Abstract
BACKGROUND
This report and a companion report describe a validation of the ability of serum proteomic profiling via SELDI-TOF mass spectrometry to detect prostatic cancer. Details of this 3-stage process have been described. This report describes the development of the algorithm and results of the blinded test for stage 1.
METHODS
We derived the decision algorithm used in this study from the analysis of serum samples from patients with prostate cancer (n = 181) and benign prostatic hyperplasia (BPH) (n = 143) and normal controls (n = 220). We also derived a validation test set from a separate, geographically diverse set of serum samples from 42 prostate cancer patients and 42 controls without prostate cancer. Aliquots were subjected to randomization and blinded analysis, and data from each laboratory site were subjected to the decision algorithm and decoded.
RESULTS
Using the data collected from the validation test set, the decision algorithm was unsuccessful in separating cancer from controls with any predictive utility. Analysis of the experimental data revealed potential sources of bias.
CONCLUSION
The ability of the decision algorithm to successfully differentiate between prostate cancer, BPH, and control samples using data derived from serum protein profiling was compromised by bias.
Multiple laboratories have reported that the spectral patterns of mass spectrometric examinations of specimens of serum can be used to identify patients with several types of tumors (1–7). Based on work at Eastern Virginia Medical School (EVMS)11 and the Fred Hutchinson Cancer Research Center, a diagnostic profile of 9 spectral peaks was reported that could be used to identify patients with prostate cancer (PCa) based on evaluation of serum using SELDI-TOF mass spectrometry. These studies suggested that an accuracy >90% in classification of patients with cancer from controls could be expected (8). Such results would be very important because the current screening methods for PCa using serum concentrations of prostate-specific antigen (PSA) do not detect the majority of prostate cancers, including high-grade tumors (9).
Extensive controversy has accompanied the use of mass spectrometric techniques for identification of early cancers (10–16); there is concern for false discovery and unwarranted generalizability (17) arising from the analysis of data using methods such as SELDI-TOF mass spectrometry and other multiplex assays for the detection of disease (18, 19). In response to such controversy and because of the desire to evaluate the potential clinical utility of such methodologies, the National Cancer Institute Early Detection Research Network (EDRN) decided that a rigorous validation study should be undertaken. The validation study (20) was divided into 3 stages and was specifically targeted at evaluation of the previously published EDRN study for the detection of PCa (1). In stage 1, a group of 6 institutions reported that SELDI-TOF mass spectrometry instruments at separate sites could be accurately standardized over a 3-month period and could be used to accurately classify previously studied PCa patient and control sera using known spectral features (21). Also in stage 1, the intention was to test if the same algorithm could discriminate between cancer and noncancer samples derived from independent and geographically distinct nonoverlapping populations. The development of the original algorithm and the independent testing are the subject of this report. We present this study and a companion report as a model for biomarker validation studies.
Materials and Methods
SAMPLE SELECTION
We identified 194 PCa patients, 216 patients with benign prostatic hyperplasia (BPH), and 1326 healthy control patients from samples collected at EVMS under a protocol approved by the EVMS Institutional Review Board and sent to the EDRN Data Management and Coordinating Center. Some patient samples were excluded for the following reasons: PCa—race not African-American or white (n = 2), age >80 years (n = 4), 1st available sample collected after biopsy (n = 3), and insufficient volume (n = 4); BPH—race not African-American or white (n = 42), age >80 years (n = 28), and insufficient volume (n = 3); control—race not African-American or white (n = 48) and age >80 years (n = 64). All nonexcluded PCa (n = 181) and BPH (n = 143) sera were selected for inclusion in the study. In addition, sera from men with no history of PCa or BPH were identified as normal controls and were selected to match the approximate age and race distribution of men with PCa and BPH. For PCa and BPH disease conditions, the age frequency distribution over 5-year intervals was constructed by race. For each age/race group, controls were selected at random, with frequency matching the larger of the case-specific age/race frequencies. The number of normal controls selected was 220.
For the validation test set, 4 institutions provided 84 samples, 42 PCa and 42 normal controls, to be evaluated with the classifiers developed from the training data. Two institutions contributed 14 samples each and 2 contributed 28 samples each. Each institution provided an equal number of PCa and normal control samples. All samples were collected following strict standard operating procedure (20). Balance between PCa and normal control sample collection within each contributing bio-repository produced consistent sample collection methods for case and control samples despite imbalance in the number of samples from different centers.
We divided all samples into 18 aliquots of 30 µL and distributed the aliquots evenly to 6 laboratories having SELDI-TOF mass spectrometry systems that had been optimized/standardized as reported (21). The 6 participating sites were the University of Texas Health Center at San Antonio (CTRC), University of Pittsburgh Cancer Institute (UPCI), Johns Hopkins University Medical Center (JHU), Center for Prostate Disease Research (CPDR), University of Alabama Birmingham (UAB), and EVMS. Each laboratory received 252 aliquots. In addition, 36 aliquots from a pooled reference serum sample were sent to each laboratory for quality control monitoring. Each laboratory spotted the 252 specimen and 36 pooled serum samples on 36 IMAC-3 ProteinChips® (Ciphergen Biosystems), with either 3 case and 4 control or 4 case and 3 control aliquots per chip along with 1 reference sample. The position of each aliquot was randomized separately for each laboratory, so PCa and normal control aliquots occurred in every well with equal probability. Sample replicates (aliquots) were treated as individuals in randomization. Each laboratory ran the 36 chips in 3 bio-processors. Each laboratory used separate calibrations to convert time-of-flight to mass/charge (m/z) values.
MASS SPECTROMETRY ANALYSIS
All serum processing steps were performed robotically with a Biomek 2000 Workstation liquid handling robot (Beckman Instruments) exactly as described by Semmes et al. (21).Mass accuracy was calibrated externally using the All-in-1 peptide molecular weight standard (Ciphergen Biosystems).
DATA PROCESSING
Spectral degradation was apparent for some samples. Before further analysis, all spectra were examined and assessed regarding spectral quality. A logistic regression was fitted with visual assessment of spectrum degradation as the response and 3 spectrum-specific predictor variables: a) standard deviation of spectral values, b) autocorrelation of spectrum values measured by the Durbin-Watson statistic, and c) maximum spectral intensity. The model for spectral degradation yielded specificity 95% for sensitivity 95%. Because the probability model for spectral degradation is based on measurable criteria, we used the modeled probability to exclude spectra from classifier development and testing. Classifying all samples with modeled probability of spectrum degradation greater than P = 0.1 as degraded, 51 spectra (4.53%) were removed from the training data sets and 12 (2.47%) were removed from the test set. In most cases, only a single replicate was eliminated from any sample. However, all replicates for 2 control group samples were excluded, leaving n = 152 control samples for classifier development.
A test data set of 30% of samples in each group was chosen at random, and we used the remaining 70% to construct a disease status classifier. We identified signal peak locations using methods described in Yasui et al. (22). We subjected validation spectral data to baseline subtraction and total ion content normalizations and then sent them to the Data Management and Coordinating Center for subsequent analysis. Peak intensities over the interval m/z* (1 ± 0.002) were constructed for all 2570 m/z values returned by the Yasui algorithm. We used a larger window for these secondary test data to account for slight differences between the SELDITOF mass spectrometry instruments in the participating laboratories.
In addition, we used wavelet decomposition of the mass spectrometry signal to identify and measure peak intensities. Wavelet decomposition, which measures local rate of change in intensity rather than local intensity, has been shown to be effective at peak identification on shoulders of dominant peaks (23).
DATA ANALYSIS
Using boosted logistic regression, we constructed 2 classifiers separating PCa from normal controls with the training data from EVMS. The first classifier used median peak intensity across replicates within samples at each peak alignment value as candidate predictor variables. To construct the second classifier, peak intensities were ranked within spectra and ranks binned into 100 levels. We computed median bin values across replicates within samples for each aligned m/z value. These median bin values were used as candidate predictor variables for the second classifier. The second method would be more robust if between-spectrum variability were large even after normalization and baseline subtraction.
Boosting models were fitted using a 10-fold cross-validation stopping rule. We divided groups into 10 sets each, balanced with respect to the number of observations in each set. For each 10% set, the remaining 90% of the data were used to construct a boosting classifier with K terms (or K iterations), K = 1, 2, 3, …. For each of the 10 divisions of the data, we computed the misclassification error rate for the 10% of samples that were not used to construct the classifier and averaged the misclassification error rate over the 10 sets at every boosting iteration. The average misclassification error rate across the 10 cross-validation sets was used to determine the number of iterations (M) necessary to achieve a best model. The boosted logistic regression model was then computed employing all data in the training set, stopping after M iterations.
For evaluation, we scored the 30% test set data using both classifiers, with positive values of the boosting linear predictor indicating cases and negative values indicating controls. For the 30% test set, we computed sensitivity and specificity of the classifier. The validating test data were scored with the same classifiers. We also examined the sensitivity and specificity for data generated from each laboratory.
Results
CONSTRUCTION OF CLASSIFIER FOR DISCRIMINATING PROSTATE CANCER
After peak selection, we subjected the data to 2 classifier development approaches: the first used median peak intensities and the second used median binned peak ranks. The boosting cross-validation error rates decreased through 3 iterations, with a final cross-validation error rate of 25% for the classifier constructed from median peak intensities. The experimental m/z values for the 3 peaks included in the classifier were 7775.93, 3651.38, and 3246.57, listed in order of entry into the classifier. In cross-validation, we observed sensitivity 71% and specificity 79%. The classifier constructed from median binned peak ranks required only 2 iterations to achieve a minimum cross-validation error rate of 23%. The m/z values for the 2 peaks included in the classifier were 5943.44 and 3449.77. Cross-validation of this set yielded sensitivity 63% and specificity 89%. It was expected that the cross-validation error rates were somewhat optimistic. When the 2 classifiers were used to predict status in the 30% test data set, the misclassification error rates were 27% and 28%, respectively. For the classifier constructed using median intensities, we observed sensitivity 59% and specificity 85%. The classifier constructed from median binned peak ranks had sensitivity 57% and specificity 82%.
ANALYSIS OF DATA FOR SOURCES OF BIAS
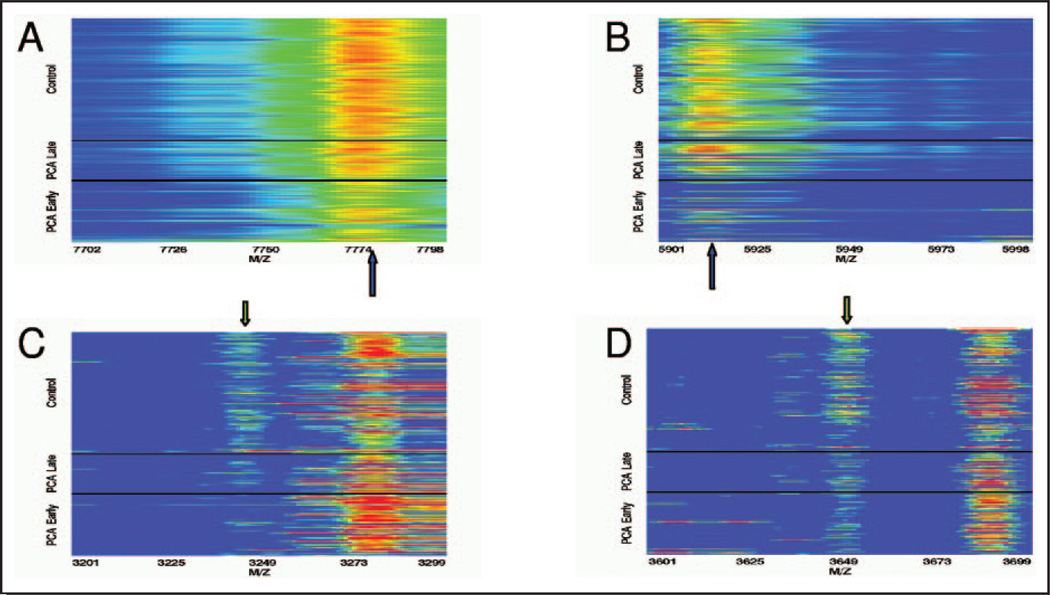
Postexperimental analysis can reveal hidden bias by evaluating the overall detail of collected data. To identify potential bias, we constructed spectral intensity heat maps with spectra arranged with respect to sample characteristics such as case status and specimen collection date within case status. In this analysis, we observed differences in mass spectroscopy profiles between prostate cancer cases collected before and after 1996. Heat maps of the mass spectroscopy profiles around primary peaks 7775.93 and 5943.44, which were dominant features for classification, suggested that the PCa cases collected before 1996 have considerably different spectral profiles from those collected in 1996 or later (Fig. 1A and B). For the 1st peaks that enter into each classifier, PCa cases collected after 1996 appear to have spectral profiles more similar to normal control samples, which were all collected after 1995. We also observed that overall higher intensities were associated with normal control and recent PCa cases compared with older PCa cases. The fact that older cases contained lower intensities and were all cancers was strong evidence of sample bias. Interestingly, this was not the case for the secondary peaks (Fig. 1C and D). Peaks that entered the classifiers in the 2nd or 3rd boosting iteration exhibited little difference between PCa samples collected before and after 1996. When we compiled all potential confounding aspects of the sample collection (see Table 1), we uncovered some disparities in time of storage (reflected as date of collection) and the number of freeze-thaws.
Fig. 1. Serum spectra profiles in the vicinity of decision peaks.
A. Primary peak for the classifier based on median intensities. B. Primary peak for the classifier based on median binned rank intensities. C. Secondary peak for the classifier based on median intensities. D. Secondary peak for the classifier based on median binned rank intensities. The arrows indicate the peak of interest. PCa and normal control serum specimens with early and late collection periods for the PCa specimens are indicated. Specimens are ordered by collection date within each group.
Table 1.
Known characteristics of the serum specimens used for training the classifier.
| Characteristic | Dx Group | ||
|---|---|---|---|
| Normal | PCa | BPH | |
| n | 220 | 181 | 143 |
| Mean age, years (SD) | 62.5 (6.6) | 60.7 (6.7) | 65.8 (5.8) |
| Year blood collected, % | |||
| 1980–1989 | 0.0 | 8.3 | 0.0 |
| 1990–1995 | 1.4 | 52.5 | 90.2 |
| 1996–2001 | 98.6 | 39.2 | 9.8 |
| Race, % | |||
| White | 82.7 | 79.6 | 93.0 |
| African-American | 17.3 | 20.4 | 7.0 |
| Neoadjuvant treatment, % | |||
| 0 | 100.0 | 85.6 | 99.3 |
| 1 | 0.0 | 11.6 | 0.0 |
| Data missing | 0.0 | 2.8 | 0.7 |
| Source, % | |||
| DTU Clinic | 0.0 | 88.4 | 0.0 |
| Sentara Hospital | 100.0 | 8.8 | 94.4 |
| Data missing | 0.0 | 2.8 | 5.6 |
| Estimated freeze-thaw, % | |||
| 1 or 2 | 63.2 | 25.4 | 57.3 |
| >2 | 35.4 | 68.7 | 22.2 |
| Data missing | 1.4 | 5.9 | 20.4 |
EVALUATION OF CLASSIFICATION ROBUSTNESS
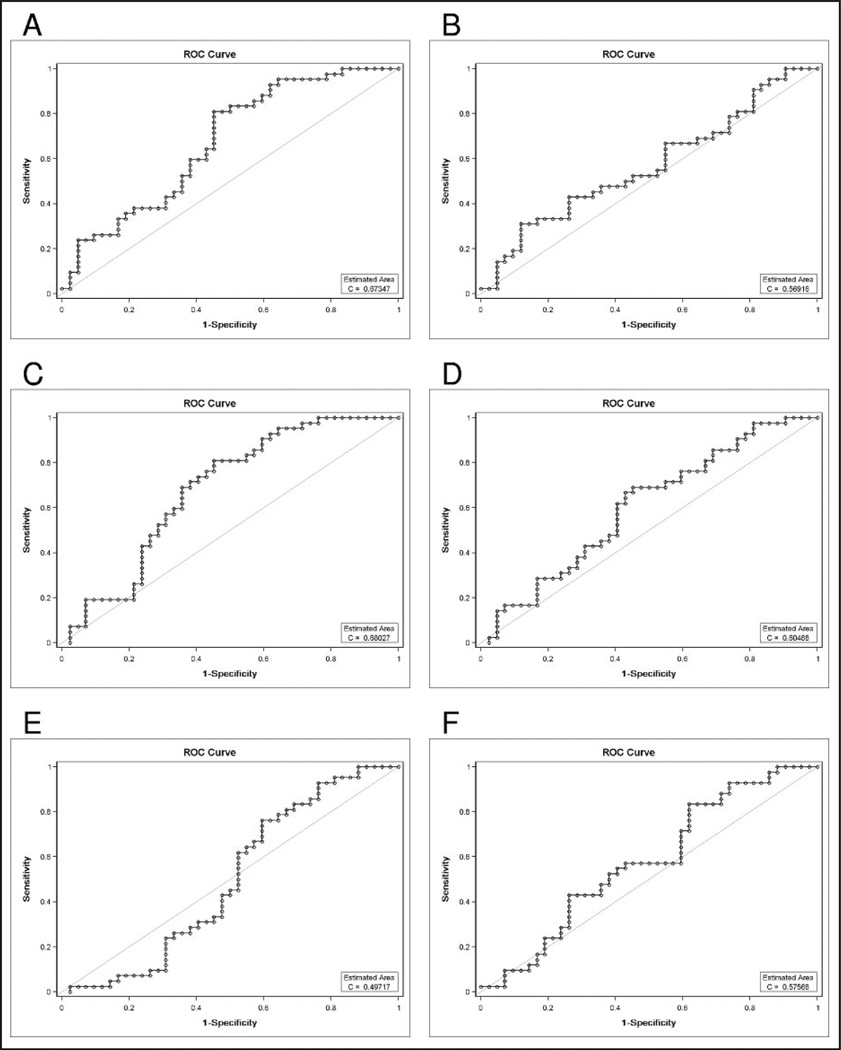
Similarity in secondary peak intensity values between pre- and post-1996 PCa samples suggests there might be some ability to discriminate between PCa and normal control samples in the independent 84-sample test set collected from the 4 biorepositories. Therefore, we performed all subsequent analysis both with and without the pre-1996 data. We will refer to the initial data set as study A and after removal of the pre-1996 spectra as study B. We performed the same classifier construction approaches on study A and study B; because of space limitations, the results of study B are included as supplemental data. Fig. 2 displays ROC curves showing the utility of the classifier constructed from median intensities in predicting cancer status in study A. For Pittsburgh, the best point along the ROC curve produces 58.3% correct classification. Both EVMS and CTRC achieve 67.9% correct prediction at the best point along the ROC curve. Across the 6 laboratories, the average maximum correct prediction probability is 62.8%. The median intensity classifier from study A has significant ability to predict cancer status only for the 2 laboratories EVMS and CTRC. ROC curves for the median binned rank classifier approach in study A (see Supplemental Data Fig. 2) demonstrate similar classifier function, with a mean across the 6 laboratories of 64.6%.
Fig. 2. ROC curves for the study A median intensity boosting classifier based on peak intensities obtained using the Yasui method for predicting prostate cancer status in 42 PCa and 42 normal control serum specimens collected from 4 biorepositories and processed by SELDI-TOF-MS instruments at 6 EDRN laboratories: EVMS (A), UAB (B), CTRC (C), CPDR (D), UPCI (E), and JHU (F).
ROC curves constructed for the 4 classifiers obtained when we restrict sample collection to the post-1996 time period indicate no improvement in predictive utility for the models tested, except for 1 model employing median binned rank intensity values for peak locations and peak intensities measured through wavelet detail functions (see Supplemental Data Fig. 3).
MULTILABORATORY TESTING OF THE CLASSIFIER; AGREEMENT BETWEEN LABORATORIES
We next examined the across-laboratory agreement for each classifier as applied to the test set. Again, we analyzed the data with and without the pre-1996 data (see Supplemental Data Tables 1 and 2). Laboratory agreement among the 6 sites in predicting case status is shown in Table 2. Agreement exceeds 80% in all but 1 instance (agreement between laboratories at JHU and CPDR was 78.6%). For the median intensity classifier, the association of cancer status prediction across laboratories was significant at P <0.05 (Fisher exact χ2). There was significant association at P <0.05 for the prediction of cancer status across all but 2 laboratory pairs (UAB with CTRC and UAB with JHU) when using the median binned peak ranks classifier. The high agreement between laboratories is confounded by the poor predictive ability of the models, which places constraints on the number of samples for which the prediction can differ. Both classifiers predicted the majority of samples as controls (see Table 3).
Table 2.
Percent agreement between sites in classification of 84 phase 1C samples.
| Site | Site | |||||
|---|---|---|---|---|---|---|
| EVMS | UAB | CTRC | CPDR | UPCI | JHU | |
| EVMS | — | 97.6a (90.9) | 100.0a (93.1) | 81.0b (75.4) | 97.6a (90.9) | 81.0b (75.4) |
| UAB | 85.7a (78.0) | — | 97.6a (90.9) | 83.3a (74.1) | 95.2a (88.8) | 81.0a (74.1) |
| CTRC | 82.1a (71.4) | 84.5 (81.2) | — | 81.0b (75.4) | 97.6a (90.9) | 81.0b (75.4) |
| CPDR | 86.9a (71.4) | 89.3a (81.2) | 85.7a (73.8) | — | 83.3a (74.1) | 78.6a (65.0) |
| UPCI | 88.1a (67.7) | 81.0b (75.9) | 82.1a (69.7) | 86.9a (69.7) | — | 81.0a (74.1) |
| JHU | 86.9a (74.3) | 89.3 (85.5) | 88.1a (77.1) | 90.5a (77.1) | 86.9a (72.4) | — |
Agreement on boosted logistic regression classifier fitted to peak intensities above the diagonal. Values below the diagonal are agreement based on boosted logistic regression classifier employing peak rank values. Expected probability of agreement in parentheses.
P <0.01
P <0.05.
Table 3.
Marginal probability expressed as a percent (number of serum specimens) classified as prostate cancer of 84 samples split equally between case and control.
| Model | Site | |||||
|---|---|---|---|---|---|---|
| EVMS | UAB | CTRC | CPDR | UPCI | JHU | |
| Real boosting with intensities | 3.6 (3) | 6.0 (5) | 3.6 (3) | 22.6 (19) | 6.0 (5) | 22.6 (19) |
| Real boosting with ranked peaks | 19.0 (16) | 4.8 (4) | 15.5 (13) | 21.4 (18) | 15.5 (13) | 10.7 (9) |
The classifiers were constructed from training data sets.
INTERSTUDY ANALYSIS FOR THE PRESENCE OF M/Z PEAKS THAT DISPLAY CONSISTENT DISCRIMINATORY VALUE
The classifiers constructed for the training data may select candidate predictors that are not truly predictive (type I error) and may fail to select markers that are predictive (type II error). Without a priori knowledge regarding which classifiers to investigate among more than a thousand candidates, the probability of committing either type I or type II error is high. Accordingly, we investigated the predictive utility in the validation study data for a set of candidate markers with the best marginal predictive utility in the training data.
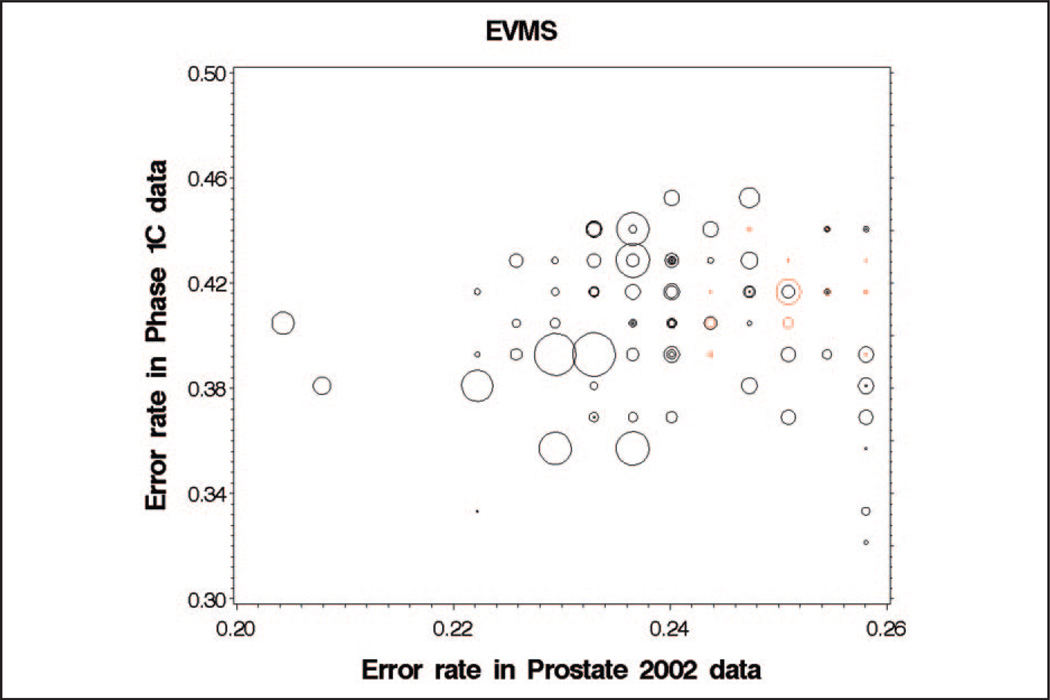
We plotted training data prediction error rates against validation study error rates, allowing sample-specific cut points for classifying diseased and nondiseased groups. Markers that have inconsistent effect-direction (e.g., mean in the PCa group is higher than mean in the normal controls in the training data but lower in the data of the confirmation study) are plotted in red (Fig. 3). Markers that have consistent effect-direction are plotted in black. We examined 96 candidate markers with the best predictive utility in the training data. To account for differential sample selection and laboratory effects, sample-specific cut points were allowed for separating PCa from normal controls. In addition to plotting consistent and inconsistent markers with different color symbols, symbol size is constructed proportional to marker mean intensity in the training data (Fig. 3).
Fig. 3. Classification error rate in the validation data for 96 peak locations with the lowest classification error rate in the Prostate 2002 training data.
Mean peak intensity in the training data is indicated by the size of each bubble, with higher intensity peaks having larger bubbles. Black bubbles indicate peak features where the effect direction is consistent between training and validation data (cases have higher intensity than controls in both data sets or cases have lower intensity than controls in both data sets). Red bubbles indicate peaks where the effect direction is inconsistent.
Among the 96 markers with best predictive utility, the number of markers with inconsistent effect-direction was approximately 15 (EVMS 14, UAB 15, CPDR 14, CTRC 18, UPITT 15, JHU 15). If we assume that results for the 6 laboratories in the validation sample are independent of one another, the number of markers that have 5 or 6 consistent observations across the validation laboratories have an expected value of 10.5 (χ2 458.77, df=1, P <0.0001). We observed 76 markers with 5 (n = 11) or 6 (n = 65) consistent observations that were significantly different from the expected value of 10.5. Assuming that 5 of 6 laboratories with consistent direction is indicative of a consistent effect, observed consistency is still much greater than chance (χ2 32.67, df=1, P <0.0001). Among the 54 markers with best performance in the training data, 53 were consistent across 5 or 6 laboratories. The number of markers that were inconsistent across training and evaluation data sets was between 1 and 2 (EVMS 0, UAB 3, CPDR 1, CTRC 2, UPITT 1, JHU 1).
Discussion
The algorithm described in this study is identical to the one used in our early analysis of analytical reproducibility (21). In that study, we demonstrated that the algorithm could correctly differentiate between PCa and control samples when those samples were derived from the same patient cohort used to develop the algorithm. Over the intervening time period, we maintained instrument output optimization by weekly calibration to 3 serum reference peaks as described (21). Thus, the low probability of correctly predicting prostate cancer among the 84 test set samples indicates that the initial samples collected for training the classifier differed markedly from the 84 samples collected for evaluating consistency of case assignment. Further investigation of the samples collected for developing the classifier revealed that the collection period for PCa samples was considerably different from the collection period for normal control samples. Of the 127 PCa samples used to construct the classifier, 78 (61.4%) were collected before 1996. In contrast, only 1 of the normal control samples (0.7%) was collected before 1996. This storage bias was obvious in retrospect. However, when designing discovery studies aimed at distinguishing between early less aggressive cancer vs late/aggressive cancer, this storage bias may be difficult to avoid. Specifically, in the case of prostate cancer, the incidence of advanced disease (Gleason >8) has dramatically reduced to the point that it is difficult to accrue significant numbers of high-grade disease at a single clinical site. Other disease models can be expected to present different but equally cryptic challenges as clinical treatment options change.
Our analysis uncovered possible sources of storage time variability that arose from different collection protocols. Specifically, many samples collected before 1996 were derived from in-house PSA testing conducted at EVMS. In the intervening years, the standardization of laboratory tests for PSA resulted in a reduction in the number of serum samples derived from testing in-house. This change resulted in the reduction of freeze-thaw cycles in the majority of samples post-1996 from 1 cycle to 0 cycles. Also of critical impact to this study was the decline in the numbers of patients with advanced vs early PCa, a trend being experienced wherever PSA testing is aggressively employed. Thus, researchers can expect that increased demand for sample numbers will be especially affected by decreasing sample availability and variations in the storage process. As we learn more about the ideal conditions for both collection and storage of samples, even more changes may be introduced, and these changes might introduce new bias. These are critical issues often overlooked in the biomarker discovery process that are likely to be the single greatest reason most biomarker discoveries fail to be validated.
Differences associated with sample age may result from serum degradation associated with storage time or freeze-thaws. As has been described before, these and other preanalytical variables can greatly affect the peptide/protein content of samples (24–30). At peaks used in the 2 classifiers developed for this study, PCa samples collected after 1996 appeared to be more similar to normal controls. Also of consideration is the possibility that clinical protocols such as a decision to collect serum after voiding or fasting may become standardized without notification to the research group. A particularly insidious issue is the struggle with “improving” or “standardizing” collection protocol based on knowledge gained regarding sample stability. Although a close relationship between the clinic and the research laboratory helps reduce some concerns, even careful practice cannot prevent variability. Thus, it is recommended that poststudy data analysis such as was performed here be an integral component of biomarker discovery and validation. In fact, the collective experiences of many laboratories leading human biomarker discovery efforts has led to calls for both experimental standards (31) and uniformity in sample preparation (32).
Because our global analysis of the data suggested potential discriminating elements in the post-1996 data, we decided to construct a new decision algorithm. Additionally, our study population identified for stage 2 of the overall validation process was derived from multiple laboratory sites, imposing stricter sample storage criteria including a requirement for post-2002 collection date. Accordingly, such a study was initiated and these results are reported in the companion article.
Acknowledgments
Grant/funding Support: This work was supported by the Early Detection Research Network, National Cancer Institute Grant CA084986 (to O.J.S.) and Grants CA86402 (to I.M.T.), CA84968 (to W.L.B), CA86368 (to Z.F.), CA86359 (to W.E.G.), CA85067 (to O.J.S.), and CA86323 (to A.P.).
Footnotes
Nonstandard abbreviations: EVMS, Eastern Virginia Medical School; PCa, prostate cancer; PSA, prostate-specific antigen; EDRN, National Cancer Institute Early Detection Research Network; BPH, benign prostatic hyperplasia; CTRC, University of Texas Health Center at San Antonio; UPCI, University of Pittsburgh Cancer Institute; JHU, Johns Hopkins University Medical Center; CPDR, Center for Prostate Disease Research; UAB, University of Alabama Birmingham.
Financial Disclosures: None declared.
References
- 1.Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- 2.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 3.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 4.Petricoin EF, 3rd, Ornstein DK, Paweletz CP, Ardekani A, Hackett PS, Hitt BA, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst. 2002;94:1576–1578. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- 5.Rosty C, Christa L, Kuzdzal S, Baldwin WM, Zahurak ML, Carnot F, et al. Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res. 2002;62:1868–1875. [PubMed] [Google Scholar]
- 6.Vlahou A, Laronga C, Wilson L, Gregory B, Fournier K, McGaughey D, et al. A novel approach toward development of a rapid blood test for breast cancer. Clin Breast Cancer. 2003;4:203–209. doi: 10.3816/cbc.2003.n.026. [DOI] [PubMed] [Google Scholar]
- 7.Vlahou A, Schellhammer PF, Mendrinos S, Patel K, Kondylis FI, Gong L, et al. Development of a novel proteomic approach for the detection of transitional cell carcinoma of the bladder in urine. Am J Pathol. 2001;158:1491–1502. doi: 10.1016/S0002-9440(10)64100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu Y, Adam BL, Yasui Y, Ward MD, Cazares LH, Schellhammer PF, et al. Boosted decision tree analysis of surface-enhanced laser desorption/ionization mass spectral serum profiles discriminates prostate cancer from noncancer patients. Clin Chem. 2002;48:1835–1843. [PubMed] [Google Scholar]
- 9.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 10.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 11.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 12.Diamandis EP. Peptidomics for cancer diagnosis: present and future. J Proteome Res. 2006;5:2079–2082. doi: 10.1021/pr060225u. [DOI] [PubMed] [Google Scholar]
- 13.Diamandis EP. Validation of breast cancer biomarkers identified by mass spectrometry. Clin Chem. 2006;52:771–772. doi: 10.1373/clinchem.2005.064972. author reply 2. [DOI] [PubMed] [Google Scholar]
- 14.Grizzle W, Semmes O, Bigbee W, Zhu L, Malik G, Oelschlager D, Manne B. The need for the review and understanding of SELDI/MALDI mass spectroscopy data prior to analysis. Cancer Informatics. 2005;1:86–97. [PMC free article] [PubMed] [Google Scholar]
- 15.Hortin GL, Jortani SA, Ritchie JC, Jr, Valdes R, Jr, Chan DW. Proteomics: a new diagnostic frontier. Clin Chem. 2006;52:1218–1222. doi: 10.1373/clinchem.2006.067280. [DOI] [PubMed] [Google Scholar]
- 16.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 18.Grizzle W, Semmes O, Bigbee W, Malik G, Miller E, Manne B, et al. Use of mass spectrographic methods to identify disease processes. In: Patrinos G, Ansorg W, editors. Molecular Diagnosis. Vol. 17. 2005. pp. 211–222. [Google Scholar]
- 19.Sharp V, Utz PJ. Technology insight: can autoantibody profiling improve clinical practice? Nat Clin Pract Rheumatol. 2007;3:96–103. doi: 10.1038/ncprheum0404. [DOI] [PubMed] [Google Scholar]
- 20.Grizzle WE, Adam BL, Bigbee WL, Conrads TP, Carroll C, Feng Z, et al. Serum protein expression profiling for cancer detection: validation of a SELDI-based approach for prostate cancer. Dis Markers. 2003;19:185–195. doi: 10.1155/2004/546293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 22.Yasui Y, McLerran D, Adam BL, Winget M, Thornquist M, Feng Z. An automated peak identification/ calibration procedure for high-dimensional protein measures from mass spectrometers. J Biomed Biotechnol. 2003;2003:242–248. doi: 10.1155/S111072430320927X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randolph TW, Yasui Y. Multiscale processing of mass spectrometry data. Biometrics. 2006;62:589–597. doi: 10.1111/j.1541-0420.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- 24.Banks RE, Stanley AJ, Cairns DA, Barrett JH, Clarke P, Thompson D, Selby PJ. Influences of blood sample processing on low-molecularweight proteome identified by surface-enhanced laser desorption/ionization mass spectrometry. Clin Chem. 2005;51:1637–1649. doi: 10.1373/clinchem.2005.051417. [DOI] [PubMed] [Google Scholar]
- 25.Drake SK, Bowen RA, Remaley AT, Hortin GL. Potential interferences from blood collection tubes in mass spectrometric analyses of serum polypeptides. Clin Chem. 2004;50:2398–2401. doi: 10.1373/clinchem.2004.040303. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh SY, Chen RK, Pan YH, Lee HL. Systematical evaluation of the effects of sample collection procedures on low-molecular-weight serum/plasma proteome profiling. Proteomics. 2006;6:3189–3198. doi: 10.1002/pmic.200500535. [DOI] [PubMed] [Google Scholar]
- 27.Karsan A, Eigl BJ, Flibotte S, Gelmon K, Switzer P, Hassell P, et al. Analytical and preanalytical biases in serum proteomic pattern analysis for breast cancer diagnosis. Clin Chem. 2005;51:1525–1528. doi: 10.1373/clinchem.2005.050708. [DOI] [PubMed] [Google Scholar]
- 28.Timms JF, Arslan-Low E, Gentry-Maharaj A, Luo Z, T’Jampens D, Podust VN, et al. Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem. 2007;53:645–656. doi: 10.1373/clinchem.2006.080101. [DOI] [PubMed] [Google Scholar]
- 29.Traum AZ, Wells MP, Aivado M, Libermann TA, Ramoni MF, Schachter AD. SELDI-TOF MS of quadruplicate urine and serum samples to evaluate changes related to storage conditions. Proteomics. 2006;6:1676–1680. doi: 10.1002/pmic.200500174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West-Nielsen M, Hogdall EV, Marchiori E, Hogdall CK, Schou C, Heegaard NH. Sample handling for mass spectrometric proteomic investigations of human sera. Anal Chem. 2005;77:5114–5123. doi: 10.1021/ac050253g. [DOI] [PubMed] [Google Scholar]
- 31.Mischak H, Apweiler R, Banks RE, Conaway M, Coon J, Dominiczak A, et al. Clinical proteomics: a need to define the field and to begin to set adequate standards. Proteomics Clin App. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 32.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]