Summary
Apolipoproteins are key structural elements of lipoproteins and critical mediators of lipid metabolism. Their detergent-like properties allow them to emulsify lipid or exist in a soluble lipid-free form in various states of self-association. Unfortunately, these traits have hampered high-resolution structural studies needed to understand the biogenesis of cardioprotective high-density lipoproteins (HDL). We derived a crystal structure of the core domain of human apolipoprotein (apo)A-IV, an HDL component and important mediator of lipid absorption. The structure at 2.4 Å depicts two linearly connected 4-helix bundles participating in a helix swapping arrangement that offers a clear explanation for how the protein self-associates as well as clues to the structure of its monomeric form. This also provides a logical basis for antiparallel arrangements recently described for lipid-containing particles. Furthermore, we propose a “swinging door” model for apoA-IV lipid association.
Lipid transport in blood is accomplished by lipoproteins, nanometer-scale emulsions of lipid and protein that solve the fundamental oil-in-water solubility problem. One lipoprotein class, high-density lipoproteins (HDL), has been intensively studied due to its association with various cardioprotective functions (recently reviewed in (Meurs et al., 2010)). HDL proteins, called apolipoproteins, include (apo)A-I, apoA-II, apoA-IV, and many less abundant species (Vaisar et al., 2007;Gordon et al., 2010b;Gordon et al., 2010a). Many of these can interconvert between a soluble lipid-free state and the lipid-bound form, with the lipid-free form able to interact with the cell surface ATP binding cassette transporter (ABCA1) to produce HDL particles (Rust et al., 1999; Lawn et al., 1999; Brooks-Wilson et al., 1999; Remaley et al., 2001a). Unfortunately, the details of this process are poorly understood, largely because the basic structures of both the lipid-free and -bound forms of most HDL apolipoproteins have not been clearly determined.
In terms of lipid-bound HDL apolipoproteins, much of our structural understanding has come from the study of well-defined recombinant HDL (rHDL) particles created in vitro (reviewed in (Davidson et al., 2007)). The heaviest focus has been on apoA-I, which adopts a ‘double-belt’ orientation in which two apoA-I molecules wrap around a patch of phospholipid bilayer in an antiparallel fashion (Koppaka et al., 1999; Segrest et al., 2000). The overall tenets of this model have been supported by numerous experimental studies, including chemical cross-linking that confirmed the general inter-molecular alignments of the resident apoA-I molecules (i.e registry)(Davidson et al., 2003;Bhat et al., 2005;Silva et al., 2005b). Several refinements to the model have been proposed that share the basic registry and antiparallel orientation (Bhat et al., 2007;Martin et al., 2006;Wu et al., 2007). Despite this understanding, fundamental questions remain as to how the lipid-free forms transition from a structurally unknown lipid-free state to this antiparallel arrangement in lipoprotein particles. Unfortunately, progress on a high-resolution structural understanding of the lipid-free forms of HDL proteins has been slow. Prior to last year, only three credible high-resolution structures for human lipid-free apolipoproteins had been reported (Wilson et al., 1991;Sivashanmugam et al., 2009;Borhani et al., 1997). These have been helpful for understanding some aspects of apolipoprotein function, though critical questions remain. The difficulties in determining these structures can be attributed to their tendency to form heterogeneous oligomeric arrays in the lipid-free state. The functional role of this self-association, as well as its structural basis, has been questioned for nearly 30 years. Recently, Mei and Atkinson reported the crystal structure of the N-terminal 184 residues of lipid-free human apoA-I (Mei et al., 2011). The structure showed a half-circle dimer with two helical bundles connected by shared antiparallel helices. This “helix swapping” motif offered important structural clues to the mechanism for apoA-I self association and also a logical mechanism for lipoprotein particle formation. However, the applicability of this motif and mechanism to other members of the exchangeable apolipoprotein family is not known.
Human apolipoprotein (apo)A-IV is the largest apolipoprotein (MW of 46 kDa). It is made in the gut and has been associated with lipid absorption and chylomicron assembly (Stan et al., 2003). As a prominent component of HDL, apoA-IV has also been proposed to be a mediator of reverse cholesterol transport (Emmanuel et al., 1994;Duverger et al., 1994;Main et al., 1996;Remaley et al., 2001b), an antioxidant (Qin et al., 1998), a satiety factor (Tso et al., 2001), a mediator of gut inflammation (Vowinkel et al., 2004), and has recently been linked to β-amyloid clearance in the brain (Cui et al., 2011). ApoA-IV shares several structural characteristics with apoA-I and other exchangeable apolipoproteins, including putative amphipathic α-helical repeats that are critical for lipid binding and self-association (Segrest et al., 1994). Here, we describe the X-ray crystal structure of a stable core domain of human apoA-IV. Although the apoA-IV structure is consistent with the dimer arrangement observed in the N-terminal apoA-I structure, including a similar helix-swapping motif, significant structural differences are apparent in the composition of the helical bundles. Analysis of the structure suggests plausible molecular explanations for the structure of monomeric apoA-IV and may provide important hints for how apoA-IV may bind lipids.
Results
X-ray structure of dimeric apoA-IV64-335
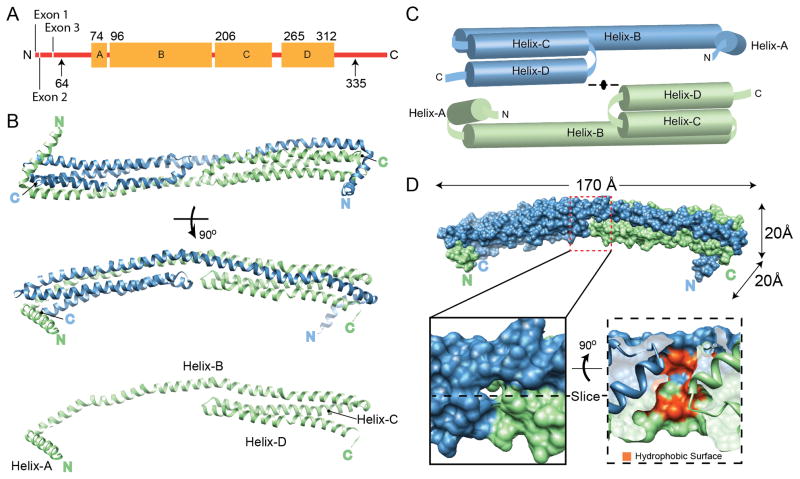
Numerous attempts to crystallize full-length apoA-IV have thus far failed to provide diffraction quality crystals. However, we previously showed that lipid-free apoA-IV exhibits a compartmentalized structure with a large, well-folded central core and less organized N- and C-termini that likely interact (Tubb et al., 2007). Using limited proteolysis, we localized the core domain to residues 64-335 (Tubb et al., 2008), and isolated dimers of this modified protein formed diffraction quality crystals. The crystal structure of this domain was determined to 2.4 Å resolution by MAD with two molecules in the asymmetric unit (Table 1). The model consisted of residues 74-312 but lacks interpretable electron density for 10 and 23 residues at the N- and C-termini, respectively (Figure 1A). The structure revealed a long rod-like dimer (170 Å L × 20 Å H × 20 Å W) consisting of two 4-helix bundles stacked end-to-end in opposing orientations (Figure 1B). There is a progressive right-handed twist that turns the structure by 90° over the length of the rod. The helical hydrophobic faces are directed toward the center of the bundles as seen in other exchangeable apolipoproteins (Wilson et al., 1991;Wang et al., 2002). The monomers, each consisting of 4 helices (labeled A–D), are related by 2-fold symmetry across an axis perpendicular to the rod (Figure. 1C). The N-terminal Helix-A (74-94) runs perpendicular to the rod and caps the outer end of the helical bundle. Helix-B (96-204) is extremely long, spanning the entire length of the dimer. Following an 180° turn, Helix-C (206-255) and Helix-D (265-312) form antiparallel helical arms of ~80 Å in length and complete the other half of the bundle. The loops connecting Helix-C and D (256-264, C–D loop) in each monomer oppose each other at the center of the rod, causing a break in the hydrophobic core. This results in a large cavity (>700 Å3) (Dundas et al., 2006) that is lined at the basin with exposed hydrophobic residues from Helix-B of each monomer and topped with small polar residues from the C–D loop (Figure 1D and S1). The N- and C- termini from opposing chains are in proximity at each end of the rod, consistent with an intermolecular interaction that we proposed previously (Tubb et al., 2007). Each 4-helix bundle in the dimer is composed of three helices from one chain with the fourth stretching across from the opposing chain. This interlocking interaction offers an immediate explanation for apoA-IV dimerization with the interface burying ~5,900 Å2 of predominately hydrophobic surface on each monomer. In fact, relative to the quaternary interfaces of other protein assemblies, the ratio of buried surface area at the dimer interface to total solvent accessible surface area is exceptionally high (Ponstingl et al., 2004).
Table 1.
Data collection, phasing and refinement statistics for MAD (SeMet)
| Native1 | SeMet1 | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6122 | |||
| Cell dimensions | ||||
| a, b, c (Å) | 70.8, 70.8, 512.4 | 71.2, 71.2, 513.4 | ||
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | ||
| Peak (E1) | Inflection (E2) | Remote (E3) | ||
| Wavelength (Å) | 1.0332 | 0.9794 | 0.9796 | 0.9494 |
| Resolution (Å) | 19.8–2.4 (2.53-2.4) | 2.8 (2.9-2.8) | 2.8 (2.9-2.8) | 2.8 (3.0-2.8) |
| Rmerge | 0.084 | 0.081 | 0.082 | 0.083 |
| Mn(I/sd) | 19.4 (1.9) | 16.5 (4.5) | 16.4 (4.3) | 17.3 (3.9) |
| Total reflection | 485,811 | 220,658 | 220,938 | 242,804 |
| Unique reflection | 31,135 | 20,321 | 20,304 | 20,353 |
| Completeness (%) | 99.2 (99.3) | 99.5 (99.8) | 99.5 (99.8) | 99.6 (99.8) |
| Redundancy | 15.4 (15.9) | 9.2 (9.5) | 9.1 (9.5) | 9.1 (9.5) |
| Refinement | ||||
| Resolution (Å) | 18.8–2.4 | |||
| Rwork (%) / Rfree2(%) | 20.0/22.9 | |||
| No. atoms | 3,959 | |||
| Protein | 3,937 | |||
| Water | 22 | |||
| B-factors | ||||
| Protein | 95.7 | |||
| Water | 93.5 | |||
| Rmsd | ||||
| Bond lengths (Å) | 0.010 | |||
| Bond angles (°) | 1.12 | |||
| Ramachandran plot | ||||
| Favored (%) | 99.37% | |||
| Outliers (%) | 0.0% | |||
Two crystals were used for data collection, one native and one SeMet.
Values in parentheses are for highest-resolution shell as defined in the resolution row.
Rfree is defined as 5% of initial total number of reflections. Rmsd bond lengths and angles are the deviations from ideal values.
Mn(I/sd) is defined as <merged<Ih>/sd(<Ih>) > ~= signal/noise
Figure 1.
Structure of apoA-IV64-335. (A) Linear diagram of human apoA-IV. Arrows indicate the extent of the protein used for crystallization (64-335). The residues that were visible in the structure (74-312) are shown with the four major helical domains (A–D) indicated as orange boxes. (B) The structure, rotated through 90°, is depicted as blue and green ribbons interacting through a helical domain swap mechanism. The green monomer is also shown in the absence of the blue monomer. (C) Schematic representation of the dimer pulled apart. (D) Space filling representation of the dimer with molecular dimensions indicated. A cavity at the center of the structure is highlighted, showing the exposure of hydrophobic core residues in red (see also Figure S1). See also Figure S8.
Structure validation
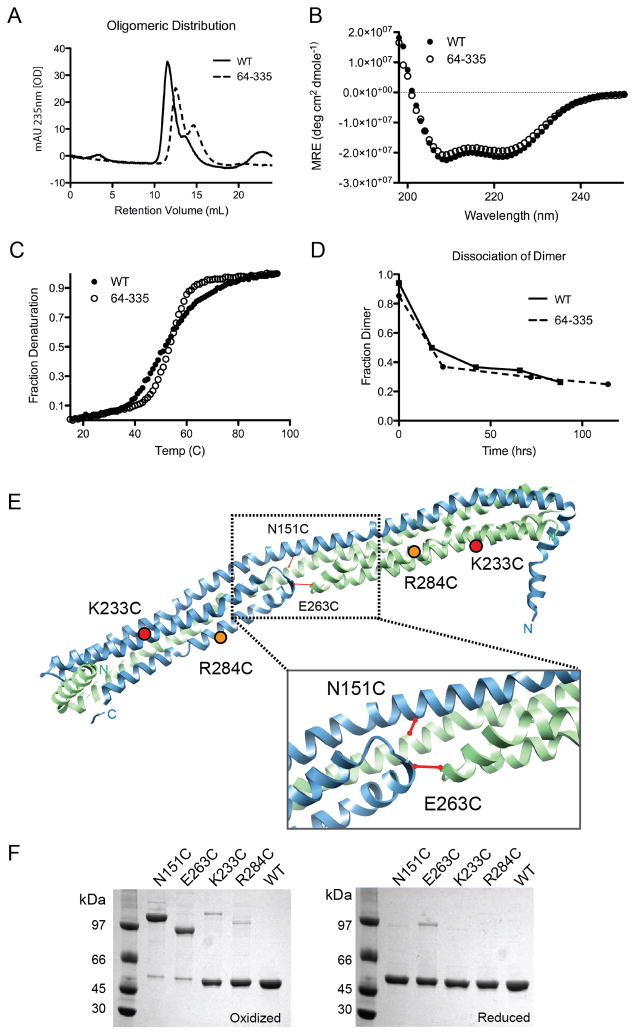
Since crystallization typically occurs at high protein concentrations and apolipoproteins tend to adopt interconverting oligomeric species, we set out to verify that the crystallization-state matched the solution-state. In solution, apoA-IVWT exists as a mixture of monomers and dimers (Pearson et al., 2004), as does apoA-IV64-335 (Figure 2A). The helix sharing structure we observed in apoA-IV64-335 and the large amount of hydrophobic surface buried at the dimer interface suggest high dimer stability with a correspondingly slow rate of dissociation. In fact, when we purified the apoA-IV64-335 dimer by size exclusion chromatography (SEC) and then reapplied the pooled dimer to the column, little to no dissociation into monomer occurred at room temperature or below (Figure S2A). This was verified by sedimentation velocity (Figure S2C). These observations demonstrated that the apoA-IV64-335 dimer is a stable species, thus making it possible for us to characterize it in solution. Circular dichroism (CD) spectroscopy of the isolated dimeric apoA-IV64-335 estimated the helical content at 83% (Figure 2B). The visible crystal structure of apoA-IV64-335 is about 87% helical. These values are in remarkable agreement, especially if one assumes that the invisible regions at the termini are largely random coil. Furthermore, lysine residues cross-linked in dimeric apoA-IV64-335 by Bis(Sulfosuccinimidyl) suberate (BS3) (Table S1) were consistent with the structure and well within the 23 Å distance cutoff of BS3. These data gave confidence that the visible crystal structure was a highly plausible model for the solution structure of dimeric apoA-IV64-335.
Figure 2.
Ability of apoA-IV64-335 to model apoA-IVWT. (A) Isolated apoA-IVWT and apoA-IV64-335 dimer incubated at 37°C in PBS for 24hrs and analyzed by SEC (dimers at 11–12 ml, monomers at 14–15 ml). (B) Far UV CD spectra of dimeric apoA-IVWT and apoA-IV64-335 at 0.85μM at 25°C. (C) Thermal denaturation of both forms as monitored by CD at 222nm. (D) Rate of dimer disassociation over time measured by SEC for both apoA-IVWT and apoA-IV64-335. (E) Introduction of single Cys point mutations (N151C, E263C, K233C and R284C) to test the dimer model. Predicted disulfide bond formation in WT apoA-IV based on the crystal structure is indicated with a solid line. (F) SDS-PAGE of point mutations of apoA-IVWT designed in (D) that were purified under reducing conditions (right) and then oxidized to allow disulfide bond formation (left), stained with coomassie blue. See also Figure S2 and Table S1.
Satisfied that the structure model recapitulates soluble apoA-IV64-335, we next evaluated how well apoA-IV64-335 served as a structural surrogate for apoA-IVWT. We found that, despite differences at the termini, the two proteins had remarkably similar characteristics. For example, both versions exhibited highly stable dimers that could be isolated by SEC and studied (Figure S2B and S2C). They exhibited similar CD spectra and thermal denaturation profiles with a Tm of 53°C ±0.5 (Figure 2B and 2C), though the WT protein exhibited slightly less cooperatively. In addition, at 37°C, the isolated dimers of both forms dissociated with similar kinetics and reached a similar monomer/dimer ratio at equilibrium (60 and 40% respectively, Figure 2D). This suggests that the majority of the amino acids responsible for the stability and self-association of apoA-IVWT are present and functional in apoA-IV64-335. Finally, Lys residues found to be cross-linkable in apoA-IVWT (Tubb et al., 2008) were highly plausible in the crystal structure of apoA-IV64-335. In total, these data indicate that the apoA-IV64-335 crystal structure is an appropriate template for making structural predictions in the full-length protein.
We further tested this by strategically introducing Cys into apoA-IVWT (which normally lacks them) and examining whether a disulfide-bonded dimer could form. Two ‘symmetrical’ amino acids coming into close contact in the center of the dimer were identified that would potentially form intermolecular disulfide linkages in the apoA-IV64-335 dimer (Figure 2E). In the first mutant, apoA-IVN151C, a Cys was introduced in the center of Helix-B at a predicted point of direct alignment across the dimer interface. In apoA-IVE263C, the Cys was placed in the C-D loop at a point of predicted alignment at the tips of the two helical arms. Figure 2F shows that after oxidation both apoA-IVN151C and apoA-IVE263C existed primarily as disulfide linked dimers in the lipid-free state when analyzed by non-reducing PAGE, consistent with the crystal structure (note: the apparent MW of the mutant dimers is likely different because of gel migration effects related to the location of the disulfide bond (Thomas et al., 2006)). Under reducing conditions, all mutants reverted to monomers, confirming that the MW differences observed in the non-reduced PAGE were due to the disulfides. In contrast, apoA-IVK233C and apoA-IVR284C, where the introduced Cys are predicted to be 94 and 71 Å apart in the crystal structure, remained mostly monomeric under oxidizing conditions (Figure 2F). Collectively, these data indicate that the crystal structure is consistent with that of soluble dimeric apoA-IV64-335 and most importantly, full-length apoA-IV.
Distinct segmentation of apoA-IV
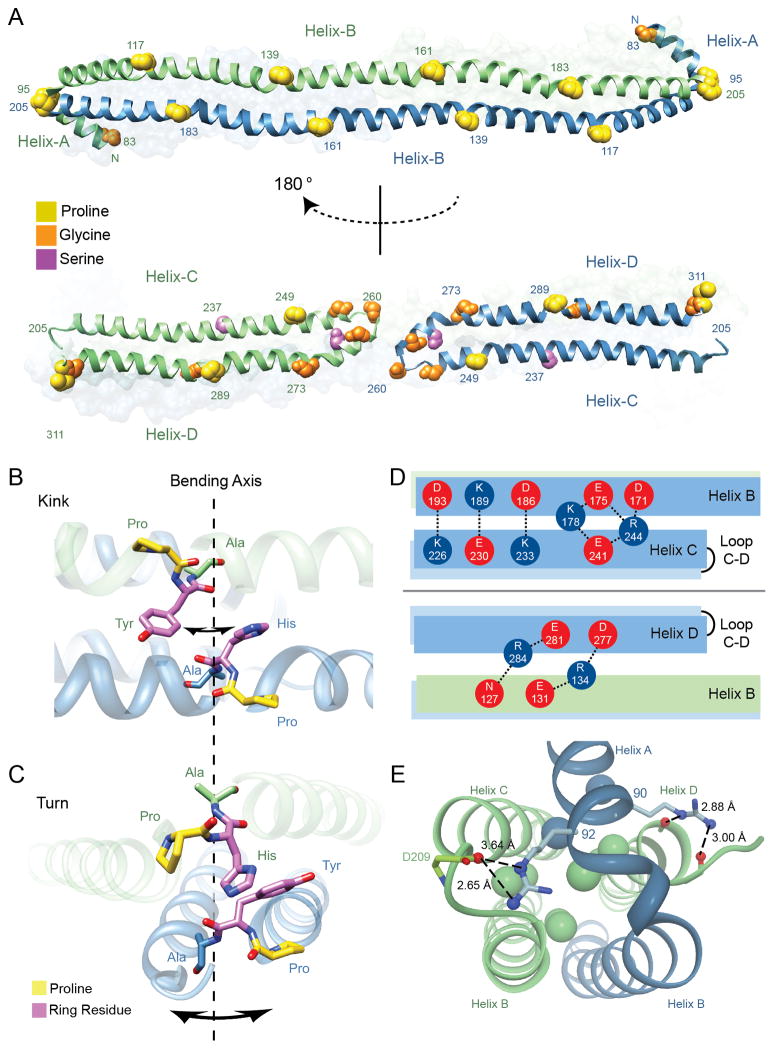
A well-recognized theme from sequence analyses of the exchangeable apolipoproteins is the repeating motif of 22 a.a. amphipathic α-helices (Segrest et al., 1994). Consistent with this paradigm, our structure shows that each monomer is composed of amphipathic helical segments separated by ‘kinks’ that are associated with helix-breaking residues, most commonly proline. Remarkably, the kinks align with the opposing monomer along the shared Helix-B and within each monomer between Helix C and D, despite their antiparallel orientation (Figure 3A). These residues are highly conserved across species (Figure S3), indicating a key structural and functional role. Six pairs of proline residues align vertically and partition the B helices into five even segments of 22 residues each (Figure 3A). A set of paired proline residues (Pro 95 and Pro 205) caps the B helices at either end. The helical arms composed of Helix C and D are also segmented, but differently than in Helix-B. Here two prolines in the helical arm domain (249 in Helix-C, and 289 in Helix-D) do not align with each other, but each aligns with a highly conserved glycine or serine residue (Figure 3A and 3S). The segmentation of Helix-C and D is flanked at one end by the turn that connects the two helices, which consists of three conserved glycine residues and Pro 311 of Helix-D at the other end. This partitions the helical arm into three segments that vary in length from 11–20 residues. Taken together, the core domain of the apoA-IV64-335 dimer is divided into 11 paired helical segments, five that are intermolecular and 6 that are intramolecular (Figure 3A).
Figure 3.
Molecular characteristics of the apoA-IV64-335 dimer. (A) Alignment of helix disrupting ‘kinks’ reveals paired segmentation. Proline, glycine and serine residues are represented by spheres and colored accordingly. (top) Helix-B from both green and blue chains run anti-parallel and conserved proline residues from each helix are in a shifted alignment, dividing the B helices into five paired segments of 22 a.a. in length. (bottom) Segmentation is also observed between Helix-C and D, where proline residues align vertically with glycine and serine. Additional glycine residues that form the Helix-C to D turn are also displayed with the central glycine (Gly 260) annotated. (B) Illustration of the P(H/Y)A motifs that align vertically across Helices-B looking down onto the two stacked helices (hydrophobic core at the top and polar exterior on the bottom). Shown are residues PHA (161-163) and PYA (139-141) from opposing B helices. The potential axis of bending and direction is indicated with arrows around dashed line. (C) Illustration of paired P(H/Y)A motifs that align in turns (95-97 linking Helix-A to B) and (205-207 linking opposing Helix-B to C). Orientation is looking down the length of the dimer rod. (D) Hydrogen bonding and salt bridges at the interface of Helix-B and helical arms (Helix-C and D). (top) Intramolecular interactions are shown between acidic (red) and basic (blue) residues looking down on the dimer rod parallel to the plane made up by the stacked B-helices. (bottom) Intermolecular interactions are shown looking up in the same plane (i.e. rotated 180°). (E) Helix-A caps the four-helix bundle at each opposing dimer end. Shown are interactions of two conserved arginine residues (90 and 92) within Helix-A that interact at the top of the bundle. Interior leucine residues are shown as spheres and colored in accordance with each monomer. See also Figures S3, S4 and S8, and Table S2.
Since prolines typically destabilize α-helix extension, it was surprising to find several distributed throughout the remarkably long Helix B, inducing only minor directional kinks (Table S2). Of the proline residues that do not cause a turn, the mean kink angle was 16° (±4°), which is less than the average reported value of 26° (Barlow et al., 1988). Certainly crystal-packing could be partly responsible for these observations, as described in further detail (Deng et al., 2012), and in Figure S4. Upon further examination of these kinks, their alignment across the helical interface revealed an interesting pattern. While the prolines were clearly associated with the kinks, they are offset from each other by approximately half of a helical turn toward the N-terminus of each helix. The actual axis of each bend was centered on a conserved alanine residue located two amino acids C-terminal to each proline (Figure 3B). The intervening residue was always a tyrosine or histidine, both large ringed residues, creating a P(Y/H)A structural motif. When two of these motifs are aligned in an antiparallel fashion, as occurs throughout the length of the shared B helices, the alanines come into direct alignment. The side chains of the alanine residues point into the bundle while those of the ringed residues are positioned on the outside (Figures 3B, 3C and S8). The disparity in atomic volumes between the ringed residues and the alanines indicates that further bending of the helix at the kink position would only be favorable in one direction. Therefore, the aligned P(Y/H)A motif appears to create a ‘door hinge’ mechanism by which the kink can only bend inward toward the hydrophobic core. As bending progresses, the small-side chains of the alanines allow them to pinch together. Movement in the opposite direction (i.e. bending away from the hydrophobic core) appears unlikely due to steric hindrances and unfavorable energetics, requiring a flip in the carbonyl geometry of each proline. The P(Y/H)A motifs situated at the ends of the bundle (residues 95-97 and 205-207) provide examples of a fully bent hinge (Figure 3C). Remarkably, all the P(Y/H)A kinks in the molecule appear designed to bend toward the hydrophobic core, irrespective of their location. This directional hinging may be important for structural transitions during lipid binding (see below).
Interhelical interactions and bundle stability
During lipid binding, the helical bundles of apolipoproteins must partially or completely transition from protein:protein interactions to protein:lipid interactions. Therefore, we examined features of the structure that offer insight into bundle stability, such as hydrophobic interactions, hydrogen bonds and salt bridges. Surprisingly, only a modest number of hydrogen bonds were observed across the chains of the dimer as compared to interfaces of other oligomeric assemblies (Figure S7). More significant were the number of salt bridges, specifically at the interface of Helix-B and C, which were clustered in two patches and an intermolecular patch that links Helix-B to D (Figure 3D). Although these patches form networks of hydrogen bonds/salt bridges, their contribution to stabilizing the bundle may be less than optimal. Due to the spatial arrangement of the charged side chains, comparable stabilizing interactions can form across helices as well as within the helix. This competition can be envisioned to reduce the free energy contribution of these interactions to the bundle stability (Figure 3D). Therefore, the hydrogen bonds and salt bridges observed in the bundle appear more favorable for aligning the helices than for stabilizing the bundle. At the N-terminus, the amphipathic Helix-A caps the ends of the bundle and is involved in a number of interhelical interactions. The hydrophobic face of Helix A wedges between Helix-C and D of the opposing molecule and is anchored by interactions coordinated through two conserved arginine residues (Arg 90 and 92) (Figure 3E and S3). This splits the ends of Helix-C and D, widening the bundle and creating a small cavity in the hydrophobic core filled with ten leucines. The loose packing of hydrophobic residues in this area could decrease the overall stability of the bundle. These features may offer an explanation for the overall low melting temperature and stability of apoA-IV (Weinberg et al., 1985a).
Mechanism for self-association
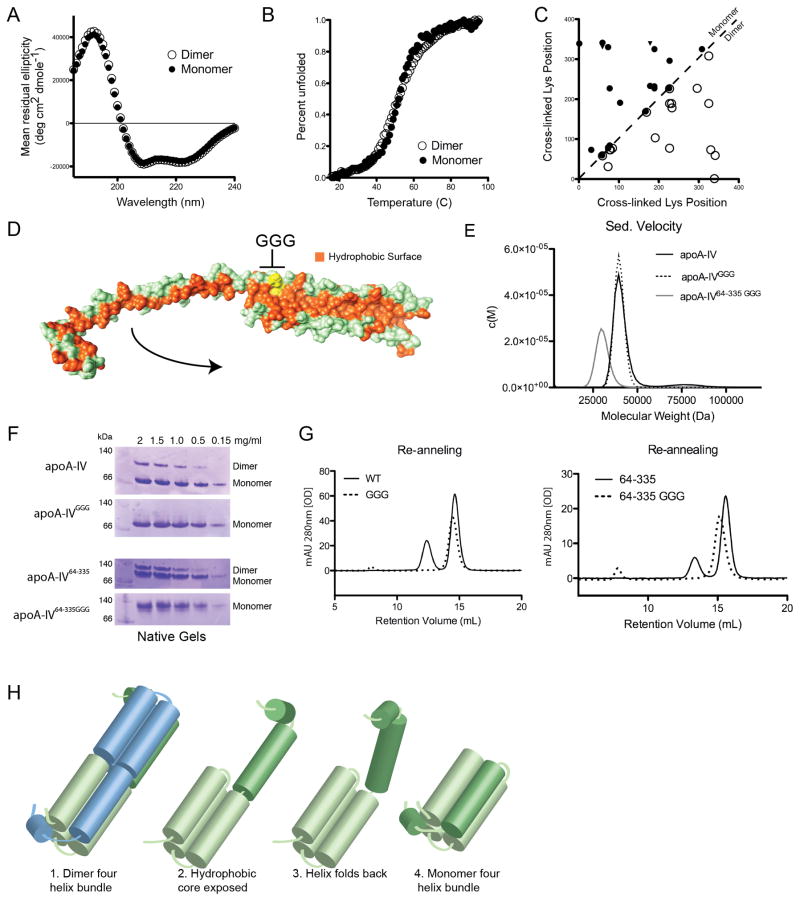
Unlike apoA-I and apoE, apoA-IV s high oligomeric stability afforded us the first opportunity to study the conformation of an apolipoprotein monomer and dimer in isolation. Figure 4A shows that the far UV CD spectra of the apoA-IVWT monomer and dimer were essentially identical, indicating a similar degree of secondary structure. Upon thermal denaturation, the two forms exhibited similar unfolding curves with equivalent Tm values of 53°C (Figure 4B). Finally, our cross-linking studies showed only minor differences in the linkage pattern between the two forms (Figure 4C). Taken together, these data strongly indicate that the overall protein fold of the monomer and dimer are highly related and make similar molecular contacts irrespective of the presence of one or two molecules.
Fig 4.
Transition of apoA-IV dimer to monomer. (A) Far UV circular dichroism (CD) spectra of the SEC isolated dimer and monomer forms. (B) Thermal denaturation of isolated dimer and monomer as monitored by CD. (C) A plot of cross-linking patterns derived from the isolated apoA-IVWT monomer (upper left, filled symbols) and dimer (lower right, open symbols). The X and Y-axes show the sequence number of the Lys residue involved in a specific cross-link (both intra- and intermolecular). Cross-links indicated with a filled triangle were unique to the monomer. The high similarity of the cross-linking patterns on both sides of the unity line dividing the plot is clear, indicating overall similarity in the tertiary folds of the two forms. (D) Placement of the glycine hinge introduced into Helix-B. Depicted is one component of the dimer with the polar surface in green and the exposed hydrophobic core in orange. Introduced glycines are shown in yellow. The arrow indicates how Helix-B might fold back onto the bundle to bury the exposed hydrophobic residues to form a monomer. (E) Sedimentation velocity molecular weight distribution plot of monomer apoA-IVWT (40.4 ±4 kDa), apoA-IVGGG (39.8 ±3 kDa) and apoA-IV64-335GGG (30.6 ±3.9 kDa). (F) Native gels showing the redistribution of monomer and dimer of apoA-IV variants that were subjected to thermal denaturation and subsequently re-annealed. Concentrations of protein are indicated for each lane. (G) Thermally denatured then re-annealed samples at 1.5 mg/ml from (F) analyzed by SEC. (H) Proposed model of transition from dimer to monomer where, in the absence of a second molecule, the extended Helix-B folds back on itself, completing a monomolecular four-helix bundle. See also Figure S5.
Based on these observations and the domain swapping apparent in our crystal structure, we envisioned a straightforward model for the monomeric form. Consider the hypothetical separation of the dimer to produce an elongated monomer, such as shown in Figure 4D. If a hinge is present near the middle of the long Helix-B, the N-terminal half could bend back toward the helical bundle and lie in the hydrophobic trough normally occupied by the same helix from an opposing monomer (Figure 4H). This action can be likened to closing the extended blade of a pocket knife. In this configuration, the molecular contacts, as measured by cross-linking, will be similar to those found in the dimer - except that those involving the N-terminal half of Helix-B would be intra-molecular instead of inter-molecular in the dimer. This should not dramatically change the helicity of the monomer vs. the dimer, consistent with the CD data, nor should it affect the thermal denaturation properties of the two forms on a molar basis. Thus, we reasoned that placing turn-favoring residues in the middle of Helix-B might shift the oligomeric equilibrium in favor of the monomer by allowing Helix-B to bend back instead of domain-swapping with another apoA-IV molecule. A candidate site for such a turn is at the kink sequence following Pro161 (Figure 4D). A GOR4 secondary structure prediction of this region showed a high propensity for helix formation but, by substituting three glycines (GGG) in place of HAD immediately after Pro 161, random coil is predicted between residues 158-164. This mutation was performed in both apoA-IVWT and apoA-IV64-335. In sedimentation velocity experiments, isolated monomers of both apoA-IVGGG and apoA-IV64-335GGG exhibited MW consistent with their respective monomer MW (Figures 4E and S5). Isolated monomers of both GGG mutants were thermally denatured, 2 min at 100°C, at various concentrations and then allowed to re-anneal for 10 min at room temperature. A native PAGE analysis (Figure 4F) showed that apoA-IVWT distributed between monomer and dimer in a concentration dependent manner, with dimer favored at higher concentrations (Note: on native gels, apoA-IV tends to run larger than predicted by its MW, possibly due to its extended conformation). By contrast, apoA-IVGGG completely failed to form dimers even at the highest concentration. The results were identical for apoA- IV64-335 and its corresponding GGG mutant. The same conclusions were obtained when SEC, rather than native PAGE, was used as the readout for extent of dimerization (Figure 4G). These results strongly argue that introduction of a GGG “hinge” shifts the oligomerization equilibrium of apoA-IV towards the monomer, consistent with the structural scheme shown in Figure 4H.
Particle formation
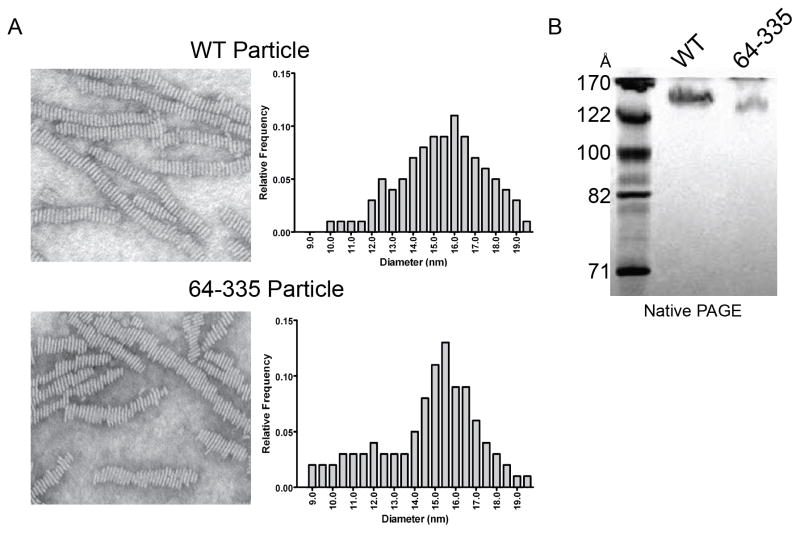
Most apolipoproteins can form rHDL via a number of mechanisms, including spontaneous association with short chain phospholipids such as dimyrstoyl phosphatidylcholine (DMPC). Using the conditions in Methods, apoA-IVWT produced DMPC-containing particles with a hydrodynamic diameter of 140 Å as measured by native PAGE. ApoA-IV64-335 also produced an rHDL particle, though slightly smaller at about 132 Å (Figures 5A, 5B, and S6). The chemical compositions of both particles are shown in Table S3. Furthermore, EM images (Figure 5A) showed that these particles are disc-shaped and can stack into characteristic structures called rouleux, a well noted behavior for apoA-I and other exchangeable apolipoproteins (Nichols et al., 1983). The particle diameters as determined by EM were larger than most estimated by native PAGE for both apoA-IVWT and apoA-IV64-335 (16 and 15.5 nm respectively), but both particles were similar in size and morphology. This suggests that the core four-helical bundle domain identified in the structure of apoA-IV64-335 exhibits similar particle forming characteristics as apoA-IVWT, including overall particle size.
Figure 5.
Particle formation comparison of apoA-IVWT and apoA-IV64-335. (A) Negative stain electron microscopy images of reconstituted lipoprotein DMPC particles generated with apoA-IVWT (top) and apoA-IV64-335 (bottom), with their respective particle diameter measurement distribution. (B) Native PAGE gel of reconstituted lipoprotein DMPC particles generated with apoA-IVWT and apoA-IV64-335, Coomassie stained. See also Figure S6 and Table S3.
Discussion
Compared to fully soluble proteins, only a handful of apolipoprotein structures have been solved by X-ray crystallography or nuclear magnetic resonance. The current study adds a new structure to this knowledge base that we believe offers significant new insight into long held questions concerning self-association and lipoprotein assembly that may be applicable to the larger family of exchangeable apolipoproteins and their role in HDL biogenesis.
Mechanism for self-association
The physiological role and the mechanism for apolipoprotein self-association have long been questioned, but the molecular basis for the interactions have only just started to be resolved with the recent crystal structure of lipid-free apoA-I (Mei et al., 2011). Both dimeric apoA-I and apoA-IV are bona fide examples of significant 3D domain swaps that provide a clear mechanism for how the proteins self-associate. This departs substantially from previously proposed mechanisms that invoked interaction between charged or hydrophobic patches between independently folded monomers (Silva et al., 2005a;Weinberg et al., 1985b). In the case of apoA-IV, the monomer and dimer forms both exist in stable low energy states that interconvert slowly enough so that they can be studied in isolation. We found that both the dimer and monomer exhibit highly similar structures as determined by CD and chemical cross-linking. Indeed, our introduction of a flexible region in the center of the long, swapped helix resulted in a form that remained monomeric (Figure 4H). This provides the first detailed picture of an apolipoprotein monomer/dimer relationship, but its applicability to other apolipoproteins, including apoA-I, remains to be determined.
Lipoprotein particle biogenesis
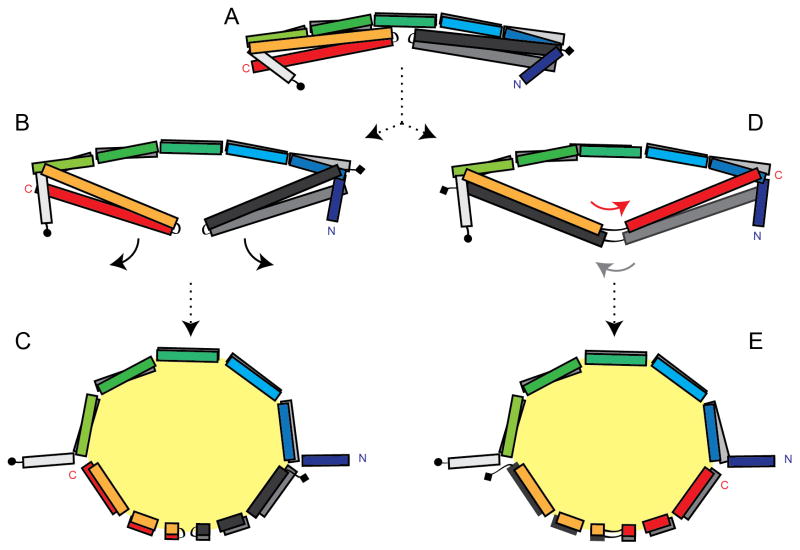
As stated in the Introduction, the most well supported model for the structure of nascent HDL is the double belt model in which two apolipoprotein molecules (typically apoA-I) are arranged in a ring-like, antiparallel structure that encapsulates a lipid bilayer (Li et al., 2004). A basic question in lipoprotein biology relates to the mechanism by which lipid-free proteins, presumably starting out as four-helical bundles, interact with each other and lipid to generate such an antiparallel structure. A key feature of these structures, apoA-I1-184 and apoA-IV64-335, is that their dimers already have significant portions of the participating molecules overlapping in an antiparallel manner. Combining this observation with the concept of uni-directional hinges mediated by the segmental kinks described above, we propose that an apoA-IV rod-like dimer could form a discoidal lipoprotein particle with only minor adjustments of these hinge angles (Figure 6). Beginning with the dimer rod, one can imagine inserting phospholipid molecules into the hydrophobic space located between the B helices and the helical arms composed of Helix-C and D. As lipids are added, the kink/hinges bend toward the core, allowing the helical arms or “doors” to swing away from the Helix-B backbone. Continued lipid accumulation results in a circular ring with the overall curvature mediated by the segmental kinks. One potential result is a ring with about half the circumference composed of the antiparallel Helix-B’s, making intermolecular contacts (Figures 6B and 6C). The other half is composed of the two antiparallel helical arms making intramolecular contacts. Alternatively, the helical arms may open and exchange their C-termini, resulting in a closed ring that is entirely stabilized by intermolecular interactions as in the apoA-I double belt (Figures 6D and 6E) and the scheme proposed by Mei and Atkinson. Our preliminary modeling analysis indicates that both scenarios could easily generate discoidal particles that are consistent with the particle diameters and compositions that we measured experimentally. The model does not define how phospholipid enters the hydrophobic space, however, one can speculate that lipid can access the core through the central cavity or the loose bundle packing created by Helix-A that caps the four-helix bundles. Our crystal structure should serve as an excellent guide for further mutagenesis experiments designed to sort out this issue and will be valuable for future studies designed to understand the structure of lipid-bound apoA-IV.
Figure 6.
Hypothesized model for apoA-IV rHDL. (A) A segmented apoA-IV64-335 dimer is shown. One chain is colored rainbow from N to C, where the other is colored in gray scale with (circle) being the N and (diamond) being the C terminus. The arrows indicate a proposed outward swing of the helical arm domains as lipid accumulates in the hydrophobic core. Two models are proposed that either keep the arms intact after they swing open (B–C) or the arms swap helices (D–E). See also Figure S7.
Comparison to apoA-I
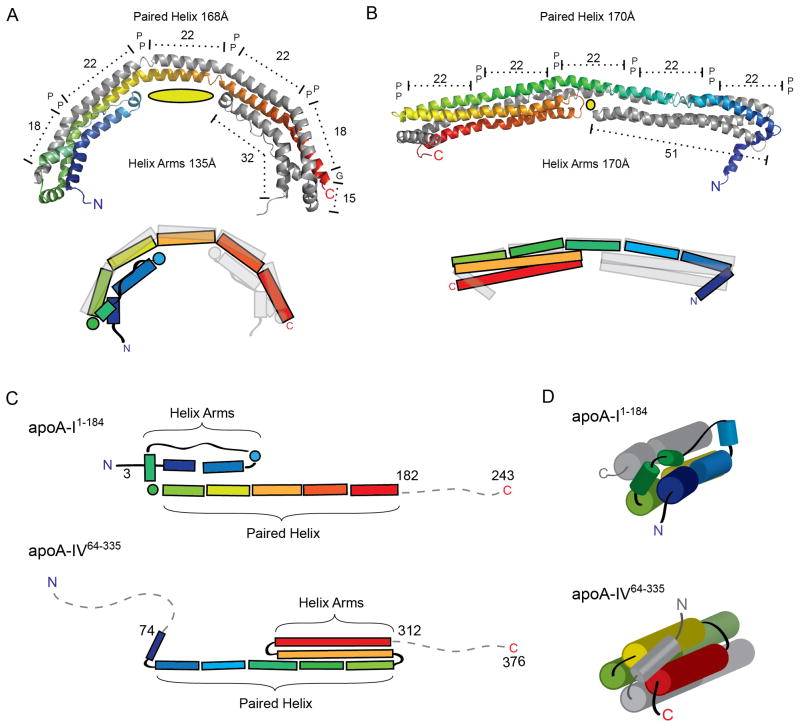
As indicated above, the dimeric structure of apoA-IV65-335 exhibited several striking similarities to the recent structure of apoA-I1-184 (Figure 7). First, residues 79-182 of apoA-I formed a long helix that participated in an antiparallel domain swapping interaction similar to apoA-IV Helix-B. Unlike the relatively straight apoA-IV structure, apoA-I was bent into a semi-circular shape owing to more extensive kink angles between helical domains of about 25° vs. 16° for apoA-IV (Table S2), nearer the average reported value of 26° (Barlow et al., 1988). Like apoA-IV, these kinks were associated with conserved Pro residues, though the P(Y/H)A motif prevalent in apoA-IV was not recapitulated in apoA-I. This provides strong evidence that domain swapping may be a common motif among exchangeable apolipoproteins that may mediate apolipoprotein self-association. Second, the apoA-I structure also features helix arms that fold across the antiparallel backbone. Interestingly, these are formed by the N-terminal 65 a.a. in apoA-I, but are composed of elements near the C-terminal end of the apoA-IV (206-312). Due to this difference, the transition from the helical arm to the paired helix is distinct for apoA-I and apoA-IV (Figure 7C), resulting in different capping mechanisms of the four-helix bundle (Figure 7D). In apoA-I1-184, the helix arm transitions into the antiparallel helix via a 10-residue helix (67-78), which caps the four-helix bundle. By contrast, in apoA-IV64-335 the paired helix transitions into the helical arm via a proline-induced turn (205-207), with the bundle being capped by Helix-A of the opposing molecule. Because of this difference in architecture, if a similar “swinging door” model is proposed for apoA-I, the helical arms become crossed in relation to the long paired helix, requiring a cross-over event in the double-belt model. Though the arrangement of the two structures is slightly different, it is surprising that varying regions of the polypeptide should give rise to apparently structurally similar features, especially given the view that the apoA-IV gene evolved from a primordial form of apoA-I (Karathanasis et al., 1986). This could be explained by a localized gene reversion event during apoA-IV evolution. Beyond the presence of the turns and amphipathic helices, we have not yet found common architectural features that predispose these residues, either in apoA-I or apoA-IV, to specifically form helical arms. However, we would point out that the missing sequences in both apoA-IV and apoA-I may interact with the bundle domains in ways that are not yet clear. Thus, the propensity for helical arm formation might not be exclusively encoded in the arms themselves. Third, the arms in apoA-I1-184 are significantly shorter, resulting in a less defined 4-helix bundle motif compared to apoA-IV64-335. As a consequence, a much larger cavity is created in the center of the rod, reducing the stability of the apoA-I1-184 helix 5 region. The authors propose that this flexible region might be a site for interaction with lecithin:cholesterol acyl transferase (LCAT). Our analysis of the corresponding region in apoA-IV64-335 suggests that there is no noticeable increase in B factor in this area with respect to other stretches of helix. Furthermore, our demonstration that apoA-IVN151C readily forms covalent dimers (Figure 2D) suggests that the helices in this region remain in close contact. This potential difference in conformational flexibility at the geometric center of each molecule may explain the remarkable difference in monomer/dimer dynamics between apoA-IV (slow) and apoA-I (fast) as well as why apoA-I is the superior activator of LCAT (Jonas et al., 1989). The availability of both of these structures will prove highly valuable for driving future studies aimed at understanding the structure/functional relationships of these and other exchangeable apolipoproteins associated with HDL. This comes at a critical time when there is a major effort within the pharmaceutical industry to develop therapies that raise HDL with the goal of enhancing protection from CVD.
Figure 7.
Structural comparison of apoA-I1-184 and apoA-IV64-335. (A–B) Ribbon (top) and schematic (bottom) representation of apoA-I1-184(A) and apoA-IV64-335(B) in their dimeric forms, each with one chain colored in rainbow from N to C terminus. Both structures reveal two long paired helices with anti-parallel orientation spanning similar distances and punctuated by proline residues. The cross chain aligned proline residues (represented as P) divide the long anti-parallel helix into two 18 and three 22 a.a paired helix regions for apoA-I1-182 and five 22 a.a paired helix regions for apoA-IV64-335. The length of the helical arms is shown, with the central cavity depicted as a yellow oval. (C) Schematic representation of both structures linearly aligned at the paired helix segment. Note the flipped orientation of the helix arm region in relation to the paired helix segment. (D) Schematic view looking down the bundle showing different linkage from the helical arms to the paired helices and how the four-helix bundle is capped in each structure.
Materials and Methods
Protein expression and purification
ApoA-IVWT, apoA-IV64-335, apoA-IVGGG, and apoA-IV64-335GGG were expressed and purified as described previously (Deng et al., 2012). Single Cys mutants of apoA-IV were immediately subjected to 5 mM DTT and kept under reducing conditions throughout the purification process. DTT was removed by dialysis (48 h at 4°C) to allow disulfide bond formation.
X-ray structure determination of apoA-IV64-335
The details of crystallization and structure determination have been previously published (Deng et al., 2012). Diffraction data were collected at the Advanced Photon Source (23ID-B GM/CA) at Argonne National Laboratory. Data on the Se-Met substituted crystals were collected at three wavelengths and utilized for MAD phasing (Table S2). The final structure has a MolProbity (Chen et al., 2010) score of 1.79 (97th percentile), with no Ramachandran outliers.
Cross-linking and mass spectrometry
Cross-linking with Bis(Sulfosuccinimidyl) suberate and subsequent analysis by electrospray mass spectrometry was performed exactly as we have described previously (Tubb et al., 2008). See supplemental methods for more detail.
Dissociation of dimer
Isolated dimers of apoA-IVWT and apoA-IV64-335 were diluted to 0.15 mg/ml in PBS and incubated at 37°C. Proteins were analyzed by SEC on a Superdex-200 column at 4°C. Monomer and dimer peaks were integrated to determine their ratio.
Circular dichroism (CD)
Immediately preceding the CD measurements, the samples were analyzed by SEC to verify the oligomeric state. Far-UV wavelength measurements were performed at 25°C with 1nm steps from 185 to 300nm using samples in 20mM NaPO4, 50mM NaF at pH 7.5. Data were analyzed using the SELCON 3 method. Thermal denaturation experiments were performed by monitoring the ellipticity at 222nm from 15°C to 95°C in 1°C increments with 2 m temperature equilibrations and 5 s averaging time. The measurements were performed in a 1 cm cuvette under constant stirring with a protein concentration of 0.37 μg/mL.
Apolipoprotein-lipid particle formation
Dimyristoyl phospatidylcholine (DMPC) (Avanti Polar Lipids, Birmingham, AL) in chloroform was dried down under nitrogen, resuspended in PBS (pH 7.3), bath sonicated for 30 s, and combined with apoA-IV at a 350:1 DMPC: apoA-IV molar ratio to a final protein concentration of 0.8 mg/ml. The lipid:protein mixture was incubated for 24 h at 24.5°C followed by a 16 h 37°C incubation. Particle homogeneity was verified by native PAGE. Composition was determined by analysis of phosphorus (Sokoloff et al., 1974) and modified Lowry assay (Markwell et al., 1978) from multiple independent rHDL preparations. Electron microscopy of rHDL particles was performed as described previously (Huang et al., 2011).
Supplementary Material
Highlights.
Structure of dimeric apoA-IV solved to 2.4 Å by x-ray crystallography.
Structure confirmed through biochemical and mutational analysis.
Understanding of apoA-IV dimerization allowed us to create a monomeric mutant.
Structural features in the dimer offer clues to the genesis of apoA-IV lipoproteins.
Acknowledgments
This work was supported by US National Institutes of Health (NIH) R01 grant HL67093 (to W.S.D.), an NIH training grant T32 HL007382 (to X.D.), an American Heart Association Pre-doctoral fellowship to M.R.T. and NIH R01 grant HL48148 (to W.G.J.). EM was carried out in the Vanderbilt University Research Electron Microscopy Resource of the Cell Imaging Core (partially supported by NIH grants CA68485, DK20593, and DK58404).
Footnotes
Accession codes. The coordinates and structure factors of Homo sapiens apolipoprotein A-IV have been deposited in the Protein Data Bank under the accession code 3S84.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barlow DJ, Thornton JM. Helix geometry in proteins. J Mol Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Huang M, He Y, Zhang S, Luo Y. Genetic ablation of apolipoprotein A-IV accelerates Alzheimer’s disease pathogenesis in a mouse model. Am J Pathol. 2011;178:1298–1308. doi: 10.1016/j.ajpath.2010.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles: A mass spectrometry study. J Biol Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- Davidson WS, Thompson TB. The structure of apolipoprotein A-I in high density lipoproteins. J Biol Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- Deng X, Davidson WS, Thompson TB. Improving the diffraction of apoA-IV crystals through extreme dehydration. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012;68:105–110. doi: 10.1107/S1744309111048706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger N, Ghalim N, Theret N, Fruchart JC, Castro G. Lipoproteins containing apolipoprotein A-IV: composition and relation to cholesterol esterification. Biochim Biophys Acta. 1994;1211:23–28. doi: 10.1016/0005-2760(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Emmanuel F, Steinmetz A, Rosseneu M, Brasseur R, Gosselet N, Attenot F, Cuine S, Seguret S, Latta M, Fruchart JC, et al. Identification of specific amphipathic alpha-helical sequence of human apolipoprotein A-IV involved in lecithin:cholesterol acyltransferase activation. J Biol Chem. 1994;269:29883–29890. [PubMed] [Google Scholar]
- Gordon S, Durairaj A, Lu J, Davidson WS. High-density lipoprotein proteomics: Identifying new drug targets and biomarkers by understanding functionality. Curr Cardio Risk Rep. 2010a;4:1–8. doi: 10.1007/s12170-009-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SM, Deng J, Lu LJ, Davidson WS. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res. 2010b;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Silva RA, Jerome WG, Kontush A, Chapman MJ, Curtiss LK, Hodges TJ, Davidson WS. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat Struct Mol Biol. 2011;18:416–422. doi: 10.1038/nsmb.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas A, Kezdy KE, Wald JH. Defined apolipoprotein A-I conformations in reconstituted high density lipoprotein discs. J Biol Chem. 1989;264:4818–4824. [PubMed] [Google Scholar]
- Karathanasis SK, Oettgen P, Haddad IA, Antonarakis SE. Structure, evolution, and polymorphisms of the human apolipoprotein A4 gene (APOA4) Proc Natl Acad Sci U S A. 1986;83:8457–8461. doi: 10.1073/pnas.83.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppaka V, Silvestro L, Engler JA, Brouillette CG, Axelsen PH. The structure of human lipoprotein A-I. Evidence for the “belt” model. J Biol Chem. 1999;274:14541–14544. doi: 10.1074/jbc.274.21.14541. [DOI] [PubMed] [Google Scholar]
- Lawn RM, Wade DP, Garvin MR, Wang X, Schwartz K, Porter JG, Seilhamer JJ, Vaughan AM, Oram JF. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J Clin Invest. 1999;104:R25–R31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen J, Mishra VK, Kurtz JA, Cao D, Klon AE, Harvey SC, Anantharamaiah GM, Segrest JP. Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J Mol Biol. 2004;343:1293–1311. doi: 10.1016/j.jmb.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Main LA, Ohnishi T, Yokoyama S. Activation of human plasma cholesteryl ester transfer protein by human apolipoprotein A-IV. Biochim Biophys Acta. 1996;1300:17–24. doi: 10.1016/0005-2760(95)00228-6. [DOI] [PubMed] [Google Scholar]
- Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- Mei X, Atkinson D. Crystal Structure of C-terminal Truncated Apolipoprotein A-I Reveals the Assembly of High Density Lipoprotein (HDL) by Dimerization. J Biol Chem. 2011;286:38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs I, Van EM, Van Berkel TJ. High-density lipoprotein: key molecule in cholesterol efflux and the prevention of atherosclerosis. Curr Pharm Des. 2010;16:1445–1467. doi: 10.2174/138161210791051022. [DOI] [PubMed] [Google Scholar]
- Nichols AV, Gong EL, Blanche PJ, Forte TM. Characterization of discoidal complexes of phosphatidylcholine, apolipoprotein A-I and cholesterol by gradient gel electrophoresis. Biochim Biophys Acta. 1983;750:353–364. doi: 10.1016/0005-2760(83)90040-1. [DOI] [PubMed] [Google Scholar]
- Pearson K, Saito H, Woods SC, Lund-Katz S, Tso P, Phillips MC, Davidson WS. Structure of human apolipoprotein A-IV: a distinct domain architecture among exchangeable apolipoproteins with potential functional implications. Biochemistry. 2004;43:10719–10729. doi: 10.1021/bi048978m. [DOI] [PubMed] [Google Scholar]
- Qin X, Swertfeger DK, Zheng S, Hui DY, Tso P. Apolipoprotein AIV: a potent endogenous inhibitor of lipid oxidation. American Journal of Physiology. 1998;274:H1836–H1840. doi: 10.1152/ajpheart.1998.274.5.H1836. [DOI] [PubMed] [Google Scholar]
- Remaley AT, Stonik JA, Demosky SJ, Neufeld EB, Bocharov AV, Vishnyakova TG, Eggerman TL, Patterson AP, Duverger NJ, Santamarina-Fojo S, et al. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem Biophys Res Commun. 2001a;280:818–823. doi: 10.1006/bbrc.2000.4219. [DOI] [PubMed] [Google Scholar]
- Remaley AT, Stonik JA, Demosky SJ, Neufeld EB, Bocharov AV, Vishnyakova TG, Eggerman TL, Patterson AP, Duverger NJ, Santamarina-Fojo S, et al. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem Biophys Res Commun. 2001b;280:818–823. doi: 10.1006/bbrc.2000.4219. [DOI] [PubMed] [Google Scholar]
- Rust S, Rosier M, Funke H, Real J, Amoura Z, Piette JC, Deleuze JF, Brewer HB, Duverger N, Denefle P, et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat Genet. 1999;22:352–355. doi: 10.1038/11921. [DOI] [PubMed] [Google Scholar]
- Segrest JP, Garber DW, Brouillette CG, Harvey SC, Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–369. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- Segrest JP, Harvey SC, Zannis V. Detailed molecular model of apolipoprotein A-I on the surface of high-density lipoproteins and its functional implications. Trends Cardiovasc Med. 2000;10:246–252. doi: 10.1016/s1050-1738(00)00078-5. [DOI] [PubMed] [Google Scholar]
- Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 2005a;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005b;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- Sivashanmugam A, Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein binding and for receptor binding activity. J Biol Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff L, Rothblat GH. Sterol to phospholipid molar ratios of L cells with qualitative and quantitative variations of cellular sterol. Proc Soc Exp Biol Med. 1974 Sep;146:1166–1172. doi: 10.3181/00379727-146-38267. [DOI] [PubMed] [Google Scholar]
- Stan S, Delvin E, Lambert M, Seidman E, Levy E. Apo A-IV: an update on regulation and physiologic functions. Biochim Biophys Acta. 2003;1631:177–187. doi: 10.1016/s1388-1981(03)00004-0. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Bhat S, Sorci-Thomas MG. The use of chemical cross-linking and mass spectrometry to elucidate the tertiary conformation of lipid-bound apolipoprotein A-I. Curr Opin Lipidol. 2006;17:214–220. doi: 10.1097/01.mol.0000226111.05060.f4. [DOI] [PubMed] [Google Scholar]
- Tso P, Liu M, Kalogeris TJ, Thomson AB. The role of apolipoprotein A-IV in the regulation of food intake. Annu Rev Nutr. 2001;21:231–254. doi: 10.1146/annurev.nutr.21.1.231. [DOI] [PubMed] [Google Scholar]
- Tubb MR, Silva RA, Fang J, Tso P, Davidson WS. A three-dimensional homology model of lipid-free apolipoprotein A-IV using cross-linking and mass spectrometry. J Biol Chem. 2008;283:17314–17323. doi: 10.1074/jbc.M800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubb MR, Silva RAGD, Pearson KJ, Tso P, Liu M, Davidson WS. Modulation of apolipoprotein A-IV lipid binding by an interaction between the N and C termini. J Biol Chem. 2007;282:28385–28394. doi: 10.1074/jbc.M704070200. [DOI] [PubMed] [Google Scholar]
- Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowinkel T, Mori M, Krieglstein CF, Russell J, Saijo F, Bharwani S, Turnage RH, Davidson WS, Tso P, Granger DN, et al. Apolipoprotein A-IV inhibits experimental colitis. J Clin Invest. 2004;114:260–269. doi: 10.1172/JCI21233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc Natl Acad Sci USA. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RB, Spector MS. Structural properties and lipid binding of human apolipoprotein A-IV. J Biol Chem. 1985a;260:4914–4921. [PubMed] [Google Scholar]
- Weinberg RB, Spector MS. The self-association of human apolipoprotein A-IV. Evidence for an in vivo circulating dimeric form. J Biol Chem. 1985b;260:14279–14286. [PubMed] [Google Scholar]
- Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, III, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.