Abstract
While mutational activation of the Epidermal Growth Factor Receptor (EGFR) features prominently in glioma and non-small-cell lung cancer (NSCLC), inhibitors of EGFR improve survival only in NCSLC. To understand how mutations in EGFR influence response to therapy, we generated glioma cells expressing either glioma- or NSCLC-derived alleles, quantifying kinase site occupancy by clinical inhibitors using novel affinity probe and kinetic methodology. At equivalent doses, erlotinib achieved lower kinase site occupancy in glioma-derived EGFRvIII, compared to NSCLC-derived EGFR mutants. Kinase site occupancy correlated directly with cell cycle arrest. EGFRvIII released erlotinib rapidly compared to wild-type EGFR, whereas NSCLC-derived mutants released erlotinib slowly.
Keywords: Erlotinib, EGFR, malignant glioma, EGFRvIII, kinase site occupancy
INTRODUCTION
The epidermal growth factor receptor (EGFR/HER1/ErbB1) is a transmembrane protein belonging to the EGFR family of receptor tyrosine kinases1. EGFR is implicated as an oncogene in a large number of cancers, driving malignancy through over-expression/amplification, activating mutation, and/or decreased turnover2. Amplification of EGFR occurs commonly in malignant glioma, the most common primary brain tumor3. These tumors typically express a truncated form of EGFR, due to the in-frame deletion of introns 2–7, resulting in a ligand-independent, constitutively active EGFR protein (EGFRvIII)4,5. EGFR amplification and mutation have also been reported in non-small cell lung cancer (NSCLC), the most aggressive form of lung cancer. Ten percent of NSCLC patients have mutations in the EGFR kinase domain, which result in activated EGFR signaling6,7.
Small molecule tyrosine kinase inhibitors (TKIs) of EGFR have been developed and tested clinically8. Erlotinib (Tarceva, OSI-774) and gefitinib (Iressa) showed poor overall responses in initial clinical trials for chemotherapy-refractory NSCLC, although a subsection of patients had dramatic responses7. Retrospective studies in responders subsequently identified mutations within the tyrosine kinase domain of EGFR, typically point mutation in exon 21 (L858R) or in-frame deletion in exon 19 (del746-750). Biochemical analyses revealed that these mutations increased EGFR activity and led to heightened sensitivity to EGFR blockade9.
In contrast, while an initial study showed increased rate of transient radiographic responses to EGFR inhibition in malignant glioma patients with EGFRvIII expression10, subsequent trials demonstrated no survival benefit11,12. The poor response to TKIs observed in glioma patients with mutationally activated EGFRvIII stands in stark contrast to the dramatic response seen in NSCLC patients with activated EGFR mutations (L858R and del746-750).
Could access to brain tissue, or cell-type differences account for this differential response to therapy? Evidence against this argument stems from observations that EGFRvIII is also found in ~5% of patients with NSCLC13, and does not correlate with response to therapy in this group. Moreover, Ji and colleagues compared human B-cells transduced with lung- or brain-cancer derived alleles of EGFR13, demonstrating EGFRvIII cells to show significant resistance to TKI treatment, relative to EGFR-L858R cells. These data suggest that the differential response of L858R and EGFRvIII is linked to mutational status of EGFR itself.
To elucidate a mechanistic basis for the differential responses to therapy observed in lung- and brain-cancer derived alleles of EGFR, we generated isogenic cell lines containing either wild type-, glioma- or NSCLC-derived alleles of EGFR. Using a novel EGFR fluorescent affinity probe to measure EGFR kinase site occupancy, we demonstrate that the differential therapeutic response across the panel of cells correlates with differential occupancy of TKI in the kinase-active site. To provide molecular insights into these differences in occupancy, we demonstrated that brain cancer-derived alleles of EGFR released erlotinib more rapidly, whereas NSCLC-derived mutants released erlotinib more slowly, in comparison with wild-type EGFR. These data provide a mechanistic basis for the differential response of lung- and brain-cancer patients to EGFR TKIs, and highlight kinase site occupancy as a prominent biomarker for efficacy.
RESULTS
Erlotinib Treatment Inhibits Growth in a Mutant EGFR-Specific Manner
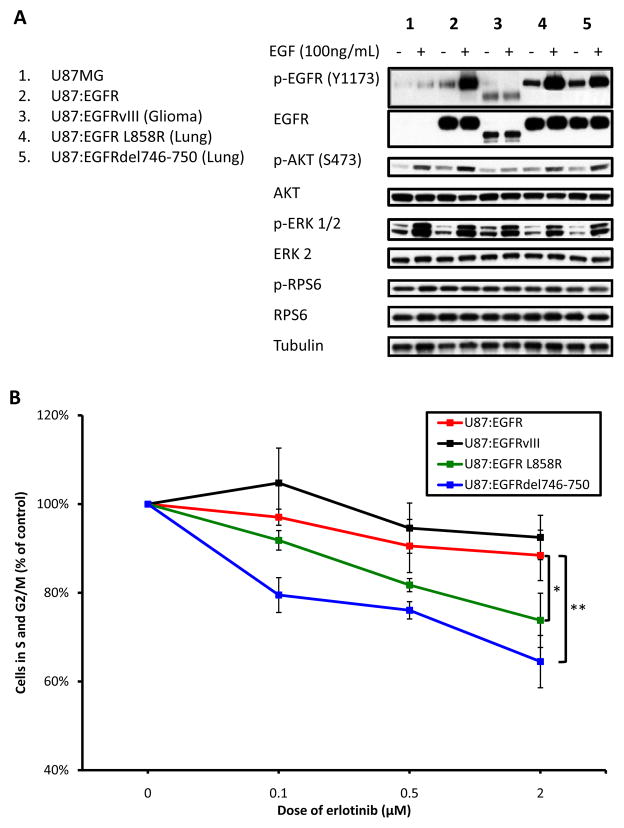
To control for cell type-specific effects, wild-type EGFR, EGFRvIII, EGFR L858R, and EGFRdel746-750 were transduced individually into glioma cell lines U87MG and LN229MG. Since PTEN may also drive resistance to EGFR TKIs14, we chose cell lines both wild-type (LN229MG) and mutant (U87MG) for PTEN. As expected5,9 the variant III, L858R, and del746-750 EGFR-mutants exhibited increased basal EGF-independent phosphorylation compared to the EGFRWT (Figure 1A). Flow cytometric analysis demonstrated the four EGFR-alleles to show differential responses to erlotinib (Figure 1b), with comparable results observed in LN229MG cells (Supplementary Figure 1). A similar trend was also observed in cell viability in both the U87MG and LN229MG EGFR-allele panels (Supplementary Figure 2). These cell-based observations were thus aligned with clinical data from patients treated trials with erlotinib.
Figure 1.
EGFR mutants exhibit differential growth inhibition in response to erlotinib. A, U87MG cells were transduced with wild type-EGFR, EGFRvIII, EGFR L858R, or EGFRdel746-750. Cells were treated or untreated with EGF 30 min prior to harvest, as shown. Cells were then lysed and analyzed by immunoblot. Signaling downstream of EGFR was analyzed using antisera to total and phospho-specific proteins shown. B, The U87MG panel was treated for 24 hours with three doses of erlotinib, as shown. Cells were analyzed for their DNA content, measured by propidium iodide staining. For each sample, the proportion of cells in S and G2/M phases was combined and graphed. Treatments were completed in triplicate with error bars representing one standard deviation (* p<0.05, ** p<0.01).
Quantifying Kinase Site Occupancy in Mutant-EGFR Alleles
We previously developed a fluorescent probe specific to the active site of EGFR, by attaching an NBD fluorophore via a PEG linker to the C7 position of PD168393, a 6-acrylamido-4-anilinoquinazoline that binds irreversibly to Cys797 of EGFR15. Despite the highly conserved nature of the kinase active site, the presence of this cysteine is rare among receptor tyrosine kinases, affording this probe, [16], high specificity for EGFR16. Our previous study validated the capacity of this probe to measure the kinase site occupancy of anilinoquinazoline derivatives in analog-sensitive (as) alleles. Here, we show that [16] also has high specificity for wild-type EGFR, as well as the glioma-and NSCLC-derived mutants (Supplementary Figure 3).
Erlotinib Achieves Allele Specific Differences in Kinase Site Occupancy in Lung-and Brain-Cancer Derived Mutants of EGFR
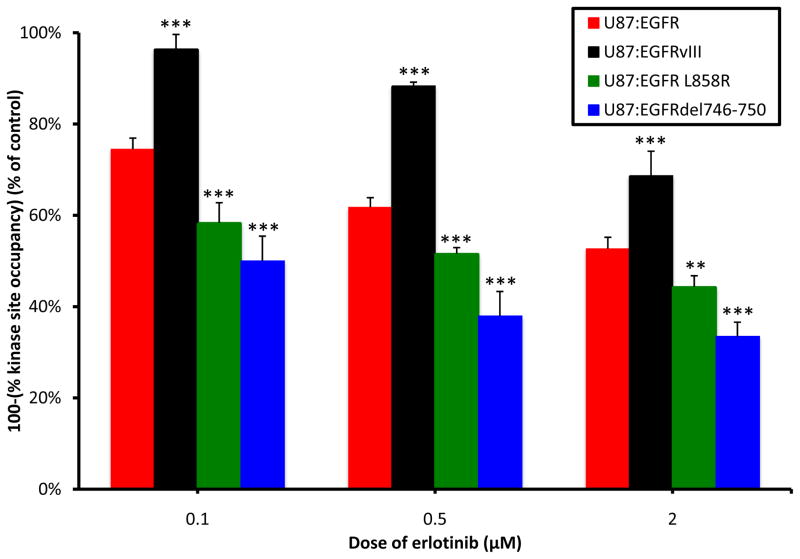
Cells were treated with erlotinib, then subjected to a short pulse-chase of the EGFR-fluorescent affinity probe ([16]) on ice. Since [16] can only bind unoccupied active site, this method quantifies open kinase site across the different mutant alleles. The binding of erlotinib to EGFR is dynamic. Thus, a fraction of erlotinib-bound EGFR will become unoccupied during the [16] pulse, and will become available for [16] binding. Therefore, [16] labeling quantifies the amount of kinase site that has remained occupied during the period of probe labeling, referred to as erlotinib’s kinase site occupancy. In both drug-treated U87 and LN229 panels, erlotinib achieved significantly higher levels of kinase-site occupancy in NSCLC-derived alleles of EGFR, compared with EGFRvIII (Figure 2, Supplementary Figure 4).
Figure 2.
EGFR alleles differ in levels of kinase site occupancy achieved after erlotinib treatment. The U87MG panel was treated overnight with erlotinib at doses shown, and stimulated for 30 minutes with 100ng/mL EGF. Cells were subjected to a 25-minute pulse-chase with 60μM [16], then lysed and separated by SDS-PAGE. Gels were scanned on a Typhoon fluorescence imager using a 488nm excitation laser. Levels of fluorescence correspond to the amount of kinase active site that is unbound by erlotinib (100-(% kinase site occupancy)), and thus is available for probe binding. The fluorescence intensity for each treatment was quantified by densitometry and scaled as a percent of the EGF-stimulated control lane. Results were completed in triplicate with error bars representing one standard deviation (** p<0.01, *** p<0.005).
Kinase Site Occupancy is a Biomarker for Efficacy
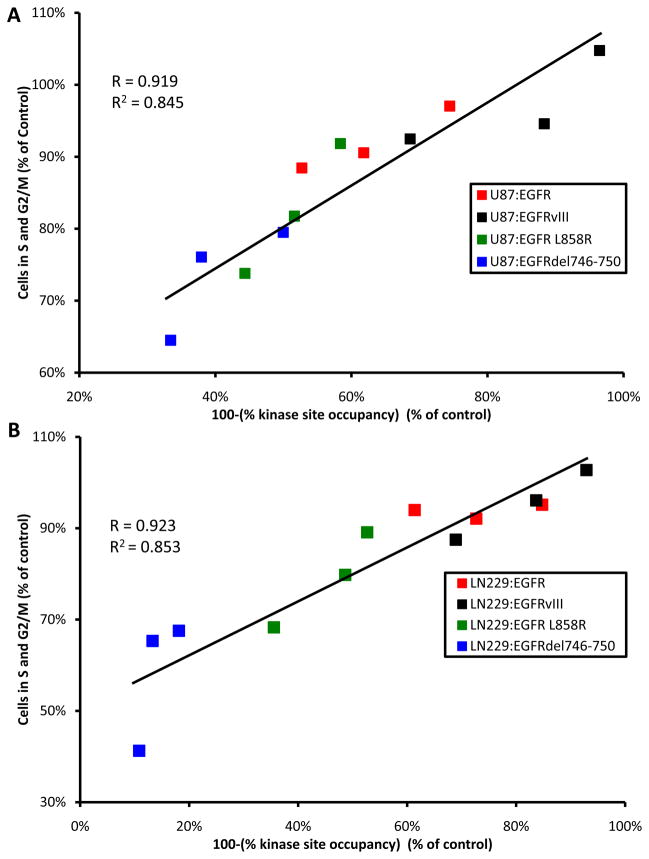
Calculated levels of kinase site occupancy mirrored the trend of erlotinib’s efficacy observed in patients. Kinase site occupancy was also closely aligned with cell cycle arrest achieved by erlotinib across the panels. The correlation coefficient of open kinase site and percent dividing cells was identical, 0.92, for both the U87MG and LN229MG EGFR-allele panels, (Figure 3A and B). These data suggest kinase site occupancy as a biomarker for the differential efficiency of erlotinib across tumor-derived, activated alleles of EGFR. In addition, different mutationally activated alleles of EGFR all showed identical trends between kinase site occupancy and proliferation in two different cell lines (Supplementary Figure 5). Thus, data in Figure 3 and Supplementary Figures 5 demonstrate that allele-specific differences in kinase occupancy are the major arbitrator distinguishing differential sensitivity to erlotinib.
Figure 3.
Kinase site occupancy is a better biomarker than abundance of p-EGFR, for efficacy of erlotinib. A, For each EGFR-allele and at each dose of erlotinib, 100-(% kinase site occupancy) was assigned the x-coordinate. The proportion of cells in S and G2/M phases was combined and graphed on the y-coordinate. We found a strong (R=0.916) correlation between kinase site occupancy and efficacy for the U87 panel. B, The same analysis was applied to the LN229 panel, where a correlation of R=0.923 was found between kinase site occupancy and efficacy.
Antiproliferative Effects of Erlotinib Correlate Poorly with Abundance of p-EGFR
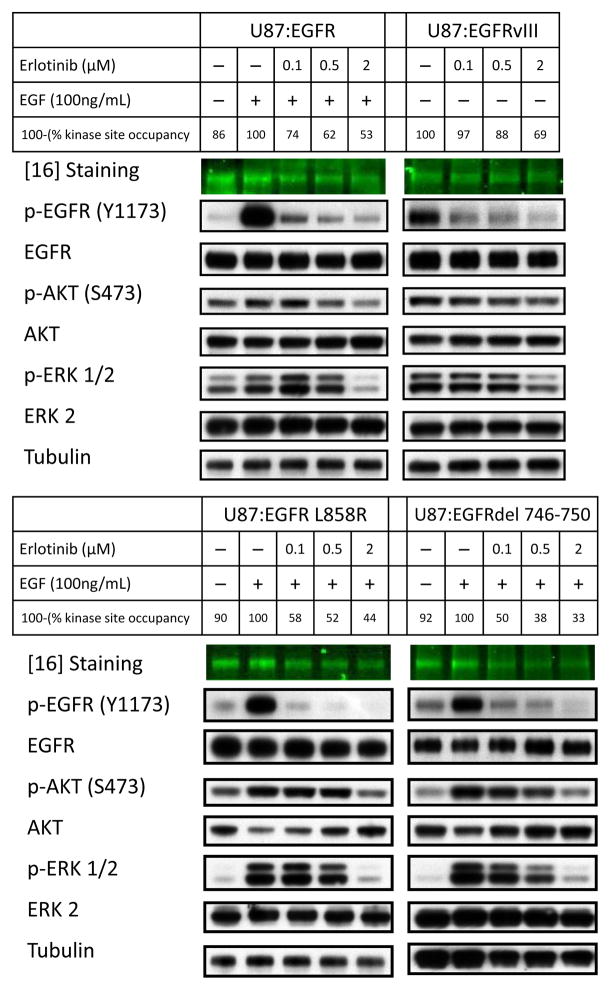
Using the reversible EGFR inhibitor erlotinib in the panel of wild-type and mutant alleles of EGFR, we assessed the relationship between kinase site occupancy and downstream signaling (Figure 4). Immunoblot analysis of the U87MG panel revealed a marked difference between kinase site occupancy and abundance of p-EGFR as measured at Y1173 and global phosphorylation of EGFR as measured by 4G10 anti-tyrosine antibody (Supplementary Figure 6). Analysis of the western blots using fluorescently-coupled secondary antibodies and densitometry revealed coefficients of 0.71 and 0.50 for the correlation of kinase site occupancy with p-EGFR (Y1173) and p-Tyr (4G10), respectively. Weak correlations were also measured between antiproliferative efficacy and abundance of p-EGFR (Y1173) and p-Tyr (4G10), with correlation coefficients of 0.68 and 0.52, respectively (Supplementary Figure 7, Supplementary Figure 8). The weak overall correlation between p-EGFR levels and efficacy was due to differences in the cell cycle response of each allele, at similar abundances of p-EGFR (Supplementary Figure 9), visualized by the differences in the trend lines for each allele. These observations suggest that p-EGFR levels are a poor biomarker for erlotinib’s efficacy across EGFR-alleles.
Figure 4.
Antiproliferative effects of erlotinib correlate poorly with abundance of p-EGFR. The U87MG panel was treated with three doses of erlotinib, as shown, then pulsed with 100 ng/mL EGF prior to harvesting. Phospho- and total protein levels were visualized by western blotting. While low dosages of erlotinib efficiently block p-EGFR (Y1173) in all cell lines, levels of p-ERK 1/2 (T202/Y204) were decreased in NSCLC-derived alleles, compared with brain-cancer derived EGFRvIII, paralleling the antiproliferative response in Figure 1B. U87MG cells are mutant for PTEN, disconnecting signaling between EGFR and PI3K. Thus, the differential abundance of p-AKT (S473) was less apparent in this experiment, as compared with PTENWT LN229 cells (Supplemental Figure 10). Levels of kinase site occupancy more closely follow abundance of phosphorylated downstream molecules.
The abundance of p-EGFR also did not accurately reflect abundance of downstream pathway targets p-AKT and p-ERK1/2. In contrast, levels of kinase site occupancy correlated more accurately with levels of p-ERK1/2, and moderately with levels of p-AKT, though clearly, this correlation was imperfect (Figure 4). Similar results were observed in both U87MG and LN229MG EGFR-allele panels, arguing that these effects were both independent of PTEN-status, and not specific to a particular allele of EGFR. The abundance of p-AKT and p-ERK 1/2 was particularly sensitive to erlotinib in NSCLC-derived mutants, as compared with glioma-derived EGFRvIII, shown clearly in the PTENWT LN229 panel (Supplementary Figure 10). Studies in U87 and LN229 cells expressing a mutant form of EGFR that is resistant to erlotinib (EGFR T790M)17,18, suggest that this effect is not due to any off-target effects of erlotinib (Supplementary Figure 11). This observation demonstrates that kinase site occupancy accurately reflects oncogenic signaling through downstream molecules.
Differences in Kinetics of Erlotinib Binding and Release Underlie Differential Erlotinib Occupancy Observed in Glioma- Versus NSCLC-Derived Mutants of EGFR
To probe the basis for differential kinase site occupancy, we analyzed the kinetics of erlotinib binding to EGFR. Erlotinib-EGFR binding follows a simple equilibrium reaction, with EGFR existing in either erlotinib-bound or erlotinib-unbound states at all times. However, this reaction is difficult to probe in a cellular setting without altering either EGFR or erlotinib in a way that would also change their relative interactions.
Exploiting the fact that the fluorescent probe ([16]) binds all studied EGFR-alleles irreversibly and with a higher affinity than erlotinib, we used [16] to analyze the kinetics of EGFR binding to erlotinib across the panel of EGFR-alleles. EGFR binds irreversibly to [16] through the covalent linkage of Cys797 to [16]. Thus the reaction of Cys797 with [16] acts as a sink for EGFR, preventing it from taking part in the equilibrium reaction with erlotinib. Since [16] has a higher affinity than erlotinib for the active site of EGFR, [16] will, over time, replace erlotinib within the active site. Therefore, the rate with which [16] exchanges with erlotinib can be used as a tool for studying the kinetic interaction between EGFR and erlotinib (detailed in Supplementary Figure 12, legend).
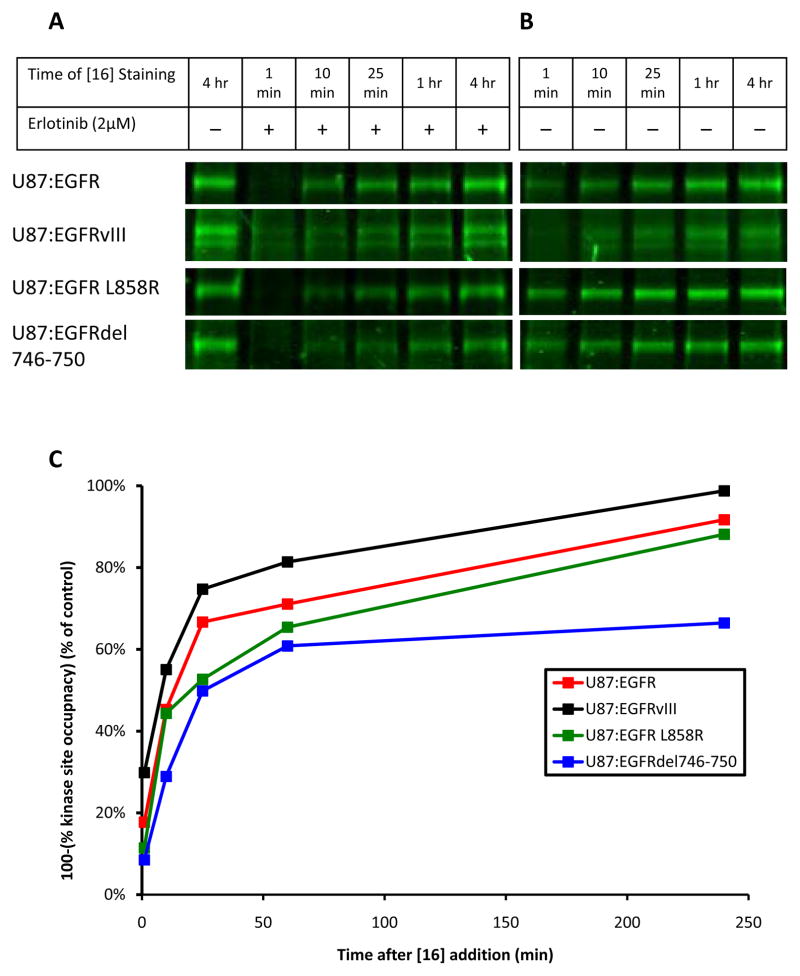
Analyzing these kinetics (Supplementary Figure 12), we found a gradual replacement of erlotinib by [16], over time, represented by an increase in [16] binding to EGFR (Figure 5A and B). Mirroring our previous experiments, the rate replacement in the glioma-derived EGFRvIII was greater than that of the wild-type allele. In contrast, NSCLC-derived EGFR L858R and EGFRdel746-750 both showed slower rates of replacement (Figure 5C). Analysis of the clinical EGFR inhibitor, gefitinib, confirmed that these results were not erlotinib specific (Supplementary Figure 13).
Figure 5.
Kinetics of erlotinib binding/unbinding differ across EGFR-alleles. A and B, The U87 panel was treated with 2μM erlotinib (A) or DMSO (B) for 24 hours and then pulsed with 60μM [16] for 1 minute, 10 minutes, 25 minutes, 1 hour, or 4 hours. In the drug treated experiment (A), the control was untreated and pulsed with [16] for 4 hours. As erlotinib cycles out of EGFR, the active site is irreversibly bound by [16], preventing erlotinib re-binding. Over time, this occurs with all EGFR-bound erlotinib. The rate with which this replacement occurs is related to the speed with which erlotinib is unbound by EGFR. The control (B) established the rate at which [16] alone binds each EGFR-allele (k 2 in Equation (2)). C, Analysis of (A) and (B) by densitometry allowed for the quantification of [16] binding over time in the presence or absence of erlotinib. The level of [16] staining of a single EGFR-allele (as a % of the 4hr control lane) determined in (A) was divided by the level of [16] staining of that same EGFR-allele (as a % of the 4hr control lane) determined in (B), allowing for the tracking of kinase site occupancy for each EGFR-allele over time. These data determine that erlotinib replacement occurs more quickly in glioma-derived EGFRvIII than in NSCLC-derived EGFR-mutants.
To quantify these observations, we determined the time taken for half of the EGFR within the cell to be bound by [16], t1/2 (Table 1). The values of t1/2 represent the relative time during which erlotinib occupies the active site of each EGFR-allele, as compared to the wild-type. The inverse of t1/2 is also related to the speed (Vrelease,Erl) with which erlotinib moves in and out of the active site of each EGFR-allele. These measurements establish the basis for the differential kinase occupancy demonstrated in Figures 2–4, with erlotinib cycling in and out of the active site of EGFRvIII rapidly in comparison with EGFRWT. In contrast, erlotinib moves in and out of the active site of NSCLC-derived alleles of EGFR much more slowly in comparison with EGFRWT. Analogous results were reached using gefitinib (Supplementary Table 1).
Table 1.
| (1) |
The derived function was used to calculate the t1/2, the time after which one half of the EGFR active site has been bound by [16]. The values of t1/2 represent the relative speed with which each allele of EGFR releases erlotinib. The relative t1/2 was calculated by scaling all values relative to the t1/2 of the wild-type kinase. The inverse of this value provides the rate, relative to EGFRWT, with which the mutant EGFR-alleles release erlotinib, Vrelease,Erl, and is related to erlotinib’s period of occupancy of each kinase site.
| Kinetic Constants of EGFR Variants Panel with Erlotinib | |||
|---|---|---|---|
| Calc. t1/2 (min) | Rel. t1/2 | Vrelease, Erl | |
| U87:EGFR | 10.75 | 1.00 | 1.00 |
| U87:EGFRvIII | 5.89 | 0.55 | 1.83 |
| U87:EGFR L858R | 18.16 | 1.69 | 0.59 |
| U87:EGFRdel746-750 | 25.30 | 2.35 | 0.43 |
DISCUSSION
Although TKIs of EGFR are now in widespread clinical use, therapeutic efficacy varies greatly among tumor types and associated EGFR alleles19. In this report, we describe a method for the determination of efficacy by measuring kinase site occupancy, the level of total protein bound by an active site inhibitor, through use of an active-site specific fluorescent affinity probe. Erlotinib and gefitinib, small molecule inhibitors of EGFR, achieved higher levels of kinase site occupancy in lung-cancer-derived mutants of EGFR, as compared with a commonly occurring glioma-derived allele. Kinase site occupancy correlated directly with cell cycle arrest. These data suggest kinase site occupancy as a biomarker for efficacy.
We reported previously that in cells treated with an irreversible EGFR-inhibitor, kinase site occupancy reflected the abundance of both p-EGFR and of its downstream oncogenic signaling through AKT and ERK 1/215. In this report, using reversible clinical inhibitors, erlotinib and gefitinib, the abundance of p-EGFR was reduced to nearly basal levels at very low doses, while much higher doses were required to reduce its oncogenic signaling and decrease growth. Furthermore, levels of kinase site occupancy were aligned better with the abundance of p-AKT and p-ERK 1/2, than with abundance of p-EGFR. That this disconnect was observed upon reversible, but not irreversible EGFR inhibition, suggested that the kinetics of reversible inhibitor cycling underlies therapeutic efficacy.
In our kinetic analyses, all three mutant kinases differed dramatically from wild-type EGFR in the rate with which erlotinib moved in and out of the active site, quantified by the constants t1/2 and Vrelease,Erl. As a result of these differential kinetics, glioma-derived EGFRvIII required higher concentrations of erlotinib to achieve similar levels of kinase site occupancy. Therefore, increased doses of erlotinib were required to decrease downstream signaling in glioma-derived EGFRvIII than in EGFRWT, and lower in lung-derived EGFR L858R and EGFRdel746-750.
How do these data explain the disconnect observed between the abundance of p-EGFR and growth inhibition? We propose that at all studied doses, the half-life with which erlotinib occupies the active site of EGFR is sufficient to prevent significant ATP catalysis and autophosphorylation of tail tyrosine residues. However, the period of occupancy required to reduce oncogenic signaling of downstream molecules is longer, and is only reached at doses of erlotinib or gefitinib sufficient to rapidly reoccupy the EGFR active site and maintain high levels of kinase site blockade.
Our study argues that wild-type EGFR and EGFRvIII are viable small molecular therapeutic targets, and that erlotinib fails to produce a survival advantage in malignant glioma because it fails to achieve sufficient levels of kinase site occupancy in glioma-derived EGFR-alleles. Use of irreversible EGFR inhibitors, and combinatorial blockade of both EGFR and of key downstream outputs, represent important areas of investigation to improve overall pathway inhibition.
METHODS
Cell Culture
U87MG and LN229MG cell lines were obtained from ATCC (Manassas, VA) and were authenticated using STR DNA fingerprinting at UCSF, using the Promega Powerplex 1.2 platform. pcDNA3.1 plasmids containing human full-length EGFR or EGFRvIII cDNA were a gift from Dr. C. David James (UCSF) and plasmids containing EGFR L858R or EGFRdel746-750 were a gift from Dr. Susumu Kobayashi, (Harvard Medical School, MA). The EGFR constructs were digested with XhoI and SalI and ligated into pWZLhygro vector and transduced into U87MG and LN229MG cells using retrovirus.
Cells were maintained in phenol red-free Dulbecco’s Modified Eagle Medium High Glucose (DME H-21) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Low serum media for signaling experiments contained 1% FBS. Cells were stored at 37°C in a 5% CO2 incubator. For western blot analysis, cells were plated in 6-well plates at full serum for 24 hours, then changed to low serum media for 24 hours before being treated in the same media for an additional 24 hours. For flow cytometric analysis, cells were plated in 6-well plates at full serum for 24 hours, then treated in the same media for an additional 24 hours. For viability assays, cells were plated at 2 ×103 cells per well in a 24-well plate and treated once for three days. Media was changed prior to each treatment. Cell viability was determined using at WST-1 assay (Roche, Indianapolis, IN), according to manufacturer’s instructions.
EGFR Tyrosine Kinase Inhibitors and EGFR-Specific Fluorescent Probe
Erlotinib tablets (Genentech, South San Francisco, CA)were purchased, ground to powder and dissolved in aqueous HCl. The aqueous phase was extracted with ethyl acetate. The combined organic extracts were dried over sodium sulfate and concentrated toyield pure erlotinib, which was dissolved at 10mM in DMSO for storage at −20°C. Working dilutions of erlotinib were made immediately prior to use by serial dilution in low-serum media. The EGFR-specific fluorescent probe, [16] (MW: 837g/mol), was also dissolved to 10mM in DMSO and protected from light in storage at −20°C. The working dilution was made by diluting the stock concentration 1:10 in an 85:15 PBS:DMSO mixture, and spinning at top speed in a table-top centrifuge for 10 minutes to remove the precipitate.
Western Blotting
Six-well plates were pulsed with100ng/mL human recombinant EGF (Roche), when applicable, for 30 minutes, then washed with ice cold PBS. Protein was harvested from cultured cells using cell lysis buffer (Cell Signaling, Boston, MA) supplemented with complete protease inhibitor cocktail (Roche, Indianapolis, IN). Equal amounts of protein, as determined by a BCA Protein Assay (Thermo Scientific, Rockford, IL), were loaded into a 4–12% SDS-polyacrylamide gel for electrophoresis and transferred to PVDF membrane. Membranes were blocked in 5% non-fat milk dissolved in TBS-Tween 20 for 1 hour, then incubated overnight at 4°C in primary antibody in 5% bovine serum albumin. Mouse anti-phospho-tyrosine (4G10) was obtained from Upstate Biotechnology (Waltham, MA). Rabbit anti-ERK 2, anti-EGFR and anti-phospho-EGFR (Y1173) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-AKT, anti-phospho-AKT (S473), anti-p44/42 MAPK, anti-rpS6, and anti-phospho-rpS6 (S235/S236) were obtained from Cell Signaling. Mouse anti-β-tubulin was obtained from Millipore (Billerica, MA). Antibodies were detected with HRP-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Calbiochem, Gibbstown, NJ) followed by enhanced chemiluminescence (ECL, GE Healthcare, Piscataway, NJ) or with DyLight 680 dye-coupled anti-rabbit secondary antibodies (Thermo Scientific, Rockford, IL) and imaged using a LI-Cor Odyssey Imaging System (LI-Cor Biosciences, Lincoln, NE).
Fluorescent Gels
Six-well plates were pulsed with EGF and washed with ice cold PBS (as described above), then pulsed with 60μM fluorescent probe for 25 minutes on ice. Cells were then harvested and run on a gel (as described above). Gels were rinsed in a solution of 15% methanol and 5% Transfer Buffer (Invitrogen, Carlsbad, CA) for 20 minutes, then scanned on a Typhoon fluorescence imager (Molecular Dynamics) using a 488nm laser and a 560nm low pass emission filter. The fluorescent intensity was measured using ImageJ software (NIH, 2008). The net band signal was determined by subtracting the fluorescent intensity of the gel below the band from the fluorescent intensity of the band. The band intensity of the control was normalized to 100%, and all subsequent band intensities scaled accordingly.
Flow Cytometry
Cells were washed with PBS then harvested using 0.25% Trypsin. All media and PBS were collected for analysis. Cells were pelleted and resuspended in PBS without Ca2+ and Mg2+ ions, then permeabilized with ice cold 70% ethanol, and then placed on ice for at least 30 minutes. Cells were then washed once with PBS then resuspended in PBS containing RNaseA (Invitrogen, 1:100) and 10μg/mL propidium iodide (Invitrogen). Cells were sorted using FACSCalibur (Becton, Dickinson and Company) and analyzed for their level of propidium iodide staining using ModFit LT 3.2 (Verity). Ten thousand live cell events were collected per treatment.
Analysis of the Kinetics of the Binding of Erlotinib to EGFR-alleles
Cells were plated as described above for western blotting and treated for 24 hours with 2μM erlotinib or 1μM gefitinib to allow the EGFR-drug reaction to reach equilibrium. Cells were then harvested after 1 minute, 10 minutes, 25 minutes, 1 hour, or 4 hours of treatment with 60μM [16] on ice. A single control lane that was not treated with drug was treated with [16] for 4 hours, allowing for comparison with the non-treating [16]-binding assay. The experiment was repeated with 24 hours of DMSO treatment as a control, which determined the differences in [16]-binding to each EGFR-allele. The level of [16] staining of a single drug-treated EGFR-allele (as a % of the 4hr control lane) was divided by the level of [16] staining of that same EGFR-allele following DMSO-treatment (as a % of the 4hr control lane), allowing for the tracking of kinase site occupancy for each EGFR-allele over time, corrected for differences in the kinetics of [16]-binding for each EGFR-allele.
To determine the t1/2 of erlotinib and gefitinib replacement by [16], the experimental kinetic data was best fit to an equation of the form of equation (1) using the solver function of Microsoft Excel to minimize the sum of the differences between the calculated value of [16]-binding and the experimental values.
| (1) |
Using these experimentally determined equations, the t1/2 was determined for each EGFR-allele. The relative value of t1/2 was determined by dividing the calculated value of the t1/2 of each EGFR-allele by that of the wild-type.
Statistical Analysis
For statistical comparison, a Student’s one-tailed t-test were used, with P values of <0.05 considered significant.
Supplementary Material
SIGNIFICANCE.
These data suggest kinase site occupancy as a biomarker for efficacy of EGFR inhibitors, that rapid binding and release of erlotinib in glioma-derived EGFRvIII opposes blockade of downstream signaling, and that slower cycling of erlotinib within the active site of NSCLC-derived mutants underlies their improved clinical response.
Acknowledgments
We are grateful to Trever Bivona and Neil Shah for critical review of the manuscript and Igor Vivanco, H. Ian Robins, Tim Cloughesy and Ingo K. Mellinghoff for sharing unpublished data.
GRANT SUPPORT
This work was supported by grants from Accelerate Brain Cancer Cure, The Burroughs Wellcome Fund, the Frank A. Campini Foundation, the Howard Hughes Medical Institute, NIH K08NS065268, NIH P50CA097257, and the Samuel Waxman Cancer Research Foundation.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
We have no potential conflicts of interest to disclose.
Author contributions: KJB, TN, KS, and WAW designed experiments and analyzed the data. KJB, TN, SH, AG, and QWF performed experiments. KJB, TN, and WAW wrote the manuscript. All authors were involved in the interpretation of data and manuscript content.
References
- 1.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 2.Zandi R, Larsen AB, Andersen P, Stockhausen M-R, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19:2013–23. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Frederick L, Wang X-Y, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–7. [PubMed] [Google Scholar]
- 4.Sagawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci. 1990;87:8602–6. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H-JS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji X-D, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Bio Chem. 1997;272:2927–35. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 8.Arteaga CL. ErbB-targeted therapeutic approaches in human cancer. Exp Cell Res. 2003;284:122–30. doi: 10.1016/s0014-4827(02)00104-0. [DOI] [PubMed] [Google Scholar]
- 9.Yun C-H, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, et al. Structures of lung-cancer derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–27. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 11.Van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MCM, Kros JM, Carpentier AF, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–74. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaekcle KA, Uhn JH, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma mutliforme: North central cancer treatment group study N0177. J Clin Oncol. 2008;26:5603–9. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci. 2006;103:7817–22. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumor suppression. Nature. 2011;476:163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blair JA, Rauth D, Kung C, Yun C-H, Fan Q-W, Rode H, et al. Structure-guided development of affinity probes for tyrosine kinases using chemical genetics. Nat Chem Bio. 2007;3:229–38. doi: 10.1038/nchembio866. [DOI] [PubMed] [Google Scholar]
- 16.Singh J, Dobrusin EM, Fry DW, Haske T, Whitty A, McNamara DJ. Structure-based design of a potent, selective, and irreversible inhibitor of the catalytic domain of the erbB receptor subfamily of protein tyrosine kinases. J Med Chem. 1997;40:1130–5. doi: 10.1021/jm960380s. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciardiello F, Trotora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.