In the past 15 years, nuclear magnetic resonance (NMR) spectroscopy has emerged as one of the principle techniques of structural biology (1, 2). It not only is capable of solving protein structures to atomic resolution but also has the unique ability to accurately measure the dynamic properties of proteins and to probe the process of protein folding (3, 4). However, a major drawback of macromolecular NMR is its size limitation caused by two technical barriers. First, larger molecules have slower tumbling rates and shorter NMR signal relaxation times. This reduces the sensitivity of the complicated pulse sequences that often use long delays for the necessary coherence transfer steps. The increased molecular weight also introduces more complexity to a given spectrum, simply because there are more NMR-active nuclei and, therefore, more interactions among them. The current size limit of protein NMR is ≈35 kDa, but recent advances in both hardware and experimental design promise to allow the study of much larger proteins (2). The future is even brighter with the development of novel strategies for isotopic labeling of proteins that are synergistic with the new NMR techniques. One such strategy is the segmental protein isotopic labeling scheme described by Xu et al. in this issue of the Proceedings (5).
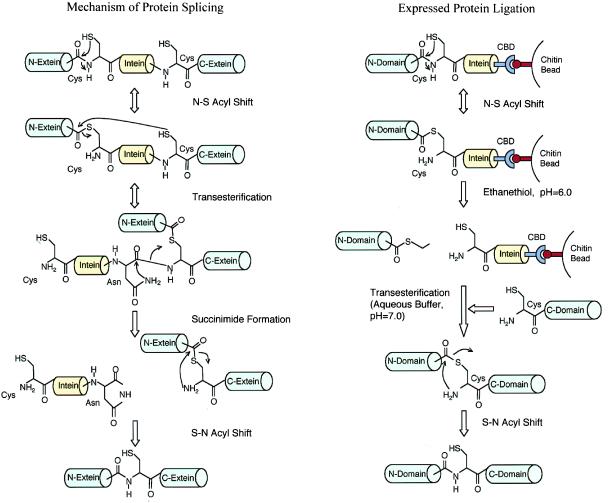
Xu et al. have successfully ligated together two independently expressed, folded protein domains under mild conditions in vitro, making it possible to selectively label a given fragment or domain of modular proteins (5). They have done so by taking advantage of protein splicing (6, 7). A small number of proteins are made as precursors that contain motifs called inteins. During the maturation process, inteins are excised from these precursors, and the two resulting fragments or exteins then rejoin. This process has been called protein splicing presumably because it is conceptually similar to RNA splicing. Much has been learned about the mechanism of protein splicing (Fig. 1). As a first step, the N-terminal cysteine of the intein attacks the C-terminal amide bond of the N-extein, resulting in an N-S acyl shift and the formation of a thioester at the N-terminal splice site. In a transesterification reaction, the N-extein is ligated to the N-terminal cysteine of the C-extein. The sidechain amide of a conserved Asn residue at the C terminus of the intein then attacks the main chain amide bond to form a succinimide group, excising the intein. The ligated exteins undergo an S-N acyl rearrangement to form a native amide bond in the final step.
Figure 1.
Mechanism of protein splicing and expressed protein ligation (see text for details).
Certain intein mutants are defective in the cleavage of the C-terminal splice site but still capable of cleavage at the N terminus, leading to accumulation of the thioester intermediate between the intein and N-extein (8). Recently, the properties of these mutant inteins have attracted much attention from protein chemists. For example, commercial protein expression vectors have been constructed to allow the production of protein of interest as an intein-chitin-binding domain fusion, which can be purified on a chitin affinity column (9). Because of mutations within the intein, the fusion protein is trapped as a thioester between the protein of interest and the intein. The intein-chitin-binding domain then can be cleaved off by the addition of reducing agents, such as DTT. Based on similar principles, Muir et al. have developed a technique termed “expressed protein ligation” (10, 11). Instead of reducing the thioester intermediate with DTT, they showed that the thioester generated by the splicing process can react with peptides containing a cysteine at their N-termini. Using this technique, they ligated synthetic peptides to recombinant proteins through native peptide bonds (10, 11).
Xu et al. have now taken this approach one step further (5). They postulated that similar principles should work for the ligation of two folded proteins. To accomplish this, they made a simple yet elegant extension to their existing scheme (Fig. 1). In their earlier work with peptide ligation, they mixed the thioester form of the fusion protein on chitin beads directly with peptides. For the reactions to go to completion, they used high concentrations of peptides. Because it is difficult to obtain a similarly high molar concentration of proteins, the ligation reaction between two proteins would have been inefficient. Their solution to the problem was to incubate the beads with a small thiol compound, ethanethiol, which led to the liberation of an ethyl-thioester derivative into solution. The ethyl-thioester reacted efficiently with a second protein. Using this seemingly simple modification, they were able to join together the SH3 and SH2 domains of the Abelson tyrosine kinase with a remarkable 70% yield. Furthermore, the final products were characterized thoroughly by using analytical techniques such as mass spectrometry to confirm their chemical structures. They also obtained SH3-SH2 proteins with only the SH2 portion labeled with 15N. Comparison of the 1H/15N heteronuclear single quantum correlation (HSQC) spectrum of the SH3-15N-SH2 with that of uniformly 15N-labeled SH3-SH2 confirmed that the SH2 domain retained its tertiary fold after the ligation reaction. As the authors pointed out, this strategy permits three protein domains to be joined together in vitro.
Evans et al. also developed a method to trap the thioester intermediate of a different intein with 2-mercaptoethanesulfonic acid (12), although they only attempted the ligation of the trapped thioester with synthetic peptides in their semisynthesis of RNase A and HpaI enzymes. Other strategies for generating regioselective isotopically labeled proteins in vitro include a trans-splicing scheme reported by Yamazaki et al., in which two protein fragments each containing part of the intein and the target protein were individually expressed (13). These two fragments then were mixed to allow for the excision of the intein in a trans-splicing process. A serious limitation of this method, however, is its requirement of a protein denaturation-refolding sequence. It is also not as versatile as that presented by Xu et al. because it cannot be extended to ligate three protein fragments.
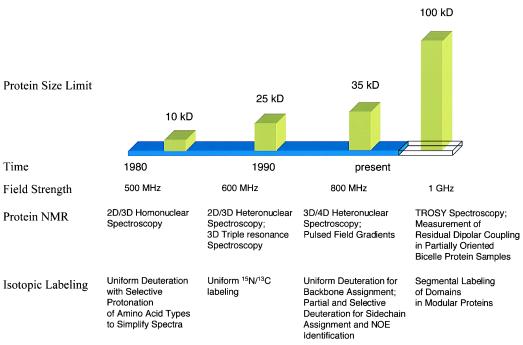
During its relatively short history, protein NMR already has undergone several transformations that have extended its size limit (Fig. 2) (1). These transformations were brought about by technical advances in NMR spectroscopy and by progress in protein labeling schemes. The assignment of resonance and identification of nuclear Overhauser effects (NOEs) initially were accomplished by analyzing two-dimensional homonuclear spectra, limiting the size of proteins suitable for NMR studies under 10 kDa because of spectral complexity. The subsequent availability of uniformly 15N/13C-labeled proteins produced in bacteria led to the development of the so-called triple resonance experiments in the late 1980s and early 1990s. These multidimensional heteronuclear experiments establish sequential connections of backbone resonance on the basis of the larger and more uniform heteronuclei through-bond couplings instead of through-space NOEs that vary greatly in intensity depending on the local conformation. In addition, the NMR signals have been spread effectively into additional 15N and/or 13C dimensions, alleviating spectral degeneracy. These advances, together with the later incorporation of pulsed field gradients, extended the range of proteins suitable for NMR to 25 kDa. More recently, 2H labeling of proteins has contributed greatly to the field of protein NMR. By substituting the nonexchangeable protons with deuterons, the relaxation time of heteronuclear signals are prolonged, resulting in narrowed linewidth and a dramatic increase in resolution and sensitivity. This has increased the current size limit of protein NMR to 35 kDa (14–18).
Figure 2.
Advances in NMR spectroscopy and isotopic labeling have extended the size limit of protein NMR (see text for details).
In the last year or two, the availability of higher field magnets (1H frequency of 800 MHz) has allowed new experiments that take advantage of chemical shift anisotropy and residual dipolar coupling. The first such experiment is transverse relaxation-optimized spectroscopy (TROSY) (2, 19–21). It is known that both chemical shift anisotropy and dipolar coupling contribute to line broadening. At higher fields, chemical shift anisotropy becomes larger and subtracts the contribution of dipolar coupling to relaxation in one of the four multiplets formed by scalar coupling in a two-spin system. For an amide 1H-15N group, the complete cancellation of chemical shift anisotropy and dipolar coupling happens at a field equivalent to a proton frequency of 1 GHz. TROSY-type experiments using 800-MHz spectrometer show significantly narrower linewidth and higher sensitivity, and the TROSY pulse sequence is now being incorporated into triple resonance experiments. Because the sensitivity gain of TROSY experiments seems to be independent of molecular weight, this technique promises to push the size of proteins suitable for NMR studies up to 100 kDa (2, 19–21). Another approach that has the potential to extend the size limit of NMR is the measurement of residual dipolar coupling in partially oriented proteins dissolved in lipid bicelle medium (22, 23). The N-H and C-C vector orientation relative to the magnetic anisotropy tensor can be deduced from the magnitude of residual dipolar coupling, thus providing valuable structural information. Because measurement of 1H-15N and 13C-13C dipolar couplings can be performed on perdeuterated samples, this technique should be applicable to proteins in the 50-kDa range. However, larger molecular weight also means an increased number of peaks in a given spectral range, making it much more difficult to resolve overlapping signals. This is where the segmental protein labeling scheme developed by Xu et al. comes into play (5).
As the authors alluded to in their discussion, there are many potential applications for in vitro ligation of protein domains, the most important of which may be its ability to generate proteins that are selectively labeled on certain segments. These samples are extremely valuable for NMR structural analyses, especially in light of the recent technological advances in NMR spectroscopy. As mentioned above, both TROSY-type experiments and the use of semi-liquid crystalline protein samples will help extend the size limit, yet neither is able to solve the problem of increased spectral complexity accompanying larger proteins. On the other hand, the approach by Xu et al. allows a 100- to 150-residue segment of a large protein to be selectively isotopically labeled (5). As a result, the rest of the protein becomes largely “invisible” in the isotopically dispersed spectra, thus reducing spectral complexity and signal overlaps. This will greatly facilitate the structure determination of a protein domain in the context of the full length protein. In principle, the structures of different segments in the protein can be solved one at a time, and their quaternary arrangement then can be determined by a relatively small number of interdomain NOEs.
Segmental isotopic labeling also can be combined with other selective labeling methods. For example, Rosen, Gardner, and Kay (18, 24) recently developed an elegant labeling scheme that allows selective protonation of methyl groups of alanines, valines, leucines, and isoleucines in an otherwise perdeuterated protein molecule. They also devised a set of NMR experiments to correlate the methyl resonance with that of the backbone amide protons (18). Introduction of the protonated methyl groups into perdeuterated proteins permits the observation of NOEs involving backbone amide proton and the protonated methyl groups while retaining the advantage of slower relaxation rate and sharper linewidth compared with nondeuterated protein samples. In model calculations, they found that NH-NH, NH-methyl, and methyl-methyl NOE restraints alone are sufficient to yield low resolution structures (18, 25). Therefore, incorporation of methyl-protonated and otherwise 2H/15N/13C-labeled protein segments into the full length protein through in vitro protein ligation will further simplify the NMR spectra and increase the possibility of obtaining a tertiary fold of any given domain in larger proteins.
In addition to facilitating NMR structure determination, the segmental labeling method also will be beneficial to NMR experiments that measure protein dynamics and to the Structure-Activity Relationship-by-NMR approach (3, 26). Another important application of in vitro chemical ligation is the synthesis of proteins that cannot be successfully expressed in bacteria or other hosts as full length because of either insolubility or spontaneous degradation in vivo. Although the expression of intein-containing proteins only was attempted in bacteria, it should be straightforward to incorporate the intein coding sequence into the popular baculoviral expression vectors, which will allow this type of fusion proteins to be produced in insect cells. This will further expand the applicability of expressed protein ligation.
Despite its versatility, the approach described by Xu et al. has its limitations. In the present form, it can only be applied to the synthesis of modular proteins that comprise of multiple, independently folded domains. It also requires prior knowledge about the boundaries of these domains. On the other hand, many proteins larger than 30 kDa contain more than one discernable domain (27). For example, a large number of intracellular signaling proteins are composed of modules such as SH2, SH3, PH, or PTB domains (28). Other notable examples include transcriptional factors and extracellular matrix proteins. For less well characterized systems, there are several methods available for identifying the domain boundaries within a modular protein. An independently folded protein domain often is encoded by one or two exons in the gene, and domain boundaries sometimes coincide with the exon–intron boundaries. As more genomes are being sequenced and more proteins are being identified, sophisticated sequence alignment algorithms undoubtedly will identify more and more homologous protein motifs that are present in otherwise functionally distinct proteins. These motifs often represent intact structural domains. Finally, domain boundaries can be determined empirically by limited proteolysis followed by mass spectrometry analysis. This approach is now used widely by x-ray crystallographers to eliminate less well ordered fragments outside of the protein cores, thereby facilitating crystallization.
Throughout its history, protein NMR has benefited tremendously from the availability of isotopic labeled samples. Exciting progress continues to be made in both NMR spectroscopy and protein engineering. The development of new labeling schemes such as that described by Xu et al., combined with advances in NMR procedures, promises to push the size limit of protein NMR to 100 kDa. As these techniques become more mature and widely practiced, the young and resilient field of protein NMR will undergo yet another major transformation in the foreseeable future.
Footnotes
The companion to this Commentary begins on page 388.
References
- 1.Wagner, G. (1997) Nat. Struct. Biol.4, Suppl., 841–844. [PubMed]
- 2.Wüthrich, K. (1998) Nat. Struct. Biol.5, Suppl., 492–495. [DOI] [PubMed]
- 3.Kay, L. E. (1998) Nat. Struct. Biol.5, Suppl., 513–517. [DOI] [PubMed]
- 4.Dobson, C. M. & Hore, P. J. (1998) Nat. Struct. Biol.5, Suppl., 504–507. [DOI] [PubMed]
- 5.Xu R, Ayers B, Cowburn D, Muir T W. Proc Natl Acad Sci USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perler F B, Xu M Q, Paulus H. Curr Opin Chem Biol. 1997;1:292–299. doi: 10.1016/s1367-5931(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 7.Shao Y, Kent S B. Chem Biol. 1997;4:187–194. doi: 10.1016/s1074-5521(97)90287-8. [DOI] [PubMed] [Google Scholar]
- 8.Xu M Q, Perler F B. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 9.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 10.Muir T W, Sondhi D, Cole P A. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severinov K, Muir T W. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 12.Evans T C, Jr, Benner J, Xu M Q. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H. J Am Chem Soc. 1998;120:5591–5592. [Google Scholar]
- 14.Clore G M, Gronenborn A M. Curr Opin Chem Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 15.Clore G M, Gronenborn A M. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 16.Kay L E, Gardner K H. Curr Opin Struct Biol. 1997;7:722–731. doi: 10.1016/s0959-440x(97)80084-x. [DOI] [PubMed] [Google Scholar]
- 17.Kay L E. Biochem Cell Biol. 1997;75:1–15. doi: 10.1139/o97-023. [DOI] [PubMed] [Google Scholar]
- 18.Gardner K H, Kay L E. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 19.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. Proc Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czisch M, Boelens R. J Magn Reson. 1998;134:158–160. doi: 10.1006/jmre.1998.1483. [DOI] [PubMed] [Google Scholar]
- 22.Tjandra N, Bax A. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 23.Prestegard, J. H. (1998) Nat. Struct. Biol.5, Suppl., 517–522. [DOI] [PubMed]
- 24.Rosen M K, Gardner K H, Willis R C, Parris W E, Pawson T, Kay L E. J Mol Biol. 1996;263:627–636. doi: 10.1006/jmbi.1996.0603. [DOI] [PubMed] [Google Scholar]
- 25.Gardner K H, Rosen M K, Kay L E. Biochemistry. 1997;36:1389–1401. doi: 10.1021/bi9624806. [DOI] [PubMed] [Google Scholar]
- 26.Shuker S B, Hajduk P J, Meadows R P, Fesik S W. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 27.Campbell, I. D. & Downing, A. K. (1998) Nat. Struct. Biol.5, Suppl., 496–499. [DOI] [PubMed]
- 28.Kuriyan J, Cowburn D. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]