Abstract
Nanodiscs are a new class of model membranes that are being used to solubilize and study a range of integral membrane proteins and membrane-associated proteins. Unlike other model membranes, the Nanodisc bilayer is bounded by a scaffold protein coat that confers enhanced stability and a narrow particle size distribution. The bilayer diameter can be precisely controlled by changing the diameter of the protein coat. All these properties make Nanodiscs excellent model membranes for single molecule fluorescence applications. In this chapter, we describe our work using Nanodiscs to apply total internal reflection fluorescence microscopy (TIRFM), fluorescence correlation spectroscopy (FCS) and Förster resonance energy transfer (FRET) to study the integral membrane protein cytochrome P450 3A4 and the membrane-binding proteins islet amyloid popypeptide (IAPP) and α-synuclein, respectively. The monodisperse size distribution of Nanodiscs enhances control over the oligomeric state of the membrane protein of interest, and also facilitates accurate solution-based measurements. Nanodiscs also comprise an excellent system to stably immobilize integral membrane proteins in a bilayer without covalent modification, enabling a range of surface-based experiments where accurate localization of the protein of interest is required.
Keywords: Nanodiscs, Fluorescence correlation spectroscopy, Total internal reflection fluorescence microscopy, Förster resonance energy transfer, Nile Red, cytochrome P450, islet amyloid polypeptide, α-synuclein
1. Introduction
Phospholipid bilayer Nanodiscs (Bayburt et al., 2002; Nath et al., 2007a; Bayburt and Sligar, 2009; Ritchie et al., 2009) are an emerging model membrane system for the study of membrane-associated proteins. Nanodiscs consist of a phospholipid bilayer surrounded by a protein coat formed of membrane scaffold protein (MSP) and are derived from nascent (discoidal) high-density lipoprotein (HDL) particles. Nanodiscs are more stable and monodisperse than conventional model membranes such as liposomes, bicelles, and micelles, making them a very appealing model system for a range of biochemical and biophysical experiments with integral and peripheral membrane proteins. Given the importance of membrane proteins in so many biological and pharmacological questions, there has been an understandable interest in novel Nanodisc technology and a number of exciting developments in membrane protein biochemistry over the past few years (Alami et al., 2007; Boldog et al., 2006; Morrissey et al., 2008).
Concurrent with the growing use of Nanodiscs, there has been a rise in the application of single molecule fluorescence techniques to a range of biological problems, including movement of motor proteins (Park et al., 2007; Peterman et al., 2004), ribosome dynamics (Blanchard et al., 2004) and enzyme catalysis (Henzler-Wildman et al., 2007; Lu et al., 1998) that have provided fundamentally new mechanistic insights and a new appreciation for the role of stochasticity and nonlinear dynamics in a range of biological processes. Several groups have recently reported the application of single molecule fluorescence to integral membrane protein incorporated in Nanodiscs (Nath et al., 2008b) or HDL particles (Kuszak et al., 2009; Whorton et al., 2007). In this chapter, we present detailed protocols from our published work, as well as new methods and results using Nanodiscs to study peripheral membrane-binding proteins, in the hope that this will prove useful to other investigators of membrane proteins.
2. Nanodiscs and HDL particles
Nanodisc technology was developed in the early part of this decade by the group of Stephen G. Sligar at the University of Illinois, Urbana-Champaign, and builds on the considerable body of knowledge contributed by numerous groups about the structure and characterization of HDL particle biology. While the biology of HDL particle formation, maturation and maintenance has been reviewed elsewhere (Atkinson and Small, 1986; Ohashi et al., 2005), we will briefly describe parts of the process relevant to Nanodisc preparation. HDL is involved in reverse cholesterol transport from various tissues to the liver. Apopliprotein A-I (ApoA-I) can bind serum phospholipids to form a discoidal particle with two protein monomers wrapped around a lipid bilayer ~10 nm in diameter (Segrest et al., 1999); as many as five varieties of this species have been proposed, each with slightly different lipid:protein ratios or protein packing around the bilayer. Subsequent loading of cholesterol and cholesteryl esters into the discoidal HDL particles by lecithin:cholesterol acyltransferase (LCAT) transforms them into mature, spheroidal HDL particles. HDL maturation and cholesterol uptake has recently been studied by single particle fluorescence (Sanchez et al., 2007).
Nanodisc formation is achieved by combining detergent-solubilized lipids and MSP in appropriate molar ratios, essentially determined by the optimal fluid-phase surface area occupied by the lipids in a bilayer and the diameter of the desired Nanodisc (Ritchie et al., 2009). The length of the protein coat defines the diameter of the Nanodisc particle, resulting in a relatively narrow size distribution that makes them especially well-suited to single molecule experiments. Additionally, the Sligar lab has assembled a library of MSP constructs of varying lengths, all of which are ultimately derived from human ApoA-I, that can be used to modulate Nanodisc size with high fidelity (Denisov et al., 2004). In recent work, Hoeprich and co-workers (Baker et al., 2009) have developed apolipoprotein E4-based scaffold proteins that can be used to solubilize membrane proteins in particles up to 20 nm in diameter, further extending the versatility of Nanodisc-type model membranes.
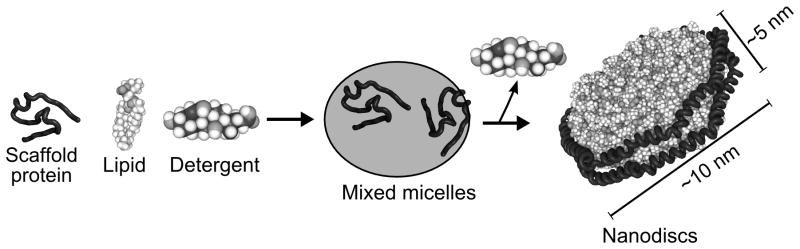
Table 1 contains example lipid ratios for common combinations of MSP constructs and lipids. Nanodisc self-assembly is triggered upon detergent removal from the mixed micelles, by dialysis or using hydrophobic adsorbents such as Bio-Beads SM-2 (Biorad, Hercules, CA) or Amberlite XAD-2 (Sigma Aldrich, St. Louis, MO) (Fig. 1). Protocol 2.1 describes the fabrication of Nanodiscs without an integrated target protein.
Table 1.
Lipid-protein ratios for Nanodisc reconstitution
| Diameter (nm) | Lipid composition | Protein (relative amount1) | Lipid 1 (relative amount1) | Lipid 2 (relative amount1) |
|---|---|---|---|---|
| 9.7 | 100% POPC | MSP1D1 (1) | POPC (65) | - |
| 9.7 | 100% DPPC | MSP1D1 (1) | DPPC (90) | - |
| 9.7 | 40%:60% POPC:DOPG | MSP1D1 (1) | POPC (26) | DOPG (39) |
| 12.9 | 100% POPC | MSP1E3D1 (1) | POPC (130) | - |
| 12.9 | 40%:60% POPC:POPS | MSP1E3D1 (1) | POPC (52) | POPS (78) |
The value in parentheses is the desired molar excess of the relevant species with respect to scaffold protein concentration. For example, 40%:60% POPC:DOPG Nanodiscs made with MSP1D1 require a reconstitution mixture consisting of 1:26:39 MSP1D1:POPC:DOPG, plus an appropriate concentration of detergent. More details can be found at the Sligar group website, http://sligarlab.life.uiuc.edu or in Ritchie et al. (2009).
Figure 1.
Schematic of Nanodisc self-assembly. Scaffold proteins, lipid and detergent are combined in appropriate ratios to form mixed micelles. Integral membrane proteins can also be included at this stage, as long as they can be solubilized in a suitable detergent. Detergent removal from the mixed micelles triggers self-assembly of the Nanodisc particle. Approximate dimensions for Nanodiscs created with MSP1 or MSP1D1 are shown. (The model, kindly provided by Dr. S.C. Harvey of Georgia Tech, is of a discoidal HDL particle with ApoA-I[Δ1-33] (Segrest et al., 1999). The images are not to scale.)
2.1. POPC-MSP1D1 discs
An appropriate volume of lipid dissolved in chloroform (e.g. 200 μl of 20 mg/ml POPC) in a glass test tube is dried to a thin film under a gentle stream of nitrogen, and then dried overnight in a vacuum dessicator or for 2 h in a lyophilizer. To incorporate functionalized lipids, an appropriate mixture is used instead (e.g. 190 μl of 20 mg/ml POPC + 8 μl of 20 mg/ml biotinyl-cap-DPPE for a 5% biotinylated bilayer).
The lipid film is dissolved in 200 μl of a buffer of 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 40 mM sodium cholate. Gentle vortexing and sonication may be necessary to completely solubilize the lipid; the resulting solution should look completely clear.
An appropriate volume of MSP (e.g. 560 μl of 150 μl MSP1D1) in 20 mM Tris-HCl, pH 7.4, 100 mM NaCl (Buffer A) is added, and the mixture is incubated with gentle shaking close to the transition temperature of the lipid (4°C for POPC) for 30 to 90 min. Note that the molar ratios of MSP:lipid:detergent are approximately 1:65:100, and the final concentration of detergent is slightly above its critical micelle concentration (6 mM for cholic acid); the latter consideration facilitates both mixed micelle formation and easy removal of detergent either by dialysis or adsorbent.
Bio-Beads are washed with 2 volumes of methanol and 4 volumes of Buffer A. Approximately 0.3 g of these wet Bio-Beads are added to the mixture from step 3. (A saturated suspension of Bio-Beads has a density of ~1 g/ml, so an appropriate volume can be transferred using a disposable pipet if this is more convenient.)
The mixture with Bio-Beads is incubated close to the transition temperature of the lipid (4°C for POPC) for 4 to 12 h, and the Bio-Beads are then separated using a small gravity column or careful pipeting.
The size and monodispersity of the Nanodisc prep is verified using size exclusion chromatography on a Superdex 200 10/300 (GE Healthcare, Piscataway NJ).
The dimensions, bilayer properties and biophysical behavior of Nanodiscs have been extensively characterized (Denisov et al., 2004; Grinkova et al., 2004), and generally correspond to what is known of HDL structure. By size-exclusion chromatography and native PAGE, the hydrodynamic diameter is close to 10 nm for particles made with ApoA-I or the scaffold protein constructs MSP1 (ApoA-I[Δ1–43]) or MSP1D1 (ApoA-I[Δ1–54], also referred to as MSP1T2 in some papers). The constructs MSP1E3 and MSP1E3D1, in which three additional 22-mer helical segments have been inserted into the sequences of MSP1 and MSP1D1 respectively, produce larger particles ~12.5 nm in diameter. Small-angle X-ray scattering, atomic force microscopy and transmission electron microscopy all indicate disc-shaped particles with similar diameters and a thickness of ~5 nm depending on the lipid composition (Denisov et al., 2004). Finally, differential scanning calorimetry measurements indicate that the influence of the scaffold protein on the bilayer is such that the lipids display a broad phase transition, less like liposomes and more like biological membranes (Denisov et al., 2005).
Nanodiscs have been used to study integral membrane proteins such as G-protein coupled receptors (Leitz et al., 2006), cytochrome P450s (CYPs) (Davydov et al., 2007; Denisov et al., 2007; Kijac et al., 2007; Nath et al., 2007b) and bacterial chemoreceptors (Boldog et al., 2006; Boldog et al., 2007), as well as peripheral membrane-associated proteins such as coagulation factors (Morrissey et al., 2008; Shaw et al., 2007). The constrained, monodisperse size of the Nanodisc bilayer has greatly facilitated close study of the stoichiometry of membrane protein complexes and their functional importance (Bayburt et al., 2006; Boldog et al., 2006; Boldog et al., 2007). In one study of bacterial transmembrane chemoreceptors, the authors were able to modulate the oligomerization state of the target Tar protein by varying the stoichiometry of Tar per Nanodisc (Boldog et al., 2006). They found that the minimal structural unit of Tar is a homodimer, and while homodimers were properly modified in response to ligand binding, further downstream signaling required a higher-order oligomer: a trimer of dimers. This level of control over oligomerization in a membrane is difficult to achieve with traditional model membranes.
3. Single Molecule Techniques and Applications to Membrane Proteins
As this volume of Methods in Enzymology is devoted to single molecule techniques, we will provide only a very brief overview of the methods we use; the other chapters in this volume or standard textbooks (Gell et al., 2006; Lakowicz, 2006) provide a much more detailed discussion. We have used Nanodiscs in experiments with fluorescence correlation spectroscopy (FCS), Förster resonance energy transfer (FRET) and total internal reflection fluorescence microscopy (TIRFM).
In FCS (Magde et al., 1972), the fluctuations in fluorescence from a dilute solution of particles diffusing through a small (~1 fL) focal volume are autocorrelated. Any processes that change fluorescence cause a change in the calculated autocorrelation function, and the rates of these processes determine the timescale at which autocorrelation decays. The most common contribution to autocorrelation decay is simply the diffusion of particles into and out of the confocal volume: measurement of the diffusion time through a given volume enables an estimation of the diffusion time of a particle, and hence (by the Stokes-Einstein equation) its hydrodynamic radius. Although FCS is not purely a single molecule technique, it can be used on the single molecule level and it is frequently discussed in the same category as strictly single molecule fluorescence methods.
FRET is the non-radiative transfer of energy that occurs via a transition dipole interaction between two fluorophores, a donor and an acceptor (Roy et al., 2008). The efficiency of transfer (ETeff) is proportional to the inverse 6th power of the distance: for a given pair of fluorophores, a low ETeff corresponds to fluorophores that are further apart than those with a high ETeff. For our purposes, FRET is used to report on protein conformation and thus both the donor and acceptor fluorophore will be placed on a single protein. As with FCS, fluorescence emission is measured from the molecules as they diffuse through the focal volume. The sample is diluted so that only a single labeled protein is present in the observation volume and the relative emission collected from the donor and acceptor fluorophores is used to calculate ETeff for each molecule.
In TIRFM (Axelrod, 1981), a laser beam is totally internally reflected at a glass surface (using either a prism or a high numerical-aperture oil objective). This generates an evanescent electromagnetic field that decays exponentially from the surface of the slide, and thus specifically excites the fluorophores in the sample within 100 to 200 nm of the surface. If particles of interest are attached to the glass slide at sufficiently low surface concentrations that they are separated by at least the diffraction limit, then fluorescence from single particles can be individually observed. The three techniques mentioned above are not mutually exclusive, and can be used in combination: for example, FCS-FRET (Kim et al., 2002) can provide the rates of conformation fluctuations of molecules in solution, while TIRFM-FRET (Kozuka et al., 2006) reports on the conformational states of surface-attached molecules.
Single molecule fluorescence has been used extensively to study membrane-binding peptides and integral membrane proteins, interacting with a range of model membranes and mimetics including micelles, unilamellar vesicles and supported bilayers, or even in living cells (Brunger et al., 2009; Garcia-Saez and Schwille, 2007). To cite just a few recent examples from the literature, single-particle tracking has been used to study the mechanism of protein recruitment to supported bilayers (Knight and Falke, 2007) and the formation of signaling complexes in living cells (Murakoshi et al., 2004). FCS can measure the difference in diffusion rates between a free protein and a liposome-bound state, enabling measurements of membrane affinity with high sensitivity (Rhoades et al., 2006). Single molecule FRET has been used to study conformational dynamics of intrinsically disordered proteins bound to detergent micelles (Ferreon et al., 2009) as well as liposome-bound transmembrane motors (Diez et al., 2004).
Compared to other model membranes, Nanodiscs are unsuitable for investigations of membrane leakage or diffusion within a bilayer. They are, however, advantageous for solution-based techniques that would benefit from a monodisperse membrane size distribution, such as FCS-based binding measurements. Because the Nanodisc bilayer is relatively small and its size can be accurately controlled, Nanodiscs are also an excellent system for the characterization of oligomers and complexes of membrane proteins in situ. Finally, their usefulness is most evident in the unique ability to stably immobilize membrane proteins in a discrete spatial location without any covalent modification or encapsulation of the protein (Rhoades et al., 2003). This enables long-term observation of single membrane proteins in something very close to a native lipid environment. In the following sections, we describe our results and protocols using Nanodiscs to study three different protein systems by TIRFM, FCS and FRET.
4. Cytochrome P450 3A4 and its Allosteric Behavior
Cytochrome P450 3A4 (CYP3A4) is the major drug-metabolizing enzyme in humans, responsible for the clearance of ~50% of pharmaceutical compounds (Thummel and Wilkinson, 1998). It is a member of the cytochrome P450 (CYP) superfamily of heme-thiolate monooxygenases, capable of catalyzing reactions including hydroxylation, heteroatom oxidative dealkylation and epoxidation on a broad range of different substrates (Davydov and Halpert, 2008; Guengerich, 2001). CYP3A4 is an integral membrane protein with an N-terminal helical anchor and other hydrophobic membrane-associated regions, and was one of the first proteins successfully incorporated in Nanodiscs (Baas et al., 2004).
CYP3A4 incorporated into Nanodiscs is stable, monomeric and soluble, and this complex has enabled a range of experiments on CYP3A4 biophysics that were previously impossible or impractical, or suffered from complications caused by the tendency of CYP3A4 to aggregate or oligomerize. Experiments with CYP3A4-Nanodiscs include equilibrium ligand binding (Baas et al., 2004; Nath et al., 2007b), stopped-flow kinetic measurements (Davydov et al., 2005), pressure-perturbation spectroscopy (Davydov et al., 2007) and turnover kinetics measurements with Nanodiscs containing a complex of CYP3A4 an accessory oxidoreductase required for normal catalysis (Denisov et al., 2007). It has been suggested that interactions between two or more CYPs in a membrane can affect catalytic activity and ligand binding (Fernando et al., 2007; Subramanian et al., 2009). CYP3A4-Nanodiscs provide a minimal system to study CYP3A4 behavior in a native-like membrane, but without interference from other membrane proteins. Nanodiscs have enabled valuable new insights into CYP3A4 mechanisms.
One particularly challenging aspect of CYP3A4 enzymology is that it displays atypical allosteric kinetics with a large fraction of its substrates. CYP3A4 metabolizes thousands of substrates, ranging in size from ~150 Da (e.g. acetaminophen) to ~1200 Da (cyclosporin), and with other physico-chemical properties just as diverse. Because it needs to accommodate such a broad group of substrates, CYP3A4 is capable of binding one, two or three substrate molecules simultaneously. If the different complexes have altered Vmax values (or different affinities for additional substrate molecules), the functional result is a deviation from standard Michaelis-Menten kinetics.
Although not classical multi-subunit allosterism in the sense of Monod-Wyman-Changeux (Monod et al., 1965) or Koshland-Nemethy-Filmer (Koshland et al., 1966) models, CYP3A4 does display quite a high degree of cooperativity both with a single chemical species acting homotropically, and with heterotropic cases involving distinct substrates and effectors (Atkins, 2004). This is clinically relevant: in part because of CYP3A4 allosterism, many combinations of drugs can alter each other’s metabolism by inhibition or activation, in ways that are difficult to predict solely from in vitro data for each drug alone. These drug-drug interactions can lead to unfavorable pharmacokinetics, where one or more of a patient’s drugs are outside their therapeutic window: if clearance by CYP3A4 or other drug-metabolizing enzymes is activated, then the circulating drug concentration might be too low to be efficacious; if clearance is inhibited, then the drug could build up to toxic levels. This has generated significant pharmacological interest in preventing adverse drug-drug interactions, and in harnessing favorable drug-drug interactions by co-administering adjuvants that extend the duration of the therapeutic window (Zeldin and Petruschke, 2004).
Many steroids and small polycyclic aromatic molecules act as CYP3A4 effectors: at low concentrations, they boost the metabolism of other substrates, and their own turnover often displays sigmoidal kinetics, implying that they boost their own metabolism. There has been a growing consensus in the past few years that these effectors bind to a peripheral site (outside the active site) and thereby modulate CYP3A4 structure and/or dynamics, or protein-protein interactions within the membrane, so as to increase the affinity of substrate binding at the active site. There is strong evidence from X-ray crystallography (Williams et al., 2004) that progesterone can bind outside the active site. As reviewed in (Davydov and Halpert, 2008) and (Denisov et al., 2009), a large body of biophysical and enzymological characterization (Denisov et al., 2007; Isin and Guengerich, 2007; Lampe and Atkins, 2006; Roberts and Atkins, 2007; Tsalkova et al., 2007) focuses on the mechanism of effectors such as testosterone, progesterone and α-naphthoflavone (ANF).
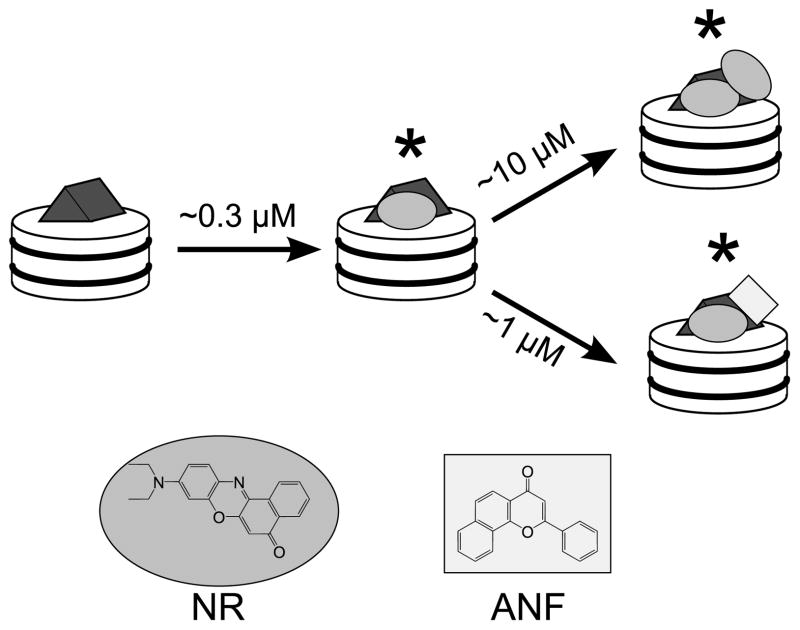
Our studies of allosteric mechanisms were greatly facilitated by the discovery that the membrane dye Nile Red (NR) was not only a highly active substrate of CYP3A4, but also an allosteric reporter (Lampe et al., 2008; Nath et al., 2008a). Using a combination of equilibrium binding experiments monitored by fluorescence and absorbance spectroscopy, and catalytic assays, we found that CYP3A4 can bind one or two NR molecules, with higher affinity for the active site than the peripheral effector site (in contrast to archetypal effectors such as ANF). NR fluorescence is greatly enhanced upon binding CYP3A4, and the doubly-bound species displays much brighter and red-shifted fluorescence than the 1:1 complex. Finally, because NR preferentially binds the active site and ANF preferentially binds the peripheral effector site, CYP3A4 can easily form a trimeric complex with both NR and ANF bound. This heterotrimeric species is spectrally distinct from both the singly- and doubly-occupied NR-CYP3A4 complexes (Fig. 2). NR fluorescence thus provides a rich set of probes to investigate different aspects of homotropic and heterotropic cooperativity displayed by CYP3A4.
Figure 2.
Our model of Nile Red (oval) and α-naphthoflavone (rectangle) binding to CYP3A4 (gray prism) incorporated into a model membrane such as Nanodiscs (white cylinder), based on data from previous work (Nath et al., 2008; Nath et al., 2007). NR binds with high affinity to the active site, and with lower affinity to the peripheral effector site. In contrast, ANF binds with high affinity to the effector site, and can form a heterocomplex with NR. All three species marked with an asterisk are fluorescent and can be spectrally resolved.
We applied single molecule TIRFM to the CYP3A4-NR-ANF system as described in a recent paper (Nath et al., 2008b), and were able to measure residence times of NR in the CYP3A4-Nanodisc complex based on the marked increase in NR fluorescence intensity when bound to either the CYP3A4 active site or the Nanodisc bilayer. Here we describe in detail the methods used in these experiments. CYP3A4 incorporated into Nanodiscs containing biotinylated lipids as described in Protocol 4.1. Surface attachment of CYP3A4-Nanodiscs using biotin-avidin chemistry is described in Protocol 4.2.
4.1. Incorporation of CYP3A4 in Nanodiscs
MSP1D1 is treated with AcTEV protease (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions to remove the N-terminal hexahistidine tag. Aliquots are stored at −80°C. The resulting protein is denoted MSP1D1(−).
A suitable amount of lipid (e.g. 190 uL of 20 mg/ml POPC + 8 uL of 20 mg/ml biotinyl-cap-DPPE for a 5% biotinylated bilayer) is dried to a thin film under a N2 stream and stored overnight in a vacuum desiccator.
CYP3A4 is expressed as previously described (Nath et al., 2007b). CYP3A4 (5 μM in 2 ml of 100 mM potassium phosphate pH 7.4 with 20% glycerol) is solubilized with 20 μl of a 10% (v/v) solution of Emulgen-913 (Kao Chemicals, Osaka, Japan) for a final detergent concentration of 0.1% (v/v). The mixture is incubated with gentle shaking at room temperature for 1 hour.
CYP3A4 is exchanged into Buffer A (from Protocol 2.1) with 0.1% E-913 using Amicon centrifugal filter columns (30 kDa cut-off; Millipore, Billerica, MA) until the final glycerol concentration is less than 3%. (Higher levels of glycerol interfere with the disc formation process.) At the final centrifugation step, the sample is concentrated to a final volume of ~1 ml.
Lipid films are resuspended in 400 μl of Buffer A with 100 mM cholate by gentle vortexing and sonication.
MSP1D1(−) (580 μl of 150 μM solution) is added to the lipid-detergent solution.
The solubilized CYP3A4 sample from step 4 is added. The mixture now contains CYP3A4:MSP1D1(−):lipid in approximate 0.1:1:63 molar ratio with ~20 mM cholate. This corresponds to an approximate four-fold excess of empty discs without CYP3A4, thereby disfavoring incorporating multiple CYP3A4 molecules into a single Nanodisc.
The mixture is incubated with gentle shaking at 4°C (Tm of lipid) for 1 hour.
Wet Bio-Beads (2 g) are added to the mixture and incubated with gentle shaking at 4°C for at least 4 h.
Bio-Beads are removed using a gravity flow column, and the sample is further dialyzed three times against 1 L changes of Buffer A for 2 h each to remove residual detergent.
Nickel-affinity chromatography (Ni-NTA Superflow resin, Qiagen, Valencia, CA) is used to separate CYP3A4-Nanodiscs from plain Nanodiscs. CYP3A4-Nanodiscs are eluted with Buffer A with 300 mM imidazole, pH 7.4.
The CYP3A4-Nanodiscs are dialyzed overnight against Buffer A. Monodispersity of CYP3A4-Nanodiscs is verified using size exclusion chromatography on a Superdex 200 10/300 (GE Healthcare, Piscataway NJ). If the sample is not homogenous, fractions corresponding to the size of the CYP3A4-Nanodisc complex should be collected and pooled. For long term storage, add glycerol to 10% and store at −80°C.
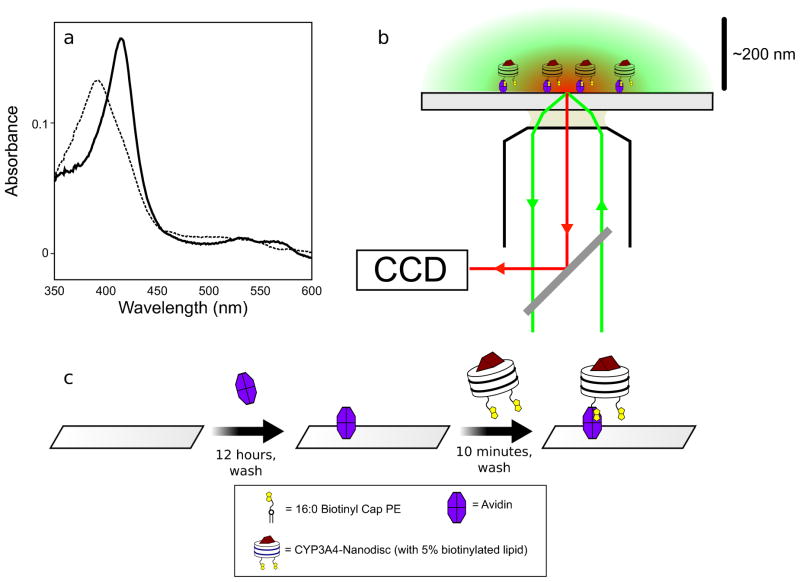
A successful preparation of monomeric CYP3A4-Nanodiscs will show approximately equal absorbance at 280 nm and 417 nm. (There can be some variability due to minor changes in the resting spin state of the CYP3A4 heme, which affects absorbance at 417 nm.) The addition of saturating bromocriptine will induce a complete conversion to the low-spin state with an absorbance peak at 388 nm, as shown in Fig. 3a.
Figure 3.
Setup of a TIRFM experiment using CYP3A4-Nanodiscs. (a) Bromocriptine (8 μM) induced an almost complete shift in the heme absorbance signal of CYP3A4-Nanodiscs from low-spin (solid line) to high-spin (high spin). After verifying appropriate size and homogeneity by size-exclusion chromatography, this experiment serves as a convenient check that a preparation of CYP3A4-Nanodiscs is properly functional. (b) Schematic of a through-objective TIRFM experiment. Excitation laser light (green) is guided through one side of an oil objective so that it is incident at a glass slide at the critical angle. This generates an evanescent field that decays, selectively exciting fluorophores within a few hundred nanometers of the slide surface. Emitted light (red) from these particles is passed through a dichroic mirror to a CCD camera. c. Schematic of the surface attachment procedure using biotin-avidin crosslinking described in Protocol 4.2.
4.2. Surface attachment of CYP3A4-Nanodiscs
Glass coverslips (22 mm x 22 mm, #1 thickness, Fisher Scientific, Pittsburgh, PA) are cleaned by sonication for 30 min in 10% (v/v) 7x cleaning solution (MP Biomedical, Solon, OH) followed by another 30 min of sonication in Milli-Q water (i.e., deionized and 0.22 μm-filtered in a Milli-Q Integral system, Millipore, Billerica, MA). As an alternative, coverslips can be cleaned by 10 min incubation in Piranha solution, made by gradual addition of hydrogen peroxide (30%) to concentrated H2SO4 to a final ratio of 1:3. Piranha-cleaned slides must be thoroughly rinsed with MilliQ water before use. Clean coverslips were dried overnight at 60°C.
Flowcells are assembled by stacking two cleaned coverslips separated by two parallel thin strips of Parafilm (Pechiney Plastic Packaging, Chicago, IL) and heating for 15 s at ~200°C, just until the Parafilm melts and secures the two coverslips together.
Flowcells are rinsed with two volumes of 50% NH4OH and 15 volumes of MilliQ water, and then incubated overnight at 4°C with a 1 mg/ml aqueous solution of avidin (Sigma Aldrich, St. Louis, MO).
Flowcells are rinsed with 10 volumes of Buffer A (from Protocol 2.1), and then Nanodiscs or CYP3A4-Nanodiscs containing 5% biotinylated lipids are added at a concentration of 30 nM.
After 10 min, flowcells are rinsed with 2 more volumes of Buffer A to remove any unbound particles, and then Nile Red (~50 nM in Buffer A) is added (Fig. 3c). Loaded flowcells are immediately transferred to the microscope for TIRFM visualization.
Our TIRFM instrumentation (Fig. 3b) is based on an Olympus IX-71 inverted microscope (Olympus America, Center Valley, PA). A 561 nm, 50 mW diode-pumped solid state continuous-wave laser (Newport Corp, Irvine, CA) is passed through a neutral density filter (OD was varied from 0.04 to 2.0), then through a 5:1 beam expander. The beam is then focused into the back aperture of a 60x/1.45 N.A. oil objective (Olympus) and the entry of the beam into the back aperture is adjusted to achieve total internal reflection, creating a ~75 μm diameter evanescent field. Emitted fluorescence is passed through a 585 nm long-pass filter (Chroma Technology Corp., Rockingham, VT) and then to an iXon back-thinned EMCCD camera (Andor USA, South Windsor, CT) for detection. Images are captured using Andor Solis software. EM gain is varied from 30 dB to 280 dB, and exposure time is varied from 6 ms to 100 ms in an effort to maximize sensitivity while still capturing rapid binding events. Trajectories are collected for 1000 frames each (i.e. 6 s to 100 s in length).
5. Image Filtering by Singular Value Decomposition
Trajectories with short exposures (6 to 30 ms) showed high levels of background noise (Fig. 4a). One popular approach to improve sensitivity is to average neighboring frames (Ulbrich and Isacoff, 2007), but in our case this runs the risk of discarding rapid binding events and thereby biasing the distribution of residence times. The residence time of NR in membranes has been reported to be ~15 ms (Gao et al., 2006), and so rapid events are expected to be especially relevant when characterizing binding to the Nanodisc bilayer. Therefore, we chose to filter images with exposure times <50 ms using singular value decomposition (SVD), a popular technique in image processing (Hendler and Shrager, 1994; Protter and Elad, 2009).
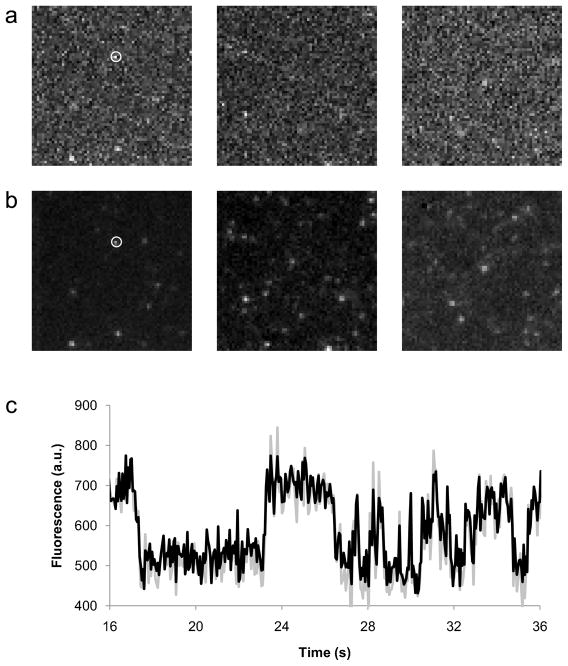
Figure 4.
Nile Red binding CYP3A4-Nanodiscs as observed by TIRFM. (a) Three unprocessed single frames from TIRF trajectories of NR binding CYP3A4-Nanodiscs. (b) The same three frames after noise reduction performed by singular value decomposition as described in Protocol 5.1. c. Comparison of unprocessed (gray) and filtered (black) time traces from the particle indicated by the white circle in a, showing how the SVD process preserves information on bright periods.
SVD is related to principal components analysis (PCA), and similarly seeks to identify common trends in a set of data. When considering a set of TIRFM images, all of the pixels corresponding to the diffraction-limited image of a single particle will, on average, display higher covariance (i.e. changes in intensity correlated with each other) than random noise, which is uncorrelated. Conceptually, SVD can be thought of as the determination of the eigenvectors and eigenvalues of the covariance matrix of a data set. Each eigenvector signifies an observed trend in the data, and the corresponding eigenvalue represents the magnitude or significance of that trend. In the context of SVD, the eigenvalues of the covariance matrix are called ‘singular values’. Higher singular values generally reflect meaningful trends in the data while lower singular values correspond to random, uncorrelated noise. If the SVD is inverted while neglecting the lower singular values, the reconstructed data set will lack much of the noise present in the original data. The key parameter in SVD-based filtering is the number of singular values to retain in the reconstructed image: too many, and the reconstructed image will still contain noise; too few, and significant ‘meaningful’ information will be lost. Protocol 5.1 is a pseudocode description of our SVD filtering procedure. Sample source code in Python for SVD filtering is available at http://www.yale.edu/rhoadeslab. Fig. 4b shows the reduction in noise achieved by SVD, while Fig. 4c that SVD-filtered intensity traces from single particles retain information on rapid binding events.
5.1. SVD-based Image Filtering Pseudocode
Convert each frame of a trajectory into a one-dimensional array, so that a 256×256 pixel image, for example, becomes a 65536-value vector, with the value at each position corresponding to the brightness of a particular pixel.
Assemble the resulting arrays from all the frames of a trajectory into a matrix D, so that for example a 1000-frame trajectory of 256×256 pixel images becomes a 65536×1000 matrix. (Note: matrix operations of this size can be very memory-intensive, so to improve performance it may be necessary to spatially divide the original into, for example, four 1000-frame 128×128 pixel trajectories and process each one separately.)
Perform SVD on the matrix D. Many efficient implementations exist for various programming environments; we used the linalg.svd() function of Scientific Python (Jones et al., 2001-). SVD produces three matrices U, S and V such that USVT = D. U (65536×1000) contains all the eigenvectors of the covariance matrix (i.e. common trends in the images) and V (1000×1000) shows how they change over the course of the trajectory. S (1000×1000) contains the singular values along its diagonal (and zeroes everywhere else), reflecting the relative contribution of each trend to the information in the original trajectory.
Choose a number n greater than the estimated number of particles visible in the trajectory, and discard all columns of U and S, and all rows of VT, whose index is greater than n. Some trial and error in setting the value of n is necessary for each trajectory: if n is too large, then the filtered trajectory will still contain much of the noise present in the original data. However, if n is too small, the filtered trajectory will lack significant transitions from the original data (i.e. the data will be oversmoothed), and meaningful information will be lost.
Reconstruct the filtered trajectory by multiplying the truncated versions of U, S and VT to generate R (65536×1000).
Convert each column of R to an image with the same dimensions as the originals (256×256).
Assemble the images into a trajectory of the same length as the original (1000 frames).
For further analysis, we averaged intensities from a 3×3 pixel area around well-separated diffraction limited spots and manually measured the extent of bright periods, which represent bound NR. The resulting distribution of residence times was fit to exponential decays to determine the off-rate of NR under different conditions. As described in our earlier paper (Nath et al., 2008b), we measured a residence time for NR in Nanodiscs of ~30 s−1, in good agreement with the measurements of residence times in lipid bilayers made by others (Gao et al., 2006). Using CYP3A4-Nanodiscs, we found a new, slower (~1.5 s−1) phase which we ascribed to NR dissociation from the CYP3A4 active site. Miconazole, a competitive inhibitor, blocked NR binding to CYP3A4 and eliminated the slow phase. The presence of 5 μM ANF, which should predominantly bind the peripheral effector site while leaving the active site accessible, further slowed the putative off-rate (~0.3 s−1). This would seem to suggest that CYP3A4 effectors may act by increasing the residence time of substrates in the enzyme active site. ANF did not affect off-rates from plain Nanodiscs, suggesting the observed effect is mediated by CYP3A4 and not the bilayer.
A common consideration with single-molecule data of this type is whether they contain artifacts due to photophysical effects. Intermittent photoblinking associated with the triplet state should occur on a submillisecond timescale for oxazine dyes like NR in aerobic solutions (Vogelsang et al., 2009), so they are unlikely to contribute to the observed bright and dark periods. Photobleaching of bound NR can also contribute to the observed decay in bright periods. To test the effect of this phenomenon, we compared data collected with different illumination powers (Fig. 5) and found relatively minor differences.
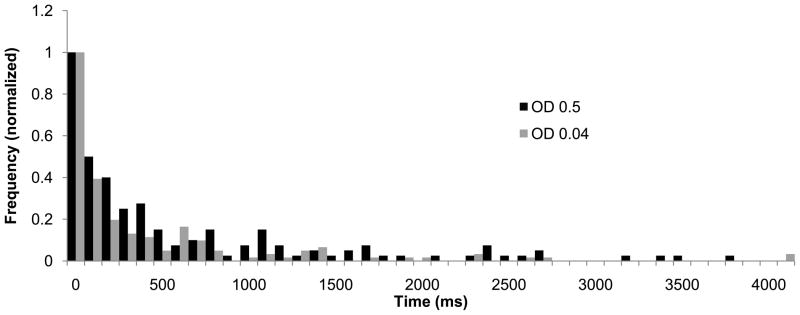
Figure 5.
Dwell-time histograms measured using neutral density filters of OD 0.04 (grey bars) and OD 0.5 (black) bars in the excitation beam path. The higher laser power corresponded to an apparent decrease in the residence time of NR in the CYP3A4-Nanodisc active site of ~25%, which we attribute to photobleaching of the fluorophore. Although this is a relatively minor change relative to the effect of ANF (a four-fold increase in residence time), we used the lower laser power for our measurements to minimize the contribution of photobleaching.
6. Islet Amyloid Polypeptide binding to Nanodiscs
Islet amyloid polypeptide (IAPP; also called amylin) is an intrinsically disordered amyloidogenic peptide strongly implicated in the pathogenesis of Type II diabetes (Hebda and Miranker, 2009). IAPP is a peptide hormone cosecreted with insulin by pancreatic β-cells in healthy individuals, but readily forms amyloid fibers in vitro. Preamyloid oligomeric intermediates may be significant effectors of β-cell death, and thereby contribute to the progression of type II diabetes. Understanding what factors mediate pathogenic misfolding of IAPP is therefore of deep interest for the design of therapeutics.
IAPP binds anionic membranes with high affinity. Membrane binding catalyzes the formation of predominantly α-helical oligomers and also accelerates amyloid (cross-β) fibril formation (Knight et al., 2006; Knight and Miranker, 2004). In turn, membrane-bound oligomers appear to contribute to toxicity by inducing membrane leakage. Studies of this complicated thermodynamic landscape, with membrane partitioning linked to the cooperative formation of two different types of oligomer, are fortunately simplified by the fact that the rat IAPP homolog, rIAPP, also forms membrane-bound oligomers but does not form amyloid fibrils (Knight et al., 2006).
Several groups (Ling et al., 2009; Mishra et al., 2009; Nanga et al., 2008) are attempting to characterize structural features of different states in this pathway using NMR and infrared spectroscopy. However, our current thermodynamic understanding of IAPP-membrane interactions relies primarily on the ensemble spectroscopic techniques of circular dichroism and thioflavin T fluorescence. FCS-based binding measurements with Nanodiscs may provide a more sensitive and detailed way to dissect the thermodynamics of IAPP-membrane interactions.
For the case of a single freely-diffusing species observed by FCS, the autocorrelation decay is fit with the following equation:
Here N is the average number of molecules of the species in the confocal volume, τD is the diffusion time of the species, and s is a measure of the prolateness of the observation volume (the ratio of Z-axial radius to XY-radius). For a system with two components possessing distinct diffusion times, such as a mixture of free and lipid-bound labeled protein, the equivalent equation is:
Here A is the fraction of free protein, τf is the diffusion time of the free protein and τb is the diffusion time of the lipid-bound species. It should be noted that this equation assumes the free and bound states of the protein have equal brightness. The diffusion time of the bound species will correspond to that of plain Nanodiscs, which can be determined with high precision by virtue of their monodisperse size distribution.
The FCS-based binding approach enables sensitive direct measurements of free and bound concentrations, while requiring orders of magnitude less material than ensemble experiments. This facilitates precise measurements of high-affinity binding events, which can be difficult when binding partners are present at high concentrations. As a proof of principle, we describe FCS measurements of rIAPP binding to Nanodiscs containing 60% DOPG:40% POPC.
6.1. FCS measurement of rIAPP binding Nanodiscs
Tetramethylrhodamine-succinimidylester (TAMRA-SE) is used to label rIAPP at the ε-amino position of Lys1, the sole lysine residue in the rIAPP sequence. TAMRA-SE (0.2 mg in 100 μl DMSO) is added to rIAPP (1 mg in 1 ml Buffer A from Protocol 2.1) and the mixture is incubated with gentle shaking in the dark for 4 h at room temperature. Excess free dye is separated from labeled rIAPP using a Sephadex G-25 desalting column (GE Healthcare, Piscataway, NJ). Efficient labeling is verified by reverse-phase HPLC on a C-18 column (Grace Vydac, Deerfield, IL).
Nanodiscs are prepared as in Protocol 2.1, except that the lipid mixture is 100 μl of 20 mg/ml and 100 μl of 25 mg/ml DOPG.
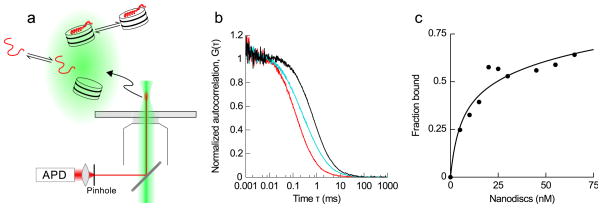
FCS measurements are performed on an Olympus IX-71 inverted microscope with 60x/1.2 N.A. water objective. The output of a 561 nm, 50 mW diode-pumped solid state continuous-wave laser (Newport Corp, Irvine, CA) is adjusted with neutral density filters to a measured power of ~5 μW just prior to entry to the microscope. Emitted fluorescence is collected through a 585 nm long-pass filter (Chroma Technology Corp., Rockingham, VT) and a 50 μm optical fiber (Oz Optics, Ottawa, Canada) to an avalanche photodiode (PerkinElmer, Waltham, MA) coupled to a Flex03-LQ-12 correlator (Correlator.com, Bridgewater, NJ) (Fig. 6a).
To measure the Nanodisc diffusion time, 10 autocorrelation traces of 10 nM NR in the presence of 10 μM Nanodiscs are collected, averaged and fit with the single-component FCS equation to determine τb. Under these conditions, the only contribution to observed fluorescence is from bound NR, which therefore provides an accurate measurement of Nanodisc τD. Alternatively, this calibration can be performed on Nanodiscs containing 1% rhodamine-DOPE.
30 autocorrelation traces of 10 nM TAMRA-rIAPP are collected, averaged and fit to the single-component FCS to determine τf.
30 autocorrelation traces of 10 nM TAMRA-rIAPP are collected at a range of Nanodisc concentrations (Fig. 6b). Each set is averaged and fit to the two-component FCS equation with N and A (free fraction) as the only variables. A is plotted against Nanodisc concentration to generate a binding curve (Fig. 6c). Typical standard errors of A from this fitting approach are on the order of 0.5% to 1% of the free fraction (i.e. ranging from about 0.0015 to 0.006 over the course of the titration). Given that the uncertainty in τf and τb values is also about 1%, the overall uncertainty in the value of the bound fraction is about 2%.
Figure 6.
IAPP binding Nanodiscs as observed by FCS. (a) Schematic of an FCS experiment. Excitation light (green) is focused into a sample, and emitted light (red) from a ~1 fL observation volume is directed through an optical fiber (that serves as a pinhole) to an avalanche photodiode (APD) and autocorrelated. Any change in the intensity of fluorescence manifests as a decay in autocorrelation that provides information on the rate of the underlying process. In our case, the relevant processes are the diffusion of labeled rIAPP in free and Nanodisc-bound states (not shown to scale). (b) Autocorrelation traces of free rIAPP (red) and Nanodiscs with trace NR (black), showing the rightward shift expected with larger hydrodynamic size and longer diffusion time. In cyan is a mixture of labeled rIAPP and Nanodiscs under conditions with about half of the peptide bound, showing the intermediate two-component autocorrelation decay. (c) Binding isotherm obtained as Nanodiscs containing 60% DOPG:40% POPC were titrated into 10 nM labeled rIAPP. Performing this experiment at a range of different peptide concentrations will allow further study of the complex binding cooperativity observed in rIAPP-membrane interactions (Knight et al., 2006).
The resulting binding isotherm has an apparent KD of ~50 nM, corresponding to a membrane partition coefficient of 2×105. This is relatively close to the equivalent partition coefficient for 100% DOPG membranes of 5×104, measured using circular dichroism spectroscopy (Knight et al., 2006). It must be strongly emphasized that IAPP-membrane binding is a complex thermodynamic process, with separate parameters governing membrane partition, nucleation and growth of membrane-bound oligomers. A meaningful understanding of the system requires global analysis of data collected over a range of different protein and lipid concentrations. The improved sensitivity afforded by single molecule techniques enables experiments over a much broader concentration range than the ~10 μM regime necessary for the ensemble techniques we previously used (Knight et al., 2006), and so should lead to a more complete and accurate understanding of IAPP-membrane interactions.
7. α-Synuclein conformations on Nanodiscs
α-Synuclein (αS) is the primary protein constituent of cytoplasmic Lewy bodies and Lewy neurites that are pathologically linked to Parkinson’s Disease (PD) (Goedert, 2001; Ueda et al., 1993). Although αS is strongly implicated in disease progression (Cookson, 2005) its precise role in PD is unclear. The native function of αS is also poorly understood, although evidence suggests that it may play a role both in maintaining neuronal plasticity and in the regulation of synaptic vesicle recycling (George et al., 1995; Lotharius and Brundin, 2002) and thus that binding to membranes may be important to its function.
αS is intrinsically disordered in solution (Weinreb et al., 1996) but undergoes a conformational change to an α-helical structure upon association with negatively charged membranes (Davidson et al., 1998; Jo et al., 2000). Two contrasting models have been proposed for the conformation of membrane-bound αS, either an extended, continuous helix (Bussell and Eliezer, 2003; Georgieva et al., 2008; Jao et al., 2004; Jao et al., 2008) or two anti-parallel, noninteracting helices (Borbat et al., 2006; Chandra et al., 2003; Drescher et al., 2008; Ulmer et al., 2005), with an unstructured loop region in between. We recently used single molecule FRET to study αS membrane-bound conformations using lipid vesicles and SDS micelles (Trexler and Rhoades, 2009). We observed that αS displays either an extended helix on 50 nm and 100 nm lipid vesicles, and a hairpin conformation on the much more highly curved detergent micelles, in good agreement with the findings of other recent single molecule (Ferreon et al., 2009) and EPR (Georgieva et al., 2008) studies. Here, the use of Nanodiscs as a model membrane allows us to decouple the effects of membrane size and curvature: unlike vesicles and micelles, it is possible to modulate Nanodisc size by appropriate choice of MSP while retaining a flat bilayer (Denisov et al., 2004).
The double cysteine mutant, αS T33C/T72C, labeled with a donor and acceptor fluorophore, can differentiate between bent and extended helical conformations. Here, we use this αS mutant to report on the helical-conformation of αS bound to Nanodiscs made with the extended scaffold protein MSP1E3, which have a bilayer ~13 nm in diameter. αS T33C/T72C was expressed, purified, and labeled essentially as previously described (Rhoades et al., 2006; Trexler and Rhoades, 2009).
7.1. smFRET measurement of αS bound to Nanodiscs
Nanodiscs are prepared as described in Protocol 2.1, except with the following mixture of scaffold protein and lipid: MSP1E3 (265 μl of 155 μM) and POPS (200 μl of 20 mg/ml).
Single molecule FRET measurements are performed on an Olympus IX-71 inverted microscope with 60x/1.2 N.A. water objective. The output of a 488 nm, 50 mW diode-pumped solid state continuous-wave laser (Newport Corp, Irvine, CA) is adjusted with neutral density filters to a measured power of ~10 μW just prior to entry to the microscope. Fluorescence is split using a 585 nm dichroic mirror (Chroma Technology Corp., Rockingham, VT) and fiber-coupled (100 μm fibers, Oz Optics, Ottawa, Canada) to avalanche photodiodes (PerkinElmer, Waltham, MA).
Samples contain 90 pM double-labeled αS T33C/T72C, the low concentration ensuring that each observed burst represents a single particle passing through the observed volume. The Nanodisc concentration is 600 nM, an excess that enhances the probability of having at most one protein bound per disc.
-
Data are collected in 1 ms time bins and processed using MATLAB scripts as described in detail in our previous work (Trexler and Rhoades, 2009). For each burst of photons associated with a Nanodisc-bound protein, the energy transfer efficiency (ETeff) value is calculated using the following formula:
Here Ia and Id are intensity in the acceptor and donor channel, respectively, corrected for background counts. β accounts for bleed-through of donor photons to the acceptor channel, while γ accounts for differences in quantum yield and detection efficiency of the donor and acceptor fluorophores and was experimentally determined for our instrument as 1.2. ETeff values were then compiled into a histogram for analysis.
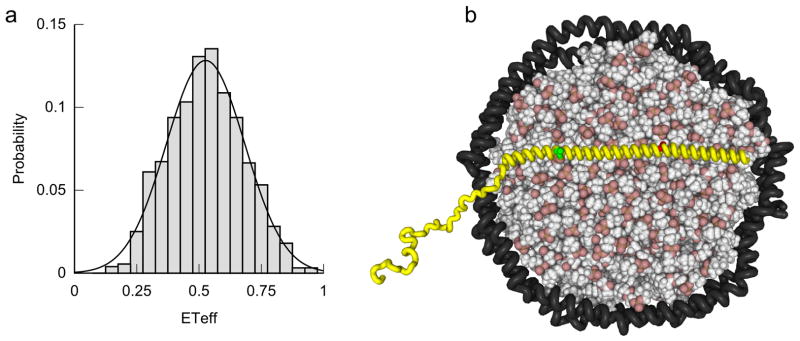
When bound to the Nanodisc bilayer, αS T33C/T72C showed a peak ETeff of 0.53 (Fig. 7a), close to that measured using 100 nm POPS vesicles (0.57), and considerably lower than the corresponding value for SDS micelles (0.72) (Trexler and Rhoades, 2009). This observation suggests that Nanodiscs induce an extended helix conformation in αS (Fig. 7b) similar to that induced by much larger vesicles, but unlike the hairpin conformation observed with detergent micelles ~4 nm in diameter. Membrane curvature may be an important determinant of αS conformation, and thus may be relevant to both native function and PD-associated pathology.
Figure 7.
Conformation of αS bound to Nanodiscs as observed by FRET. (a) FRET histogram collected for double-labeled αS T33C/T72C bound to MSP1E3D1-POPS Nanodiscs shows a single species with a peak energy transfer efficiency close to that observed for an extended helix on large unilamellar vesicles. (b) Model for αS (in yellow) extended helix conformation when bound to a Nanodisc, scaled appropriately to account for the larger diameter of particles made with MSP1E3D1 scaffold protein. Red and green spheres denote sites of labeling: a hairpin conformation, such as that observed with SDS micelles, would bring the dyes closer together and increase energy transfer efficiency.
8. Summary
In this chapter, we discussed how Nanodiscs may be used in popular single-molecule fluorescence techniques. Nanodiscs are a powerful complement to conventional model membranes, especially well-suited to single-molecule approaches. Solution-based single-molecule experiments may be facilitated by the relatively precise control over particle size afforded by different MSP constructs. Nanodiscs may prove even more useful in surface-based experiments: they provide a system to stably immobilize single membrane proteins in a native-like bilayer without covalent modification.
Acknowledgments
We very gratefully acknowledge Prof. S.G. Sligar and members of his research group for helpful advice and generous gifts of Nanodisc scaffold proteins. Funding was provided by NIH grants GM-32165 to W.M.A. and E.R., and GM-084391 to E.R. and A.D.M., and by the Ellison Medical Foundation to E.R.
Abbreviations
- HDL
high-density lipoprotein
- MSP
membrane scaffold protein
- FCS
fluorescence correlation spectroscopy
- TIRFM
total internal reflection fluorescence microscopy
- FRET
Förster resonance energy transfer
- NR
Nile Red
- ANF
α-naphthoflavone
- CYP
cytochrome P450
- IAPP
islet amyloid polypeptide
- POPC
palmitoyloleoylphosphatidylcholine
- POPS
palmitoyloleoylphosphatidylserine
- DOPG
dioleoylphosphatidylglycerol
- LCAT
lecithin:cholesterol acyltransferase
- PD
Parkinson’s disease
- SVD
singular value decomposition
- PCA
principal components analysis
- TAMRA-SE
tetramethylrhodamine-succinimidylester
References
- Alami M, Dalal K, Lelj-Garolla B, Sligar S, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO Journal. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins W. Implications of the allosteric kinetics of cytochrome P450s. Drug Discov Today. 2004;9:478–484. doi: 10.1016/S1359-6446(04)03072-7. [DOI] [PubMed] [Google Scholar]
- Atkinson D, Small D. Recombinant Lipoproteins - Implications for Structure and Assembly of Native Lipoproteins. Annu Rev Biophys Bio. 1986;15:403–456. doi: 10.1146/annurev.bb.15.060186.002155. [DOI] [PubMed] [Google Scholar]
- Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89:141–5. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas B, Denisov I, Sligar S. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–28. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Baker S, Hopkins R, Blanchette C, Walsworth V, Sumbad R, Fischer N, Kuhn E, Coleman M, Chromy B, Letant S, Hoeprich P, Adams M, Henderson P. Hydrogen Production by a Hyperthermophilic Membrane-Bound Hydrogenase in Water-Soluble Nanolipoprotein Particles. J Am Chem Soc. 2009;131:7508–7509. doi: 10.1021/ja809251f. [DOI] [PubMed] [Google Scholar]
- Bayburt T, Sligar S. Membrane protein assembly into Nanodiscs. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayburt T, Grinkova Y, Sligar S. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–22. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2002;2:853–856. [Google Scholar]
- Blanchard S, Kim H, Gonzalez R, Puglisi J, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog T, Grimme S, Li M, Sligar S, Hazelbauer G. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–14. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog T, Li M, Hazelbauer G. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–35. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- Borbat P, Ramlall T, Freed J, Eliezer D. Inter-helix distances in lysophospholipid micelle-bound alpha-synuclein from pulsed ESR measurements. J Am Chem Soc. 2006;128:10004–10005. doi: 10.1021/ja063122l. [DOI] [PubMed] [Google Scholar]
- Brunger A, Weninger K, Bowen M, Chu S. Single-molecule studies of the neuronal SNARE fusion machinery. Annu Rev Biochem. 2009;78:903–28. doi: 10.1146/annurev.biochem.77.070306.103621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell R, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- Chandra S, Chen X, Rizo J, Jahn R, Sudhof T. A broken alpha-helix in folded alpha-synuclein. J Biol Chem. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- Cookson M. The biochemistry of Parkinson’s disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- Davidson W, Jonas A, Clayton D, George J. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- Davydov D, Baas B, Sligar S, Halpert J. Allosteric mechanisms in cytochrome P450 3A4 studied by high-pressure spectroscopy: pivotal role of substrate-induced changes in the accessibility and degree of hydration of the heme pocket. Biochemistry. 2007;46:7852–64. doi: 10.1021/bi602400y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov D, Fernando H, Baas B, Sligar S, Halpert J. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: Heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov D, Halpert J. Allosteric P450 mechanisms: multiple binding sites, multiple conformers or both? Expert Opin Drug Met. 2008;4:1523–1535. doi: 10.1517/17425250802500028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov I, Baas B, Grinkova Y, Sligar S. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–76. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- Denisov I, Frank D, Sligar S. Cooperative properties of cytochromes P450. Pharmacol Therapeut. 2009 doi: 10.1016/j.pharmthera.2009.05.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov I, Grinkova Y, Lazarides A, Sligar S. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- Denisov I, McLean M, Shaw A, Grinkova Y, Sligar S. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez M, Zimmermann B, Borsch M, Konig M, Schweinberger E, Steigmiller S, Reuter R, Felekyan S, Kudryavtsev V, Seidel C, Graber P. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat Struct Mol Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- Drescher M, Veldhuis G, van Rooijen B, Milikisyants S, Subramaniam V, Huber M. Antiparallel arrangement of the helices of vesicle-bound alpha-synuclein. J Am Chem Soc. 2008;130:7796–7797. doi: 10.1021/ja801594s. [DOI] [PubMed] [Google Scholar]
- Fernando H, Davydov D, Chin C, Halpert J. Role of subunit interactions in P450 oligomers in the loss of homotropic cooperativity in the cytochrome P450 3A4 mutant L211F/D214E/F304W. Arch Biochem Biophys. 2007;460:129–140. doi: 10.1016/j.abb.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreon A, Gambin Y, Lemke E, Deniz A. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci U S A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Mei E, Lim M, Hochstrasser R. Probing lipid vesicles by bimolecular association and dissociation trajectories of single molecules. J Am Chem Soc. 2006;128:4814–4822. doi: 10.1021/ja058098a. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez A, Schwille P. Single molecule techniques for the study of membrane proteins. Appl Microbiol Biot. 2007;76:257–266. doi: 10.1007/s00253-007-1007-8. [DOI] [PubMed] [Google Scholar]
- Gell C, Brockwell D, Smith A. Handbook of single molecule fluorescence spectroscopy. Oxford University Press; Oxford ; New York: 2006. [Google Scholar]
- George J, Jin H, Woods W, Clayton D. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron. 1995;15:361–372. doi: 10.1016/0896-6273(95)90040-3. [DOI] [PubMed] [Google Scholar]
- Georgieva E, Ramlall T, Borbat P, Freed J, Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: Long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- Grinkova Y, Denisov I, Bayburt T, Sligar S. Structure and composition of self-assembled discoidal phospholipid bilayer nanoparticles formed with different membrane scaffold proteins. Biophys J. 2004;86:252A–252A. [Google Scholar]
- Guengerich F. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–50. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Hebda J, Miranker A. The interplay of catalysis and toxicity by amyloid intermediates on lipid bilayers: insights from type II diabetes. Annu Rev Biophys. 2009;38:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- Hendler R, Shrager R. Deconvolutions based on singular-value decomposition and the pseudoinverse - a guide for beginners. J Biochem Bioph Meth. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson M, Petsko G, Karplus M, Hubner C, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- Isin E, Guengerich F. Multiple sequential steps involved in the binding of inhibitors to cytochrome p450 3A4. J Biol Chem. 2007;282:6863–6874. doi: 10.1074/jbc.M610346200. [DOI] [PubMed] [Google Scholar]
- Jao C, Der-Sarkissian A, Chen J, Langen R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci U S A. 2004;101:8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C, Hegde B, Chen J, Haworth I, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci U S A. 2008;105:19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo E, McLaurin J, Yip C, St George-Hyslop P, Fraser P. alpha-synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275:34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P. SciPy: Open Source Scientific Tools for Python. 2001 http://scipy.org.
- Kijac A, Li Y, Sligar S, Rienstra C. Magic-angle spinning solid-state NMR spectroscopy of nanodisc-embedded human CYP3A4. Biochemistry. 2007;46:13696–703. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Nienhaus G, Ha T, Orr J, Williamson J, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–9. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J, Falke J. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys J. 2007;96:566–82. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J, Hebda J, Miranker A. Conserved and cooperative assembly of membrane-bound alpha-helical states of islet amyloid polypeptide. Biochemistry. 2006;45:9496–9508. doi: 10.1021/bi060579z. [DOI] [PubMed] [Google Scholar]
- Knight J, Miranker A. Phospholipid catalysis of diabetic amyloid assembly. J Mol Biol. 2004;341:1175–1187. doi: 10.1016/j.jmb.2004.06.086. [DOI] [PubMed] [Google Scholar]
- Koshland D, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Kozuka J, Yokota H, Arai Y, Ishii Y, Yanagida T. Dynamic polymorphism of single actin molecules in the actin filament. Nat Chem Biol. 2006;2:83–6. doi: 10.1038/nchembio763. [DOI] [PubMed] [Google Scholar]
- Kuszak A, Pitchiaya S, Anand J, Mosberg H, Walter N, Sunahara R. Purification and functional reconstitution of monomeric mu-opioid receptors: allosteric modulation of agonist binding by Gi2. J Biol Chem. 2009;284:26732–41. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz JR. Principles of fluorescence spectroscopy. Springer; New York ; Berlin: 2006. [Google Scholar]
- Lampe J, Atkins W. Time-resolved fluorescence studies of heterotropic ligand binding to cytochrome P450 3A4. Biochemistry. 2006;45:12204–12215. doi: 10.1021/bi060083h. [DOI] [PubMed] [Google Scholar]
- Lampe J, Fernandez C, Nath A, Atkins W. Nile Red is a fluorescent allosteric substrate of cytochrome P450 3A4. Biochemistry. 2008;47:509–16. doi: 10.1021/bi7013807. [DOI] [PubMed] [Google Scholar]
- Leitz A, Bayburt T, Barnakov A, Springer B, Sligar S. Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques. 2006;40:601–12. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- Ling Y, Strasfeld D, Shim S, Raleigh D, Zanni M. Two-dimensional Infrared Spectroscopy Provides Evidence of an Intermediate in the Membrane-catalyzed Assembly of Diabetic Amyloid. J Phys Chem B. 2009;113:2498–2505. doi: 10.1021/jp810261x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotharius J, Brundin P. Impaired dopamine storage resulting from alpha-synuclein mutations may contribute to the pathogenesis of Parkinson’s disease. Hum Mol Genet. 2002;11:2395–2407. doi: 10.1093/hmg/11.20.2395. [DOI] [PubMed] [Google Scholar]
- Lu H, Xun L, Xie X. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- Magde D, Webb W, Elson E. Thermodynamic Fluctuations in a Reacting System—Measurement by Fluorescence Correlation Spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- Mishra R, Sellin D, Radovan D, Gohlke A, Winter R. Inhibiting Islet Amyloid Polypeptide Fibril Formation by the Red Wine Compound Resveratrol. ChemBioChem. 2009;10:445–449. doi: 10.1002/cbic.200800762. [DOI] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux J. On nature of allosteric transitions - a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Morrissey J, Pureza V, Davis-Harrison R, Sligar S, Ohkubo Y, Tajkhorshid E. Blood clotting reactions on nanoscale phospholipid bilayers. Thromb Res. 2008;122(Suppl 1):S23–6. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci U S A. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanga R, Brender J, Xu J, Veglia G, Ramamoorthy A. Structures of Rat and Human Islet Amyloid Polypeptide IAPP(1–19) in Micelles by NMR Spectroscopy. Biochemistry. 2008;47:12689–12697. doi: 10.1021/bi8014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Atkins W, Sligar S. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007a;46:2059–69. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- Nath A, Fernández C, Lampe J, Atkins W. Spectral resolution of a second binding site for Nile Red on cytochrome P4503A4. Arch Biochem Biophys. 2008a;474:198–204. doi: 10.1016/j.abb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A, Grinkova Y, Sligar S, Atkins W. Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J Biol Chem. 2007b;282:28309–28320. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- Nath A, Koo P, Rhoades E, Atkins W. Allosteric Effects on Substrate Dissociation from Cytochrome P450 3A4 in Nanodiscs Observed by Ensemble and Single-Molecule Fluorescence Spectroscopy. J Am Chem Soc. 2008b;130:15746–15747. doi: 10.1021/ja805772r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi R, Mu H, Wang X, Yao Q, Chen C. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. QJM - Int J Med. 2005;98:845–856. doi: 10.1093/qjmed/hci136. [DOI] [PubMed] [Google Scholar]
- Park H, Toprak E, Selvin P. Single-molecule fluorescence to study molecular motors. Q Rev Biophys. 2007;40:87–111. doi: 10.1017/S0033583507004611. [DOI] [PubMed] [Google Scholar]
- Peterman E, Sosa H, Moerner W. Single-molecule fluorescence spectroscopy and microscopy of biomolecular motors. Annu Rev Phys Chem. 2004;55:79–96. doi: 10.1146/annurev.physchem.55.091602.094340. [DOI] [PubMed] [Google Scholar]
- Protter M, Elad M. Image sequence denoising via sparse and redundant representations. IEEE T Image Proc. 2009;18:27–35. doi: 10.1109/TIP.2008.2008065. [DOI] [PubMed] [Google Scholar]
- Ritchie T, Grinkova Y, Bayburt T, Denisov I, Zolnerciks J, Atkins W, Sligar S. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–31. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc Natl Acad Sci U S A. 2003;100:3197–202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades E, Ramlall T, Webb W, Eliezer D. Quantification of alpha-synuclein binding to lipid vesicles using fluorescence correlation spectroscopy. Biophys J. 2006;90:4692–4700. doi: 10.1529/biophysj.105.079251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Atkins W. Energetics of heterotropic cooperativity between alpha-naphthoflavone and testosterone binding to CYP3A4. Arch Biochem Biophys. 2007;463:89–101. doi: 10.1016/j.abb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Tricerri M, Gratton E. Interaction of high density lipoprotein particles with membranes containing cholesterol. J Lipid Res. 2007;48:1689–1700. doi: 10.1194/jlr.M600457-JLR200. [DOI] [PubMed] [Google Scholar]
- Segrest J, Jones M, Klon A, Sheldahl C, Hellinger M, De Loof H, Harvey S. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- Shaw A, Pureza V, Sligar S, Morrissey J. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–63. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson C, Tracy T. CYP2D6-CYP2C9 Protein-Protein Interactions and Isoform-Selective Effects on Substrate Binding and Catalysis. Drug Metab Dispos. 2009 doi: 10.1124/dmd.109.026500. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel K, Wilkinson G. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- Torres T, Levitus M. Measuring conformational dynamics: a new FCS-FRET approach. J Phys Chem B. 2007;111:7392–400. doi: 10.1021/jp070659s. [DOI] [PubMed] [Google Scholar]
- Trexler A, Rhoades E. alpha-Synuclein Binds Large Unilamellar Vesicles as an Extended Helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalkova T, Davydova N, Halpert J, Davydov D. Mechanism of interactions of alpha-naphthoflavone with cytochrome p450 3A4 explored with an engineered enzyme bearing a fluorescent probe. Biochemistry. 2007;46:106–119. doi: 10.1021/bi061944p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero D, Kondo J, Ihara Y, Saitoh T. Molecular-cloning of cDNA-encoding an unrecognized component of amyloid in Alzheimer-disease. Proc Natl Acad Sci U S A. 1993;90:11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich M, Isacoff E. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer T, Bax A, Cole N, Nussbaum R. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- Vogelsang J, Cordes T, Forthmann C, Steinhauer C, Tinnefeld P. Controlling the fluorescence of ordinary oxazine dyes for single-molecule switching and superresolution microscopy. Proc Natl Acad Sci U S A. 2009;106:8107–8112. doi: 10.1073/pnas.0811875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb P, Zhen W, Poon A, Conway K, Lansbury P. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- Whorton M, Bokoch M, Rasmussen S, Huang B, Zare R, Kobilka B, Sunahara R. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Cosme J, Vinkovic D, Ward A, Angove H, Day P, Vonrhein C, Tickle I, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- Zeldin R, Petruschke R. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemoth. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]