNA expression and function into the neural crest gene regulatory network.
VI. Epigenetic Control of the Neural Crest State
Epigenetic contributions to the control of the NC-GRN are an emerging area of focus in the field. The regulation of higher order chromatin structure via histone modifications and association of chromatin remodelers that catalyse those modifications, as well as modifications of the DNA proper, is undoubtedly of high significance to understanding the formation of neural crest progenitors and their subsequent development as chromatin state dynamics will have direct consequences for the recruitment of the transcriptional activation or repression machineries. In building our understanding of how the expression of neural crest specifiers is initiated, it will be important to take into account variables such as the presence of histone variants, modification of histones, the role of ATP-dependent chromatin remodeling factors, and their effects on chromatin structure in prospective progenitor cells. Indeed, given the unusual increase in developmental potential that underlies the formation of the neural crest precursor population, and the fundamental role that epigenomics plays in the regulation of stemness more generally, this level of regulation in neural crest progenitors is likely to be of central importance.
The best-characterized histone modifications are post-translational modifications of histone tails by methylation, acetylation, phosphorylation, and ubiquitination (Berger, 2007). Additional histone modifications include sumoylation, ADP ribosylation, and deamination, and the non-covalent proline isomerization (Gibney and Nolan, 2010). Histone modifications contribute to the control of gene expression by recruiting chromatin modifiers and transcriptional activators or repressors. Histone methylation, in particular, is a widely studied modification. For example, methylation of histone H3 subunit on the fourth and twenty seventh lysine residue (H3K4me3 and H3K27me3) is catalyzed by histone methyl transferases of the Trithorax (TrxG) and Polycomb group and plays a central role in flagging the active and repressed loci, respectively (Barski et al., 2007; Pan et al., 2007; Liu and Xiao, 2011). H3K4me3, together with H3K36me3 are frequently considered transcriptional activation marks (predominantly found in promoter or body of the gene, respectively) whereas H3K9me3 and H3K27me3 are considered to be repressive marks the latter considered to be a key signal for Polycomb-mediated repression (Simon and Kingston, 2009). Genome wide analysis of histone methylation states in early Xenopus embryos during gastrulation confirmed H3K4me3 and H3K27me3 are marks for active and repressed genes respectively (Akkers et al., 2009). Similarly, in 24 hpf (hours post-fertilization) zebrafish embryos, H3K4me3 and H3K4me1 marks are found at putatively active gene targets (Aday et al., 2011). Recent analysis of embryonic stem cells emphasized the importance of these histone marks, as well as histone acetylation, in identifying active and repressed genes as well as distant active sites of regulatory activity (Rada-Iglesias et al., 2011). This study further suggested that H3K27me3 marked genes were in a poised state in advance of developmental roles in gastrulation, mesoderm formation and neurulation. Additionally they showed that poised enhancers could drive spatially and temporally correct patterns of reporter expression in zebrafish despite the absence of clear sequence conservation (Rada-Iglesias et al., 2011). Epigentic signatures can thus be utilized for efficient identification of functional enhancer regions of developmentally important genes.
Jumonji family histone demethylases, have recently been shown to play an essential role in neural crest development, highlighting the importance of epigenetic regulation in these cells. This study provided evidence that a member of this family, JmjD2A, mediates demethylation of H3K9me3 that is required for activation of neural crest specifier genes Sox9, Sox10, FoxD3, and Snail2 (Strobl-Mazzulla et al., 2010). Consistent with such a role, JmjD2A is expressed in the neural plate but then downregulated in migrating neural crest cells (Strobl-Mazzulla et al., 2010). Another recent study has shown that CHD7 (chromodomain helicase DNA-binding domain), an ATP-dependent chromatin remodeler related to the Drosophila trithorax-group factor Kismet, is essential for the formation and migration of neural crest cells (Bajpai et al., 2010). This study found that CHD7 association with distant enhancer elements is essential for activation of numerous neural crest specifiers including Sox9, Twist and Slug. In neural crest cells induced from human ES cells, CHD7 was also found to associate with PBAF a SWI/SNF family chromatin-remodelling complexes, and occupy an neural crest specific Sox9 enhancer as well as a regulatory element upstream of Twist.
Histone acetyl transferases (HATs) acetylate histones whereas histone deacetylases (HDACs) remove these groups (Carrozza et al., 2003; Hsieh et al., 2004). Histone acetylation increases the accessibility of DNA to transcription factors and promotes gene transcription, while deacetylation of histones results in a more compact chromatin confirmation resulting in silencing of gene expression (Jenuwein and Allis, 2001). HDAC8 has been shown to be essential for cranial neural crest cells to form the craniofacial skeleton (Haberland et al., 2009). Mice deficient for HDAC8 show derepression of important regulatory factors including Otx2 and Lhx1 and other homeobox genes normally not expressed in cranial neural crest cells. The most remarkable aspect of this work is the finding that broadly expressed factors such as Class I HDACs can have such highly specific developmental functions (Haberland et al., 2009). While we have barely scratched the surface in understanding the contributions of chromatin regulatory mechanisms to the formation and development of neural crest cells, it is clear that is will be an important and fruitful area of future investigation. Understanding the epigenetic landscape of neural crest progenitors should shed important light on the acquisition of stem cell characteristics in general, and the mechanisms that led to the evolution of vertebrates.
Conclusion
Our current insights into neural crest development are based mainly on gene expression analyses using in situ hybridization and immunohistochemistry, combined with perturbation analysis of individual genes and signaling pathways. These powerful approaches have provided the enormous amounts of data that seed our present understanding of the neural crest regulatory network. System wide approaches are beginning to be employed to identify additional neural crest regulatory factors, their targets, and epigenetic marks characteristic of these cells from their induction through their differentiation. Fully deciphering the role and regulation of signaling pathways and transcription factors that are key players in the GRN will require understanding the contributions of epigenetics regulation as well as post-transcriptional/translational modifications. Indeed, a central challenge to understanding complex developmental processes such as neural crest development on a systems-wide level is to understand how the function of each protein in the network is controlled, individually and coordinately. This is of particular importance given the reiterative usage of many of these key factors in the neural crest gene regulatory network and the links that many of these factors have to the acquisition of stem cell characteristics and developmental potency.
Figure 2.
Summary of regulatory inputs leading to the formation of the neural crest progenitor population. The neural plate border region receive signals from neural and non-neural ectoderm, and underlying mesoderm, to establish a zone of competence at the neural plate border that expresses border specifiers including Pax3/7, Dlx3/5, Zic1, Msx1, AP2α.. Neural plate border specifiers function together with extracellular signals to induce the expression of neural crest specifiers, including Pax3/7, Id, Snail1/2, Sox9/10, FoxD3, Twist, several of which have links to the establishment of stem cell attributes in multiple systems. Post-transcriptional, post-translational and epigenetic regulatory mechanisms play key roles in both the establishment of the zone of competence at the neural plate border, and the induction of the neural crest progenitor population within the border region. P-phosphorylation, SU-SUMOylation, Ub-ubiquitination.
Highlights.
-
3)
We review our current understanding of the formation of neural crest progenitors
-
4)
Neural crest progenitors have attributes of stem cells and their formation is controlled by factors linked to pluripotency
-
5)
Neural crest progenitors arise in a zone of competence at the neural plate border established by border specifier factors
-
6)
Neural crest specifiers are used reiteratively and thus subject to context-dependent control mechanisms
-
7)
Post-translational and epigenetic mechanisms contribute to formation of neural crest stem cells
Acknowledgments
The authors thank Elsy Buitrago Delgado, Keith Hultman, Kara Nordin and Ritika Giri for comments on the manuscript. We apologize for work that could not be included due to space constraints. Relevant work in the authors’ labs is supported by NIH RO1CA114058, RO1GM077288 and R21DE022150 (CL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acloque H, Ocana OH, Matheu A, Rizzoti K, Wise C, Lovell-Badge R, Nieto MA. Reciprocal repression between Sox3 and snail transcription factors defines embryonic territories at gastrulation. Dev Cell. 2011;21:546–558. doi: 10.1016/j.devcel.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol. 2011;357:450–462. doi: 10.1016/j.ydbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci. 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet JP. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroffio A, Dupin E, Le Douarin NM. Common precursors for neural and mesectodermal derivatives in the cephalic neural crest. Development. 1991;112:301–305. doi: 10.1242/dev.112.1.301. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser SE. Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature. 1988;335:161–164. doi: 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Sieber-Blum M, Cohen AM. Clonal analysis of the avian neural crest: migration and maturation of mixed neural crest clones injected into host chicken embryos. J Comp Neurol. 1980;193:423–434. doi: 10.1002/cne.901930209. [DOI] [PubMed] [Google Scholar]
- Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, Cho JY, Dobias SL, Yi SE, Lyons K, Bell JR, Arora K, Warrior R, Maxson R. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development. 2004;131:5153–5165. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Chiang EF, Pai CI, Wyatt M, Yan YL, Postlethwait J, Chung B. Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev Biol. 2001;231:149–163. doi: 10.1006/dbio.2000.0129. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Sementchenko VI, Watson DK, Trojanowska M. Ets1 is an effector of the transforming growth factor beta (TGF-beta ) signaling pathway and an antagonist of the profibrotic effects of TGF-beta. J Biol Chem. 2002;277:20399–20408. doi: 10.1074/jbc.M200206200. [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci USA. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, Ward A, Kelsh RN. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol. 2008;8:105. doi: 10.1186/1471-213X-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Endo Y, Osumi N, Wakamatsu Y. Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development. 2002;129:863–873. doi: 10.1242/dev.129.4.863. [DOI] [PubMed] [Google Scholar]
- Fafeur V, Tulasne D, Queva C, Vercamer C, Dimster V, Mattot V, Stehelin D, Desbiens X, Vandenbunder B. The ETS1 transcription factor is expressed during epithelial-mesenchymal transitions in the chick embryo and is activated in scatter factor-stimulated MDCK epithelial cells. Cell Growth Differ. 1997;8:655–665. [PubMed] [Google Scholar]
- Fang X, Cai Y, Liu J, Wang Z, Wu Q, Zhang Z, Yang CJ, Yuan L, Ouyang G. Twist2 contributes to breast cancer progression by promoting an epithelial-mesenchymal transition and cancer stem-like cell self-renewal. Oncogene. 2011;30:4707–4720. doi: 10.1038/onc.2011.181. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Gessert S, Bugner V, Tecza A, Pinker M, Kuhl M. FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev Biol. 2010;341:222–235. doi: 10.1016/j.ydbio.2010.02.031. [DOI] [PubMed] [Google Scholar]
- Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity (Edinb) 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Girard M, Goossens M. Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett. 2006;580:1635–1641. doi: 10.1016/j.febslet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004;131:347–359. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, Tam WL, Mani SA, van Oudenaarden A, Weinberg RA. Slug and sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldin CE, LaBonne C. SoxE factors as multifunctional neural crest regulatory factors. Int J Biochem Cell Biol. 2010;42:441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol Dev. 2000;2:3–5. doi: 10.1046/j.1525-142x.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Coustry F, Stephens S, Eberspaecher H, Takigawa M, Yasuda H, de Crombrugghe B. Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 2008;36:3011–3024. doi: 10.1093/nar/gkn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol Biol Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhou X, Lefebvre V, de Crombrugghe B. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9’s ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol Cell Biol. 2000;20:4149–4158. doi: 10.1128/mcb.20.11.4149-4158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, Iwasaki H, Akashi K, Morimoto A, Hitzler JK, Pestina TI, Jackson CW, Tanaka R, Chong MJ, McKinnon PJ, Inukai T, Grosveld GC, Look AT. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer Cell. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- Ito K, Sieber-Blum M. In vitro clonal analysis of quail cardiac neural crest development. Dev Biol. 1991;148:95–106. doi: 10.1016/0012-1606(91)90320-3. [DOI] [PubMed] [Google Scholar]
- Ji Z, Degerny C, Vintonenko N, Deheuninck J, Foveau B, Leroy C, Coll J, Tulasne D, Baert JL, Fafeur V. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene. 2007;26:395–406. doi: 10.1038/sj.onc.1209789. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- Jones NC, Trainor PA. Role of morphogens in neural crest cell determination. J Neurobiol. 2005;64:388–404. doi: 10.1002/neu.20162. [DOI] [PubMed] [Google Scholar]
- Karafiat V, Dvorakova M, Krejci E, Kralova J, Pajer P, Snajdr P, Mandikova S, Bartunek P, Grim M, Dvorak M. Transcription factor c-Myb is involved in the regulation of the epithelial-mesenchymal transition in the avian neural crest. Cell Mol Life Sci. 2005;62:2516–2525. doi: 10.1007/s00018-005-5297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Bronner-Fraser M. To proliferate or to die: role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes Dev. 2005;19:744–755. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, Bapat SA. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Molecular mechanisms of neural crest formation. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–190. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Lander R, Nordin K, LaBonne C. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J Cell Biol. 2011;194:17–25. doi: 10.1083/jcb.201012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Boldrini R, Dominici C, Donfrancesco A, Yokota Y, Inserra A, Iavarone A. Id2 is critical for cellular proliferation and is the oncogenic effector of N-myc in human neuroblastoma. Cancer Res. 2002;62:301–306. [PubMed] [Google Scholar]
- Le Douarin NM, Dupin E. Cell lineage analysis in neural crest ontogeny. J Neurobiol. 1993;24:146–161. doi: 10.1002/neu.480240203. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C. The Neural Crest. 2 Cambridge University Press; Cambridge: 1999. [Google Scholar]
- Lee YH, Aoki Y, Hong CS, Saint-Germain N, Credidio C, Saint-Jeannet JP. Early requirement of the transcriptional activator Sox9 for neural crest specification in Xenopus. Dev Biol. 2004;275:93–103. doi: 10.1016/j.ydbio.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–3278. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W, Vernon AE, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005;132:1831–1841. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Liu Y, Huang T, Zhao X, Cheng L. MicroRNAs modulate the Wnt signaling pathway through targeting its inhibitors. Biochem Biophys Res Commun. 2011;408:259–264. doi: 10.1016/j.bbrc.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Manzanares M, Blanco MJ, Nieto MA. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc Natl Acad Sci USA. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta. 2009;1789:363–374. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maczkowiak F, Mateos S, Wang E, Roche D, Harland R, Monsoro-Burq AH. The Pax3 and Pax7 paralogs cooperate in neural and neural crest patterning using distinct molecular mechanisms, in Xenopus laevis embryos. Dev Biol. 2010;340:381–396. doi: 10.1016/j.ydbio.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev Biol. 1996;177:580–589. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Alvarez C, Blanco MJ, Perez R, Rabadan MA, Aparicio M, Resel E, Martinez T, Nieto MA. Snail family members and cell survival in physiological and pathological cleft palates. Dev Biol. 2004;265:207–218. doi: 10.1016/j.ydbio.2003.09.022. [DOI] [PubMed] [Google Scholar]
- Mayor R, Guerrero N, Young RM, Gomez-Skarmeta JL, Cuellar C. A novel function for the Xslug gene: control of dorsal mesendoderm development by repressing BMP-4. Mech Dev. 2000;97:47–56. doi: 10.1016/s0925-4773(00)00412-3. [DOI] [PubMed] [Google Scholar]
- McKeown SJ, Lee VM, Bronner-Fraser M, Newgreen DF, Farlie PG. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev Dyn. 2005;233:430–444. doi: 10.1002/dvdy.20341. [DOI] [PubMed] [Google Scholar]
- Menendez L, Yatskievych TA, Antin PB, Dalton S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc Natl Acad Sci USA. 2011;108:19240–19245. doi: 10.1073/pnas.1113746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Fletcher RB, Harland RM. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–3124. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev Cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Montero JA, Giron B, Arrechedera H, Cheng YC, Scotting P, Chimal-Monroy J, Garcia-Porrero JA, Hurle JM. Expression of Sox8, Sox9 and Sox10 in the developing valves and autonomic nerves of the embryonic heart. Mech Dev. 2002;118:199–202. doi: 10.1016/s0925-4773(02)00249-6. [DOI] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev Dyn. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Morales AV, Perez-Alcala S, Barbas JA. Dynamic Sox5 protein expression during cranial ganglia development. Dev Dyn. 2007;236:2702–2707. doi: 10.1002/dvdy.21282. [DOI] [PubMed] [Google Scholar]
- Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- Moury JD, Jacobson AG. The origins of neural crest cells in the axolotl. Dev Biol. 1990;141:243–253. doi: 10.1016/0012-1606(90)90380-2. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P, Brock G, Appana S, Webb C, Greene RM, Pisano MM. MicroRNA gene expression signatures in the developing neural tube. Birth Defects Res A Clin Mol Teratol. 2011;91:744–762. doi: 10.1002/bdra.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson ML, Kang HS, Lee GM, Blaszczak AG, Lau DK, McIntosh LP, Graves BJ. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Natl Acad Sci USA. 2010;107:10026–10031. doi: 10.1073/pnas.0915137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina N, Tong L, Bronner ME. Ancestral network module regulating prdm1 expression in the lamprey neural plate border. Dev Dyn. 2011;240:2265–2271. doi: 10.1002/dvdy.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland JW, Raible DW. Signals derived from the underlying mesoderm are dispensable for zebrafish neural crest induction. Dev Biol. 2004;276:16–30. doi: 10.1016/j.ydbio.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Romano LA, Runyan RB. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223:91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Barks JL, Iademarco MF, Fisher RJ, Dean DC. An intricate arrangement of binding sites for the Ets family of transcription factors regulates activity of the alpha 4 integrin gene promoter. J Biol Chem. 1994;269:15652–15660. [PubMed] [Google Scholar]
- Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc Natl Acad Sci USA. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D, Tanaka Y, Endo Y, Osumi N, Okamoto H, Wakamatsu Y. Regulation of Slug transcription in embryonic ectoderm by beta-catenin-Lef/Tcf and BMP-Smad signaling. Dev Growth Differ. 2005;47:471–482. doi: 10.1111/j.1440-169X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Development and evolution of the migratory neural crest: a gene regulatory perspective. Curr Opin Genet Dev. 2006;16:360–366. doi: 10.1016/j.gde.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D, Kalcheim C. Regulation of the onset of neural crest migration by coordinated activity of BMP4 and Noggin in the dorsal neural tube. Development. 1999;126:4749–4762. doi: 10.1242/dev.126.21.4749. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development. 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. The genesis of avian neural crest cells: a classic embryonic induction. Proc Natl Acad Sci USA. 1996;93:9352–9357. doi: 10.1073/pnas.93.18.9352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber-Blum M, Cohen AM. Clonal analysis of quail neural crest cells: they are pluripotent and differentiate in vitro in the absence of noncrest cells. Dev Biol. 1980;80:96–106. doi: 10.1016/0012-1606(80)90501-1. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Dy P, Mitra S, Lefebvre V. Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J Cell Biol. 2004;164:747–758. doi: 10.1083/jcb.200312045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Hillgartner S, Wegner M. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res. 2008;36:5427–5440. doi: 10.1093/nar/gkn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol. 2010;42:437–440. doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell. 2010;19:460–468. doi: 10.1016/j.devcel.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Taylor KM, LaBonne C. Modulating the activity of neural crest regulatory factors. Curr Opin Genet Dev. 2007;17:326–331. doi: 10.1016/j.gde.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Teng L, Labosky PA. Neural crest stem cells. Adv Exp Med Biol. 2006;589:206–212. doi: 10.1007/978-0-387-46954-6_13. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompers DM, Foreman RK, Wang Q, Kumanova M, Labosky PA. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Dev Biol. 2005;285:126–137. doi: 10.1016/j.ydbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Trentin A, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Self-renewal capacity is a widespread property of various types of neural crest precursor cells. Proc Natl Acad Sci USA. 2004;101:4495–4500. doi: 10.1073/pnas.0400629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–6452. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17:141–153. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- Vesuna F, Lisok A, Kimble B, Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–1328. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichalkovski A, Gresko E, Hess D, Restuccia DF, Hemmings BA. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29:3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23:213–216. doi: 10.1016/s0968-0004(98)01211-0. [DOI] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–6538. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Yang J, Mani SA, Weinberg RA. Exploring a new twist on tumor metastasis. Cancer Res. 2006;66:4549–4552. doi: 10.1158/0008-5472.CAN-05-3850. [DOI] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, Chang SY, Lee OK, Wu KJ. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]