In embryonic stem (ES) cell lines generated from human embryos determined through preimplantation genetic diagnosis to carry the fragile X mutation, the FMR1 gene is expressed in undifferentiated cells but undergoes transcriptional silencing following ES cell differentiation (Eiges et al., 2007). Here, we generated induced pluripotent stem (iPS) cell lines from fibroblasts of individuals carrying the fragile X mutation. Despite successful reprogramming of the somatic cells to pluripotency, the FMR1 gene remained inactive, and carried DNA methylation and histone modifications indicative of inactive heterochromatin. These data highlight critical differences between ES and iPS cells in modeling Fragile X disorder.
Pluripotent stem cells are potentially an important tool to model human genetic disorders. Human embryonic stem cells can recapitulate early stages of human development, and they can also differentiate into cells from the three embryonic germ layers (Schuldiner et al., 2000; Thomson et al., 1998). Thus, human pluripotent stem cells can be used to analyze the effect of specific mutations on the differentiation of various cell types and on early developmental processes that are otherwise inaccessible for research.
In the past few years several diseases have been modeled in pluripotent stem cells, either by direct gene mutagenesis or by deriving ES cells from embryos determined by preimplantation genetic diagnosis (PGD) to carry genetic mutation (Eiges et al., 2007; Urbach et al., 2004 reviewed in Lengerke and Daley, 2009). Recently, human pluripotent stem cells have been derived from somatic cells by introduction of defined factors (Lowry et al., 2008; Park et al., 2008c; Takahashi et al., 2007; Yu et al., 2007). These induced pluripotent stem (iPS) cells show remarkable similarity to human ES cells (Lowry et al., 2008; Park et al., 2008c; Takahashi et al., 2007; Yu et al., 2007). By reprogramming somatic cells from patients, one may isolate pluripotent cells that harbor disease-specific mutations (Park et al., 2008a); The reprogramming of somatic cells into pluripotent cells raised the question whether iPS cells will be able to replace human ES cells in basic research as well as in clinical applications (Belmonte et al., 2009). We are now in a unique position to compare disease phenotypes manifest in ES cells to those seen in iPS cells.
Fragile X (FX) syndrome is the most common form of inherited mental retardation (Crawford et al., 2001; Rousseau et al., 1992). It is caused by the absence of expression of the fragile X mental retardation 1 (FMR1) gene (O'Donnell and Warren, 2002). The vast majority of FX patients do not express FMR1 due to CGG triplet repeat expansion in the 5' untranslated region of the gene (Pearson et al., 2005; Verkerk et al., 1991). Full expansion of the CGG repeat usually coincides with hypermethylation of the repeat region and its upstream promoter (Oberle et al., 1991), and with chromatin modifications such as histone H3 tail deacetylation, histone H3K9 methylation and histone H3K4 demethylation (Coffee et al., 1999). Until recently, early events in FMR1 silencing could not be characterized due to the lack of an appropriate animal model (Bontekoe et al., 1997; Lavedan et al., 1997), but human ES cells have now been derived from FX blastocysts determined through pre-implantation genetic diagnosis (PGD) (Eiges et al., 2007). In the undifferentiated FX-ES cells, the full expansion of the CGG triplet repeat is not sufficient to inactivate the expression of the FMR1 gene and gene silencing occurs only upon differentiation (Eiges et al., 2007). Evidence from chorionic villus samples supports a similar conclusion that transcriptional silencing of the FMR1 gene occurs with human development (Sutcliffe et al., 1992; Willemsen et al., 2002). Current data suggest that upon cell differentiation the mutated FMR1 gene recruits specific histone modifications followed by DNA methylation, which silence its transcription (Eiges et al., 2007; Pietrobono et al., 2005).
In the current study we have isolated iPS cell lines from three FX affected males, and compared the regulation of FMR1 transcription to that of human FX-ES cells. Fibroblasts from four-year old and 28-year old individuals, as well as fetal-lung fibroblasts from a 22 week-old affected fetus with FX syndrome were reprogrammed in culture according to published protocols (Park et al., 2008b; Takahashi et al., 2007). The efficiency of reprogramming of the FX-fibroblasts was similar to that of the wt-fibroblasts (0.0056% and 0.0024% respectively), as determined by counting the number of Tra-1-60 positive colonies. Multiple FX-iPS cell clones were analyzed (seven from the first, two from the second and two from the third patient). The iPS cell clones demonstrated typical characteristics of pluripotent stem cells: morphology similar to that of ES cells and expression of alkaline phosphatase, Tra-1-60, OCT4, SOX2, NANOG, SSEA3, SSEA4, and Tra-1-81 (Supplementary Figure 1A); silencing of retroviral transgenes (Supplementary Figure 1B, and data not shown); reactivation of genes indicative of pluripotency (Supplementary Figure 1C,D and data not shown); and maintenance of a normal diploid karyotype (Supplementary Figure 1E). By hierarchical clustering and scatter plot analysis of DNA microarray results we observed that the iPS cells cluster together with human ES cells and apart from their cell of origin (Supplementary Figure 2A,B). The cells generated embryoid bodies (Supplementary Figure 2C-I) that expressed markers of endoderm (Supplementary Figure 2C-II&III), mesoderm (Supplementary Figure 2A-IV&V) and ectoderm (Supplementary Figure 2C-VI) as demonstrated by immunostaining, and also by RT-PCR (data not shown). The iPS cell lines also differentiated in vivo into teratomas that manifest elements of all embryonic germ layers (Supplementary Figure 2D). Thus, these human iPS cell lines met stringent criteria for pluripotency (Chan et al., 2009; Daley et al., 2009).
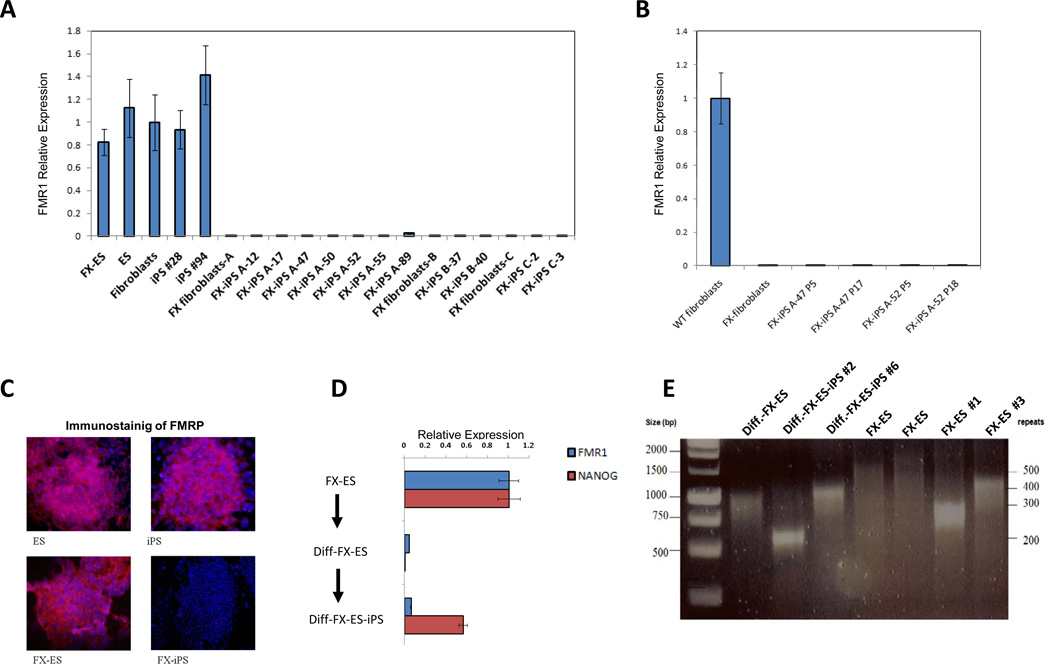
The FMR1 gene is expressed in FX-ES cells, wt-ES cells, wild type skin fibroblasts and iPS cells derived from them (Figure 1A, see also supplementary figures 1 and 2). The FMR1 gene is also expressed in wild type lung fibroblasts (MRC5), and iPS cells derived from them (data not shown). In marked contrast to all of these cells, the FMR1 gene remained transcriptionally silent in all FX-iPS cell clones derived from skin or lung FX-fibroblasts (Figure 1A). These results suggest that the differences in the expression of FMR1 between the WT and FX-iPS cells are due to the FMR1 mutation and not due to the tissue source of the original fibroblasts. The absence of FMR1 gene expression in the FX-iPS cells was observed both in early passage (P5) and at higher passages (up to P18; Figure 1B), indicating a stable phenotype. The FX-iPS cells also lacked expression of the FMR1 protein by immunostaining (Figure 1C).
Figure 1. FMR1 expression in FX-iPS cells.
A. FMR1 transcript expression as analyzed by real-time PCR in FX-ES cells, normal ES cells, normal fibroblasts, two normal iPS cell clones, FX-fibroblasts from three different patients (A, B, C), and eleven FX-iPS cell clones derived from the FX-fibroblasts. B. Comparison of FMR1 expression levels in two FX-iPS cell clones at low passage (p5) and high passage (p17 and p18). C. Immunostaining for FMRP in ES, iPS, FX-ES and FX-iPS cells using goat anti human FMRP antibody and Hoechst 33258 for nuclear staining. D. FMR1 and NANOG expression levels in FX-ES cells, Diff-FX-ES cells (a population of cells differentiated from FX-ES cells), and Diff-FX-ES-iPS cells (iPS cells derived from differentiated FX-ES cells). E. Analysis of CGG repeat number in Diff-FX-ES, Diff-FX-ES-iPS clones #2 and #6, two samples of FX-ES cells, and two subclones of the FX-ES cells. Note that the Diff.-FX-ES and the FX-ES represent cell populations that are heterogeneous with respect to CGG repeat number (Eiges et al, 2007), whereas in the Diff-FX-ES-iPS and the FX-ES subclones a dominant band indicates the more homogenous population expected in subclones. The size of the DNA marker bands in base pairs and the number of the CGG repeats are shown at the left and right sides of the gel, respectively. For further characterization of the undifferentiated iPS clones and their pluripotency see also supplementary figures 1,2.
Downregulation of the FMR1 gene occurs upon differentiation of human FX-ES cells, concomitant with downregulation of pluripotency-associated genes such as Nanog (Eiges et al., 2007 and figure 1D). Interestingly, when differentiated cells derived from FX-ES cells were reverted to pluripotency by introduction of the reprogramming factors Oct4/ Sox2/ c-Myc/ Klf4, they also failed to reactive FMR1 expression (Figure 1D). The CGG repeat length in the iPS cells derived from the differentiated FX-ES cells is in the same range as the FX-ES cells and the two subclones of FX-ES cells (Figure 1E). The data suggests that the silencing of the FMR1 in the FX-iPS cells is not due to expansion of the CGG repeats above that observed in FX-ES cells, where the gene is still active. The data also indicates that the reprogramming process has no major effect on the instability of the CGG repeats, as the reprogrammed cells have a similar number of repeats as their parental fibroblast (supplementary Figure 2E).
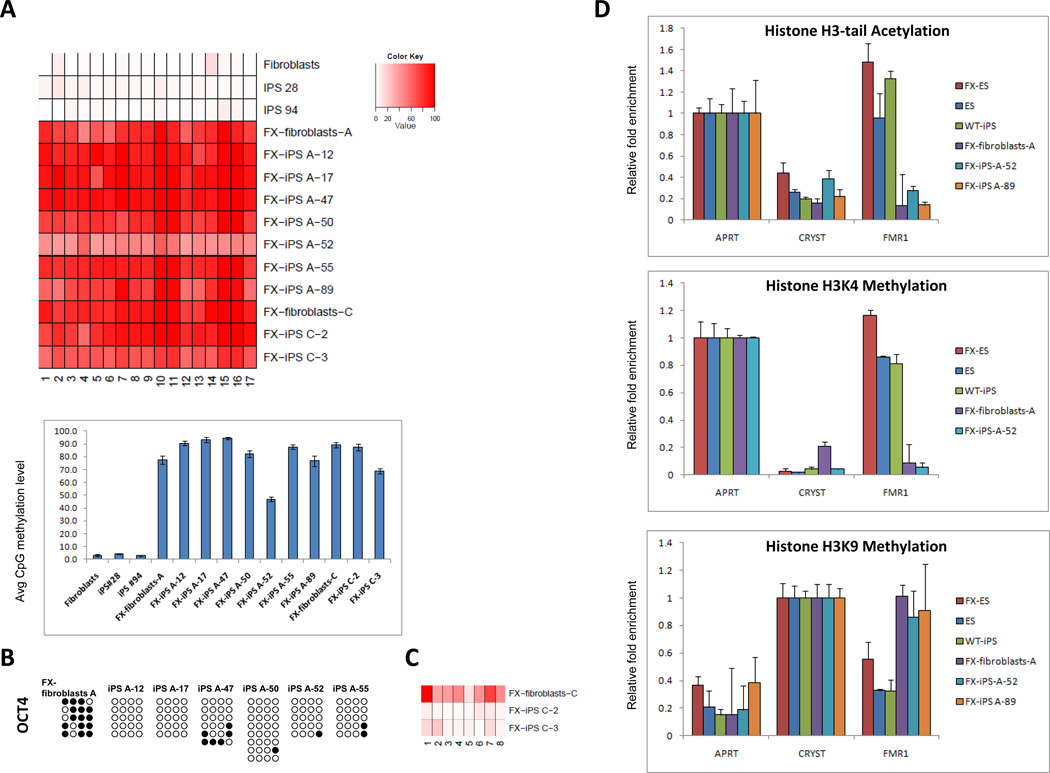
To examine the molecular basis for the silencing of FMR1 transcription in FX-iPS cells, we analyzed DNA methylation of the FMR1 promoter using bisulfite treatment followed by pyrosequencing in fibroblasts and in their derived iPS cell lines. Pyrosequencing is an accurate and reliable method to determine the degree of methylation at several CpGs in close proximity with high quantitative resolution (Brakensiek et al., 2007; Dupont et al., 2004; Shaw et al., 2006; Tost and Gut, 2007; Wong et al., 2006). While the FMR1 locus consistently lacked methylation in normal fibroblasts and their derivative iPS cell lines, 8 of 9 FX-iPS clones analyzed were highly methylated at levels comparable to the original fibroblasts; one iPS clone (FX-iPS A-52) showed an intermediate methylation level, but still much higher than the WT- fibroblasts and iPS cells (Figure 2A). DNA methylation at the FMR1 locus persisted in the FX-iPS cell clones despite the complete lack of methylation at the promoters for OCT4 (Figure 2B,C) and NANOG (not shown). The FX-iPS cell clones thus show a persistence of DNA methylation at the FMR1 locus even after reprogramming.
Figure 2. Epigenetic modifications in FX-iPS cells.
A. Pyrosequencing analysis of the FMR1 promoter in wt-fibroblasts, two clones of wt-iPS cells (iPS 28, iPS 94), FX-fibroblasts-A, and seven derivative iPS cell lines (FX-iPS A-12, 17, 47, 50, 52, 55, 89), FX-fibroblasts-C and two derivative iPS cell lines (FX-iPS C-2, C-3). Upper panel – methylation level at CpG position. Bottom panel – average methylation level of all CpG sites for each sample. B. Bisulfite sequencing analysis of the OCT4 promoter in FX-fibroblasts-A and FX-iPS cell clones derived from them. Open circles, unmethylated CpGs; Black circles, methylated CpGs. C. Pyrosequencing analysis of the OCT4 5’UTR in FX-fibroblasts-C and FX-iPS cell clones derived from them. D. Histone modifications at the FMR1 locus in FX-iPS cells: ChIP analysis of histone H3-tail acetylation and H3K4 and H3K9 methylation in FX-iPS cells. Real-time PCR was performed on bound and input sonicated DNA fragments using primers for the FMR1 promoter. Adenine phosphoribosyl transferase (APRT) and Crystalline (CRYST) served as positive and negative controls, respectively. Values were normalized to the appropriate positive control and shown with their respective standard errors.
We further explored histone modifications associated with transcriptionally active (H3 tail acetylation and lysine 4 methylation) and repressed (lysine 9 methylation) chromatin states associated with FMR1 silencing in somatic cells of FX individuals (Coffee et al., 2002; Coffee et al., 1999; Pietrobono et al., 2005). We carried out chromatin immunoprecipitation (ChIP) experiments to analyze these modifications in two different FX-iPS cell lines (# 52 and #89). In contrast to the active chromatin marks on the FMR1 locus in FX-ES cells, normal ES cells, and normal iPS cells, the FMR1 locus in the FX-iPS cells is methylated at H3K9 and lacks histone acetylation and H3K4 methylation (Figure 2D). Our data suggest that in FX-iPS cells, the mutated FMR1 gene has chromatin modifications that are consistent with its transcriptionally silent state (See Supplementary Figure 2E).
Our results thus imply that the mutant FMR1 locus in FX-iPS cell lines is resistant to activation by the iPS cell reprogramming protocol. Although reversion of aberrant methylation, heterochromatin formation, and loss of FMR1 expression has not been studied during passage of the mutant allele through the germ line in FRAXA families, conflicting data has been reported for the capacity to reactivate the silenced FMR1 gene in vitro. Two groups have shown that transfer of the X chromosome from FX patient cells into mouse embryonal carcinoma cells (Wohrle et al., 2001) or fusion of FX cells with normal fibroblasts (Stoyanova et al., 2004) result in DNA demethylation and reactivation of the FMR1 gene, whereas a third group’s attempt to reactivate the FMR1 gene by cell fusion with mouse embryonic carcinoma cells failed to erase the aberrant methylation of the mutant FMR1 gene (Burman et al., 1999a). Similarly, de novo methylation of an unmethylated mutant FMR1 gene did not occur when inserted via microcell-mediated chromosome transfer into a mouse embryonal carcinoma cell line (Burman et al., 1999b). Treatment of somatic FX-fibroblasts with the demethylating agent 5-azacytidine has been shown to reactivate FMR1 expression (Tabolacci et al., 2005). Apparently, under some circumstances the epigenetic marks that maintain silencing of a mutant FMR1 gene can be erased; however the exact conditions that enable the reactivation of the FMR1 gene remain unclear. Although we cannot exclude the possibility that reprogramming cells from a different tissue, or by a different method might reactivate FMR1 gene expression over time in FX-iPS cells, the FMR1 locus seems to be highly resistant to the reprogramming process, according to current standard practice.
Human ES and iPS cells offer significant advantages for regenerative medicine (Amabile and Meissner, 2009; Muller et al., 2009; Nishikawa et al., 2008), and the modeling of human genetic disorders (Muller et al., 2009; Nishikawa et al., 2008; Park et al., 2008a), although potential limitation such as chromosomal abnormalities (Baker et al., 2007; Lefort et al., 2008; Maitra et al., 2005; Spits et al., 2008) have to be considered before using the cells as a disease model. Our data highlight a significant difference between FX-ES and FX-iPS cells with regard to their expression of the FMR1 gene. The mutated FMR1 gene is expressed in FX-ES cells and transcriptionally silenced upon differentiation, while in FX-iPS cells the FMR1 locus remains inactive, and is not reset by the reprogramming process to the transcriptionally active state. It is thus possible that other disorders related to epigenetic defects, including triplet repeat and imprinting disorders, may likewise evade the reprogramming process. Although FX-iPS cells do not model the differentiation-dependent silencing of the FMR1 gene, as shown for FX-ES cells, they may remain valuable for analyzing the role of FMR1 in neural cells. In FX-iPS cells, like in FX-neurons and in contrast to normal human iPS cells the FMR1 gene is methylated, its chromatin is in a closed conformation, and the gene is not expressed. Therefore, differentiation of FX-iPS cells into neurons may nonetheless facilitate the study of FMR1 in neural cells. Until a deeper understanding of the potential differences between iPS and ES cells is delineated, the study of both iPS cells from patients and human ES cells carrying the same mutation (either from PGD embryos or by genetic manipulation) might, whenever possible, be the optimal approach to model human genetic disorders through cell culture. Finally, the distinction between FX-ES and FX-iPS cells at the FMR1 locus might be a particular example of a more general phenomenon of epigenetic differences between human ES cells and iPS cells, which highlights the need for more studies to clarify the similarity and differences between ES cells and iPS cells.
Supplementary Material
Acknowledgments
We would like to thank Tamar Lev-Golan and Inbal Caspi for technical assistance. A.U. is an EMBO fellow. N.B. is the Herbert Cohn Chair in Cancer Research. This research was partially supported by funds from the Israel Science Foundation (Grant number 227/06), and the European Community (ESTOOLS, Grant number 018739). We gratefully acknowledge support for this project provided by a grant from the Legacy Heritage Fund of New York. G.Q.D. was supported by grants from the National Institutes of Health, the Howard Hughes Medical Institute, and the Manton Center for Orphan Disease Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amabile G, Meissner A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends in molecular medicine. 2009;15:59–68. doi: 10.1016/j.molmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nature biotechnology. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- Belmonte JC, Ellis J, Hochedlinger K, Yamanaka S. Induced pluripotent stem cells and reprogramming: seeing the science through the hype. Nat Rev Genet. 2009;10:878–883. doi: 10.1038/nrg2700. [DOI] [PubMed] [Google Scholar]
- Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA. FMR1 premutation allele (CGG)81 is stable in mice. Eur J Hum Genet. 1997;5:293–298. [PubMed] [Google Scholar]
- Brakensiek K, Wingen LU, Langer F, Kreipe H, Lehmann U. Quantitative high-resolution CpG island mapping with Pyrosequencing reveals disease-specific methylation patterns of the CDKN2B gene in myelodysplastic syndrome and myeloid leukemia. Clinical chemistry. 2007;53:17–23. doi: 10.1373/clinchem.2007.072629. [DOI] [PubMed] [Google Scholar]
- Burman RW, Popovich BW, Jacky PB, Turker MS. Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Human molecular genetics. 1999a;8:2293–2302. doi: 10.1093/hmg/8.12.2293. [DOI] [PubMed] [Google Scholar]
- Burman RW, Yates PA, Green LD, Jacky PB, Turker MS, Popovich BW. Hypomethylation of an expanded FMR1 allele is not associated with a global DNA methylation defect. American journal of human genetics. 1999b;65:1375–1386. doi: 10.1086/302628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature biotechnology. 2009 doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Ceman S, Warren ST, Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. American journal of human genetics. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell stem cell. 2009;4:200–201. doi: 10.1016/j.stem.2009.02.009. author reply 202. [DOI] [PubMed] [Google Scholar]
- Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Analytical biochemistry. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell stem cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Lavedan CN, Garrett L, Nussbaum RL. Trinucleotide repeats (CGG)22TGG(CGG)43TGG(CGG)21 from the fragile X gene remain stable in transgenic mice. Hum Genet. 1997;100:407–414. doi: 10.1007/s004390050525. [DOI] [PubMed] [Google Scholar]
- Lefort N, Feyeux M, Bas C, Feraud O, Bennaceur-Griscelli A, Tachdjian G, Peschanski M, Perrier AL. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nature biotechnology. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- Lengerke C, Daley GQ. Disease models from pluripotent stem cells. Annals of the New York Academy of Sciences. 2009;1176:191–196. doi: 10.1111/j.1749-6632.2009.04962.x. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- Muller LU, Daley GQ, Williams DA. Upping the Ante: Recent Advances in Direct Reprogramming. Mol Ther. 2009 doi: 10.1038/mt.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nature reviews. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annual review of neuroscience. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M, Mandel J. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nature protocols. 2008b;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008c;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- Pietrobono R, Tabolacci E, Zalfa F, Zito I, Terracciano A, Moscato U, Bagni C, Oostra B, Chiurazzi P, Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Human molecular genetics. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Heitz D, Mandel JL. The unstable and methylatable mutations causing the fragile X syndrome. Human mutation. 1992;1:91–96. doi: 10.1002/humu.1380010202. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton DA, Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, Lowe D, Field JK, Risk JM. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. British journal of cancer. 2006;94:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der Elst J, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nature biotechnology. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- Stoyanova V, Rossetti S, L VANU, Oostra BA, Hoogeveen AT. Loss of FMR1 hypermethylation in somatic cell heterokaryons. Faseb J. 2004;18:1964–1966. doi: 10.1096/fj.04-2499fje. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Human molecular genetics. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tabolacci E, Pietrobono R, Moscato U, Oostra BA, Chiurazzi P, Neri G. Differential epigenetic modifications in the FMR1 gene of the fragile X syndrome after reactivating pharmacological treatments. Eur J Hum Genet. 2005;13:641–648. doi: 10.1038/sj.ejhg.5201393. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nature protocols. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem cells (Dayton, Ohio) 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- Wohrle D, Salat U, Hameister H, Vogel W, Steinbach P. Demethylation, reactivation, and destabilization of human fragile X full-mutation alleles in mouse embryocarcinoma cells. American journal of human genetics. 2001;69:504–515. doi: 10.1086/322739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HL, Byun HM, Kwan JM, Campan M, Ingles SA, Laird PW, Yang AS. Rapid and quantitative method of allele-specific DNA methylation analysis. BioTechniques. 2006;41:734–739. doi: 10.2144/000112305. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.