Abstract
Biodegradable core-shell structured nanoparticles with a poly(β-amino-ester) (PBAE) core enveloped by a phospholipid bilayer shell were developed for in vivo mRNA delivery, with a view toward delivery of mRNA-based vaccines. The pH-responsive PBAE component was chosen to promote endosome disruption, while the lipid surface layer was selected to minimize toxicity of the polycation core. Messenger RNA was efficiently adsorbed via electrostatic interactions onto the surface of these net positively-charged nanoparticles. In vitro, mRNA-loaded particle uptake by dendritic cells (DCs) led to mRNA delivery into the cytosol with low cytotoxicity, followed by translation of the encoded protein in these difficult-to-transfect cells at a frequency of ~30%. Particles loaded with mRNA administered intranasally in mice led to the expression of the reporter protein luciferase in vivo as soon as 6 h after administration, a timepoint when naked mRNA given i.n. showed no expression. At later timepoints, luciferase expression was detected in naked mRNA-treated mice, but this group showed a wide variation in levels of transfection, compared to particle-treated mice. This system may thus be promising for non-invasive delivery of mRNA-based vaccines.
Keywords: Vaccine delivery, gene delivery, messenger RNA, polymeric nanoparticles, lipids
Introduction
Gene-based vaccination was born in 1990 after Wolff et al. demonstrated the local uptake and expression of exogenously injected plasmid DNA and in vitro transcribed messenger RNA (mRNA),1 followed shortly thereafter by the demonstration that injected nucleic acids could promote immune responses to encoded antigens.2,3 Both DNA and mRNA-based vaccines share many advantages over alternative protein, peptide, or live vector-based vaccine strategies in terms of safety, manufacturability and suitability for long-term storage, and the ability to promote broad cytotoxic T-cell responses.4,5 Since the first demonstrations, DNA-based vaccines have undergone extensive preclinical and clinical testing, while mRNA-based therapies remained less thoroughly investigated. However, DNA vaccines have failed to show potency in large-animal models and humans, in discord from results in small-animal studies,6 which may reflect in part the difficulty of overcoming not only the barrier posed by the plasma membrane of cells but also the need to transport DNA through the nuclear membrane of non-dividing cells. Vaccines based on mRNA, by contrast, require nucleic acid delivery only to the cytosol, thereby allowing the transfection of quiescent and post-mitotic cells that comprise the majority of target cells in vivo.7,8
To deliver macromolecules intracellularly, synthetic vectors are preferred over approaches based on viral vectors for their low cost, ease of large-scale production and potential for improved safety.9–12 To date, strategies reported for non-viral delivery of mRNA for vaccines and gene therapy applications include injection of naked mRNA,1,4 polyplexes,13,14 lipoplexes or liposome-entrapped mRNA,15,16 bolistic delivery via gene gun,17,18, particulate carrier-mediated delivery19 and electroporation.20–22 Recently, clinical trials in which human patients were vaccinated with naked or protamine-complexed mRNA against tumor antigens via intradermal injections were completed, demonstrating feasibility, lack of toxicity and promising responses based on clinical and immunological read-outs.23,24 However, more efficient transfection in vivo, and the ability to deliver these nucleic acids non-invasively and/or to mucosal sites, would be expected to enhance the prospects of this strategy for vaccination.
A variety of polymer and/or lipid-based drug delivery systems with the capability to disrupt endosomes have been developed that might be applicable for mRNA vaccine delivery25–29, but systems that achieve efficient cytosolic delivery or transfection in vivo with minimal cytoxicity are still sought. For vaccine applications, cytosolic delivery of mRNA into dendritic cells (DCs), immune cells that play a key role in the initiation of adaptive immune responses30, is desired but the transfection of these cells poses a significant challenge as synthetic agents generally achieve only 10–35% transfection of these cells in vitro.31–33 We previously reported a pH-responsive core-shell nanoparticle system prepared by sequential emulsion polymerization of a secondary-amine-containing monomer (diethlyaminoethyl methylacrylate) forming a pH-responsive core, followed by a second monomer (aminoethyl methacrylate) forming a hydrophilic corona.34,35 The core-shell particle structure enabled the physical and compositional segregation of the functions for the particle into an endosome-disrupting pH-responsive core and a shell whose composition could be separately tuned to facilitate particle targeting, cell binding, and/or drug binding. These particles efficiently delivered associated protein or oligonucleotide cargos to the cytosol of dendritic cells through endosomal disruption mediated by the core polymer via the proton-sponge effect, while maintaining low cytotoxicity by sequestering the cationic charge and hydrophobicity of the polymer core within a more hydrophilic polymeric shell. However, a limitation of this proof-of-concept system is its lack of biodegradability, which hinders clinical translation.
Here we adapted our core-shell design approach to a fully degradable system comprised of a pH-responsive poly(β-amino-ester) (PBAE) core and a phospholipid shell. As a first step toward enhanced mRNA vaccines, we show that negatively-charged mRNA can be adsorbed via electrostatic interactions onto the surface of these cationic nanoparticles, protecting the nucleic acids from degradation in serum. Lipid-enveloped PBAE particles disrupted endosomes and delivered mRNA into the cytosol of DCs with minimal cytotoxicity, leading to in vitro transfection of a DC clone at levels comparable to the best reports for DC transfection from the literature. Importantly, mRNA-loaded lipid-enveloped particles also promoted in vivo transfection following non-invasive intranasal delivery, suggesting their potential utility for mRNA vaccine formulations.
Materials and Methods
Materials
The poly(β-amino ester) poly-1 with a number averaged molecular weight of ~10 kDa was synthesized as previously reported.36 Poly(lactide-co-glycolide) (PLGA) with a 50:50 lactide:glycolide ratio and molecular weight of ~46 kDa was purchased from Lakeshore Biomaterials (Birmingham, AL). The lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy-(polyethyleneglycol) -2000] (ammonium salt) (DSPE-PEG), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy-(polyethyleneglycol)-2000-N′-carboxyfluorescein] (ammonium salt) (DSPE-PEG-CF) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (DOPE-rhodamine) were purchased from Avanti Polar Lipids (Alabaster, AL). Poly(vinyl-alcohol) (PVA) with a molecular weight of ~78 kDa was purchased from Polysciences Inc. (Warrington, PA). Calcein, DAPI and sodium acetate buffer were purchased from Sigma Chemical Co. (St. Louis, MO). Triton X-100 (molecular biology grade) was from Promega Corp. (Madison, WI). Poly (L-aspartic acid) (sodium salt) was from Alamanda Polymers (Huntsville, AL). 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD), Annexin V Alexa 350 conjugate, LysoTracker Green, ribogreen and SYBR Green II RNA gel stain were purchased from Invitrogen (Eugene, OR). Poly I:C was purchased from InvivoGen (San Diego, CA). RNAse inhibitor was purchased from Roche (Indianapolis, IN). D-Luciferin (potassium salt) was from Caliper Life Sciences (Hopkinton, MA). Cy3 Label ITR nucleic acid labeling kit was purchased from Mirus Bio LLC (Madison, WI). All materials were used as received unless otherwise noted.
Synthesis and Characterization of Lipid-coated PBAE Nanoparticles
Lipid-coated nanoparticles with a poly-1 core were synthesized via two different processes: a double emulsion/solvent evaporation approach or a solvent diffusion/nanoprecipitation strategy. For double emulsion synthesis, we adapted a previous approach we used for preparing lipid-enveloped PLGA particles37: 30 mg of poly-1 (or PLGA for pH-insensitive control particles) and 2 mg of the phospholipids DOPC, DOTAP, and DSPE-PEG in a 7:2:1 molar ratio were co-dissolved in 1 ml of dicholoromethane (DCM). PBS (200 μl) was then added to the mixture on ice during a 1 min sonication step at 7 W using a probe tip sonicator (Misonix XL2000, Farmingdale, NY) to form a first emulsion. The primary emulsion was then dispersed into 6 ml of distilled, deionized nuclease-free water with sonication at 12 W for 5 min, before leaving on a shaker for 18 h at 25 °C to evaporate the organic solvent. For nanoprecipitation synthesis, the same quantities of DOPC, DOTAP, and polymer were co-dissolved in 4 ml of ethanol and added drop-wise to 40 ml of distilled, deionized nuclease-free water, followed by gentle stirring for 5 h to evaporate ethanol. For particles prepared by nanoprecipitation, we found that adding DSPE-PEG in the organic phase together with DOPC, DOTAP and polymer reduced particle yield significantly, so DSPE-PEG was introduced into the lipid coating via a post-insertion process: DSPE-PEG lipid was added at 1 mM to 0.5 mg/ml particles in distilled, deionized nuclease-free water and the suspension was stirred for 16 h at 25 °C. The particles were collected and washed once via centrifugation, resuspended in fresh water and stored at 4 °C until use. Lipid-free PVA-stabilized particles were prepared by similar processes except the organic emulsion or solution containing polymer only was dispersed into a 2% w/vol PVA aqueous solution.
A fraction of each particle batch was dried in a vacuum oven to determine the particle concentration (mg/ml) by measuring the dry mass. Dynamic light scattering (DLS) and zeta potential measurements were used to determine the particle size and surface charge using a ZetaPALS dynamic light scattering detector (Brookhaven Instruments). To estimate the percentage of PEG-lipid incorporated and the resultant mol% of PEG-lipid present in the lipid surface coating, lipid-enveloped particles were prepared by nanoprecipitation as described above with 1 mol% DOPE-rhodamine added as a tracer for measuring the total amount of lipid incorporated into the particles, before the post-insertion of a DSPE-PEG lipid labeled with carboxyfluorescein (DSPE-PEG-CF). The lipids were then stripped from the particle surface by treatment with 2% triton X-100 for 15 min and the supernatant was measured for both rhodamine and fluorescein signals to quantitate the amount of PEG lipid incorporated.
To investigate the surface structure of lipid-coated PBAE particles by cryoelectron microscopy (cryoEM), particles were embedded in ice by blotting a particle suspension (3 uL) on a 1.2/1.3 μm holey carbon-coated copper grid (Electron Microscopy Sciences) and immediately freezing the sample in liquid ethane using a Leica plunge-freezing machine. Samples were transferred to a cryogenic holder and imaged using a JEOL 2200FS transmission electron microscope at 185 μA emission current and 40 000× magnification.
Analysis of Endosomal Disruption
The dendritic cell clone DC2.4 (gift from Prof. Kenneth Rock) was plated at 1.2 × 105 cells/well in Lab-Tek chambers (Nunc) for 18h, and then calcein (150 μg/ml, 0.24 mM) was added to the cells with or without 75 μg/ml of lipid-coated nanoparticles containing either a poly-1 or PLGA core in RPMI 1640 complete medium (10% fetal bovine serum (FBS), 5mM L-glutamine, 10mM HEPES, and penicillin/streptomycin) for 1 h at 37 °C. After washing with medium to remove extracellular calcein/particles, the cells were imaged live under a confocal microscope (Zeiss LSM 510) at 63x. To quantify the percentage of cells displaying cytosolic/nuclear distribution of calcein, cells were detached with Trypsin/EDTA, resuspended in flow cytometry buffer (1% BSA, 0.1% NaN3 in Hank’s balanced salt solution, pH 7.4) and analyzed by flow cytometry, gating on the cell population expressing high mean fluorescence intensity beyond that exhibited by cells treated with calcein alone.
Cytotoxicity Assay
Cytotoxicity of the particles was assessed by incubating DC2.4 cells (6 × 105 cells/well in 12 well plates seeded 18 h prior to experiments) with 50, 75 or 100 μg/ml of lipid-coated PBAE particles with or without PEG-lipid incorporation, or PBAE particles stabilized with poly-vinyl-alcohol (PVA) as surfactant, in complete medium for 1 or 12 h at 37 °C. After washing with medium to remove extracellular particles, the cells were detached with Trypsin/EDTA, resuspended in complete medium and rested for 1 h at 37 °C. The cells were then stained with DAPI and annexin V according to manufacturer’s recommendation to identify cells undergoing apoptosis and necrosis. The percentage of live cells was quantified via flow cytometry (BD LSR II) by counting cells that were negative for both stains.
Messenger RNA
Synthetic mRNA was prepared as described previously.38,39 Briefly, linearized plasmid DNA bearing the T7 promoter, an open reading frame (GFP, 811 nucleotides or firefly luciferase, 1714 nucleotides), and poly-A tail was used as a template for in vitro transcription using the Message Machine T7 Ultra Transcription kit (Ambion). The mRNA product was precipitated with LiCl, resuspended in nuclease-free water, and quantitated with a NanoDrop spectrophotometer. RNA size, purity and integrity were ascertained with an Agilient Bioanalyzer 2100. An RMA cell line electroporated with these mRNA transcripts was also used to confirm the functionality of these mRNA in vitro (data not shown). mRNA was labeled with a fluorescent Cy3 tag using a Label IT® nucleic acid labeling kit (Mirus Bio LLC) according to the manufacturer’s protocol.
RNA Loading and Release
Poly I:C or mRNA were loaded onto the surface of lipid-coated PBAE particles by first diluting the particle suspension to 1.5 mg/ml in nuclease-free water, then adding 200 μl of the diluted suspension dropwise to 100 μl containing 4 μg of RNA under gentle vortexing. The vial was then incubated at 4 °C for 2 h on a rotator to allow RNA adsorption. Bound RNA was determined indirectly by measuring fluorescence of either Cy3-labeled or ribogreen-stained RNA remaining in the supernatant after centrifugation of the particles using a fluorescence plate reader (SPECTRAmax, Molecular Devices Corp.). RNA bound to the particles was determined by subtracting the quantity of RNA detected in the supernatant from the quantity measured in identically-treated control vials containing RNA solutions but no particles. To assess the binding kinetics and capacity of the particles, the particle concentration was fixed at 1 mg/ml while the binding time and RNA amount was varied as described in the text. Binding efficiency was defined as the percentage of RNA initially present in solution that bound to the particles. Loading was defined as the amount of RNA bound (μg) per mg of particles. To determine the release kinetics of bound RNA from particles in RPMI 1640 culture medium containing 10% FBS, aliquots of particle-adsorbed poly I:C (70 μg particles containing 1.87 μg poly I:C) were resuspended in 140 μl of serum-containing-media and incubated at 37 °C under gentle mixing. At each designated time-point, an aliquot was removed and particles were washed once with nuclease free water before resuspending in a digestion buffer (100 mM sodium acetate, 2% triton X-100 and 1 mg/ml Poly (L-aspartic acid)) to dissolve the particles and disrupt any lipid-RNA or poly-1-RNA complexes. The amount of RNA remaining on the particles was then determined by measuring the fluorescence following staining with ribogreen as above, and the amount of RNA released was calculated by subtracting the amount of remaining RNA from the original amount.
RNA Degradation Protection Assay
1 μg of mRNA (encoding for GFP) either naked or loaded onto PEGylated PBAE nanoparticles was incubated in the presence of 2 or 10% FBS in nuclease free water for 5 min or 1 h at 37 °C. After the incubation period, 100U of RNAse inhibitor was added to quench degradation and the samples were analyzed by electrophoresis on a 1% agarose gel at 75V for 1 h. The gel was visualized following staining with SYBR Green II RNA gel stain according to manufacturer’s protocol.
In Vitro Transfection of DCs
To test whether mRNA loaded onto particles was able to escape from endosomes into the cytosol, DC2.4 cells were incubated in the presence of a pH-sensitive fluorescent indicator LysoTracker Green (1 μM) and 1 μg of Cy3-labeled mRNA, either naked mRNA in serum-free Opti-mem media or the same quantity of labeled mRNA loaded onto PBAE particles (75 μg particles/ml) in RPMI medium containing 10% FBS. After 1 h, the cells were washed and imaged live by confocal microscopy.
In vitro transfection of DC2.4 cells was also visualized via fluorescent confocal microscopy using Cy3-labeled mRNA encoding for green fluorescent protein (GFP). Because mRNA loading on particles was limited by particle aggregation at high mRNA:particle ratios, conditions for transfection studies were determined by identifying the maximum concentration of particles that could be incubated with DCs in vitro without giving rise to overt cytotoxicity (75 μg particles/ml), and maximizing the amount of the mRNA that could be loaded onto this quantity of particles without inducing particle aggregation. This was determined to be ~4–8 μg of mRNA per 300 μg particles. mRNA-coated lipid-coated PBAE particles with or without PEGylation (75 μg/ml), loaded with 1 μg of mRNA, were diluted into RPMI complete medium containing 10% FBS and incubated for 12 h with 6 × 105 cells seeded in 12-well plates 18 h prior to experiments. mRNA uptake and transfection efficiency were further assessed by flow cytometry, following rinsing with fresh medium and trypsinization, to quantify the percentage of cells positive for Cy3 and/or GFP, compared with control cells incubated with naked mRNA. Particle uptake efficiency was quantified simultaneously by labeling particles with a lipophilic tracer DiD and measuring the fraction of cells positive for the tracer.
In Vivo Transfection
Animals were cared for in the USDA-inspected MIT Animal Facility under federal, state, local and NIH guidelines for animal care. Lipid-coated PBAE particles were tested in vivo by administering 4 μg of luciferase-encoding mRNA adsorbed to 150 μg particles intranasally in 20 μl serum-free RPMI medium to anesthetized C57BL/6J mice. Particle fluorescence and luciferase expression were tracked via bioluminescence imaging following injection of 300 μl of 15 mg/ml luciferin i.p. in anesthetized mice (Xenogen IVIS Spectrum Imager).
Statistical Analysis
One-way ANOVA followed by Bonferroni’s Multiple Comparison Test was applied to determine the significance of the difference between the percentage of cells displaying a cytosolic/nuclear distribution of calcein with different treatments, as well as the fluorescence and bioluminescence radiance detected in mice of different treatment groups.
Results and Discussion
Design, Synthesis, and Characterization of Lipid-coated PBAE particles
We previously showed that core-shell structured hydrogel nanoparticles composed of a pH-responsive proton sponge core and a hydrophilic crosslinked shell could be loaded with anionic proteins or oligonucleotides by electrostatic adsorption of these cargos to the particle surfaces.35 Uptake of these core-shell particles by cells led to disruption of acidifying endolysosomes and efficient delivery of the cargo molecules into the cytosol of cells with minimal toxicity.34,35 However, these particles were prepared from vinyl monomers and are effectively non-biodegradable. To translate this proof of concept to a resorbable materials system for in vivo mRNA delivery, we sought to prepare biodegradable core-shell particles with the structure schematically outlined in Figure 1A: The particle core is composed of a pH-sensitive biodegradable poly(β-amino ester) (PBAE), chosen to promote endolysosomal disruption on particle uptake by cells. We focused on the PBAE poly-1 (Figure 1A) first synthesized by Lynn et al. and studied for a number of biomedical applications such as drug and DNA delivery.26,40–42,21,43–45 We sought to surround the poly-1 core by a phospholipid shell, with the aims of limiting contact of the hydrophobic, cationic polymer core with cellular constituents prior to degradation and providing a tunable surface to mediate binding of mRNA to the particle surfaces.
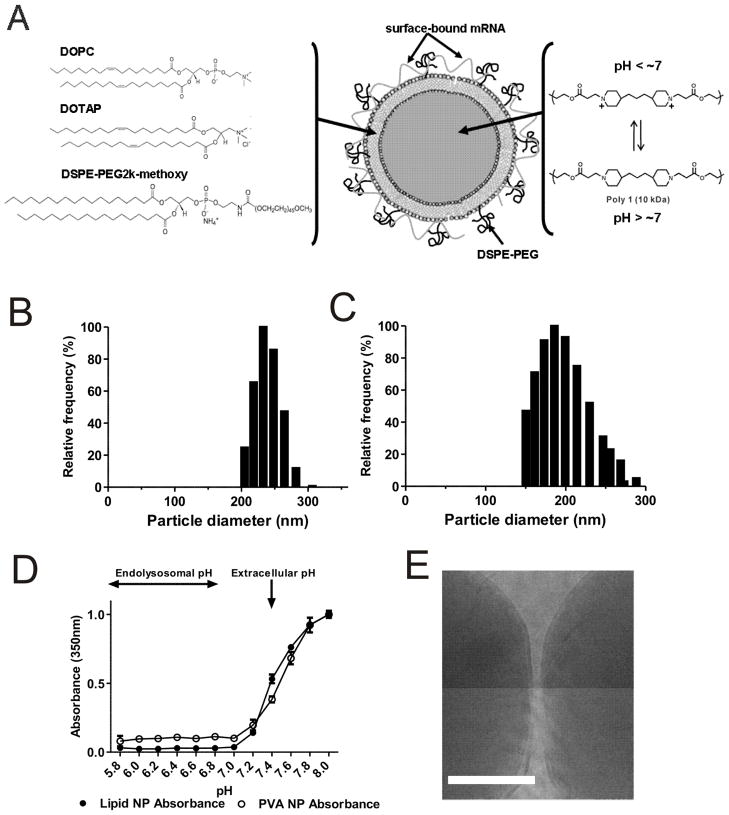
Figure 1.
Design and characterization of lipid-enveloped PBAE particles. (A) Schematic of structure and composition of lipid-coated PBAE particles and mRNA cargo association. (B, C) Size histograms of particles prepared via nanoprecipitation (B) or emulsion/solvent evaporation (C) methods. (D) Measurement of absorbance of particle suspensions as a function of solution pH for both lipid and PVA stabilized particles. (E) Representative cryoEM image of lipid-enveloped PBAE particles, showing the presence of the lipid coating at the particle surfaces (scale bar = 50 nm).
Apropos of this goal, we recently carried out a detailed study of the structure of nanoparticles formed by an emulsion/solvent-evaporation process, where an organic phase containing poly(lactide-co-glycolide) (PLGA) polymer co-dissolved with phospholipids was emulsified in water, followed by evaporation of the organic solvent to form solid polymer particles.37 In this process, the lipids act as surfactants and self-assemble to form a tightly-apposed lipid bilayer envelope around each particle.37 We tested whether variations on this synthesis strategy could be applied to form lipid-enveloped poly-1 particles: First, we prepared particles by co-dissolving poly-1 with a mixture of lipids (DOPC:DOTAP 70:20 mol:mol) in dichloromethane (DCM), followed by emulsification of this organic phase in water and evaporation of the organic solvent. We also tested an approach designed to avoid the need for toxic DCM, based on nanoprecipitation:46,42,47 PBAE and lipids were co-dissolved in ethanol, and then dispersed into excess water. In this process, solid PBAE nanoparticles form as the solvent rapidly diffuses out of the polymer droplets into the surrounding aqueous phase, leading to the precipitation of the polymer core with lipids stabilizing the particle surface. For both synthesis strategies, particles were separated from free liposomes by centrifugation. To promote colloidal stability of particles prepared by either method, poly(ethylene glycol)-phospholipid conjugates (DSPE-PEG) were introduced into the bilayer surfaces of the particles during lipid self-assembly (for the double emulsion process) or by post-insertion48–50 (for nanoprecipitation), resulting in 6–10 mol% PEG-lipid in the lipid shell. Notably, as discussed below, PEGylation did not affect electrostatic adsorption of polynucleotides onto the highly charged particle surfaces; but PEGylation did make the particles resistant to aggregation and easier to resuspend during centrifugation/washing steps.
Particles prepared by the emulsion/solvent evaporation or nanoprecipitation processes had similar size distributions as measured by dynamic light scattering (Figure 1B, C), and similar net positive surface charges indicated by their zeta potentials of 40 ± 9 mV and 42 ± 8 mV in deionized water, respectively. As a control non-pH-responsive core polymer, we also prepared lipid-enveloped PLGA nanoparticles by the emulsion/solvent evaporation process, which had mean diameters of 445 ± 67 nm. Functionally, PBAE-core particles prepared by either synthesis method exhibited indistinguishable endosomal disruption capacities, RNA loading, and in vitro and in vivo transfection in subsequent assays, and thus we report on data collected with particles prepared by both methods below.
Poly-1 is a weak polyelectrolyte that is water insoluble at elevated pH but ionizes and dissolves in aqueous solutions below pH ~7. This selective solubility has been exploited to promote cytosolic delivery of drug cargos following uptake by cells.51 Theoretically, once internalized by cells, particles containing this polymer can trigger disruption of the endosomal membrane by at least two (non-exclusive) mechanisms: (i) via a strong osmotic pressure gradient generated across the endosomal membrane as the solid PBAE particles dissolve into individual polymer chains52; (ii) via an osmotic pressure buildup due to buffering of acidification by the charged polymer backbone and subsequent counterion buildup in the endosome.53,54 We first verified the pH-responsiveness of “naked” PBAE particles (lacking lipids and stabilized only by PVA) by measuring the 350 nm absorbance of particle suspensions in 100 mM phosphate buffers of different pHs, using this absorbance as a surrogate measure of particle light scattering. As shown in Figure 1D, intact particles gave highly scattering milky suspensions at pH > 7.3, but became transparent solutions at lower pHs modeling the acidic environment found within endosomes. Consistent with earlier studies of poly-1 PBAE microparticles51, particle dissolution was rapid (within 5 min) upon exposure to acidic buffers. The same trend was observed for particles prepared with lipid as surfactant, showing that the lipid coating did not alter the dissolution properties of the particles. The presence of intact particles at pH > 7 and disappearance of particles at acidic pH was confirmed by confocal microscopy (data not shown). We thus expect that following acidification of endolysosomal compartments, mRNA on the particles would be quickly released into the cytosol upon rapid build up of osmotic pressure triggered by particle dissolution, thereby minimizing contact with acidic conditions inside endolysosomes.
To provide direct evidence for the assembly of the lipid coating at the surface of the particles, we also imaged samples by cryoelectron microscopy. Similar to our prior findings with PLGA particles prepared using lipids as a surface modifier37, we saw that PBAE particles prepared by the emulsion/solvent evaporation process had electron dense lipid bands coating the surfaces of the particles (Fig. 1D). Table 1 summarizes the size, polydispersity indices (PDI) and zeta potentials of “naked” PBAE particles (PVA-stabilized particles lacking lipid coating), lipid-coated PBAE particles with and without PEGylation, and PEGylated particles following mRNA adsorption (~12–14 μg mRNA/mg particles loaded, discussed below), all measured in deionized water, the medium used for particle synthesis and mRNA loading. A positive zeta potential was measured in all cases and is likely attributable to some degree of ionization of the PBAE core in deionized water (pH ~ 5.5). In contrast with the pH-dependent solubility profile measured in Figure 1D in high ionic strength buffers (100 mM phosphate buffers), particles remained stable in deionized water despite its acidic pH. We hypothesize this reflects the lack of sufficient ionic strength to fully ionize the core, which would otherwise lead to particle dissolution. At very low ionic strength, polyelectrolyte ionization can be dramatically suppressed relative to the expected equilibrium at high ionic strength.55–58 However, when particle zeta potentials were measured in high ionic strength basic pH buffers where no ionization of the core is expected, the zeta potentials of the particles were reduced accordingly (10 ± 4 mV and −7 ± 3 mV for non-PEGylated and PEGylated particles at pH 8, respectively).
Table 1.
Size and zeta potentials of PBAE nanoparticles determined by dynamic light scattering.
| Sample | Effective Diameter (nm) | PDI | Zeta Potential in deionized water (mV) |
|---|---|---|---|
| PVA NP | 300 ± 50 | 0.141 ± 0.08 | 32 ± 8 |
| Non-PEGylated lipid-coated NP | 250 ± 45 | 0.120 ± 0.06 | 41 ± 10 |
| PEGylated lipid-coated NP | 230 ± 40 | 0.100 ± 0.05 | 42 ± 8 |
| mRNA-loaded PEGylated lipid-coated NP | 280 ± 70 | 0.182 ± 0.10 | 40 ± 7 |
Endosomal Escape by Lipid-coated PBAE Particles
To test the ability of lipid-enveloped PBAE particles to disrupt endosomes, calcein, a membrane-impermeable fluorophore, was used as a tracer to monitor the stability of endosomes59 following particle uptake by dendritic cells (DCs), a key cellular target of interest for vaccine delivery.34,60 As shown in Figure 2A, DCs incubated with calcein alone showed a punctate distribution of fluorescence indicative of endolysosomal compartmentalization of internalized dye. In contrast, cells co-incubated with calcein and fluorescently-tagged lipid-enveloped PBAE nanoparticles exhibited calcein fluorescence throughout the cytosol and nucleus (at uniform high levels in both the cytosol and nucleus due to free diffusion of cytosolic dye throughout intracellular compartments), suggesting escape of calcein from intracellular vesicles following co-internalization of extracellular fluid containing both dye and particles (Figure 2B, C). Calcein entry into the cytosol triggered by the presence of nanoparticles required the PBAE core, as calcein remained in an endosomal distribution in cells co-incubated with calcein and lipid-enveloped PLGA particles (Figure 2D).
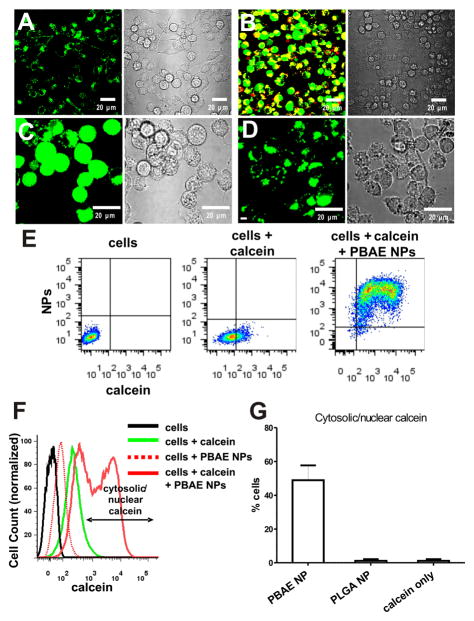
Figure 2.
pH-responsive lipid-enveloped PBAE particles chaperone the delivery of the membrane-impermeable dye molecule calcein into the cytosol of dendritic cells. DC2.4 cells were incubated for 1 h with calcein alone or calcein and lipid-coated poly-1 or PLGA particles, washed to remove unbound particles, then imaged live by confocal microscopy (A–D) or analyzed by flow cytometry (E–G). Confocal images of DC2.4 cells incubated with calcein alone (A) or calcein and lipid-coated PBAE particles (B) (LHS = fluorescence overlays: red, nanoparticles; green, calcein. RHS = bright-field images). (C, D) Cells co-incubated with calcein and either lipid-coated PBAE particles (C) or lipid-coated PLGA particles (D) (LHS = Green, calcein. RHS = bright-field images). Representative images of PEGylated particles are shown here, but a similar trend was observed with non-PEGylated particles. Flow cytometry scatter plots of particle fluorescence vs. calcein fluorescence (E) and histograms (F) for cells treated with calcein or calcein + PBAE particles compared with untreated control cells or cells incubated with particles alone. (G) Average percentage of cells exhibiting cytosolic/nuclear calcein distribution after 1 h incubation with lipid-coated PBAE particles or lipid-coated PLGA particles compared to calcein-only control. Shown are the mean ± SD from triplicate samples.
To quantitatively compare the endosomal disruption capability of PBAE-core particles vs. PLGA-core particles vs. calcein alone in the entire cell population, cells given various treatments were collected following trypsinization and their calcein fluorescence intensity was measured via flow cytometry. Cells incubated with calcein alone showed a homogeneous shift to increased green fluorescence (Figure 2E, F). In contrast, cells co-incubated with calcein and lipid-enveloped PBAE NPs showed two populations of calcein+ cells: a calceinlow and calceinhigh population (Figure 2E, F). Calcein, when trapped within endosomes, can be quenched via 2 mechanisms: self-quenching due to concentration of the dye in endolysosomes61, and quenching due to calcein’s sensitivity to pH in the endolysomal pH range62. Thus, the net fluorescence of a given quantity of internalized calcein and other pH-sensitive dyes is seen to increase on release from endolysosomes, a phenomenon reported by several groups.63,64 We used this phenomenon to quantitate the fraction of DCs showing endosome disruption on the population level, by scoring the fraction of cells exhibiting bright calcein fluorescence beyond that observed for control cells incubated with calcein alone (Figure 2F). As shown in Figure 2G, following a 1 h incubation with PBAE particles, ~50% of the cell population showed an enhanced calcein intensity in accordance with the cytosolic/nuclear distribution of calcein, compared to ~0% of cells incubated with calcein alone or calcein + PLGA particles.
To confirm that endocytosis of the nanoparticles/calcein was required for calcein delivery to the cytosol, we incubated DCs with calcein and nanoparticles at 4 °C to block endocytosis and found that neither calcein nor nanoparticles were internalized by cells up to 3 h at 4 °C (data not shown). This suggests that calcein/nanoparticle uptake and calcein entry into the cytosol of DCs required the active process of endocytosis and excluded the possibility of calcein de-quenching due to nanoparticle fusion with the plasma membrane.
Cytotoxicity of Lipid-coated PBAE Particles
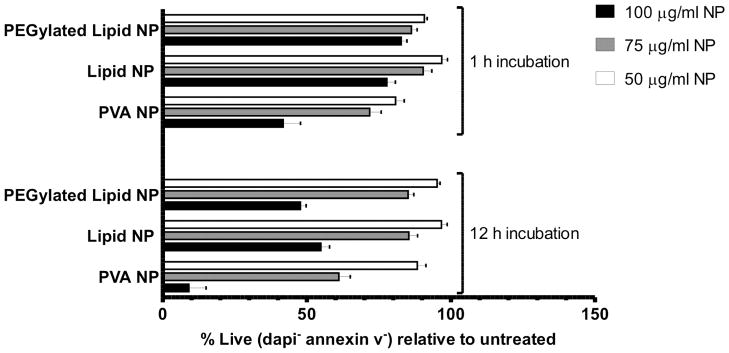
Polycations used for intracellular delivery are notorious for their toxicity;65–67 thus we next analyzed the toxicity of this lipid-enveloped particle delivery system. We incubated DC2.4 cells with lipid-enveloped poly-1 particles or “naked” PVA-stabilized poly-1 particles prepared by emulsion/solvent evaporation synthesis for 1 h or 12 h, and subsequently stained with annexin V and DAPI to identify cells undergoing apoptosis or necrosis, respectively. PVA-stabilized Poly-1 NPs appeared less toxic than some other polycations (e.g., polyethyleneimine, which is often reported to induce 50% cytotoxicity at concentrations of ~20–30 μg/ml68), but still led to ~50% cell death after only an hour incubation of cells with 100 μg/ml of the particles, and showed an LD50 between 75 and 100 μg/ml for longer incubations of 12 h. Lipid-enveloped PBAE NPs, irrespective of PEGylation, showed lower toxicity at both time-points across all tested particle concentrations (Figure 3); notably, particle concentrations that promoted robust endosome disruption by the calcein assay (75 μg/ml) showed low toxicity at both time points. Thus, the lipid shell achieved the desired effect of mitigating toxicity of the poly-1 polymer core.
Figure 3.
Assessment of nanoparticle cytotoxicity. DC2.4 cells were incubated with increasing doses of PEGylated or non-PEGylated lipid-coated poly-1 particles, or PVA-stabilized poly-1 particles for 1 h or 12 h at 37 °C, washed, detached and then stained with DAPI and annexin V to detect apoptotic and necrotic cells by flow cytometry. Shown are the percentages of live (DAPI− annexin V−) cells relative to untreated controls. Error bars represent standard deviation of triplicate samples.
RNA Loading onto Particle Surfaces
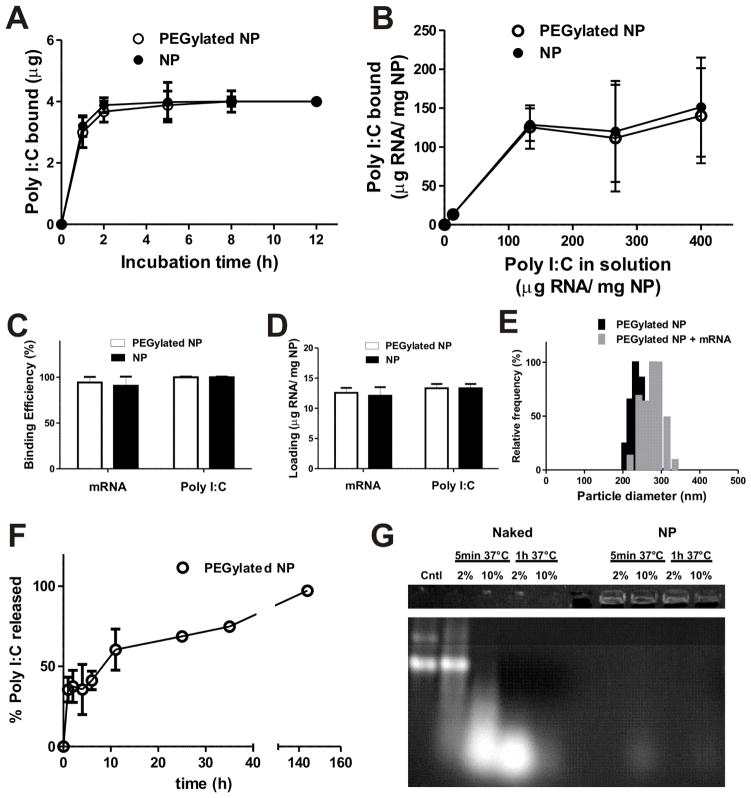
Loading of RNA cargos on these lipid-enveloped particles by adsorption post synthesis provided a means to avoid exposing RNA to harsh processing conditions. Although encapsulation of nucleic acids is often pursued in order to protect these compounds from premature enzymatic degradation in vivo, prior studies where plasmid DNA was electrostatically adsorbed to charged PLGA microparticles showed that particle-adsorbed DNA had a greatly increased half-life in vivo compared to naked plasmids69, suggesting that simple adsorption to particle surfaces may sterically protect polynucleic acids from nucleases. We hypothesized that loading of RNA onto the particle surfaces would increase bioavailability of the RNA by allowing faster release in cells, while avoiding exposure of the polynucleotides to the acidic/hydrophobic environment inside degrading particles that would be expected if we encapsulated RNA within the particle cores. To assess the efficiency of electrostatic adsorption for loading of RNA on surfaces of lipid-coated PBAE particles, we first quantified RNA binding to the particles as a function of time and RNA concentration using poly I:C, a synthetic double-stranded RNA (dsRNA) with an average size of 0.2–1 Kb, comparable in size to mRNA transcripts of interest. Poly I:C is an immunostimulatory analog of viral dsRNA and activates innate immune cells through Toll-like receptor 370,71 and the cytosolic RNA sensor MDA-572,73, which further motivated these experiments. Poly I:C was mixed with lipid-coated PBAE particles in nuclease-free water to allow RNA binding, followed by washing to remove remaining unbound RNA. We found that RNA binding equilibrated rapidly, reaching a plateau by 2 h when 4 μg of RNA was incubated with 300 μg particles in 300 μl nuclease-free water (Figure 4A). Interestingly, PEGylated particles also bound poly I:C to similar levels, suggesting that the PEG layer on the particle surface did not prevent electrostatic attraction of the RNA to the charged lipid surfaces (Figure 4A). We next measured binding of varying amounts of poly I:C added to a fixed concentration of 1 mg/ml particles, and saw that 1 mg of particles was capable of absorbing up to 133 μg of RNA before binding was saturated (Figure 4B). However, we observed that the particles exhibited increasing aggregation when higher RNA concentrations were used (not shown), likely due to the bridging of particles by RNA. To compare poly I:C and mRNA loading, we used single-stranded mRNA transcripts encoding GFP or luciferase. Fixing the mass of particles at 300 μg and RNA at 4 μg in 300 μl of nuclease free water, both poly I:C and mRNA bound to particles with over 95% efficiency (Figure 4C, D) without inducing substantial aggregation (Figure 4E), resulting in ~12–14 μg of RNA loading per mg of particles, a total loading comparable to other biodegradable particle systems where polynucleotides were incorporated by either surface loading or encapsulation in micro/nano-particles.74–76 This non-saturating level of mRNA loading on the particles did not lead to a significant change in the zeta potential of the particles in deionized water (Table 1); we believe this reflects the dominant role of the charge density present in the partially-ionized PBAE core on the zeta potential of the particles.
Figure 4.
RNA adsorption to PEGylated or non-PEGylated lipid-coated PBAE particles in water. (A) Kinetics of poly I:C adsorption to lipid-coated PBAE particles for 300 μg particles incubated with 4 μg RNA in 300 μl of water. (B) Binding of Poly I:C at varying concentrations to 300 μg particles in 300 μl of water for 2 h. (C, D) Binding efficiency (C) and RNA loading (D) of mRNA or poly I:C assessed for RNA (4 μg) added to 300 μg particles for 2 h in 300 μl of water. (E) Size histograms of particles before and after mRNA adsorption at optimized loading conditions (4 μg mRNA added to 300 μg particles in 300 μl of water). (F) Release of poly I:C from nanoparticles incubated in RPMI medium containing 10% FBS at 37 °C. (G) 1 μg mRNA (either free mRNA (Naked) or bound to PEGylated PBAE nanoparticles (NP)) was incubated with 2 or 10% FBS for 5 min or 1 h at 37 °C (or left untreated, Cntl) and analyzed by electrophoresis on a 1% agarose gel. Upper panel shows mRNA detected in the loading wells when mRNA was loaded onto particles and lower panel shows intact free mRNA (Cntl) and degraded mRNA fragments obtained when samples were exposed to serum nucleases. Naked mRNA is completely destroyed by 1 h when exposed to 10% FBS.
To assess how well adsorbed RNA would remain bound to particles upon exposure to physiological conditions, we measured the kinetics of poly I:C release from particles in RPMI containing 10% serum at 37 °C. A burst release of approximately 30% of the poly I:C desorbed within the first hour while the remaining RNA was gradually released over several days (Figure 4F), suggesting that a significant amount of RNA would likely remained bound onto the particles long enough for cellular uptake. Finally, we verified that mRNA coating did not block the ability of lipid-enveloped poly-1 particles to disrupt endosomes, by quantifying the fraction of DC2.4 cells exhibiting cytosolic/nuclear calcein following co-incubation of cells with the dye and mRNA-loaded or unloaded particles for 1 h via flow cytometry as discussed in the previous section. mRNA-loaded lipid-enveloped PBAE particles trended toward slightly reduced endosome disruption compared to unloaded particles, but this did not reach statistical significance (p >0.05) (Supporting information, Figure S1). Thus, simple electrostatic adsorption allowed for rapid loading of the particles with substantial quantities of RNA cargo, which could be retained on the lipid-coated poly-1 surface without blocking their endosome disruption activity.
A major issue for RNA-based therapeutics is their susceptibility towards nuclease degradation. To determine if mRNA loaded onto the surface of lipid-enveloped PBAE particles would be protected from nuclease degradation, we analyzed free or particle-bound mRNA by gel electrophoresis following exposure to serum containing RNA nucleases. As shown in Figure 4G, naked mRNA began to undergo degradation following as little as 5 min exposure to buffer containing 2% FBS, and was almost entirely degraded within 1 h when incubated with buffer containing 10% FBS (bright band detected at shorter fragment size after 5 min incubation and no bands after 1 h incubation) compared to untreated mRNA (cntl). In contrast, minimal degraded fragments were detected for particle-loaded mRNA, where the majority of the nucleic acid remained intact on the particles following nuclease treatment (trapped in the loading wells, top band of the gel), suggesting that mRNA adsorbed onto the particles was protected from immediate nuclease activity.
Transfection of DCs with Lipid-coated PBAE Particles In Vitro
For an mRNA vaccine, these lipid-coated PBAE particles need to deliver intact mRNA into the cytosol of antigen presenting cells such as DCs, where it can then be translated into protein antigens for processing and presentation. Dendritic cells are notoriously difficult to transfect and nonviral transfection agents generally achieve only 10–35% average transfection of these cells in vitro.31–33
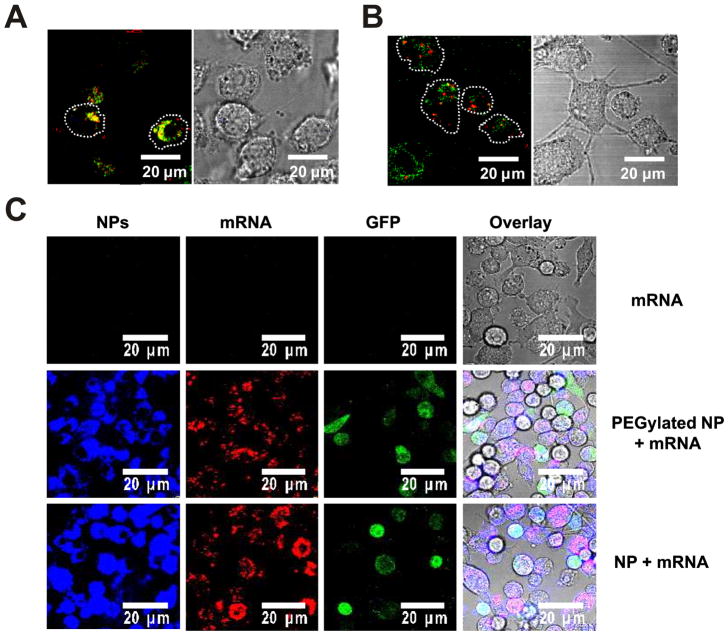
To visualize directly that mRNA loaded onto lipid-enveloped poly-1 particles was capable of escaping endosomes to access the cytosol in dendritic cells, we imaged DCs incubated with labeled mRNA and LysoTracker tracer to stain the endolysosomal compartments, and examined the intracellular trafficking of mRNA loaded onto particles compared to naked mRNA. When DCs were incubated with naked mRNA in serum-free medium, we observed colocalization of labeled mRNA with endolysosomes (Figure 5A). In contrast, particle-delivered mRNA fluorescence was detected in regions of the cell outside endolysosomes, providing direct evidence of the escape of particles and mRNA from endosomes into the cytosol (Figure 5B). Notably, when naked mRNA was incubated with DCs in complete medium containing 10% FBS, no mRNA fluorescence could be detected in the cells (data not shown), indicating the labile mRNA is readily degraded in the presence of serum nucleases without protection from the particles.
Figure 5.
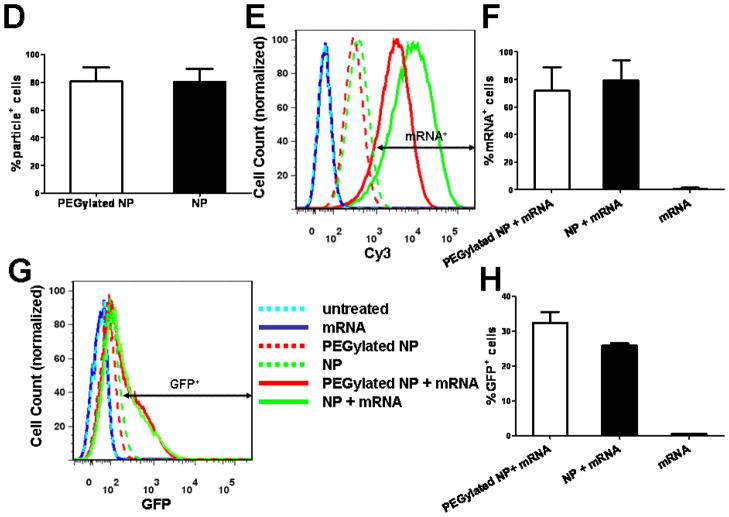
Lipid-coated PBAE particles mediate cytosolic mRNA delivery and transfection in dendritic cells in vitro. (A, B) To observe cytosolic mRNA delivery, DC2.4 cells were incubated for 1h in the presence of LysoTracker green to stain endolysosomes (green) and either naked Cy3-labeled mRNA (red) in serum-free Opti-mem (A) or Cy3-labeled mRNA loaded on particles in complete medium (10% FBS) (B). To study transfection, DC2.4 cells were incubated for 12 h in complete medium (10% FBS) with either naked mRNA (1 μg/ml) or an equivalent dose of mRNA adsorbed to PEGylated or non-PEGylated lipid-coated PBAE particles (75 μg particles/ml), then washed and imaged live by confocal microscopy at 37 °C or analyzed by flow cytometry. (C) Confocal images of DiD-labeled particles (blue), Cy3-labeled mRNA (red), and GFP expression (green) with fluorescence/bright-field overlays in DC2.4 cells. (D–H) Flow cytometry analysis of DC2.4 cells showing mean particle uptake (D), representative histograms of fluorescent mRNA uptake (E) and mean frequency of mRNA+ cells (F), representative cytometry histograms of GFP expression (G), and mean frequency of GFP+ cells (H). Shown are means ± SD from duplicates in 2 independent experiments.
We then tested transfection of DCs in vitro by lipid-enveloped poly-1 particles in the presence of serum, as a precursor to in vivo transfection studies. DC2.4 cells were incubated with Cy3-labeled mRNA encoding for green fluorescence protein (GFP); mRNA was added to the cells in soluble form or adsorbed to fluorescently-tagged lipid-enveloped NPs. Using this set of fluorescence markers, mRNA, NPs, and translated protein (GFP) were simultaneously traced in live cells. Naked mRNA is extremely labile in serum-containing medium, and in fact essentially no intact mRNA was seen to be taken up by DCs in this study when added to the cells in serum-containing medium (Figure 5C), though some uptake was detected in serum-free medium, data not shown). In contrast, mRNA loaded on poly-1 NPs was efficiently delivered into the cells (Figure 5C), supporting the contention that particle adsorption protected the RNA from serum nucleases. Further, a fraction of the cells clearly expressed GFP at the 12 h timepoint (Figure 5C). Quantifying the results via flow cytometry analysis, we found that particles were taken up by 80% of the cells and a similar fraction of the cells internalized mRNA (Figure 5D–F). Looking at protein expression, ~30% of the total cells expressed GFP (Figure 5G, H), a frequency comparable to many prior best reports of DC transfection in the literature.31–33 Irvine et. al compared transfection efficiences in human monocyte-derived DCs using a cationic peptide-plasmid DNA complex with a series of commercial nonviral agents at optimized conditions including lipofectin, lipofectamine and DOTAP lipid where they demonstrated a superior average transfection efficiency of 17% using a reporter GFP protein compared to 1–3% by commercial agents. Aswathi et. al also reported similar transfection efficiency in JAWS II dendritic cells (derived from the bone marrow of C57/BL6 mice) using a nonviral non-liposomal lipid polymer, TransIT-TKO reagent. Strobel et. al reported transfection efficacy of up to 20% when GFP RNA was delivered to monocyte-derived human DCs using commercial liposomal formulations. In contrast to our experiments where transfection was tested in the presence of serum, dendritic cells were incubated with nonviral transfection agents in serum-free medium in each of these prior studies, limiting translation of the results to conditions in vivo.
Particle uptake and GFP expression was similar for both PEGylated and non-PEGylated particles (Figure 5D, H). Notably, under these conditions providing 30% transfection, the particles were non-toxic and >95% of the cells were negative for DAPI and annexin V (data not shown).
In Vivo Transfection with Lipid-coated PBAE Particles
Finally, we tested lipid-coated PBAE particles loaded with mRNA encoding for luciferase protein in vivo, since many systems that transfect cells in vitro fail to function or have serious toxicity in vivo. Intranasal delivery is a convenient and effective route of mucosal vaccination, where the development of antigen particulate carrier systems is of considerable interest.77–80 For vaccine applications, administration via mucosal surfaces is an attractive route for inducing a protective immune response since most pathogens invade the body through mucosal surfaces. Mucosal immunization elicits both systemic and mucosal immunity81,82, while the latter is usually not achieved via parenteral administration. It has also been established that mucosal vaccines administered at one site can elicit an immune response in mucosal tissues remote from the site of initial antigen exposure, mediated by trafficking of effector immune cells between mucosal tissue compartments.83,84 Although parenteral injections of naked mRNA in vivo have been reported in both preclinical models and early clinical trials to elicit protein expression and immune responses1,4,85,23,24, it was not clear if naked mRNA administration would be effective when administered via a non-invasive route such as intranasal delivery, where the mRNA may need to traverse both the epithelial layer and the overlying mucus barrier layer to access target cells. In nasal inoculation, particulate antigens can be taken up by M-cells of nasal-associated lymphoid tissue (NALT), where they are processed and preferentially directed to the antigen-presenting cells, in contrast to soluble antigen.86–88,77,79 It has been demonstrated that particulate delivery systems, particularly those bearing cationic charges, can enhance muco-adhesiveness, reducing the clearance rate from the nasal cavity, and thereby increasing the contact time of the delivery system with the nasal mucosa, facilitating transport.77,79,80
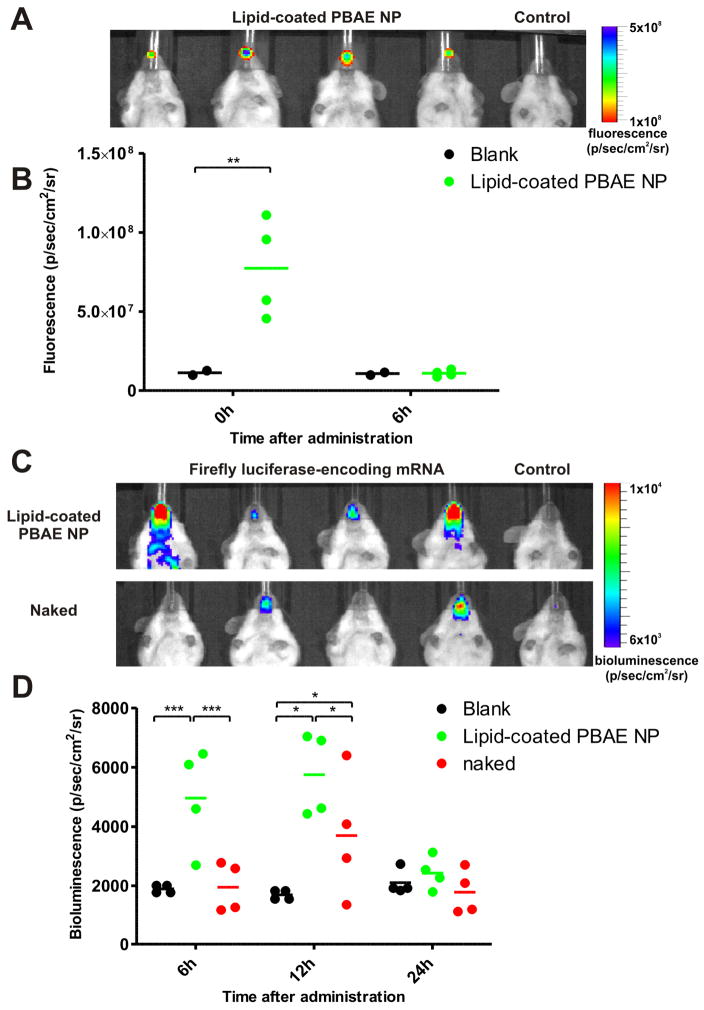
Luciferase-encoding mRNA (4 μg), in soluble form or adsorbed to 150 μg DiD-labelled NPs, was administered intranasally to anesthetized C57BL/6J mice, and the presence of particles and luciferase expression were tracked longitudinally via fluorescence and bioluminescence imaging in live animals (Figure 6). Immediately following intranasal administration, localized particle fluorescence was detected in the nasal region, but fluorescence above background could not be detected by 6 h (Figure 6A, B), suggesting that the particles are rapidly cleared from the nasal passage. However, at 6 h after administration, significant bioluminescence (p <0.01 vs. untreated animals) could be detected at the inoculation site, demonstrating successful transfection by mRNA-loaded particles, while mice treated with naked mRNA showed no signal above background (Figure 6D). Imaging at later time points revealed that the luciferase activity peaked at 12 h before declining to background levels 24 h after administration, consistent with the expected short half-life of transfected mRNA in vivo.1,85 At 12 h, although bioluminescence was detected in some mice receiving naked mRNA, expression was observed in all mice treated with particles (statistically significant above naked mRNA treatment group, p <0.05), demonstrating that lipid-enveloped poly-1 NP delivery was more effective and consistent in eliciting protein expression in vivo (Figure 6C, D). Significant variations in levels of transfection were consistently observed for naked mRNA-treated mice in multiple independent experiments, reflected in the increased variance in bioluminescence signal detected at the peak of protein expression at 12 h (st.dev.naked = 2500 vs. st.dev.NP = 1200). Generally, 1–2 mice/group always failed to respond to naked mRNA administration, possibly reflecting a fundamental limitation in the consistency of transfection efficiency of naked mRNA by this route, compared to nanoparticle administration where all mice were transfected. Though more detailed safety analyses are a topic for future study, PBAE nanoparticles did not elicit overt signs of toxicity in mice: no signs of granuloma formation or chronic inflammation were observed at i.n. or parenteral administration sites, nor were changes in the body mass or behavior of mice detected up to 3 months following particle administration (data not shown).
Figure 6.
In vivo transfection in C5BL/6J mice following intranasal administration of firefly luciferase-encoding mRNA either in soluble form or adsorbed on fluorescent lipid-enveloped PBAE particles. (A) Fluorescence image of mice immediately following intranasal particle administration. (B) Total fluorescence signal at the inoculation site at 0 and 6 h quantified from groups of particle-treated or control mice. (C) Bioluminescence images of mice 12 h following mRNA administration. (D) Bioluminescence signal at the inoculation site at 6, 12 and 24 h quantified from groups of treated and control mice. ***,p <0.001, **, p <0.01 and *, p <0.05. Data shown from one representative of two independent experiments (n = 4 mice/group).
Altogether, these data show that cytosolic delivery of mRNA is achieved by this lipid-enveloped NP delivery system both in vitro and in vivo with minimal toxicity, via the electrostatic adsorption of cargo molecules to the particle surfaces. RNA bound to these pH-sensitive particles was protected from serum nucleases and facilitated in vivo transfection. This approach provides a simple strategy to enhance mRNA delivery of interest for mRNA vaccine design, and may be useful for other instances where local gene delivery may be of therapeutic value.
Supplementary Material
Acknowledgments
We thank A. Bershteyn for assistance with cryoEM imaging and E. Horrigan for help with animal studies. This work was supported by the Institute for Soldier Nanotechnology (Dept. of Defense contract W911NF-07-D-0004) and the Ragon Institute of MGH, MIT and Harvard. X.S. acknowledges the financial support from Agency for Science, Technology and Research, Singapore. D.J.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supporting Information. Supporting figure showing mRNA coating did not block the ability of lipid-enveloped poly-1 particles to disrupt endosomes. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct Gene-Transfer into Mouse Muscle Invivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Tang DC, Devit M, Johnston SA. Genetic Immunization Is a Simple Method for Eliciting an Immune-Response. Nature. 1992;356:152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 3.Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, Dewitt CM, Friedman A, Hawe LA, Leander KR, Martinez D, Perry HC, Shiver JW, Montgomery DL, Liu MA. Heterologous Protection against Influenza by Injection of DNA Encoding a Viral Protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 4.Hoerr I, Obst R, Rammensee HG, Jung G. In Vivo Application of Rna Leads to Induction of Specific Cytotoxic T Lymphocytes and Antibodies. European Journal of Immunology. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Pascolo S. Messenger Rna-Based Vaccines. Expert Opinion on Biological Therapy. 2004;4:1285–1294. doi: 10.1517/14712598.4.8.1285. [DOI] [PubMed] [Google Scholar]
- 6.Ulmer JB. An Update on the State of the Art of DNA Vaccines. Current Opinion in Drug Discovery & Development. 2001;4:192–197. [PubMed] [Google Scholar]
- 7.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, Escande D. Polyethylenimine but Not Cationic Lipids Promotes Transgene Delivery to the Nucleus in Mammalian Cells. Journal of Biological Chemistry. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 8.Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Cell Cycle Dependence of Gene Transfer by Lipoplex Polyplex and Recombinant Adenovirus. Gene Therapy. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 9.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and Development of Polymers for Gene Delivery. Nature Reviews Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 10.Remaut K, Sanders NN, De Geest BG, Braeckmans K, Demeester J, De Smedt SC. Nucleic Acid Delivery: Where Material Sciences and Bio-Sciences Meet. Materials Science & Engineering R-Reports. 2007;58:117–161. [Google Scholar]
- 11.Ghosh P, Han G, De M, Kim CK, Rotello VM. Gold Nanoparticles in Delivery Applications. Advanced Drug Delivery Reviews. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and Strategies for Effective Delivery of Antisense and Sirna Oligonucleotides. Nucleic Acids Research. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-Mediated Rna Delivery: A Novel Approach for Enhanced Transfection of Primary and Post-Mitotic Cells. Nucleic Acids Research. 2001;29:3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A Versatile Reducible Polycation-Based System for Efficient Delivery of a Broad Range of Nucleic Acids. Nucleic Acids Research. 2005:33. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malone RW, Felgner PL, Verma IM. Cationic Liposome-Mediated Rna Transfection. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinon F, Krishnan S, Lenzen G, Magne R, Gomard E, Guillet JG, Levy JP, Meulien P. Induction of Virus-Specific Cytotoxic T-Lymphocytes in-Vivo by Liposome-Entrapped Messenger-Rna. European Journal of Immunology. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 17.Qiu P, Ziegelhoffer P, Sun J, Yang NS. Gene Gun Delivery of Mrna in Situ Results in Efficient Transgene Expression and Genetic Immunization. Gene Therapy. 1996;3:262–268. [PubMed] [Google Scholar]
- 18.Vassilev VB, Gil L, Donis RO. Microparticle-Mediated Rna Immunization against Bovine Viral Diarrhea Virus. Vaccine. 2001;19:2012–2019. doi: 10.1016/s0264-410x(00)00438-2. [DOI] [PubMed] [Google Scholar]
- 19.Zohra FT, Chowdhury EH, Akaike T. High Performance Mrna Transfection through Carbonate Apatite-Cationic Liposome Conjugates. Biomaterials. 2009;30:4006–4013. doi: 10.1016/j.biomaterials.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 20.Kyte JA, Mu L, Aamdal S, Kvalheim G, Dueland S, Hauser M, Gullestad HP, Ryder T, Lislerud K, Hammerstad H, Gaudernack G. Phase I/Ii Trial of Melanoma Therapy with Dendritic Cells Transfected with Autologous Tumor-Mrna. Cancer Gene Therapy. 2006;13:905–918. doi: 10.1038/sj.cgt.7700961. [DOI] [PubMed] [Google Scholar]
- 21.Devalapally H, Shenoy D, Little S, Langer R, Amiji M. Poly(Ethylene Oxide)-Modified Poly(Beta-Amino Ester) Nanoparticles as a Ph-Sensitive System for Tumor-Targeted Delivery of Hydrophobic Drugs: Part 3. Therapeutic Efficacy and Safety Studies in Ovarian Cancer Xenograft Model Cancer Chemotherapy and Pharmacology. 2007;59:477–484. doi: 10.1007/s00280-006-0287-5. [DOI] [PubMed] [Google Scholar]
- 22.Van Driessche A, Van de Velde ALR, Nijs G, Braeckman T, Stein B, De Vries JM, Berneman ZN, Van Tendeloo VFI. Clinical-Grade Manufacturing of Autologous Mature Mrna-Electroporated Dendritic Cells and Safety Testing in Acute Myeloid Leukemia Patients in a Phase I Dose-Escalation Clinical Trial. Cytotherapy. 2009;11:653–668. doi: 10.1080/14653240902960411. [DOI] [PubMed] [Google Scholar]
- 23.Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I, Rammensee HG, Garbe C, Pascolo S. Results of the First Phase I/Ii Clinical Vaccination Trial with Direct Injection of Mrna. Journal of Immunotherapy. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 24.Weide B, Pascolo S, Scheel B, Derhovanessian E, Pflugfelder A, Eigentler TK, Pawelec G, Hoerr I, Rammensee HG, Garbe C. Direct Injection of Protamine-Protected Mrna: Results of a Phase 1/2 Vaccination Trial in Metastatic Melanoma Patients. Journal of Immunotherapy. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 25.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. Different Behavior of Branched and Linear Polyethylenimine for Gene Delivery in Vitro and in Vivo. Journal of Gene Medicine. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 26.Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen JZ, Eisen HN, Langer R. Poly-Beta Amino Ester-Containing Microparticles Enhance the Activity of Nonviral Genetic Vaccines. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9534–9539. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park TG, Jeong JH, Kim SW. Current Status of Polymeric Gene Delivery Systems. Advanced Drug Delivery Reviews. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Wasungu L, Hoekstra D. Cationic Lipids, Lipoplexes and Intracellular Delivery of Genes. Journal of Controlled Release. 2006;116:255–264. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Remaut K, Lucas B, Raemdonck K, Braeckmans K, Demeester J, De Smedt SC. Protection of Oligonucleotides against Enzymatic Degradation by Pegylated and Nonpegylated Branched Polyethyleneimine. Biomacromolecules. 2007;8:1333–1340. doi: 10.1021/bm0611578. [DOI] [PubMed] [Google Scholar]
- 30.Steinman RM, Banchereau J. Taking Dendritic Cells into Medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 31.Irvine AS, Trinder PKE, Laughton DL, Ketteringham H, McDermott RH, Reid SCH, Haines AMR, Amir A, Husain R, Doshi R, Young LS, Mountain A. Efficient Nonviral Transfection of Dendritic Cells and Their Use for in Vivo Immunization. Nature Biotechnology. 2000;18:1273–1278. doi: 10.1038/82383. [DOI] [PubMed] [Google Scholar]
- 32.Strobel I, Berchtold S, Gotze A, Schulze U, Schuler G, Steinkasserer A. Human Dendritic Cells Transfected with Either Rna or DNA Encoding Influenza Matrix Protein M1 Differ in Their Ability to Stimulate Cytotoxic T Lymphocytes. Gene Therapy. 2000;7:2028–2035. doi: 10.1038/sj.gt.3301326. [DOI] [PubMed] [Google Scholar]
- 33.Awasthi S, Cox RA. Transfection of Murine Dendritic Cell Line (Jaws Ii) by a Nonviral Transfection Reagent. Biotechniques. 2003;35:600. doi: 10.2144/03353dd03. [DOI] [PubMed] [Google Scholar]
- 34.Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N, Irvine DJ. Cytosolic Delivery of Membrane-Impermeable Molecules in Dendritic Cells Using Ph-Responsive Core-Shell Nanoparticles. Nano Letters. 2007;7:3056–3064. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- 35.Hu YH, Atukorale PU, Lu JJ, Moon JJ, Um SH, Cho EC, Wang Y, Chen JZ, Irvine DJ. Cytosolic Delivery Mediated Via Electrostatic Surface Binding of Protein, Virus, or Sirna Cargos to Ph-Responsive Core-Shell Gel Particles. Biomacromolecules. 2009;10:756–765. doi: 10.1021/bm801199z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynn DM, Langer R. Degradable Poly(Beta-Amino Esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA. Journal of the American Chemical Society. 2000;122:10761–10768. [Google Scholar]
- 37.Bershteyn A, Chaparro J, Yau R, Kim M, Reinherz E, Ferreira-Moita L, Irvine DJ. Polymer-Supported Lipid Shells, Onions, and Flowers. Soft Matter. 2008;4:1787–1791. doi: 10.1039/b804933e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kavanagh DG, Kaufmann DE, Sunderji S, Frahm N, Le Gall S, Boczkowski D, Rosenberg ES, Stone DR, Johnston MN, Wagner BS, Zaman MT, Brander C, Gilboa E, Walker BD, Bhardwaj N. Expansion of Hiv-Specific Cd4(+) and Cd8(+) T Cells by Dendritic Cells Transfected with Mrna Encoding Cytoplasm- or Lysosome-Targeted Nef. Blood. 2006;107:1963–1969. doi: 10.1182/blood-2005-04-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ngumbela KC, Ryan KP, Sivamurthy R, Brockman MA, Gandhi RT, Bhardwaj N, Kavanagh DG. Quantitative Effect of Suboptimal Codon Usage on Translational Efficiency of Mrna Encoding Hiv-1 Gag in Intact T Cells. Plos One. 2008:3. doi: 10.1371/journal.pone.0002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little SR, Lynn DM, Puram SV, Langer R. Formulation and Characterization of Poly (Beta Amino Ester) Microparticles for Genetic Vaccine Delivery. Journal of Controlled Release. 2005;107:449–462. doi: 10.1016/j.jconrel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 41.Shenoy D, Little S, Langer R, Amiji M. Poly(Ethylene Oxide)-Modified Poly(Beta-Amino Ester) Nanoparticles as a Ph-Sensitive System for Tumor-Targeted Delivery of Hydrophobic Drugs: Part 2. In Vivo Distribution and Tumor Localization Studies. Pharmaceutical Research. 2005;22:2107–2114. doi: 10.1007/s11095-005-8343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shenoy D, Little S, Langer R, Amiji M. Poly(Ethylene Oxide)-Modified Poly(Beta-Amino Ester) Nanoparticles as a Ph-Sensitive System for Tumor-Targeted Delivery of Hydrophobic Drugs. 1. In Vitro Evaluations. Molecular Pharmaceutics. 2005;2:357–366. doi: 10.1021/mp0500420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green JJ, Langer R, Anderson DG. A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Accounts of Chemical Research. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang YH, Zugates GT, Peng WD, Holtz D, Dunton C, Green JJ, Hossain N, Chernick MR, Padera RF, Langer R, Anderson DG, Sawicki JA. Nanoparticle-Delivered Suicide Gene Therapy Effectively Reduces Ovarian Tumor Burden in Mice. Cancer Research. 2009;69:6184–6191. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, Poly(Beta-Amino Ester) Nanoparticles for Small Interfering Rna Delivery. Nano Letters. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganachaud F, Katz JL. Nanoparticles and Nanocapsules Created Using the Ouzo Effect: Spontaneous Emulsification as an Alternative to Ultrasonic and High-Shear Devices. Chemphyschem. 2005;6:209–216. doi: 10.1002/cphc.200400527. [DOI] [PubMed] [Google Scholar]
- 47.Anton N, Benoit JP, Saulnier P. Design and Production of Nanoparticles Formulated from Nano-Emulsion Templates - a Review. Journal of Controlled Release. 2008;128:185–199. doi: 10.1016/j.jconrel.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Zalipsky S, Mullah N, Harding JA, Gittelman J, Guo L, DeFrees SA. Poly(Ethylene Glycol)-Grafted Liposomes with Oligopeptide or Oligosaccharide Ligands Appended to the Termini of the Polymer Chains. Bioconjugate Chemistry. 1997;8:111–118. doi: 10.1021/bc9600832. [DOI] [PubMed] [Google Scholar]
- 49.Allen TM, Sapra P, Moase E. Use of the Post-Insertion Method for the Formation of Ligand-Coupled Liposomes. Cellular & Molecular Biology Letters. 2002;7:889–894. [PubMed] [Google Scholar]
- 50.Cheng WWK, Allen TM. Targeted Delivery of Anti-Cd19 Liposomal Doxorubicin in B-Cell Lymphoma: A Comparison of Whole Monoclonal Antibody, Fab′ Fragments and Single Chain Fv. Journal of Controlled Release. 2008;126:50–58. doi: 10.1016/j.jconrel.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Lynn DM, Amiji MM, Langer R. Ph-Responsive Polymer Microspheres: Rapid Release of Encapsulated Material within the Range of Intracellular Ph. Angewandte Chemie-International Edition. 2001;40:1707–1710. [PubMed] [Google Scholar]
- 52.Lomas H, Massignani M, Abdullah KA, Canton I, Lo Presti C, MacNeil S, Du JZ, Blanazs A, Madsen J, Armes SP, Lewis AL, Battaglia G. Non-Cytotoxic Polymer Vesicles for Rapid and Efficient Intracellular Delivery. Faraday Discussions. 2008;139:143–159. doi: 10.1039/b717431d. [DOI] [PubMed] [Google Scholar]
- 53.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in-Vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonawane ND, Szoka FC, Verkman AS. Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine-DNA Polyplexes. Journal of Biological Chemistry. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 55.Currie EPK, Sieval AB, Fleer GJ, Stuart MAC. Polyacrylic Acid Brushes: Surface Pressure and Salt-Induced Swelling. Langmuir. 2000;16:8324–8333. [Google Scholar]
- 56.Biesalski M, Johannsmann D, Ruhe J. Synthesis and Swelling Behavior of a Weak Polyacid Brush. Journal of Chemical Physics. 2002;117:4988–4994. [Google Scholar]
- 57.Biesheuvel PM. Ionizable Polyelectrolyte Brushes: Brush Height and Electrosteric Interaction. Journal of Colloid and Interface Science. 2004;275:97–106. doi: 10.1016/j.jcis.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 58.Miller AC, Bennett RD, Hammond PT, Irvine DJ, Cohen RE. Functional Nanocavity Arrays Via Amphiphilic Block Copolymer Thin Films. Macromolecules. 2008;41:1739–1744. [Google Scholar]
- 59.Straubinger RM, Hong K, Friend DS, Papahadjopoulos D. Endocytosis of Liposomes and Intracellular Fate of Encapsulated Molecules - Encounter with a Low Ph Compartment after Internalization in Coated Vesicles. Cell. 1983;32:1069–1079. doi: 10.1016/0092-8674(83)90291-x. [DOI] [PubMed] [Google Scholar]
- 60.Steinman RM. Dendritic Cells in Vivo: A Key Target for a New Vaccine Science. Immunity. 2008;29:319–324. doi: 10.1016/j.immuni.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Hamann S, Kiilgaard JF, Litman T, Alvarez-Leefmans FJ, Winther BR, Zeuthen T. Measurement of Cell Volume Changes by Fluorescence Self-Quenching. Journal of Fluorescence. 2002;12:139–145. [Google Scholar]
- 62.Tenopoulou M, Kurz T, Doulias PT, Galaris D, Brunk UT. Does the Calcein-Am Method Assay the Total Cellular ‘Labile Iron Pool’ or Only a Fraction of It? Biochemical Journal. 2007;403:261–266. doi: 10.1042/BJ20061840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Febvay S, Marini DM, Belcher AM, Clapham DE. Targeted Cytosolic Delivery of Cell-Impermeable Compounds by Nanoparticle-Mediated, Light-Triggered Endosome Disruption. Nano Letters. 10:2211–2219. doi: 10.1021/nl101157z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dacey DM, Peterson BB, Robinson FR, Gamlin PD. Fireworks in the Primate Retina: In Vitro Photodynamics Reveals Diverse Lgn-Projecting Ganglion Cell Types. Neuron. 2003;37:15–27. doi: 10.1016/s0896-6273(02)01143-1. [DOI] [PubMed] [Google Scholar]
- 65.Choksakulnimitr S, Masuda S, Tokuda H, Takakura Y, Hashida M. In-Vitro Cytotoxicity of Macromolecules in Different Cell-Culture Systems. Journal of Controlled Release. 1995;34:233–241. [Google Scholar]
- 66.Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. In Vitro Cytotoxicity Testing of Polycations: Influence of Polymer Structure on Cell Viability and Hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 67.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A Two-Stage Poly(Ethylenimine)-Mediated Cytotoxicity: Implications for Gene Transfer/Therapy. Molecular Therapy. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 68.Grayson ACR, Doody AM, Putnam D. Biophysical and Structural Characterization of Polyethylenimine-Mediated Sirna Delivery in Vitro. Pharmaceutical Research. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 69.Denis-Mize KS, Dupuis M, Singh M, Woo C, Ugozzoli M, O’Hagan DT, Donnelly JJ, Ott G, McDonald DM. Mechanisms of Increased Immunogenicity for DNA-Based Vaccines Adsorbed onto Cationic Microparticles. Cellular Immunology. 2003;225:12–20. doi: 10.1016/j.cellimm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of Double-Stranded Rna and Activation of Nf-Kappa B by Toll-Like Receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 71.Takeda K, Akira S. Toll-Like Receptors in Innate Immunity. International Immunology. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 72.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential Role of Mda-5 in Type Iifn Responses to Polyriboinosinic: Polyribocytidylic Acid and Encephalomyocarditis Picornavirus. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-Dependent Recognition of Double-Stranded Ribonucleic Acids by Retinoic Acid-Inducible Gene-I and Melanoma Differentiation-Associated Gene 5. Journal of Experimental Medicine. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh M, Briones M, Ott G, O’Hagan D. Cationic Microparticles: A Potent Delivery System for DNA Vaccines. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:811–816. doi: 10.1073/pnas.97.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang C, Ge Q, Ting D, Nguyen D, Shen HR, Chen JZ, Eisen HN, Heller J, Langer R, Putnam D. Molecularly Engineered Poly(Ortho Ester) Microspheres for Enhanced Delivery of DNA Vaccines. Nature Materials. 2004;3:190–196. doi: 10.1038/nmat1075. [DOI] [PubMed] [Google Scholar]
- 76.Blum JS, Saltzman WM. High Loading Efficiency and Tunable Release of Plasmid DNA Encapsulated in Submicron Particles Fabricated from Plga Conjugated with Poly-L-Lysine. Journal of Controlled Release. 2008;129:66–72. doi: 10.1016/j.jconrel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koping-Hoggard M, Sanchez A, Alonso MJ. Nanoparticles as Carriers for Nasal Vaccine Delivery. Expert Review of Vaccines. 2005;4:185–196. doi: 10.1586/14760584.4.2.185. [DOI] [PubMed] [Google Scholar]
- 78.Slutter B, Hagenaars N, Jiskoot W. Rational Design of Nasal Vaccines. Journal of Drug Targeting. 2008;16:1–17. doi: 10.1080/10611860701637966. [DOI] [PubMed] [Google Scholar]
- 79.Csaba N, Garcia-Fuentes M, Alonso MJ. Nanoparticles for Nasal Vaccination. Advanced Drug Delivery Reviews. 2009;61:140–157. doi: 10.1016/j.addr.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 80.Sharma S, Mukkur TKS, Benson HAE, Chen Y. Pharmaceutical Aspects of Intranasal Delivery of Vaccines Using Particulate Systems. Journal of Pharmaceutical Sciences. 2009;98:812–843. doi: 10.1002/jps.21493. [DOI] [PubMed] [Google Scholar]
- 81.Illum L, Davis SS. Nasal Vaccination: A Non-Invasive Vaccine Delivery Method That Holds Great Promise for the Future. Advanced Drug Delivery Reviews. 2001;51:1–3. [PubMed] [Google Scholar]
- 82.Holmgren J, Czerkinsky C. Mucosal Immunity and Vaccines. Nature Medicine. 2005;11:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 83.McDermont MR, JB Evidence for a Common Mucosal Immunologic System. I. Migration of B Immunoblasts in Intestinal, Respiratory and Genital Tissues. Journal of Immunology. 1979;122:1892–1898. [PubMed] [Google Scholar]
- 84.Czerkinsky C, Prince SJ, Michalek SM, Jackson S, Russell MW, Moldoveanu Z, McGhee JR, Mestecky J. Iga Antibody-Producing Cells in Peripheral-Blood after Antigen Ingestion - Evidence for a Common Mucosal Immune-System in Humans. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:2449–2453. doi: 10.1073/pnas.84.8.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Probst J, Weide B, Scheel B, Pichler BJ, Hoerr I, Rammensee HG, Pascolo S. Spontaneous Cellular Uptake of Exogenous Messenger Rna in Vivo Is Nucleic Acid-Specific, Saturable and Ion Dependent. Gene Therapy. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 86.Fernandez-Urrusuno R, Calvo P, Remunan-Lopez C, Vila-Jato JL, Alonso MJ. Enhancement of Nasal Absorption of Insulin Using Chitosan Nanoparticles. Pharmaceutical Research. 1999;16:1576–1581. doi: 10.1023/a:1018908705446. [DOI] [PubMed] [Google Scholar]
- 87.Brooking J, Davis SS, Illum L. Transport of Nanoparticles across the Rat Nasal Mucosa. Journal of Drug Targeting. 2001;9:267–279. doi: 10.3109/10611860108997935. [DOI] [PubMed] [Google Scholar]
- 88.Nagamoto T, Hattori Y, Takayama K, Maitani Y. Novel Chitosan Particles and Chitosan-Coated Emulsions Inducing Immune Response Via Intranasal Vaccine Delivery. Pharmaceutical Research. 2004;21:671–674. doi: 10.1023/b:pham.0000022414.17183.58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.