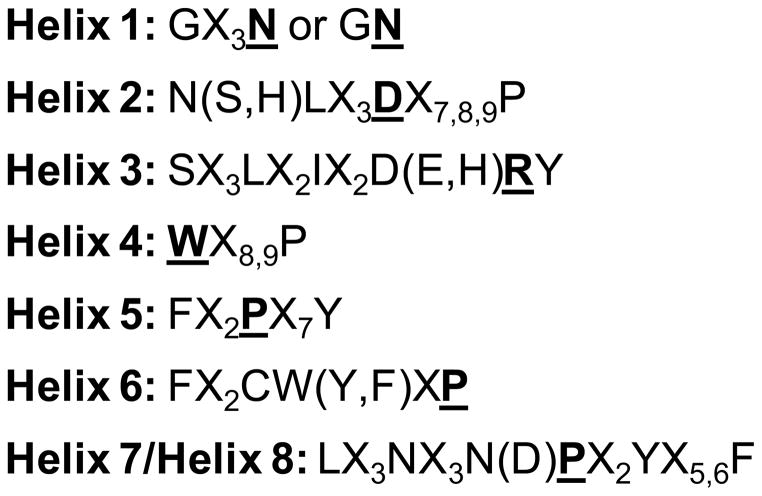Abstract
G protein-coupled receptors (GPCRs) are a large superfamily of membrane bound signaling proteins that hold great pharmaceutical interest. Since experimentally elucidated structures are available only for a very limited number of receptors, homology modeling has become a widespread technique for the construction of GPCR models intended to study the structure-function relationships of the receptors and aid the discovery and development of ligands capable of modulating their activity. Through this chapter, various aspects involved in the constructions of homology models of the serpentine domain of the largest class of GPCRs, known as class A or rhodopsin family, are illustrated. In particular, the chapter provides suggestions, guidelines and critical thoughts on some of the most crucial aspect of GPCR modeling, including: collection of candidate templates and a structure-based alignment of their sequences; identification and alignment of the transmembrane helices of the query receptor to the corresponding domains of the candidate templates; selection of one or more templates receptor; election of homology or de novo modeling for the construction of specific extracellular and intracellular domains; construction of the three-dimensional models, with special consideration to extracellular regions, disulfide bridges, and interhelical cavity; validation of the models through controlled virtual screening experiments.
Keywords: G protein-coupled receptors (GPCRs), membrane spanning helices, extracellular loops, homology modeling, de novo modeling, multiple sequence alignment, model validation, controlled virtual screening
1. Introduction
G protein-coupled receptors (GPCRs), also known as seven transmembrane (7TM) receptors, are proteins expressed on the plasma membrane that mediate the receiving of extracellular stimuli given by a variety of first messengers (1). The latter can be either endogenous molecules secreted by the body, for example neurotransmitters or hormones, or exogenous molecules of external origin, for example odorants. In humans, the superfamily of GPCRs includes over 800 members that, according to the GRAFS classification scheme, can be divided into five main families: the glutamate family (G; also class C or family III), the rhodopsin family (R; also class A or family I), the adhesion family (A; also class B or family 2, together with the secretin family), the frizzled/taste2 family (F), and the Secretin family (S, also class B or family 2, together with the adhesion family) (2). The rhodopsin family, which also comprises numerous odorant receptors, is by far the largest of the five, accounting for about 84% of the entire superfamily (2). Coupling with intracellular proteins, GPCRs transduce extracellular stimuli into biochemical signals that alter the functioning of the cell, with vast physiological and pathophysiological implications (1). Notably, GPCRs signaling can be ad hoc modulated by exogenous molecules that either stimulate the receptors in lieu of their physiological first messengers or block their stimulation. As a result of this opportunity for pharmacological intervention, GPCRs are the target of a large share of the currently marketed drugs (3) and are the object of intense studies aiming at the development of novel therapeutic strategies.
Despite the large size of the superfamily, GPCRs have traditionally been characterized by a paucity of structural information and, for many years, detailed three-dimensional (3D) structures were available only for rhodopsin. However, rhodopsin is a peculiar receptor with a very distinctive mechanism of activation: it features a covalently bound ligand, retinal, that triggers activation of the receptor upon isomerization by the action of light photons – for a synoptic perspective on the role of rhodopsin as a prototypical class A GPCR, see Costanzi et al. (4). More recently, breakthroughs in GPCR crystallography led to the solution of the structure of additional receptors, all belonging to class A. Specifically, as shown in Table 1, at the time of this writing the Protein Data Bank (www.rcsb.org), enlists structures for: bovine rhodopsin crystallized in the ground state and at early stages of the photoactivation cycle; squid rhodopsin; the unliganded opsin alone and in complex with the C-terminal peptide of the α-subunit of transducin; the β1 and β2 adrenergic receptors in complex with a variety of blockers and agonists; the adenosine A2A receptor in complex with a neutral antagonist; the CXCR4 chemokine receptor in complex with a small molecule and a cyclic peptide antagonist; and the dopamine D3 receptor (4–10). Additional structures are very likely to be solved in the near future.
Table 1.
Crystal structures of GPCRs deposited in the Protein Data Bank (www.rcsb.org) at the time of this writing.
| Receptor | PDB ID |
|---|---|
| Bovine rhodopsin, ground state | 1F88 (40), 1GZM (41), 1HZX (42), 1L9H (43), 1U19 (44), 2I35 (45), 2I36 (45), 2J4Y (46),a 3C9L (47),b 3C9M (47)b |
| Bovine rhodopsin, early stages of photoactivation | 2G87 (48), 2HPY (49), 2I37 (45), 2PED (50) |
| Squid rhodopsin, ground state | 2ZIY (34), 2Z73 (51) |
| Bovine opsin | 3CAP (52), 3DQB (53)d |
| Turkey β1 adrenergic receptor in complex with antagonists, partial agonists, and full agonists | 2VT4 (33),a,e 2Y00 (9),a,f 2Y01 (9),a,f 2Y02 (9),a,g 2Y03 (9),a,g 2Y04 (9)a,f |
| Human β2 adrenergic receptor in complex with inverse agonists, antagonists, and agonists | 2R4R (54),h,i,j 2R4S (54),h,i,j 2RH1 (27, 28),i,k 3D4S (55),i,k 3KJ6 (56),h,i,j 3NY8 (57),i,k 3NY9 (57),i,k 3NYA (57),e,k 3P0G (7),g,k.l 3PDS (8)k,m |
| Human adenosine A2A receptor in complex with an antagonist | 3EML (58)e,k |
| Human CXCR4 chemokine receptor in complex with antagonists | 3ODU (6),e,k 3OE9 (6),e,k 3OE8 (6),e,k 3OE6 (6),e,k 3OE0 (6)k,n |
| Human dopamine D3 receptor | 3PBL (10)e,k |
Thermally stable mutant receptor.
Alternative model of 1GZM
Alternative model of 2J4Y
In complex with a C-terminal peptide of the α-subunit of transducin.
In complex with an antagonist.
In complex with a partial agonist.
In complex with a full agonist.
In complex with a Fab.
In complex with an inverse agonist.
Ligand not visible.
T4-lysozime fusion protein.
In complex with a camelid antibody fragment
In complex with an irreversible agonist
in complex with a cyclic peptide antagonist.
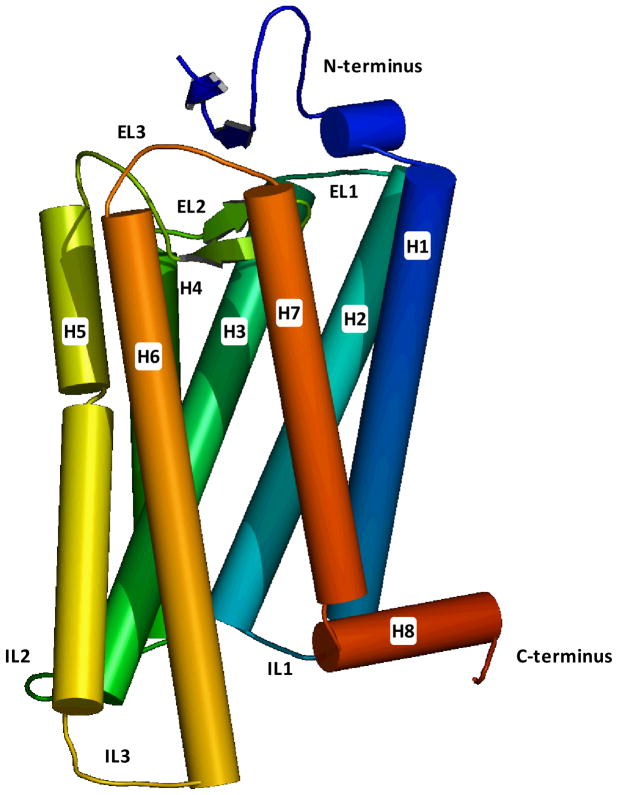
The experimentally elucidated structures confirmed the idea, initially founded on sequence analysis (4), that GPCRs are constituted by a single polypeptide chain that spans the plasma membrane seven times, with seven α-helical structures (numbered from helix 1 to helix 7) interconnected by three extracellular and three intracellular loops (ELs and ILs, numbered from EL1 to EL3 and from IL1 to IL3), as schematically shown in Figure 1 (11). The N-terminus is in the extracellular milieu. Although usually relatively short, for some receptors – notably those belonging to class B and C and to the glycoprotein hormone subfamily of class A – this region is fused to a large soluble ectodomain responsible for ligand binding. For the protease activated receptors (PAR), the N-terminus plays a very peculiar role: it functions as a tethered ligand that, when unmasked by the action of proteases, activates the receptor. The C-terminus, instead, is inside the cytoplasm. Notably, for all the receptors crystallized at the time of this writing, with the exception of the CXCR4 chemokine receptor, the portion of the C-terminal domain immediately following the junction with helix 7 has been shown to adopt an α-helical structure parallel to the plane of the membrane, dubbed helix 8. Sequence similarity suggests that many of the receptors belonging to the rhodopsin family may feature this amphipathic helix.
Figure 1.
Schematic representation of the crystal structure of bovine rhodopsin (1GZM), showing the seven transmembrane domain spanning topology characteristic of GPCRs. The structure is rendered with a continuum spectrum of colors going from blue, at the N-terminus, to red, at the C-terminus.
With such a large superfamily of pharmaceutically appealing receptors and so little structural information, homology modeling, initially based exclusively on the structure of rhodopsin, became a widespread technique to get insights into the structure-function relationships of the receptors and facilitate the discovery of chemicals capable of modulating their activity (4, 11, 12). In the most successful examples, the models were generated on the basis of biochemical and medicinal chemistry data, especially for the in silico generation of the complexes between the receptors and the small molecule ligands (13). A particularly powerful approach is the neoceptor/neoligand method developed by Jacobson and coworkers, in which receptor-ligand interactions are probed through mutagenesis experiments coupled to complementary chemical modification of the ligands (14).
In recent times, the above mentioned advancements in GPCR crystallography have significantly changed the landscape of GPCR homology modeling. First of all, multiple template strategies can now be applied to the construction of the models (11, 15, 16) – for a detailed analysis of the impact of the disclosure of new crystal structures to GPCR homology modeling, see Mobarec and coworkers (16). Moreover, comparisons between in silico and experimental models of the same receptor are now possible and can be used not only to evaluate the state of the art, but also to develop new and improved modeling strategies. In this context, soon after the β2 adrenergic receptor became the first GPCR, after rhodopsin, with a crystallographically elucidated structure, I published the first direct evaluation of the accuracy of a GPCR homology model (17). In particular, I compared the crystal structure of the β2 adrenergic receptor in complex with its inverse agonist carazolol to in silico models of the same receptor-ligand complex constructed through rhodopsin-based homology modeling followed by molecular docking. Notably, not only the structure of the receptor, but also the binding mode of the ligand and the receptor ligand interactions were approximated reasonably well by the models. A wider evaluation of the state of the art was subsequently provided by the first “community-wide assessment of GPCR structure modeling and ligand docking”, organized in coordination with the solution of the structure of the adenosine A2A receptor in complex with the neutral antagonist ZM241385 (18). This time, models of the receptor-ligand complex were submitted to the organizers of the assessment by a number of molecular modelers prior to the unveiling of the crystal structure. In line with what I had found for the β2 adrenergic receptor, this blind test revealed that the seven-helix bundle of the A2A receptor could be built with good accuracy, while the modeling of the interconnecting loops, especially the long ones, was confirmed to be problematic. The docking of the ligand revealed to be a very challenging aspect too, as testified by the wide distribution found for the accuracy of the predictions. However, the top three scoring models (submitted by Costanzi, Abagyan/Katrich and Abagyan/Lam) predicted correctly over 40% of the total number of the receptor-ligand contacts. At the time of this writing, a second community wide assessment is underway (see cmpd.scripps.edu/GPCRDock2010).
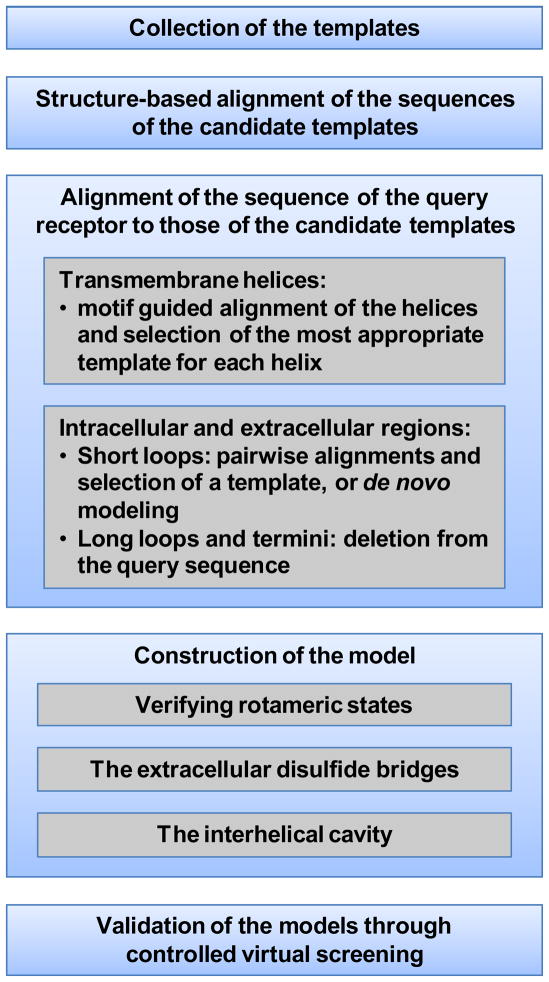
This chapter, geared towards researchers already familiar with homology modeling, provides suggestions, guidelines and critical thoughts on some of the most crucial aspect involved in the constructions of homology models of the serpentine domain of class A receptors (see Figure 2 for a schematic overview).
Figure 2.
Schematic overview of the aspects of class A GPCR modeling discussed throughout this chapter.
2. Materials
The construction and validation of homology models of GPCRs entails performing sequence alignments – including structure-based sequence alignments generating and refining 3D models, and performing docking-based virtual screening experiments. These operations can be carried out by means a variety of web servers as well as commercial and freely available software. Of note, this chapter is intended for researcher well versed with homology modeling, and does not deal with technical aspects relative to the use of specific software packages.
3. Methods
3.1 Collection of the templates
As mentioned, for a long time rhodopsin has been the only available template for the construction of homology models of class A GPCRs (4). However, this is not the case anymore, as crystal structures for a number additional of receptors have been recently solved (4–6).
Files with the coordinates of the crystallized class A GPCRs (see Table 1) can be directly downloaded in PDB format from the web site of the Protein Data Bank (www.rcsb.org). Of note, the availability of additional templates may be verified at any given moment through the “Advanced Search” feature of the web site, which allows conducting “Sequence Blast” searches based on the amino acid sequence of the query receptor, i.e. the receptor object of the modeling project.
3.2 Structure-based alignment of the sequences of the templates
Prior to the selection of the most suitable structure – or of multiple structures – to be used as template for the construction of the model of the query receptor, it is convenient to align the amino acid sequences of the candidate templates. Since structures are more conserved than sequences and since, by definition, 3D coordinates are available for all the templates, it is opportune to derive this sequence alignment through a structure-based alignment method. More specifically, it is advisable to derive the multiple sequence alignment only for the seven membrane spanning helices and, when present, for the amphipathic helix 8. In fact, it is, in these domains that the highest structural conservation is observed in GPCRs, while a much higher variability is observed in the extracellular and the intracellular regions (5).
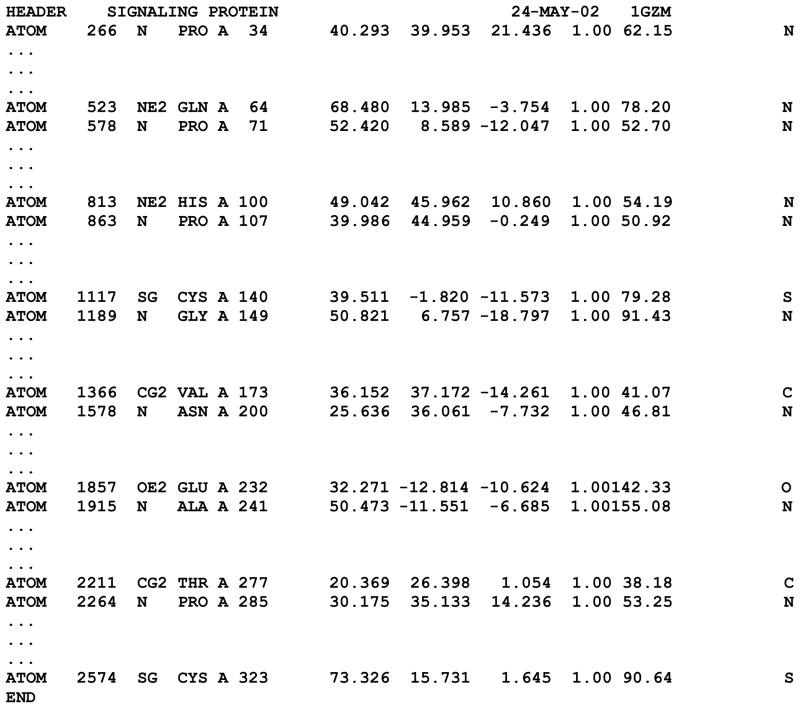
Before subjecting the PDB files to the structure-based sequence alignment, they should be appropriately edited, as several of their sections need to be expunged (see Notes 1 and 2). In particular, a PDB file often includes multiple receptor molecules contained in the unit cell, each of which with a unique chain name – for example, the β1 adrenergic receptor structure deposited with the PDB ID of 2VT4 contains four distinct instances of the receptor (chains A, B, C, and D). One of the chains should be selected to serve as a potential template for the construction of the homology model, while the others should be deleted (for a caveat on how to choose the right chain, see Note 3). A PDB file may also contain additional proteins co-crystallized with the receptor – for example, the β2 adrenergic receptor structure deposited with the PDB ID of 3R4R contains, in addition to the coordinated of the receptor (chain A), those of the light and heavy chains (chains L and H, respectively) of a co-crystallized Fab (fragment antigen binding) that recognizes the IL3 domain of the receptor. All the records pertinent to theses chains should be deleted. For the chain of interest, the ATOM records pertinent to the helical bundle of the receptor are essential for the structure-based sequence alignment and must be preserved (see Note 4). All other records, among which those relative to ligands and co-factors as well as intracellular and extracellular regions are not necessary and may be deleted. Importantly, if the crystal structure has been obtained for a fusion protein of the receptor with the T4-lysozyme, the ATOM records relative to the latter must be deleted too. By way of example, the rhodopsin structure deposited with the PDB ID of 1GZM can be reduced to what represented in Figure 3.
Figure 3.
Example of a simplified PDB file that can be used to generate a structure-based alignment of the helical bundle of the candidate templates. For each helix, the figure shows only the entries corresponding the first atom of the first residue and the last atom of the last residue, while the entries in between are indicated by suspension marks. The simplified PDB file refers to the rhodopsin structure deposited with the PDB ID of 1GZM. The segment from Pro285 to Cys323 refers to both helix 7 and helix 8.
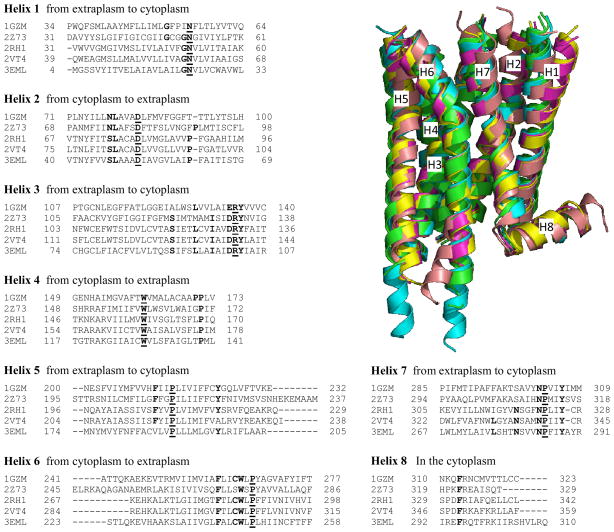
The edited PDB files of the crystallized receptors can then be used to derive a structure-based sequence alignment that, in turn, can serve as a tool for the selection of the template – or of the multiple templates – to be used for the construction of the helical bundle of the query receptor (see section 3.3). Instead, for the selection of the template for the extracellular and intracellular regions, when this is possible, pairwise alignments between each single template and the receptor to be modeled are more appropriate (see section 3.4). As a guide, a structure-based sequence alignment of the seven membrane spanning helices and the amphipathic helix 8 of bovine and squid rhodopsin, the β1 and β2 adrenergic receptors and the adenosine A2A receptors is provided in Figure 4, together with a 3D view of the resulting structural superimposition.
Figure 4.
Structure-based alignment of the sequences of the seven membrane spanning helices and the amphipathic helix 8 of bovine rhodopsin (1GZM), squid rhodopsin (2Z73), human β2 adrenergic receptor (2RH1), turkey β1 adrenergic receptor (2VT4), and adenosine A2A receptors (3EML). The most conserved residue of each helix, as defined by Ballesteros and Weinstein (see Note 5), is in bold and underlined, while additional significantly conserved residues are in bold (see Figure 5). A 3D structural superimposition is also provided, where bovine and squid rhodopsin are in green and cyan, the β1 and β2 adrenergic receptors in yellow and purple, and the adenosine A2A receptor in pink.
3.3 Alignment of the query sequence to the prealigned helical bundle of the candidate templates
The alignment of the sequence of the query receptor to the prealigned helical bundle of the candidate templates can be achieved starting with an automatic sequence alignment, performed without allowing the relative alignment of the candidate templates to change. The alignment obtained in this manner, should be subsequently subjected to a careful visual inspection and manual refinement. In particular, the correct identification of the seven membrane spanning helices of the query receptor must be verified on the basis of the presence of specific motifs, also called conservation patterns, that characterize each helix (Figure 5) (19). Of particular importance is the identification and the correct alignment of the most conserved residue of each helix (Figure 5), defined as residue X.50 according to the GPCR residue indexing system (see Note 5) (20, 21). Of note, these motifs, although frequent, are not present in the membrane spanning helices of all receptors, sometimes making the identification of a certain helix difficult. Once all the helices have been identified, the automatic alignment should be inspected and, if necessary, adjusted to ensure that the motifs of the query are aligned with those of the candidate templates. The presence of gaps in the alignment of the helices should also be avoided (however, see Note 6).
Figure 5.
Motifs relatively common in each of the seven membrane spanning helices and the amphipathic helix 8 of GPCRs. The most conserved residues of each helix, as defined by Ballesteros and Weinstein (see Note 5), are in bold and underlined; Xn indicates n contiguous non conserved residues; residues in parentheses often replace the preceding residue.
3.3.1 Single template or multiple templates?
Given that the structure of several GPCRs has been solved through X-ray crystallography, GPCR homology models can now be constructed through either a single or a multiple template strategy (16). Single template strategies involve the selection of the crystallized receptor that, overall, seems more likely to be characterized by structural similarity with the query receptor, while multiple template strategies involve the splitting of the query receptor into several domains and the subsequent selection of the most suitable template for each of these domains. In particular, once the sequences of candidate templates and query receptors have been aligned, the selection of the templates can be operated on the basis of sequence similarities, for instance through the calculation of percentages of accepted mutations (PAMs) and/or the presence of specific sequence motifs. Of note is an article published by Worth and coworkers that outlined a detailed integrated workflow for the identification of suitable templates for each of the seven membrane spanning helices and the amphipathic helix 8, based on a thorough structural analysis of the crystallized GPCRs(15). In particular, according to this scheme, the selection criteria should be based not only on sequence similarities but also on the detection of specific features and motifs detected in the sequence of the query receptor, such as the presence of specific glycine and proline residues responsible for helical kinks, or cysteine residues putatively involved in the formation of disulfide bridges (regarding the modeling of helix 7 and helix 8, see Note 7). For advice on how to construct a homology model on the basis of multiple templates, see Note 8.
3.4 The extracellular and intracellular regions: to align or not to align, that is the question
The extracellular and intracellular domains of class A GPCRs are characterized by very low sequence similarity and great length variability, which make their sequences less straightforward to align than the seven membrane spanning helices. As outlined by the published crystal structures (5, 6), the lack of sequence of similarity detected for these regions is paralleled by a correspondent significant structural diversity, which hampers their modeling by homology. Moreover, further hindering homology modeling, termini and long loops have not been solved for many of the currently crystallized receptors, while in some of the crystal structures IL3 is substituted by a fused T4-lysozyme (5). Thus, not surprisingly, molecular models of class A GPCRs are usually significantly more accurate in the helical bundle than in the extracellular and intracellular regions, if we exclude short interconnecting loops (18). Notably, besides the purely computational methods discussed in this chapter, hybrid experimental and computational approaches have also been proposed, whereby the structures of peptides mimicking the extracellular and intracellular regions of a receptor are determined experimentally, for instance through NMR spectroscopy, and subsequently merged with an in silico generated model of the helical bundle (22). Such hybrid models may offer a very powerful approach to the study of receptors that have not yet been crystallized.
3.4.1 Avoiding the alignment: de novo modeling or omission of the loop
A viable solution for the construction of short interconnecting loops can be found in de novo modeling, an approach not based on the use of a template. If this is the chosen route, the corresponding domain can be deleted from the structure of the template. Of note, if cysteine residues are present in the loop of the query receptor, special care deserves the analysis of their possible involvement in the formation of disulfide bridges on the basis of sequence analyses and experimental data (see section 3.5).
In some GPCRs, however, the considerable length of termini and some of the loops – notably IL3 – prevents an effective use of de novo modeling for their construction. It is advisable not to model the terminal regions, constructing only the portion of the receptor between the beginning of helix 1 and the end of helix 7 – or helix 8, when this thought to be present. Similarly, it is advisable not to model long loops. The omission of a domain from the model can be achieved by deleting the corresponding sequence in the query receptor (for the loops, see Note 9).
3.4.2 Aligning the loops
Despite the caveats expressed in the previous two subsections, homology modeling can be applied to the construction of interconnecting loops with a length comparable to that of the corresponding regions of the template. In this case, a sequence alignment and the selection of a template are necessary.
Due to the mentioned low sequence similarity and length variability, the alignment of the loops is better performed in a pairwise manner comparing the query receptor to one template at the time, rather than in a multiple sequence alignment context. If a loop has not exactly the same length in the template and the query receptor, a gap will have to be inserted in the sequence of the shorter one. As always in homology modeling, special care needs to be put into the positioning of such gaps, which should be driven not only by the attempt to maximize the similarity score, but also by a careful structural analysis of the template. Specifically, it is important to ensure that insertions or deletions are placed in a position compatible with the structure of the template.
If a single template strategy is chosen, it will be sufficient to align the loops of the query receptor to the corresponding loops of the template receptor chosen on the basis of the sequence similarity detected in the helical bundle. Instead, if a multiple strategy template has been chosen, once a loop of the query receptor has been separately aligned with the corresponding loop of each of the candidate templates, the template for the construction of the model can be selected according to sequence similarity or on the basis of the conservation of specific amino acids. Additionally, it is important to carefully analyze the geometric compatibility between the candidate template for the modeling of the loop and the templates chosen for the modeling of the two helices that the loop connects.
3.4.3 Special considerations concerning the second extracellular loop
EL2 connects helix 4 and helix 5 and, in the majority of class A GPCRs, is characterized by a highly conserved cysteine residue that connects it to helix 3. Modeling EL2 deserves particular attention since this loop, and in particular the portion downstream of the conserved disulfide bridged cysteine residue, is directly involved in the lining of the interhelical cavity that putatively hosts the orthosteric binding site for all members of class A GPCRs that are activated by small molecules. The crystal structures of class A GPCRs that have been solved at the time of this writing revealed that EL2 does not feature a common structure shared by all receptors (5, 6, 10), and adopts four different conformations in rhodopsin, β adrenergic, adenosine A2A, dopamine D3, and CXCR4 chemokine receptors. Specifically, in rhodopsin EL2 is characterized by a distinctive β-hairpin conformation that lays over the opening of the interhelical cavity restricting the access of water from the extracellular side, while in the β adrenergic, adenosine A2A, dopamine D3, and CXCR4 chemokine receptors it assumes a significantly more open conformation. These differences are probably attributable to the fact that, while rhodopsin features a covalently bound inverse agonist, 11-cis-retinal, that is isomerized in situ to its all-trans form by the action of a light photon and consequently triggers the activation of the receptor, the remainder of class A GPCRs are physiologically activated by diffusible agonists (4) (see Note 10).
Despite this common feature that distinguishes receptors for diffusible ligands from rhodopsin, however, a profound structural variability for EL2 has been detected among the various experimentally solved receptors, also due to the different arrays of disulfide bridges detected in their extracellular regions (5). This lack of structural conservation prevents the use of homology modeling for the construction of EL2, unless template and query receptors belong to the same subfamily, and suggests that better results could be achieved through de novo modeling, enforcing the formation of the disulfide bridges that putatively exist in the query receptor (see section 3.5). Accordingly, through a comparison of different rhodopsin-based models of the β2 adrenergic receptor, I have demonstrated that those that featured a de novo modeled EL2 resulted in lower root mean square deviations in the regions downstream of the disulfide bridge (17). In turn, this yielded the production of significantly more accurate ligand poses as a result of molecular docking (17), as well as better performances when the models were used as platforms for controlled docking-based virtual screening (23).
Alternatively to complete de novo modeling, a short portion around the conserved cysteine residue may be built by homology with one of the templates, while building the remainder of the loop de novo. Notably, I have used this strategy for the construction of C-terminal portion of EL2 in the adenosine A2A receptor model for the above-mentioned “community-wide assessment of GPCR structure modeling and ligand docking” – see supplementary information of (18) for the sequence alignment.
If the models are constructed with the intent of studying the interactions of the receptors with small molecules that bind to their interhelical cavity or conducting docking-based virtual screening experiments targeting said cavity, the segment of EL2 that really matters is the one that is downstream of the above mentioned conserved disulfide bridge that links the loop to helix 3. The remainder of the loop, if too long to allow robust de novo modeling, may be omitted (see Note 9).
3.5 Construction of the model
Once a sequence alignment has been obtained and the proper portions of query and/or template sequences have been deleted as outlined in the previous sections, a 3D model of the query receptor can be constructed through homology modeling or a combination of homology and de novo modeling – most modeling packages will directly build de novo those domains of the query receptor that are not aligned with a template.
Verifying rotameric states
Due to the availability of multiple templates, after the construction of a model, the rotameric state of each residue can be verified and adjusted in light of the whole set of crystallized receptors. Notably, if a residue of the query receptor is not conserved in the template employed to model the domain to which it belongs, nonetheless it may be conserved in one or more of the other crystallized receptors. As the structures of additional GPCRs will be solved, the number of residues of a query receptor that will be conserved in at least one of the templates will increase significantly, with obvious beneficial repercussions on homology modeling (16).
Special considerations on the extracellular disulfide bridges
As mentioned, the extracellular domains of most class A GPCRs are characterized by the presence cysteine residues involved in the formation of disulfide bridges. Among these, the disulfide bridge that connects EL2 to helix 3 is widely conserved within class A, while additional bridges, when present, are often peculiar to a specific subfamily of receptors, to which they confer a characteristic extracellular architecture functional to ligand binding. As mentioned, it is of utmost importance that the presence of cysteine residues and their putative involvement in the formation of disulfide bridges be identified prior to the construction of the model. In addition to computer-based sequence analyses, the detection and the corroboration of the presence of such bridges can be greatly assisted by biochemical data, either ad hoc generated or retrieved from the literature. For instance, mutagenesis data suggested the presence of a disuldide bridge connecting EL3 to the N-terminus of the P2Y receptors (24, 25), while they accurately predicted the presence of a disulfide bridge internal to EL2 of the β2 adrenergic receptor (26), successively confirmed by the crystal structures (27, 28). Some software for homology modeling allows the enforcement of the formation of disulfide bridges between specified pairs of cysteine residues. This feature is particularly important when the cysteine residues are not conserved in the templates or whenever using de novo loop modeling. However, if this feature is not available within the chosen software, one possible solution is the construction of many alternative loop models and the subsequent selection of those that feature the cysteine pair at a distance compatible with the formation a disulfide bridge, if present. Alternatively, the disulfide bridges can be generated after the construction of the model, for instance through molecular dynamics simulations with a harmonic restraint applied to the distance between the sulfur atoms of the bridged cysteine pairs. After the proper connection of the putative disulfide bridges, a thorough exploration of the conformations accessible to extracellular and intracellular loops, possibly in light of experimental data, is also advisable. Of note, for the extracellular loops, sometimes this operation could be better performed following the docking of a ligand (for instance, see (29)).
Special considerations on the interhelical cavity
In general, when the ligand co-crystallyzed with the template binds also to the query protein, the use the co-crystallized ligand as environment for the construction of the model significantly helps the modeling of the binding pocket and facilitates the formation of protein-ligand interactions. However, when modeling class A GPCRs, given the wide diversity found within the class and the specificity of each subfamily for a particular set of natural and synthetic ligands, in very rare cases the query receptor will share ligands with any of the available templates. Nonetheless, using the ligand co-crystallized with one of the templates as environment may still be a good practice to grant to the model a binding pocket suitable for molecular docking. Often, in fact, homology modeling procedures tend to occlude internal cavities through subtle backbone movements, especially if the construction of the model involves unrestrained energy minimizations, and through the orientation of the side chains of the residues that line the cavity towards the center of it. However, building the model of a class A GPCR around the ligand co-crystallized with one of the templates can induce artificial rotameric states to some of the residues that line the binding pocket. For example, I have shown that, when building the β2 adrenergic receptor using rhodopsin as the template and the co-crystallized retinal as the environment (17), Phe290 is prevented from adopting its natural the gauche(+) conformation by the presence of retinal (see Figure 6). Thus, after the construction of the model a thorough exploration of the rotameric states of the residues that line the binding cavity is needed. This operation can be conveniently performed after the generation of preliminary docking poses of a chosen ligand, possibly guided by experimental constraints, through a variety of differently implemented procedures dubbed “ligand-supported”, “ligand-based”, or “ligand-steered” or homology modeling (13, 30, 31).
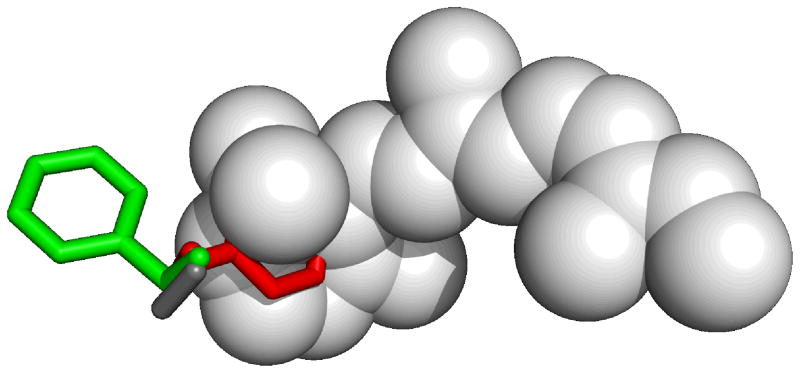
Figure 6.
As indicated by the structural superimposition shown here, Phe290 cannot adopt the right rotameric state in a rhodopsin-based model of the β2 adrenergic receptor constructed using retinal as the environment: retinal (in light gray, from 1GZM), would sterically prevent Phe290 from adopting the gauche(+) conformation revealed by the crystal structure (in red, from 2RH1) and would force it in the trans conformation (in green, from a rhodopsin-based homology model (17)). Of note, in rhodopsin, the residue corresponding to Phe290 is an alanine, namely Ala269 (in dark gray, from 1GZM).
3.6 Validation of the models through virtual screening experiments
The ultimate validation of a GPCR homology model can only derive from a direct comparison with its experimentally elucidated structure. However, such a comparison is only possible either when the model of a crystallized receptor is generated so as to probe scope and limitations of the modeling techniques, or, retroactively, when the experimental structure of a previously modeled receptor becomes available, possibly many years after the model was generated. In fact, if a computational model of a receptor is generated in order to shed light into its structure-function relationships and, possibly, to facilitate the discovery of ligands capable of modulating its activity, this very fact implies that experimental structures do not exist for the query receptor. Thus, for all intents and purposes, the only possible way to validate the usefulness of a homology model – if not necessarily its accuracy – is to test the correlation between predictions generated on its basis and experimental results. In particular, if homology models have been built with the purpose of studying receptor-ligand interactions and conducting structure-based drug discovery, the best way to validate their efficacy is to subject them to a series of controlled virtual screening experiments. These are usually performed docking at the receptor a dataset of compounds containing a number of known ligands mixed with a larger number of decoys, i.e. compounds with physico-chemical characteristics similar to those of the ligands but presumed to be inactive. Then, the ability of the screening to prioritize ligands over decoys is evaluated by monitoring enrichment factors and/or areas under the receiver operating characteristic (ROC) curve (23, 29, 31, 32). Such controlled experiments constitute very good tools not only for the selection of the initial models, but also for the control of the entire optimization process, including the refinement of loops and side-chains. Clearly, controlled virtual screening can only be performed if a significant amount of known ligands for the query receptor exists (see Note 11), while can be applied with difficulty to receptors characterized by a marked paucity of known ligands and not applied at all to orphan receptors. Moreover, it is worth keeping in mind that better virtual screening performances do not necessarily parallel higher levels of overall accuracy, and may reflect a particularly favorable arrangement, either natural or artificial, of the side chains of the residues that line the binding pocket (16, 17, 29).
Acknowledgments
This work was supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
Text editors can be conveniently used to read and edit PDB files. Alternatively, the files can be directly edited within the specialized modeling package of choice.
For a description of the PDB file format see http://www.pdb.org/docs.html.
It is not always safe to blindly opt for the first chain (usually named chain A) and discard the others. The B-factors of the various chains and their completeness are certainly important parameters on which to base the selection. Moreover, in order to choose the best chain to work with, a careful reading of the main article that describes the crystal is of utmost importance. For example, in the case of the β1 adrenergic receptor (PDB ID: 2VT4) chain B is to be preferred to chain A, since, as explained by the authors, the latter presents an anomalous 60° kink in helix 1 (33).
For a correct interpretation of the secondary structure, some programs require also the portion of the PDB file that defines it (record type: HELIX and SHEET).
The GPCR residue identifier system, devised by Ballesteros and Weinstein, is a universal way of numbering GPCR residues on the basis of reference positions that the authors identified for each of the 7 membrane spanning helices (20). Specifically, through the analysis of a sequence alignment of Class A receptors, the authors selected a reference position for each of the seven helices, chosen among those featuring one of the most conserved residues in that helix. They then defined a convention by which the identifier X.50 – where X is the helix number – is arbitrarily assigned to the reference position, while the remaining residues in the helix are numbered relatively to the reference. Later, van Rhee and Jacobson introduced a modification to the Ballesteros and Weinstein system according to which each residue is indicated with its original sequence number followed by the residue identifier, rather than solely with the residue identifier (21).
Although insertion and deletions within the seven helices are not common, structure-based alignments indicate the presence of an insertion in helix 2 of squid rhodopsin (see Figure 4) (15, 34). Moreover, the C-terminal region of helix 7, close to the hinge with helix 8, presents a deletion in some receptors, leaving only five rather than six residues between the Tyr and the Phe of the conserved NPX2YX5,6F motif (see Figure 4) (35).
The presence of either five or six intervening residues between the conserved tyrosine and phenylalanine at the hinge between helix 7 and helix 8 (see note 6) may guide the selection of the template for this region (15). Importantly, if sequence analysis does not strongly support the presence of an amphipathic helix, the sequence of the query receptor can be truncated at the end of helix 7, leaving the remainder of the receptor unmodeled.
While some homology modeling software allows the direct use of multiple templates, others require the use of a single template. A possible workaround to overcome this limitation is the generation of a hybrid template by cutting and pasting the selected portions of the various crystallized receptors into a single PDB file (on the editing of a PDB file, see also Note 1).
Some homology modeling software requires that the query be an uninterrupted protein chain. In this case, the loop (or a portion of it) can conveniently be deleted after the construction of the model. If the loop destined to be omitted from the model is particularly long, in order to avoid the expenditure of excessive computational time in its construction, it may be advisable to delete its central portion from the query sequence, thus constructing only a relatively short loop that will be subsequently removed.
As suggested by molecular modeling studies, the egression of the cleaved all-trans-retinal consequent the activation of rhodopsin and the following ingression of 11-cis-retinal into the unliganded opsin, to reform a functional rhodopsin unit, occur through openings between adjacent membrane spanning helices (36, 37). Instead, the physiological ligands of the β adrenergic receptors, as well as those of all class A GPCRs naturally activated by small molecules, are very likely to enter and exit the receptor trough the opening of the interhelical cavity towards the extracellular milieu (38).
Known ligands of the query receptor can conveniently be retrieved from the GPCR-ligand database (GLIDA, http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/) (39).
References
- 1.Pierce K, Premont R, Lefkowitz R. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Gloriam D, Fredriksson R, Schiöth H. The G protein-coupled receptor subset of the rat genome. BMC Genomics. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Costanzi S, Siegel J, Tikhonova I, Jacobson K. Rhodopsin and the others: a historical perspective on structural studies of G protein-coupled receptors. Curr Pharm Des. 2009;15:3994–4002. doi: 10.2174/138161209789824795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson MA, Stevens RC. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010 doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–40. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, Leslie AG, Schertler GF, Tate CG. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–4. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–5. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzi S. Modelling G protein-coupled receptors: a concrete possibility. Chimica Oggi-Chemistry Today. 2010;28:26–30. [PMC free article] [PubMed] [Google Scholar]
- 12.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-based virtual screening of chemical databases. II Are homology models of G-Protein Coupled Receptors suitable targets? Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 13.Moro S, Deflorian F, Bacilieri M, Spalluto G. Ligand-based homology modeling as attractive tool to inspect GPCR structural plasticity. Curr Pharm Des. 2006;12:2175–85. doi: 10.2174/138161206777585265. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson K, Gao Z, Liang B. Neoceptors: reengineering GPCRs to recognize tailored ligands. Trends Pharmacol Sci. 2007;28:111–6. doi: 10.1016/j.tips.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worth C, Kleinau G, Krause G. Comparative sequence and structural analyses of G-protein-coupled receptor crystal structures and implications for molecular models. PLoS One. 2009;4:e7011. doi: 10.1371/journal.pone.0007011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobarec J, Sanchez R, Filizola M. Modern Homology Modeling of G-Protein Coupled Receptors: Which Structural Template to Use? J Med Chem. 2009;52:5207–16. doi: 10.1021/jm9005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the beta2-adrenergic receptor. J Med Chem. 2008;51:2907–14. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michino M, Abola E, Brooks Cr, Dixon J, Moult J, Stevens R 2008 Participants, G. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8:455–63. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Rhee AM, Fischer B, van Galen PJ, Jacobson KA. Modelling the P2Y purinoceptor using rhodopsin as template. Drug Des Discov. 1995;13:133–54. [PMC free article] [PubMed] [Google Scholar]
- 20.Ballesteros JA, Weinstein H. Integrated method for the consturction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 21.van Rhee AM, Jacobson KA. Molecular architecture of G protein-coupled receptors. Drug Develop Res. 1996;37:1–38. doi: 10.1002/(SICI)1098-2299(199601)37:1<1::AID-DDR1>3.0.CO;2-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tikhonova I, Costanzi S. Unraveling the structure and function of G protein-coupled receptors through NMR spectroscopy. Curr Pharm Des. 2009;15:4003–16. doi: 10.2174/138161209789824803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilar S, Ferino G, Phatak SS, Berk B, Cavasotto CN, Costanzi S. Docking-based virtual screening for ligands of G protein-coupled receptors: Not only crystal structures but also in silico models. J Mol Graph Model. 2010 doi: 10.1016/j.jmgm.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann C, Moro S, Nicholas RA, Harden TK, Jacobson KA. The role of amino acids in extracellular loops of the human P2Y1 receptor in surface expression and activation processes. J Biol Chem. 1999;274:14639–47. doi: 10.1074/jbc.274.21.14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costanzi S, Mamedova L, Gao Z, Jacobson K. Architecture of P2Y nucleotide receptors: structural comparison based on sequence analysis, mutagenesis, and homology modeling. J Med Chem. 2004;47:5393–404. doi: 10.1021/jm049914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noda K, Saad Y, Graham RM, Karnik SS. The high affinity state of the beta 2-adrenergic receptor requires unique interaction between conserved and non-conserved extracellular loop cysteines. J Biol Chem. 1994;269:6743–52. [PubMed] [Google Scholar]
- 27.Cherezov V, Rosenbaum D, Hanson M, Rasmussen S, Thian F, Kobilka T, Choi H, Kuhn P, Weis W, Kobilka B, Stevens R. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbaum D, Cherezov V, Hanson M, Rasmussen S, Thian F, Kobilka T, Choi H, Yao X, Weis W, Stevens R, Kobilka B. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 29.Katritch V, Jaakola V, Lane J, Lin J, Ijzerman A, Yeager M, Kufareva I, Stevens R, Abagyan R. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J Med Chem. 2010;53:1799–809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evers A, Klebe G. Ligand-supported homology modeling of g-protein-coupled receptor sites: models sufficient for successful virtual screening. Angew Chem Int Ed Engl. 2004;43:248–51. doi: 10.1002/anie.200352776. [DOI] [PubMed] [Google Scholar]
- 31.Cavasotto CN, Orry AJ, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, O’Neill KA, Hine H, Burton MS, Voigt JH, Abagyan RA, Bayne ML, Monsma FJ., Jr Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J Med Chem. 2008;51:581–8. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 32.Vilar S, Karpiak J, Costanzi S. Ligand and structure-based models for the prediction of ligand-receptor affinities and virtual screenings: Development and application to the beta(2)-adrenergic receptor. J Comput Chem. 2010;31:707–20. doi: 10.1002/jcc.21346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warne T, Serrano-Vega M, Baker J, Moukhametzianov R, Edwards P, Henderson R, Leslie A, Tate C, Schertler G. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–91. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimamura T, Hiraki K, Takahashi N, Hori T, Ago H, Masuda K, Takio K, Ishiguro M, Miyano M. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J Biol Chem. 2008;283:17753–6. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci U S A. 2003;100:2290–5. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J Am Chem Soc. 2007;129:6970–1. doi: 10.1021/ja0691977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hildebrand PW, Scheerer P, Park JH, Choe HW, Piechnick R, Ernst OP, Hofmann KP, Heck M. A ligand channel through the G protein coupled receptor opsin. PLoS One. 2009;4:e4382. doi: 10.1371/journal.pone.0004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Duan Y. Ligand entry and exit pathways in the beta2-adrenergic receptor. J Mol Biol. 2009;392:1102–15. doi: 10.1016/j.jmb.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR--ligand database for chemical genomics drug discovery--database and tools update. Nucleic Acids Res. 2008;36:D907–12. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 41.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–38. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 42.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–72. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:5982–7. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–83. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 45.Salom D, Lodowski D, Stenkamp R, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros J, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–8. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–88. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenkamp RE. Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr D Biol Crystallogr. 2008;D64:902–4. doi: 10.1107/S0907444908017162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl. 2006;45:4270–3. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 49.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:12729–34. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamichi H, Buss V, Okada T. Photoisomerization mechanism of rhodopsin and 9-cis-rhodopsin revealed by x-ray crystallography. Biophys J. 2007;92:L106–8. doi: 10.1529/biophysj.107.108225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–7. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 52.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–7. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 53.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 54.Rasmussen S, Choi H, Rosenbaum D, Kobilka T, Thian F, Edwards P, Burghammer M, Ratnala V, Sanishvili R, Fischetti R, Schertler G, Weis W, Kobilka B. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 55.Hanson M, Cherezov V, Griffith M, Roth C, Jaakola V, Chien E, Velasquez J, Kuhn P, Stevens R. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bokoch M, Zou Y, Rasmussen S, Liu C, Nygaard R, Rosenbaum D, Fung J, Choi H, Thian F, Kobilka T, Puglisi J, Weis W, Pardo L, Prosser R, Mueller L, Kobilka B. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–12. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–5. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaakola V, Griffith M, Hanson M, Cherezov V, Chien E, Lane J, Ijzerman A, Stevens R. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]