Abstract
Introduction
The study of patients carrying germline EGFR mutations, which have been found in cases of familial lung adenocarcinoma, could provide unique insight into lung cancer risk and carcinogenesis in non-smokers. However, investigations into the biology of germline EGFR mutations have been hampered by the lack of an effective strategy for screening for carriers. We hypothesized that patients with lung cancers found to harbor the EGFR T790M resistance mutation prior to treatment, an uncommon occurrence, would be likely to carry underlying germline T790M mutations.
Methods
Eleven unrelated patients were identified with lung cancer harboring an EGFR T790M mutation from a 7-year institutional experience with tumor genotyping. Ten patients had benign tissue available, which was anonymously tested for presence of germline EGFR mutations.
Results
Five of 10 cases carried a germline T790M mutation (50%, CI 27%–73%). One patient’s cancer exhibited a distinctive indolent growth which has also been described in preclinical studies of T790M-mutant cancers. A second patient underwent resection of 6 separate primary lung adenocarcinomas, each carrying different sensitizing EGFR mutations as well as T790M.
Conclusions
Genotyping of lung cancers, now commonly performed to predict benefit from treatment with EGFR tyrosine kinase inhibitors, can also be used as a screening tool to identify patients at risk of carrying germline EGFR mutations. Once identified, these patients and their families can be studied prospectively in order to explore appropriate screening strategies. Further studies using existing oncogenomic data to provide insight into underlying germline genetics are warranted.
Keywords: Familial lung cancer, EGFR mutations, genetic susceptibility, T790M
Introduction
The identification of EGFR mutations in lung cancers occurring more commonly in never-smokers has added complexity to our understanding of lung cancer risk and predisposition. The most important risk factor for lung cancer development is clearly a history of smoking; indeed, studies have found that the gene most associated with lung cancer risk is one that influences nicotine metabolism.1 Yet risk factors for the development of lung cancer in never-smokers are less clear. As smoking rates decrease, lung cancer in never-smokers may come to represent a larger proportion of this deadly disease, necessitating an improved understanding of risk factors for its development.
Germline EGFR mutations, which have been identified in cases of familial lung adenocarcinoma,2, 3 have the potential to provide insight into EGFR-mutant lung cancer risk and carcinogenesis. However, because there is no effective strategy for screening for these mutations, only a handful of affected families have been identified since germline EGFR mutations were first described,2 impairing our ability to study this phenomenon. One study found no cases of germline T790M among 52 families with at least three relatives with lung cancer,4 yet 86% of this cohort had smoked and adenocarcinoma predominated in only 3 families.5 Another study screened peripheral blood from 369 never-smokers with lung adenocarcinoma and found 2 cases of germline T790M (0.5%) in this clinically selected population.6
Interestingly, EGFR T790M is also rare in unselected lung cancer patients at diagnosis,7 though T790M is common after cancers develop resistance to EGFR tyrosine kinase inhibitors (TKIs) like erlotinib.8 Now that EGFR genotyping is a standard part of the management of lung adenocarcinoma, baseline T790M mutations are occasionally identified during patient care, but the optimal management of such cases is not clear. We hypothesized that germline testing of these patients harboring baseline T790M mutations would be an effective way of screening for patients with underlying germline T790M mutations in EGFR.
Methods and Results
Through an IRB-approved mechanism, we identified 11 unrelated lung cancer patients whose tumors harbored a baseline T790M mutation, from a total of 503 patients with EGFR-mutant lung cancer treated at our institution over a 7-year period.9 Nine cases were identified incidentally through routine sequencing of EGFR or through the use of a mass spectrometry-based assay. The other two cases, known to harbor a sensitizing EGFR mutation from a mutation-specific assay, were subsequently found to harbor a concurrent pre-treatment T790M using directed testing of EGFR exon 20.10 To rule out false positive T790M results, all cases had the presence of the baselineT790M mutation confirmed with Sanger sequencing. All 11 tumors also carried a concurrent sensitizing mutation in EGFR (Table 1). Five cases were treated with EGFR TKI at some point in their course and none had a response, consistent with the expected resistance.7
Table 1.
Characteristics of all patients and germline cases*
| Characteristic | All patients (N=11) |
Germline cases (N=5) |
|
|---|---|---|---|
| Age at diagnosis |
Median (Range) | 56 (44–75) | 56 (44–73) |
| Smoking history |
Never-smoker Former/current smoker |
6 (55%) 5 (45%) |
4 (80%) 1 (20%) |
| First-degree relative with lung cancer |
Present Absent |
3 (27%) 8 (73%) |
2 (40%) 3 (60%) |
| Stage | I II III IV† |
2 (18%) 1 (9%) 0 (0%) 8 (73%) |
1 (10%) 0 (0%) 0 (0%) 4 (80%) |
| Histology | Adenocarcinoma | 11 (100%) | 5 (100%) |
|
EGFR sensitizing mutation |
Exon 19 deletion Exon 21 L858R Different mutations in different lesions† No sensitizing mutation |
4 (36%) 6 (55%) 1 (9%) 0 (0%) |
2 (40%) 2 (40%) 1 (10%) 0 (0%) |
Two of these patients have been previously reported as part of a separate study6
One patient had multiple pulmonary nodules harboring different EGFR sensitizing mutations in addition to T790M, possibly representing multiple primary lung cancers rather than stage IV disease
For the 10 patients with benign tissue available, Sanger sequencing of EGFR was performed on extracted DNA, in an anonymized fashion. Benign specimens included peripheral blood (3), benign resected lung or lymph nodes tissue (5), or other benign biopsies (skin, trachea). To rule out contamination from lung cancer cells in the benign specimens, all were confirmed to be negative for sensitizing EGFR mutations. A germline T790M mutation was identified in the benign tissue from 5 of the 10 patients (50%, CI 27%–73%; Table 1); all 5 mutations were heterozygous. Two of the 5 patients with germline T790M had a family history of lung cancer in a first-degree relative.
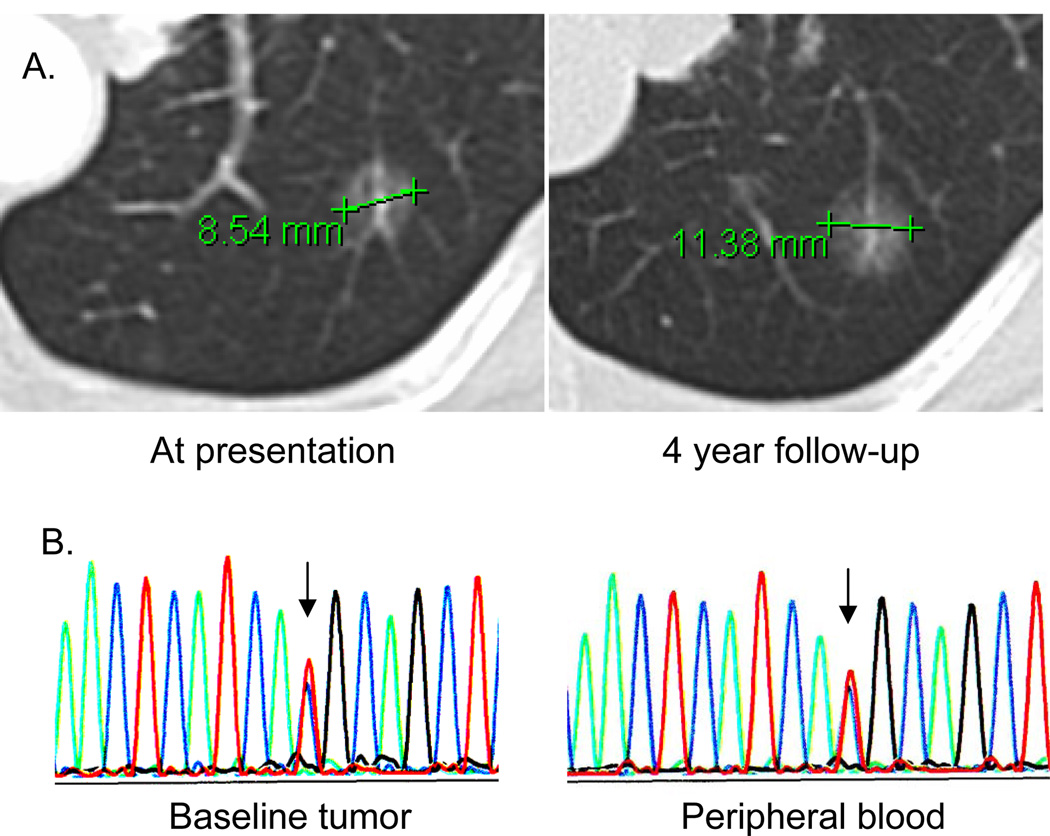
While pedigrees could not be collected due to the anonymized design of the study, one patient with a sibling with lung cancer later had presence of germline T790M confirmed in peripheral blood. This never-smoker had initially been diagnosed with advanced lung adenocarcinoma when a lung resection demonstrated multifocal disease. Because of her low tumor burden in the lungs only, the patient opted for a strategy of expectant observation. After 4 years of observation, the patient’s lung lesions grew minimally (Figure 1A); repeat biopsy again showed adenocarcinoma harboring concurrent L858R and T790M EGFR mutations despite no exposure to TKI. We suspect that the slow growth seen may be related to the indolent biology reported in cancers that acquire the T790M resistance mutation.8, 11 In retrospect, we found that the sequencing tracing from the patient’s tumor demonstrated a T790M mutant allele present in equal proportion to the wild-type allele, consistent with our subsequent finding of a germline mutation (Figure 1B). In contrast, when T790M is only detectable with a highly sensitive assay (likely indicating the T790M is present in a minor clone),10 a germline mutation may be unlikely.
Figure 1. A case of germline T790M associated with indolent lung cancer growth.
(A) A germline T790M mutation was found in one patient who presented with multifocal adenocarcinoma involving the lung. Over 4 years the patient’s lung nodules displayed minimal growth, consistent with the indolent phenotype described in other lung cancers carrying the T790M mutation.11 (B) Rebiopsy of the tumor shown (after observation only) identified an EGFR L858R mutation as well as a pre-treatment T790M mutation (left). The mutant T790M peak (red) was of equal proportion to the wild type allele (blue), consistent with the possibility of a germline mutation. After referral to clinical genetics, sequencing of peripheral white blood cell DNA (right) confirmed the germline status of the T790M mutation.
A second patient tested positive for germline T790M after resection of multiple morphologically distinct lung adenocarcinomas carrying different EGFR genotypes. This never-smoker was initially diagnosed with advanced lung adenocarcinoma after a CT showed multiple lung nodules and a biopsy found adenocarcinoma harboring a sensitizing EGFR mutation. Because of her young age, she was offered resection of her remaining lung nodes as these nodules potentially represented synchronous primary lung cancers. Six different nodules were resected, each exhibiting different adenocarcinoma morphologies. On EGFR genotyping, four carried L858R, two carried different exon 19 deletions, and all carried T790M as well. Interestingly, this patient had no family history of lung cancer, suggesting either an inherited T790M mutation with variable penetrance or a de novo germline mutation in this patient.
Discussion
We have found that patients with germline EGFR T790M mutations, which have been found in association with familial lung adenocarcinoma, can be identified through screening patients whose tumors harbor baseline EGFR T790M mutations on routine lung cancer genotyping. Estimating that 1% of EGFR-mutant lung cancers harbor T790M at diagnosis,7 we suspect that approximately 200 patients annually are diagnosed in the United States with lung cancers carrying baseline EGFR T790M. Because EGFR genotyping is now a standard part of the management of patients with non-small cell lung cancer, patients with baseline T790M will be identified as a part of routine oncology practice and could be considered for referral for germline testing. Importantly, because germline mutations are present in both benign and neoplastic cells, they should be easily detected using standard Sanger sequencing; when T790M is detected only with the use of highly sensitive assays recently developed,10, 12, 13 this would not be associated with an underlying germline mutation as this suggests that the mutation is only present in a very small proportion of the cells tested.
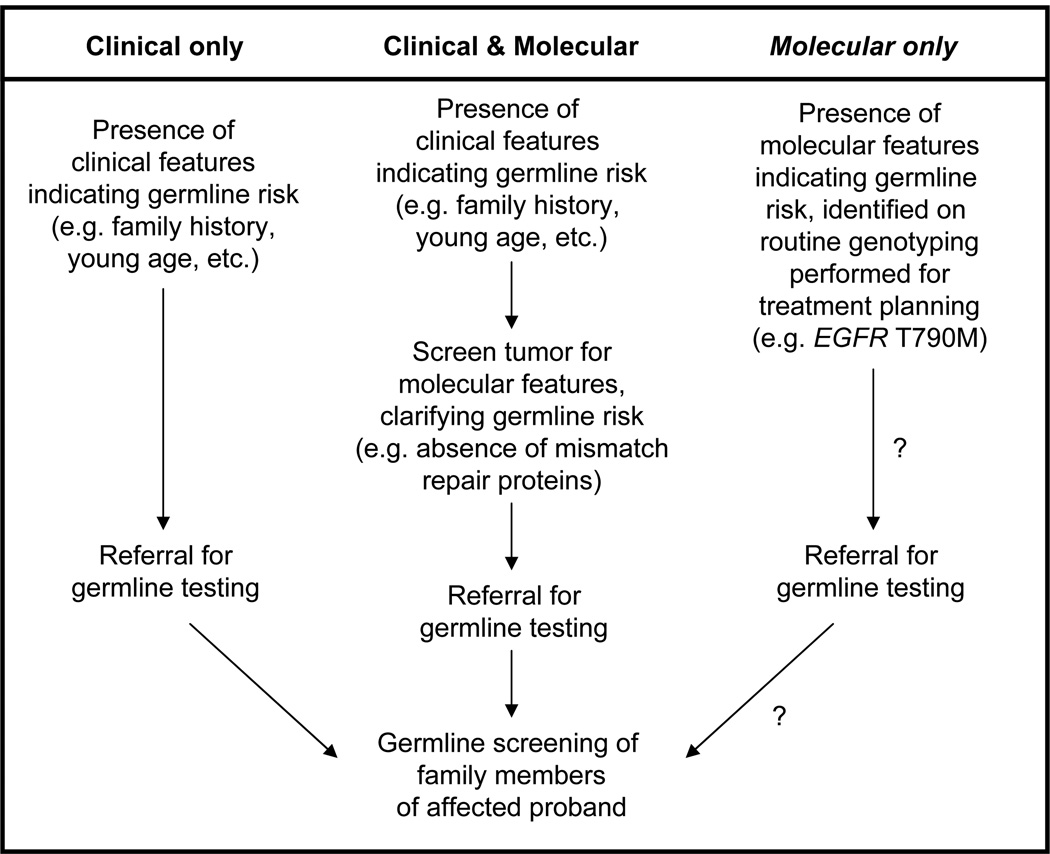
The idea of using routine tumor genotyping, performed for treatment planning, as an initial screen for patients likely to harbor germline mutations has not previously been described. While molecular characteristics of a cancer are, in some instances, used to gauge the likelihood of a germline mutation (e.g. staining for mismatch repair proteins to identify patients for Lynch syndrome screening), such testing is primarily performed when clinical characteristics raise suspicion of a hereditary disorder (Figure 2).14 However, tumor genotyping is now a standard part of the management of lung cancer (EGFR), colon cancer (KRAS), and melanoma (BRAF). Furthermore, multi-gene assays for the comprehensive genetic profiling of tumor specimens are increasingly in use,15 generating an enormous quantity of onco-genomic data. We suspect there may be additional instances where such somatic genotyping could identify unexpected germline abnormalities, similar to the incidental identification of a case of germline EGFR T790M in a recent genomic analysis of 439 cases of myelodysplastic syndrome.16 Even BRCA genotyping may come to be a component of such multi-gene assays if BRCA mutations are validated as predictive of benefit from PARP inhibitors.17 Our observation suggests that, for instance, patients either with lung tumors harboring the EGFR T790M mutation or with ovarian tumors found to harbor BRCA mutations (which can be either somatic or germline18) would be ideal candidates for subsequent germline testing.
Figure 2. A possible new strategy for the identification of “at risk” probands prior to referral for germline testing.
Traditionally, clinical characteristics such as young age, a family history of cancer, or multiple primary cancers have been used to identify patients at risk of carrying germline mutations (left). For some cancers, such as colorectal cancer, suspicion for Lynch syndrome leads first to molecular testing (staining for mismatch repair proteins) prior to germline testing (middle). We propose that, for lung cancer, merely the presence of pre-treatment T790M on EGFR genotyping performed for treatment planning could be sufficient to lead to germline testing (right).
For this analysis, we studied the most clinically relevant germline mutation in EGFR, T790M, which is interesting because of its relatively high prevalence in lung cancer patients (at least 1%7), its unique role in mediating resistance to TKIs,10, 11 and ongoing efforts to therapeutically target this mutation.8 A second germline mutation in EGFR, V843I, has also been described to be associated with familial lung cancer,3 but has not been observed as a somatic mutation in our institutional experience. Given that two variants of germline EGFR mutations have been identified to date, we suspect there may be additional germline variants yet to be discovered which may contribute to lung cancer risk in a portion of never-smokers.
In conclusion, we have found that detection of pretreatment EGFR T790M mutations on routine lung cancer genotyping signals a likelihood of an underlying germline mutation. Lung cancer patients harboring baseline EGFR T790M should be studied prospectively to better understand germline prevalence, familial penetrance, and lifetime lung cancer risk in carriers. For example, we note that only 2 of the 5 patients carrying germline T790M mutations had a family history of lung cancer. This suggests either low penetrance of the high-risk phenotype or development of de novo germline T790M mutations in a portion of patients. We favor the latter explanation given that one of the patients with no family history of lung cancer clearly exhibited a high risk phenotype with multiple synchronous EGFR-mutant lung cancers. It is clear that a registry for the prospective study of these patients and their families is needed, and is under development, in order to identify screening and counseling strategies for this rare but potentially high risk population.
Acknowledgments
Source of Funding: This work was supported in part by a Young Investigator Award from the American Society of Clinical Oncology; and from the National Cancer Institute at the National Institutes of Health (P01-CA129243).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A portion of this material was presented previously at the 14th World Conference of Lung Cancer in Amsterdam (July 4, 2011)
References
- 1.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 3.Ikeda K, Nomori H, Mori T, et al. Novel Germline Mutation: EGFR V843I in Patient With Multiple Lung Adenocarcinomas and Family Members With Lung Cancer. Ann Thorac Surg. 2008;85:1430–1432. doi: 10.1016/j.athoracsur.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Vikis H, Sato M, James M, et al. EGFR-T790M Is a Rare Lung Cancer Susceptibility Allele with Enhanced Kinase Activity. Cancer Res. 2007;67:4665–4670. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos CI, Pinney SM, Li Y, et al. A Susceptibility Locus on Chromosome 6q Greatly Increases Lung Cancer Risk among Light and Never Smokers. Cancer Res. 2010;70:2359–2367. doi: 10.1158/0008-5472.CAN-09-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an Internet-based blood collection protocol: a preliminary report. Clin Cancer Res. 2010;16:755–763. doi: 10.1158/1078-0432.CCR-09-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J-Y, Yu C-J, Chang Y-C, et al. Effectiveness of tyrosine kinase inhibitors on uncommon epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 8.Oxnard GR, Arcila ME, Chmielecki J, et al. New Strategies in Overcoming Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in Lung Cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR Exon 19 Deletions and L858R in Tumor Specimens From Men and Cigarette Smokers With Lung Adenocarcinomas. J Clin Oncol. 2011;29:2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of Lung Cancer Patients with Acquired Resistance to EGFR Inhibitors and Enhanced Detection of the T790M Mutation Using a Locked Nucleic Acid-Based Assay. Clin Cancer Res. 2011;17:1169–1180. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of Dosing for EGFR-Mutant Non–Small Cell Lung Cancer with Evolutionary Cancer Modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su K-Y, Chen H-Y, Li K-C, et al. Pretreatment Epidermal Growth Factor Receptor (EGFR) T790M Mutation Predicts Shorter EGFR Tyrosine Kinase Inhibitor Response Duration in Patients With Non–Small-Cell Lung Cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 13.Rosell R, Molina MA, Costa C, et al. Pre-treatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res. 2011;17:1160–1168. doi: 10.1158/1078-0432.CCR-10-2158. [DOI] [PubMed] [Google Scholar]
- 14.Burt RW, Barthel JS, Dunn KB, et al. Colorectal Cancer Screening. Journal of the National Comprehensive Cancer Network. 2010;8:8–61. doi: 10.6004/jnccn.2010.0003. [DOI] [PubMed] [Google Scholar]
- 15.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling Critical Cancer Gene Mutations in Clinical Tumor Samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical Effect of Point Mutations in Myelodysplastic Syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong PC, Boss DS, Yap TA, et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas T. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]