Abstract
Isocitrate dehydrogenase kinase/phosphatase (AceK) regulates entry into the glyoxylate bypass by reversibly phosphorylating isocitrate dehydrogenase (ICDH). Based on the recent complex structure of AceK-ICDH from E. coli, we have classified the structures of homodimeric NADP+-ICDHs to rationalize and predict which organisms likely contain substrates for AceK. One example is Burkholderia pseudomallei (Bp). Here we report a crystal structure of Bp-ICDH which exhibits the necessary structural elements required for AceK recognition. Kinetic analyses provided further confirmation that Bp-ICDH is a substrate for AceK. We conclude that the highly stringent AceK binding sites on ICDH are maintained only in Gram-negative bacteria.
The enzyme isocitrate dehydrogenase (ICDH) participates in the Krebs cycle converting isocitrate to α-ketoglutarate in a two-step mechanism where a dehydrogenation reaction requiring the reduction of NAD(P)+ to NAD(P)H+, is followed by a decarboxylation reaction.1 ICDH exists in all domains of life (Bacteria, Eukarya and Archaea) yet it has evolved to differ in its cofactor specificity and its oligomeric state. Based on primary sequence the family is classified into three subfamilies. Subfamily I includes archaeal and bacterial NADP+-ICDHs; subfamily II is mainly eukaryotic NADP+-ICDHs with some bacterial exceptions; and subfamily III includes hetero-oligomeric NAD+-ICDHs.2 Isocitrate dehydrogenase kinase/phosphatase (AceK) acts directly on ICDH and is the metabolic switch between the energy generating Krebs cycle and gluconeogenesis requiring glyoxylate bypass in response to nutrient availability.3 Notably, AceK only exists in certain bacteria (discussed later). AceK is a bifunctional enzyme with protein kinase, phosphatase and ATPase activities all remarkably shared at the same active site3 and is a rare example of reversible prokaryotic protein phosphorylation, acting as a “primitive” protein kinase with opposing phosphatase activity. Despite being the very founding member of prokaryotic protein phosphorylation,4,5 a full understanding of AceK has remained elusive until the structures of AceK and its complex with ICDH from E. coli were recently unveiled.6
Many protein kinases are able to use peptide substrates that are based on the sequence surrounding the phosphorylation site (P-site) albeit at a reduced affinity. Substrate specificity is often increased by docking motifs, like in MAPKs where the D domain on the MAPK target binds the docking groove on MAPK distal to the catalytic site.7,8 Some kinase-substrate interactions require scaffold proteins, particularly in intracellular signaling pathways.7 However, AceK only forms an intimate association with homodimic ICDH (Figure 1A) and cannot phosphorylate either proteolytic fragments derived from ICDH or synthetic peptides corresponding to the sequence around the phosphorylation site. There do exist kinases that require globular domains for recognition,9,10 however AceK uniquely requires the ICDH dimer.6 The substrate recognition loop (SRL; residues 484–510) of AceK deeply extends ~32 Å into the active site cleft of ICDH with a short α-helix at its tip. An extensive array of interactions between the SRL and both molecules of dimeric ICDH exist, including: hydrophobic packing, salt bridges and hydrogen bonds.6 Since AceK recognizes the tertiary structure of homodimeric ICDH to form a “productive” complex with one substrate site, a peptide is simply an inadequate interaction epitope for AceK11,12 and cannot substitute for the complexity the dimer creates.
Figure 1.
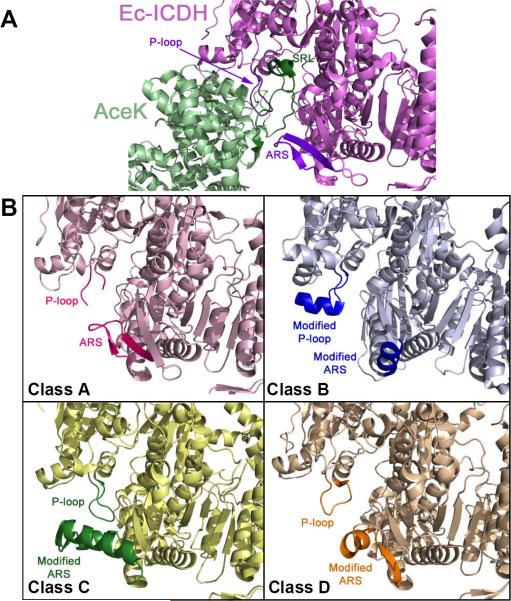
Structural classification of ICDH based on AceK binding. (A) AceK-Ec-ICDH complex structure. The SRL (green) of AceK (teal) extends into the Ec-ICDH (purple) structure. The main binding elements, ARS and P-loop (blue), for AceK on ICDH are shown. (B) Classification of ICDH based on the structural elements in the corresponding P-loop and ARS regions. For clarity only one representative structure is shown for each class (Class A: Bp-ICDH, Class B: Pc-ICDH, Class C: Bs-ICDH and Class D: Ap-ICDH).
From the complex structure of E. coli AceK-ICDH the regions of protein-protein interaction were identified. To understand possible reasons why AceK is unable to bind and act on most ICDHs a comparison and search for putative AceK binding sites on other ICDHs was undertaken. Since ICDH from E. coli (Ec), a known target of AceK, is an NADP+-dependent homodimer, we limited our comparisons to homodimeric NADP+-ICDHs whose crystal structures have been solved and deposited into the Protein Data Bank. ICDH from Bacillus subtilis (Bs), Burkholderia pseudomallei (Bp; this work), Aeropyrum pernix (Ap), Archaeoglobus fulgidus (Af), and a hypothetical ICDH from Sulfolobus tokodaii (St) are subfamily I members and Pc-ICDH (porcine heart mitochondrial), Hc-ICDH (human cytosolic), Sc-ICDH (Saccharyomyces cerevisiae), Dp-ICDH (Desulfotalea psychrophila), Tm-ICDH (Thermotoga maritima), and Mc-ICDH (mouse cytosolic) are subfamily II members. Although the sequence identity is quite low (18–23% sequence identity to Ec-ICDH) between these two subfamilies their structures are remarkably similar including the catalytic active site residues. Using DaliLite Pairwise comparison of protein structures13 the regions that likely contact AceK were evaluated. Based on these alignments and the AceK-ICDH complex structure we further classified these ICDHs according to the proposed AceK binding motifs (Figures 1B and S1). Group A is Ec- and Bp-ICDH; Group B is Pc-, Hc-, Dp-, Tm-, Sc- and Mc-ICDHs; Group C is Bs-ICDH and Group D is Ap-, Af- and St-ICDHs. Ser113 from Ec-ICDH, the target of phosphorylation by AceK,3 is structurally conserved among all ICDHs.
Only Bp-ICDH was classed with Ec-ICDH in Group A showing remarkable similarities overall and particularly with the AceK binding sites. To reveal the detailed structural elements for AceK recognition and binding, we solved the structure of Bp-ICDH at 1.65 Å resolution (see Supporting Information). Bp- and Ec-ICDH share 75% sequence identity and aligns with a Cα RMSD of 0.57 Å over 410 similar residues indicating that these enzymes are highly similar in sequence and structure (Figure S2). From the AceK-ICDH complex structure two discontinuous regions on ICDH contact AceK. These include the P-loop from monomer 1 and with the twisted antiparallel β-sheet from monomer 2 (ARS; AceK recognition segment), in the homodimeric structure of ICDH. Similar structural elements are present in Bp-ICDH (Figures 1 and S1) and like Ec-ICDH no apparent structural clashes would occur if AceK bound to Bp-ICDH (Figure S3).
Group B is comprised of subfamily II ICDHs and the most notable difference is the P-loop, which compared to other bacterial and archaeal ICDHs contains an insertion of several residues forming a short helix (Figure 1). This insertion may limit access to the active site but it would also interfere and prevent AceK binding,14 conflicting with the SRL and the helix adjacent to the SRL on AceK both sterically and through electrostatic repulsion due to the inherent negative charge of these elements. A novel self-regulatory mechanism is proposed in Hc-ICDH that mimics the phosphorylation by AceK.15 Currently no evidence exists for phosphorylation of ICDH nor has any AceK protein or homo-logues been identified in eukaryotes. During phosphorylated regulation by AceK, the phosphate moiety attached to the serine inhibits isocitrate binding to the active site. Alternatively, in the inactive form of Hc-ICDH a helix unwinds into a loop allowing Asp279 to interact with Ser94 (analogous to Ser113 of Ec-ICDH) causing inhibition of isocitrate binding without phosphorylation.15 Evidence for a self regulatory mechanism, however, is not supported by the structure of Tm-ICDH, a thermophilic bacteria,16 as such is not universal to all subfamily II members.
Bs-ICDH from B. subtilis, a Gram-positive bacterium, represents Class C and overall is 71 and 75% identical to and completely conserved near the phosphorylation site of Ec-ICDH and Bp-ICDH, respectively. However, Bs-ICDH is a poor substrate for AceK.17 Previous AceK kinetic studies reported the KM values for the kinase and phosphatase reactions to be 60- and 3450-fold greater than those for Ec-ICDH, respectively.17 Thus, Bs-ICDH is not an ideal candidate to be phosphorylated by AceK. This is not surprising given that no observations exist for the phosphorylation of Bs-ICDH in vivo nor has a corresponding Bs-AceK protein been identified.18 The key difference is the 13 residue insert (residues 246–276) in the small domain. This insert consists of a turn, a single β-strand and two α-helices, and projects 8 Å outward restricting access to the active site and would partially clash with the SRL of AceK (Figure 1).6 This region corresponds to the ARS in Ec-ICDH, which we propose is important for AceK recognition. The unique insert is also observed in Acidithiobacillus thiooxidans ICDH,19 however, it is an NAD+-dependent homodimeric ICDH but like B. subtilis no AceK proteins have been identified in this species.
Lastly, Class D is the archaeal subfamily I ICDHs and structurally very similar to Ec-ICDH with an Cα RMSD of 1.7 Å. In the region of the ARS, the second strand is substituted by an α-helix, further supporting that the ARS is a critical binding determinant for AceK. A homology search of all available archaeal genomes reveals that no ORFs encode for known or putative AceK proteins.20 Phosphorylation of the corresponding serine in archaeal ICDHs has not been observed either.21 Unfortunately, all the ICDHs structures described above, with the exception of Class A, represent those species that do not encode for any known or putative AceK as dictated by BLAST comparisons and searches of all available genomic databases. In contrast, B. pseudomallei from Class A indeed encodes AceK (Uniprot ID Q63Y16).
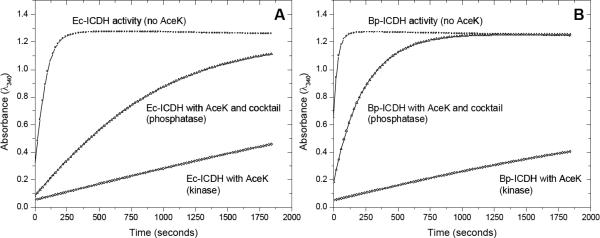
Ec-AceK phosphorylates (and dephosphorylates) Ec-ICDH and, based on our structural classification, Ec-AceK is likely to act on Bp-ICDH (Class A) with comparable efficiency. To investigate this structure-based prediction further, kinetic studies were undertaken to evaluate the ability of Bp-ICDH to be a substrate for both the kinase and phosphatase activities of AceK. The AceK kinase assay is coupled to ICDH activity whereby the reduction of NADP+ to NADPH is monitored. When AceK phosphorylates ICDH the activity of ICDH is inhibited; hence higher ICDH activity corresponds to lower AceK kinase activity.6 Both Ec- and Bp-ICDH reduce NADP+ in the absence of AceK with similar efficiency (Figure 2). When AceK is present only 3% of Ec-ICDH activity is retained and, as predicted, Bp-ICDH was similarly affected only retaining 2% activity. This loss of ICDH catalytic function infers that AceK phosphorylated both ICDH proteins. We have further shown the direct phosphorylation of both Ec- and Bp-ICDH by AceK using a dot-blot assay (Figure S4). Since AceK exhibits normal kinase function against both ICDH proteins we proceeded to evaluate the phosphatase activity. AceK initially phosphorylates ICDH which is followed by the addition of a phosphatase activator/kinase inhibitor cocktail (containing AMP and pyruvate which inhibit kinase and activate phosphatase activities of AceK22). The dephosphorylation of ICDH by AceK removes the inhibition of ICDH activity; so that higher AceK phosphatase activity leads to higher observed ICDH activity. Addition of the cocktail increased ICDH activity (indicative of phosphatase function) to 14 and 32% of the normal ICDH activity for Ec- and Bp-ICDH, respectively (Figure 2). Our structural analysis predicted that Bp-ICDH is a substrate for AceK and this is supported by the kinetics results. Both Bp- and Ec-ICDH are true substrates for this bifunctional enzyme. Therefore, our newly determined Bp-ICDH structure contains the necessary binding elements for the productive recognition by AceK and provides biochemical evidence that the P-loop and ARS are critical for this protein-protein interaction. It also illustrates the possibility of cross-species reactivity, as they are interchangeable as substrate proteins. Likewise, we predict that AceK from the two species are also interchangeable.
Figure 2.
AceK kinase and phosphatase activity against (A) Ec-ICDH and (B) Bp-ICDH. ICDH activity in the absence of AceK acts as a reference and phosphatase activity was measured in the presence of a phosphatase activator/kinase inhibitor cocktail.
A search of all known or putative AceK proteins revealed that its existence is limited to Gram-negative bacteria. The structural comparison of Bp- and Ec-ICDHs, both from Gram-negative bacteria, clearly shows conservation of the AceK binding sites, whereas in all other ICDHs these sites are modified. In addition, ICDHs from species that encode for an AceK protein show remarkable sequence conservation with no insertions (Figure S5); likely any of these ICDH proteins (like Ec- and Bp-ICDHs) would be interchangeable as substrates for AceK. It also demonstrates the highly stringent nature of this protein-protein interaction as even small structural changes eliminate this interaction.
AceK serves as the gatekeeper to the glyoxylate bypass. The bypass consists of the enzymes isocitrate lyase (ICL) and malate synthase (MS). ICL competes with ICDH for isocitrate, however ICDH has a higher affinity for this metabolite. AceK assists by temporarily shutting down ICDH allowing ICL the opportunity to convert isocitrate to glyoxylate, and then to malate by MS.23 This bypass compromises the decarboxylation steps of the Krebs cycle, which is critical for survival on two-carbon compounds preventing the loss of acetyl-CoA as CO2. Therefore, the four-carbon metabolic intermediates, succinate and malate, are continued to be produced but at the cost of generating energy. The glyoxylate bypass has been observed in bacteria, protists, plants, fungi and nematodes. The existence of this bypass in Metazoa remains controversial despite some evidence of ICL and/or MS activities.24 It is apparent that Gram-negative bacteria are the only organisms to use AceK as the on-off switch for this pathway. The genomes of B. subtilis, and other Gram-positive bacteria (e.g. Staphylococcus, Streptococcus, Enterococcus, and Clostridium) lack the entire glyoxylate bypass and its enzymes, rationalizing the absence of AceK. And despite evidence for ICL and MS in some low G+C Gram-positive bacteria, AceK remains absent in their genomes.18 Evidence does exist for the glyoxylate pathway in Archaea,25 however, with no AceK20 to turn on the bypass nor any evidence for phosphorylated regulation of ICDH,21 another regulatory mechanism must exist to funnel isocitrate into the bypass, perhaps analogous to the self-regulatory mechanism of Hc-ICDH.15
AceK is as unique to Gram-negative bacteria as are the specific structural binding elements on ICDH. Did the absence or loss of an encoded AceK protein coincide with the modification of the AceK recognition sites on ICDHs? Perhaps the differences in ICDH structure were a consequence of the lack of selective pressure to maintain these elements in the absence of AceK or the ICDH proteins evolved to modify the corresponding P-loop and ARS regions to circumvent host-pathogen interactions to protect from pathogenic Gram-negative bacteria producing AceK. Thus alterations in ICDH structure were a protective adaptation requiring alternative means to regulate ICDH under conditions of poor nutrient availability.
In summary, taking advantage of the recently determined E. coli ICDH-AceK complex structure we have re-classified ICDHs and predicted that Bp-ICDH is a substrate for Ec-AceK. Indeed, our Bp-ICDH structure displays all the essential structural elements required for Ec-AceK recognition and interaction. Our kinetic experiments further support this prediction. Based on the sequence and structural analyses, we conclude that AceK is specific for ICDHs from Gram-negative bacteria.
Supplementary Material
ACKNOWLEDGMENT
We thank the whole SSGCID team. This research was funded under Federal Contract No. HHSN272200700057C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Dept. of Health and Human Services to SSGCID and by the Canadian Institutes of Health Research (CIHR) grant to ZJ. ZJ is a Canada Research Chair in Structural Biology and a Killam Research Fellow and SPY is a recipient of a CIHR fellowship. We thank Natalie Roy and Yidai Yang for technical assistance
Footnotes
Supporting Information. Methods, Supplementary Tables and Figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Dean AM, Koshland DE., Jr. Biochemistry. 1993;32:9302–9309. doi: 10.1021/bi00087a007. [DOI] [PubMed] [Google Scholar]
- (2).Steen IH, Madern D, Karlstrom M, Lien T, Ladenstein R, Birkeland NK. J. Biol. Chem. 2001;276:43924–43931. doi: 10.1074/jbc.M105999200. [DOI] [PubMed] [Google Scholar]
- (3).Cozzone AJ. Annu. Rev. Microbiol. 1998;52:127–164. doi: 10.1146/annurev.micro.52.1.127. [DOI] [PubMed] [Google Scholar]
- (4).Garnak M, Reeves HC. Science. 1979;203:1111–1112. doi: 10.1126/science.34215. [DOI] [PubMed] [Google Scholar]
- (5).LaPorte DC, Koshland DE., Jr. Nature. 1982;300:458–460. doi: 10.1038/300458a0. [DOI] [PubMed] [Google Scholar]
- (6).Zheng J, Jia Z. Nature. 2010;465:961–965. doi: 10.1038/nature09088. [DOI] [PubMed] [Google Scholar]
- (7).Ubersax JA, Ferrell JE., Jr. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- (8).Weston CR, Lambright DG, Davis RJ. Science. 2002;296:2345–2347. doi: 10.1126/science.1073344. [DOI] [PubMed] [Google Scholar]
- (9).Dar AC, Dever TE, Sicheri F. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- (10).Komander D, Garg R, Wan PT, Ridley AJ, Barford D. EMBO J. 2008;27:3175–3185. doi: 10.1038/emboj.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Manning G, Plowman GD, Hunter T, Sudarsanam S. Trends Biochem. Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- (12).Oudot C, Cortay JC, Blanchet C, LaPorte DC, Di PA, Cozzone AJ, Jault JM. Biochemistry. 2001;40:3047–3055. doi: 10.1021/bi001713x. [DOI] [PubMed] [Google Scholar]
- (13).Holm L, Park J. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- (14).Ceccarelli C, Grodsky NB, Ariyaratne N, Colman RF, Bahnson BJ. J. Biol. Chem. 2002;277:43454–43462. doi: 10.1074/jbc.M207306200. [DOI] [PubMed] [Google Scholar]
- (15).Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. J. Biol. Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- (16).Karlstrom M, Steen IH, Madern D, Fedoy AE, Birkeland NK, Ladenstein R. FEBS J. 2006;273:2851–2868. doi: 10.1111/j.1742-4658.2006.05298.x. [DOI] [PubMed] [Google Scholar]
- (17).Singh SK, Miller SP, Dean A, Banaszak LJ, LaPorte DC. J. Biol. Chem. 2002;277:7567–7573. doi: 10.1074/jbc.M107908200. [DOI] [PubMed] [Google Scholar]
- (18).Sonenshein AL. The Krebs Citric Acid Cycle. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its closest relatives: From genes to cells. ASM Press American Society for Microbiology; Washington, DC: 2002. pp. 151–162. [Google Scholar]
- (19).Imada K, Tamura T, Takenaka R, Kobayashi I, Namba K, Inagaki K. Proteins. 2008;70:63–71. doi: 10.1002/prot.21486. [DOI] [PubMed] [Google Scholar]
- (20).Kennelly PJ. Biochem. J. 2003;370:373–389. doi: 10.1042/BJ20021547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Aivaliotis M, Macek B, Gnad F, Reichelt P, Mann M, Oesterhelt D. PLoS. One. 2009;4:e4777. doi: 10.1371/journal.pone.0004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Miller SP, Chen R, Karschnia EJ, Romfo C, Dean A, LaPorte DC. J. Biol. Chem. 2000;275:833–839. doi: 10.1074/jbc.275.2.833. [DOI] [PubMed] [Google Scholar]
- (23).Cozzone AJ, El-Mansi M. J. Mol. Microbiol. Biotechnol. 2005;9:132–146. doi: 10.1159/000089642. [DOI] [PubMed] [Google Scholar]
- (24).Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Biol. Direct. 2006;1:31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. Microbiology. 2009;155:3166–3175. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.