Abstract
To identify genes involved in osteoarthritis (OA), the most prevalent form of joint disease, we performed a genome-wide association study (GWAS) in which we tested 500,510 Single Nucelotide Polymorphisms (SNPs) in 1341 OA cases and 3496 Dutch Caucasian controls. SNPs associated with at least two OA-phenotypes were analysed in 14,938 OA cases and approximately 39,000 controls. The C-allele of rs3815148 on chromosome 7q22 (MAF 23%, 172 kb upstream of the GPR22 gene) was consistently associated with a 1.14-fold increased risk (95%CI: 1.09–1.19) for knee- and/or hand-OA (p=8×10−8), and also with a 30% increased risk for knee-OA progression (95%CI: 1.03–1.64, p=0.03). This SNP is in almost complete linkage disequilibrium with rs3757713 (located 68 kb upstream of GPR22) which is associated with GPR22 expression levels in lymphoblast cell lines (p=4×10−12). GPR22 encodes an G-protein coupled receptor with unkown ligand (orphan receptor). Immunohistochemistry experiments showed absence of GPR22 in normal mouse articular cartilage or synovium. However, GPR22 positive chondrocytes were found in the upper layers of the articular cartilage of mouse knee joints that were challenged by in vivo papain treatment or in the presence of interleukin-1 driven inflammation. GRP22 positive chondrocyte-like cells were also found in osteophytes in instability-induced OA. In addition, GPR22 is also present in areas of the brain involved in locomotor function. Our findings reveal a novel common variant on chromosome 7q22 to influence susceptibility for prevalence and progression of OA.
Introduction
Osteoarthritis (OA) (MIM165720), the most common degenerative, age-related disease of the synovial joints, is commonly characterized by cartilage degradation, formation of osteophytes and subchondral sclerosis. Unfortunately, there are no curative and hardly any symptomatic treatment options available for this disease. OA is a complex disease in which both environmental and genetic factors play an important role. Primary OA has an estimated heritability of 40% for the knee, 60% for the hip and 65% for the hand.1 Identifying the genes underlying the genetic background could give new insights into the pathophysiology of OA and could potentially lead to new drug targets. To date, genetics of OA mainly focuses on genome-wide linkage and candidate gene studies. Results of these studies have been controversial due to lack of power and replication. Currently, only two genes have been found consistently associated with OA in Caucasians: Growth Differentiation Factor 5 (GDF5)2 and Iodothyronine-Deiodinase enzyme type 2 (DIO2).3 Recently, a novel approach has become available to search for important risk genes involved in complex traits: a hypothesis-free genome-wide association study (GWAS). The first pooled-DNA GWAS for OA discovered the C-allele of the rs4140564 Single Nucleotide Polymorphism (SNP) (minor allele frequency (MAF) 8%), 75kb upstream of the Prostaglandin-Endoperoxide Synthase 2 (PTGS2) gene, to be associated with prevalent knee-OA in Caucasian women (n=387 cases and 255 controls in pooled GWAS and 1177 cases and 2372 controls in the replication studies).4 In addition, Miyamoto et al.5 identified the SNP rs7639618 in the Double Von Willebrand factor A (DVWA) gene to influence susceptibility in knee-OA (using 94 cases and 658 controls in the discovery study), but this could not be replicated in Caucasian populations with in total 2119 knee-OA cases, 2325 hip-OA cases and 3313 controls.6,7 We here report a GWAS in the population-based prospective Rotterdam Study on hip-, knee-, and hand-OA with replication in 11 additional studies with in total 14,938 cases and approximately 39,000 controls (depending on the OA-phenotype studied). Since OA can occur at different joint sites, we tried to identify SNPs that showed a consistent pattern of association across multiple joint sites. This method will likely identify more general susceptibility genes for OA rather than site-specific genes. In addition to the relation with prevalence of OA, we also examined the relationship between the genome-wide significant SNP(s) and progression of knee-OA.
Subjects and Methods
Study design
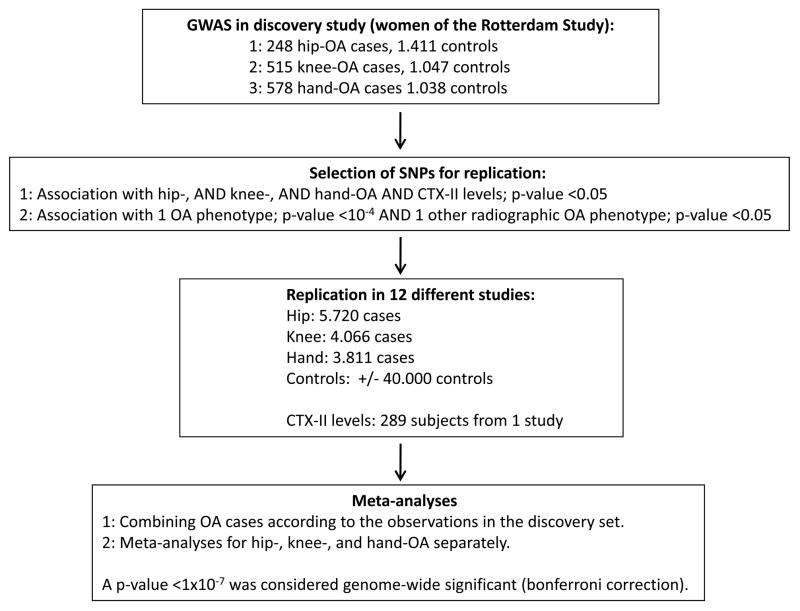
We used a two-stage design to test the association of 500,510 SNPs with hip-, knee-, and hand-OA and urinary urinary C-terminal cross-linked telopeptide of type II collagen (CTX-II) levels, a marker for cartilage degradation. Within the first stage, the SNPs were tested for association with the four OA phenotypes in women of the Rotterdam Study to identify signals that were consistent across multiple OA phenotypes. For hip-OA there was data available on genotypes and phenotypes of 248 cases and 1411 controls, for knee-OA 515 cases and 1047 controls and for hand-OA 578 cases and 1038 controls. If one of the following criteria was fulfilled, the SNP was followed up into the replication stage: 1) hip-, AND knee-, AND hand-OA AND CTX-II levels with a p-value <0.05, or 2) one of the radiographic OA phenotypes with a p-value <10−4 AND one other radiographic OA phenotype with a p-value <0.05. The direction of the association had to be the same for the different phenotypes, i.e., if a SNP was associated with an increased risk for knee OA, there also had to be an association with an increased risk of hand OA and/or hip OA. In total, 12 loci were selected on the basis of these criteria and were tested for association with OA in 11 additional studies. In total, there were 5968 hip-OA cases and 40,420 controls, 4581 knee-OA cases and 38,730 controls, and 4389-hand OA cases and 35,694 controls available for association analysis.
Phenotype definitions
For this study, radiographic definitions of OA were standardized as follows: knee OA was defined as a Kellgren/Lawrence (K/L) score >=2 (definite osteophytes and possible joint space narrowing), hand OA as 2 out of 3 hand joint groups (distal interphalangeal joints/proximal interphalangeal joints/carpometacarpal joint) affected with a K/L score >=2 (definite osteophyte) and hip OA as definite joint space narrowing. Controls were defined as having a K/L <2 for the hip and knee respectively. Subjects with hand data were a control if they did not fulfill the criteria for hand OA. Clinical studies all used total joint replacement of the hip or knee as outcome for hip- and knee-OA. The ACR criteria were used to define hand OA for the Spanish cases,8 deCODE used a definition of either squaring or dislocation of the thumb or at least two nodes at the DIP joints. In the Rotterdam Study and the Chingford Study progression of knee OA was defined as a K/L grade >=1 at baseline and an increase in at least one K/L grade during follow-up.
Study populations
Table 1 provides an overview of all populations participating in this study. An extensive description of the studies participating in this GWAS is given in the supplementary material.
Table 1.
Baseline characteristics of studies included in the GWAS meta-analysis of OA-phenotypes
| Discovery | Replication studies | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rotterdam Study | deCODE | TwinsUK | Framingham Study | C hingford Study | GARP | Spanish cases | Greek TJR cases | Oxford cohorts | Nottingham case-control | SOF | MrOS | |
| Hip OA cases/controls | 413/2474 | 1630/31654 | 33/425 | NA | 97/685 | 109/720 | 307/294 | 110/344 | 1402/871 | 929/236 | 541/1602 | 397/1540 |
| Knee OA cases/controls | 698/1893 | 1068/31654 | 53/476 | 419/1674 | 269/568 | 154/720 | 262/294 | 368/344 | 555/871 | 735/236 | NA | NA |
| Hand OA cases/controls | 832/2055 | 2725/31654 | 64/425 | NA | 286/546 | 241/720 | 241/294 | NA | NA | NA | NA | NA |
| N CTX-II | 1100 | NA | NA | NA | 289 | NA | NA | NA | NA | NA | NA | NA |
|
Case/control or cohort OA definition |
cohort radiographic |
case-control clinical |
cohort radiographic |
cohort radiographic |
cohort radiographic |
case-control radiographic/clinical |
case-control clinical |
case-control clinical |
case-control clinical |
case-control clinical |
cohort radiographic |
cohort radiographic |
| Mean age (range) | 67(55–94) | 69(19–99) | 54(37–76) | 64(29–93) | 54 (44–67) | 60(30–79) | 66(32–94) | 61(20–90) | 67(55–89) | 68(40–92) | 70(65–90) | 73(65–91) |
| Mean BMI (range) | 26(16–56) | 26(14–60) | 25(15–51) | 26(14–54) | 26 (17–47) | 27(19–47) | 31(18–53) | 26(17–34) | NA | 28(15–52) | 27(15–58) | 28(18–51) |
| % women | 59% | 58% | 100% | 56% | 100% | 63% | 66% | 72% | 55% | 62% | 100% | 0% |
| GWA platform | Illumina HumanHap 550v3 | Infinium HumanHap 300 | Infinium HumanHap 300 | Affymetrix GeneChip® Human Mapping | NA | NA | NA | NA | NA | NA | NA | NA |
GWA = genome-wide association; OA = osteoarthritis; NA = not applicable/available
Laboratory methods and quality control
GWAS Rotterdam Study (discovery study): genotyping of the samples with the Illumina HumanHap550v3 Genotyping BeadChip was carried out at the Genetic Laboratory of the Department of Internal Medicine of Erasmus Medical Center, Rotterdam, the Netherlands. The Beadstudio GenCall algorithm was used for genotype calling and quality control procedures were as described previously.9 For this study, the following quality control filters were applied: SNP call rate >= 95%, minor allele frequency >= 5%, p-value HWE >=1×10−6. After quality control 500,510 SNPs remained for association analyses. The intensity cluster plots were visually inspected for the top-hits of the Rotterdam Study and no abnormalities were discovered. Genomic inflation factors were calculated for all analyses and there was no evidence of population stratification with lambdas of 1.01 for hip- and hand-OA, 1.00 for knee-OA and 1.02 for CTX-II levels.
Replication cohorts
deCODE: all samples were assayed with the Infinium HumanHap 300 or humanCNV370 SNP chips (Illumina), containing 317,503 tagging SNPs derived from phase I of the International HapMap project. All of the SNPs tested in this report passed quality filering (a call rate >97%, a minor allele frequency >1%, not a significant distortion from HWE (p-value >10−7 on any of the three chip types used (humanHap300, humanHap300-duo and humanCNV370). Any samples with a yield <98% were excluded from the analysis. Four of the tophits, the SNPs rs3815148, rs12402320, rs4656364 and rs10248619 were not available on this platform and therefore imputed genotypes were used for analyses. Imputation of these SNPs was done using the IMPUTE software,10 but since the rs12402320 SNP is located in a region with very low LD, genotypes could not be imputed/estimated.
TwinsUK: samples were genotyped with the Infinium HumanHap 300 assay (Illumina, San Diego, USA) at the Duke University Genotyping Center (NC USA), Helsinki University (Finland) and the Wellcome Trust Sanger Institute. The Illuminus calling algorithm was used for genotype calling. After strict quality control criteria were applied as described by Richards et al. 9 there were 314075 SNPs available for analysis. Imputation was performed using the IMPUTE software (v0.2.0) to analyze the SNPS rs3815148, rs10248619 and rs4656364. Since the rs12402320 SNP is located in a region with very low LD, genotypes could not be imputed/estimated. At imputed loci, all genotypes with posterior probabilities < 0.9 were discarded and the imputed loci were filtered out using usual QC filters.
Framingham Osteoarthritis Study: samples from 9274 individuals were genotyped using the Affymetrix GeneChip® Human Mapping 500K array set and the 50K supplemental array set focused on coding SNPs and SNPs tagging protein-coding genes (Santa Clara, California) as part of the SHARE initiative. Sample exclusion criteria included call rate < 97% (n=767) and a per subject heterozygosity > ± 5 standard deviations from the mean (n=24). Finally, 2 participants were excluded for excessive Mendelian errors, resulting in 8481 participants included. Imputation was performed using MACH (version 1.0.15) to impute all autosomal SNPs using the publicly available phased haplotypes from HapMap (release 22, build 25, CEU population) as a reference population. A sample of 200 known unrelated participants with high call rates and low Mendelian errors were used to determine parameter estimates that were subsequently applied in a model for all 8481 subjects. Dosage estimates outputted by MACH were used in analysis.
Replication studies without GWA data: the MassArray iPLEX Gold from Sequenom was used for the replication of the top-hits of the Rotterdam Study except for the Study of Osteoporotic Fractures (SOF) and MrOS (Osteoporotic Fractures in Men) study which were genotyped using Taqman. To check for genotyping errors, a random set of 5% of the samples of all studies has been re-genotyped with a range of 0–0.2% discordance in genotypes. Supplementary Table 1 shows an overview of all HWE p-values, minor alleles and minor allele frequencies in the controls of all participating studies. Since the rs10248619 SNP showed deviations from HWE and had a lower minor allele frequency in the studies genotyped using Sequenom compared to the studies with GWA data, this SNP was re-genotyped using Taqman, resulting in a minor allele frequency which was comparable to the frequencies in the studies genotyped using a GWA technique. The most likely explanation for the deviation from HWE, is another SNP in the primer extension in the multiplex. The rs959396 SNP had a call rate <90% for the GARP Study and the Spanish TJR cases and therefore these two studies are not taken into account in the meta-analysis for this specific SNP. The Greek TJR samples did not perform well with the Sequenom technique due to lower DNA quality and therefore we used the Taqman to retrieve genotypes of rs10248619 and rs3815148 SNPs since this is a method which performs better with DNA of less quality. To check for genotyping errors, a random set of the samples of the Greek TJR and control samples have been re-genotyped with less than 1% discordance in genotypes.
Statistical analysis
Genome-wide association analyses: all associations with OA-phenotypes for all non-familial studies were tested using an allelic chi-square test (1df) assuming an additive effect for each SNP tested. Analyses for the Rotterdam Study, Chingford Study, Oxford cohorts, the Nottingham case-control study, the Spanish TJR and hand OA cases and the Greek TJR cases, were carried out using PLINK V1.02 software.11 CTX-II levels were log-transformed and converted to z-scores adjusted for age. The SOF and MrOS Study performed logistic regression analyses using the program SAS version 9. The program R genetics was used to calculate p-values for HWE. In the Rotterdam Study and the Chingford Study, the rs3815148 SNP was also tested for association with progression of knee-OA using a logistic regression model adjusted for gender and follow-up time in SPSS version 15.0. deCODE used a likelihood procedure described in a previous publication for the association analyses12 to calculate two-sided p-values and odds ratio for each individual allele, assuming a multiplicative model for risk, i.e., that the risk of the two alleles a person carries multiply. Some of the individuals in the Icelandic case-control groups were related to each other, causing the χ2 test statistic to have a mean >1 and median >0.6752. In addition, the inflation factor was estimated by using genomic controls by computing the median of the 300754 χ2 statistics and dividing it by 0.6752 as previously described.13 For all analyses of deCODE adjustments for the inflation factor were made.
For the TwinsUK study, robust standard errors were calculated and logistic regression was performed using the program Stata version 10 (Stata Corporation, USA) for hip-, knee- and hand-OA. The analysis used in the Framingham Osteoarthritis Study is a logistic regression using GEE (generalized estimating equations) to control for the dependency among family members. Analyses were performed using R software.
In the GARP study, standard errors were estimated from the variance between the sibling pairs (robust standard errors) to adjustfor the family relationship among sibling pairs.2 Robust standard error analyses using Stata SE8 software (Stata Corporation, USA).
Meta-analysis: a SNP-specific meta-analysis was performed for the combined phenotypes using the program Comprehensive Meta-analysis by Biostat (www.meta-analysis.com). For example, if in the discovery study a SNP was associated with knee- and hand-OA using the criteria specified in the methods section earlier, the meta-analysis was also performed for this SNP on hand- or knee-OA cases combined versus controls that had neither hand- nor knee-OA. With this method of analyses, no overlap between cases or controls existed in the analyses. If the heterogeneity metric I2 exceeded 25% a random-effects model (DerSimonian and Laird) was also used for the analysis since then moderate heterogeneity exists, otherwise only a fixed effects model (inverse variance method) was applied. The analysis was performed on the total population of all studies and was subsequently stratified for gender to reveal, if any, gender-specific associations. For family-based studies, association results of men and women combined are shown because of relatedness among men and women in the study. A p-value < 1×10−7 was considered genome-wide significant (=0.05/500,510).
Association analyses with previously published SNPs: within the Rotterdam Study, allelic chi-square tests (1df) assuming an additive model were performed for the SNPs discovered in previous studies which are mentioned in the introduction section. The rs225014 and rs12885300 SNPs of the DIO2 gene, rs4140564 75kb upstream of the PTGS2 gene, rs7639618 of the DVWA gene and rs6088813 of the GDF5 gene were tested for association with hip-, knee- and hand-OA. A p-value < 0.05 was considered significant.
mRNA expression analyses and immunohistochemistry experiments
Synovial biopsies from patients undergoing arthroscopy of the knee for meniscal and cartilage injury at the Division of Orthopedics, University Hospitals Leuven, were used for in vitro culture in Dulbecco’s Modified Eagle Medium/10% serum, using plastic adherence as described by Lories et al.14 Histological and flowcytometric analysis revealed that the synovia did not show signs of chronic inflammation. Articular chondrocytes and meniscal cells were isolated from knee cartilage and meniscus obtained from patients with osteoarthritis undergoing knee replacement surgery as described previously.15 All procedures were approved by the Ethical committee for clinical research at KU Leuven and patient informed consent was obtained. Synovial fibroblast-like cells at passage 4 and freshly isolated chondrocytes (P0) were used for quantitative PCR using Assays on Demand provided by Applied Biosystems. Immunohistochemistry was performed on mouse knee tissue after induction of papaïn, methylated bovine serum albumin induced arthritis or medial menicus instability induced arthritis as described before.16,17 Rabbit anti-GPR22 antibody was obtained from Sigma-Aldrich and used at 5 μg/ml overnight. Secondary antibody was peroxidise-conjugated goat anti-rabbit (Jackson Immunoresearch Laboratories).
Results
Association results
In the first stage, 500,510 SNPs were tested for association with osteoarthritis in women of the Rotterdam Study. Because power to pick up genome-wide associations with individual phenotypes was limited, we applied a strategy which uses the advantage of the multiple phenotypes available in our discovery study (see methods) to search for common variants showing consistent associations with OA at multiple joint sites. In total, 18 SNPs in 12 loci were identified using this approach (Table 2).
Table 2.
Identified tophits of the GWAS in the Rotterdam Study
| Radiographic OA | CTX-II | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs-number | locus | chromosome | bp-position | MAF | annotation | p-knee | p-hip | p-hand | ||||||
| Novel loci | Nearest Gene | Location | p-value | OR | p-value | OR | p-value | OR | p-value | beta | ||||
| rs10465850 | 1 | 1 | 65471791 | 0.46 | JAK1 | −347kb | 0.009 | 1.22 | 0.02 | 1.26 | 0.01 | 1.20 | 0.02 | 1.07 |
| rs12402320 | 2 | 1 | 152968030 | 0.30 | KCNN3 | intronic | 0.03 | 1.19 | 0.04 | 1.23 | 0.01 | 1.22 | 0.01 | 1.07 |
| rs4656364 | 3 | 1 | 160545479 | 0.10 | NOS1AP | intronic | 0.04 | 1.29 | 9×10−6 | 1.88 | 0.96 | 1.01 | 0.49 | 1.02 |
| rs3963342 | 4 | 4 | 43995005 | 0.33 | KCTD8 | intronic | 0.04 | 1.18 | 0.02 | 1.26 | 0.03 | 1.18 | 0.04 | 1.05 |
| rs10248619 | 5 | 7 | 50718584 | 0.21 | GRB10 | intronic | 0.002 | 1.33 | 0.03 | 1.28 | 0.04 | 1.21 | 0.01 | 1.10 |
| rs7791286 | 5 | 7 | 50824286 | 0.17 | GRB10 | intronic | 0.02 | 1.25 | 0.02 | 1.33 | 0.01 | 1.28 | 0.03 | 1.07 |
| rs3815148 | 6 | 7 | 106725656 | 0.23 | COG5 | intronic | 7×10−5 | 1.42 | 0.81 | 1.03 | 0.05 | 1.18 | 0.96 | 1.00 |
| rs1548524 | 6 | 7 | 106731799 | 0.24 | COG5 | intronic | 9×10−5 | 1.41 | 0.72 | 1.05 | 0.04 | 1.19 | 0.96 | 1.00 |
| rs997311 | 6 | 7 | 106740269 | 0.24 | COG5 | intronic | 8×10−5 | 1.41 | 0.74 | 1.04 | 0.03 | 1.20 | 0.96 | 1.00 |
| rs959396 | 7 | 9 | 37001289 | 0.38 | PAX5 | intronic | 3×10−6 | 0.69 | 0.04 | 0.81 | 0.93 | 0.99 | 0.91 | 0.95 |
| rs12352822 | 8 | 9 | 119587103 | 0.15 | TLR4 | −69kb | 2×10−5 | 0.60 | 0.001 | 0.57 | 0.30 | 0.89 | 0.19 | 0.95 |
| rs873598 | 9 | 10 | 120413174 | 0.45 | GPR10 | −68kb | 0.002 | 0.79 | 0.004 | 0.75 | 0.007 | 0.82 | 0.02 | 0.95 |
| rs880844 | 10 | 12 | 107486497 | 0.24 | ISCU | intronic | 0.02 | 1.23 | 3×10−5 | 1.56 | 0.16 | 1.13 | 0.18 | 1.05 |
| rs741542 | 10 | 12 | 107488184 | 0.24 | ISCU | 3′downstream | 0.02 | 1.23 | 3×10−5 | 1.56 | 0.16 | 1.13 | 0.18 | 1.05 |
| rs11651351 | 11 | 17 | 71478457 | 0.07 | ACOX1 | intronic | 0.02 | 0.68 | 0.02 | 0.57 | 0.03 | 0.70 | 0.01 | 0.89 |
| Known Loci | ||||||||||||||
| rs4911494 | 12 | 20 | 33435328 | 0.40 | GDF5/UQCC | coding/intron | 0.03 | 0.84 | 0.91 | 1.01 | 2×10−5 | 0.72 | 0.40 | 0.98 |
| rs6088813 | 12 | 20 | 33438595 | 0.40 | GDF5/UQCC | intronic | 0.02 | 0.84 | 0.91 | 1.01 | 1×10−5 | 0.72 | 0.40 | 0.98 |
| rs6087705 | 12 | 20 | 33464664 | 0.40 | GDF5/UQCC | 5′upstream | 0.03 | 0.84 | 0.91 | 1.01 | 1×10−5 | 0.72 | 0.40 | 0.98 |
bp= base pair; MAF = minor allele frequency; OR = odds ratio; p-values are from allele-based association analysis.
In the second stage, these 12 loci were tested for association with OA in 11 additional studies. The 12 loci were tested for association with the OA-phenotype that formed the basis for selection in the discovery phase. In total, there were 5968 hip-OA cases and 40,420 controls, 4581 knee-OA cases and 38,730 controls, and 4389 hand-OA cases and 35,694 controls available for association analysis. In Table 3 the results of the meta-analysis are shown for these 12 SNPs for the combined phenotypes. From these 18 SNPs, we observed 3 loci to be significantly associated with OA. The C-allele of the SNP rs3815148 (A>C, MAF 23%) annotated in intron 12 of the component of oligomeric golgi complex 5 (COG5) gene, was associated with knee- and/or hand-OA in the discovery study (Table 3) and also in the meta-analysis with an OR of 1.14, 95% confidence interval (CI) 1.09–1.19, a p-value of 8×10−8 and consistent results across the datasets for knee- and/or hand-OA (p-value Q-statistic for heterogeneity=0.39, I2=5%). The SNPs rs10248619 (T>C, MAF 19%, situated in the GRB10 gene) and rs6088813 (A>C, MAF 35%, situated in the promoter region of the GDF5 gene) were both associated with OA in the meta-analysis, but did not reach genome-wide significance (p=0.0004 and p=0.01 respectively). There were no gender-specific associations observed. Given its significant associations with 2 OA phenotypes in the discovery study and the meta-analysis (knee- and/or hand-OA), we further focussed on SNP rs3815148.
Table 3.
Meta-analysis results: association of the 12 top-hits with OA-phenotypes
| SNP | chromosome | minor allele | MAF | Annotation | Association with OA | CTX-II (beta) | |||
|---|---|---|---|---|---|---|---|---|---|
| nearest gene | location | OR (95%CI) | p | beta | p | ||||
| rs10465850 | 1 | T | 43% | JAK1 | −347kb | 1.02 (0.98–1.05)a | 0.42 | −0.99a | 0.82 |
| rs12402320 | 1 | T | 27% | KCNN3 | intron | 1.05 (0.99–1.12)a | 0.12 | 1.07 | 0.005 |
| rs4656364 | 1 | C | 7% | NOS1AP | intron | 1.00 (0.89–1.13)b,d | 0.95 | 1.00 | 0.97 |
| rs3963342 | 4 | C | 25% | KCTD8 | intron | 1.04 (0.97–1.12)a,d | 0.29 | 1.05 | 0.07 |
| rs10248619 | 7 | T | 19% | GRB10 | intron | 1.09 (1.04–1.14)a | 5×10−4 | 1.05a | 0.26 |
| rs3815148 | 7 | C | 23% | COG5 | intron | 1.14 (1.09–1.19)c | 8×10−8 | 0.98a | 0.37 |
| rs959396 | 9 | G | 38% | PAX5 | intron | 0.94 (0.86–1.04)b,d | 0.26 | 0.95 | 0.01 |
| rs12352822 | 9 | G | 10% | TLR4 | −69kb | 0.99 (0.86–1.15)b,d | 0.93 | 1.01a | 0.89 |
| rs873598 | 10 | T | 49% | GPR10 | −68kb | 0.98 (0.91–1.06)a,d | 0.65 | 0.95 | 0.03 |
| rs741542 | 12 | T | 24% | ISCU | 3′downstream | 1.03 (0.91–1.16)b,d | 0.65 | 1.02 | 0.43 |
| rs11651351 | 17 | T | 7% | ACOX1 | intron | 0.89 (0.74–1.08)a,d | 0.24 | 0.91a | 0.33 |
| rs6088813 | 20 | C | 35% | GDF5 | intron | 0.91 (0.84–0.98)c,d | 0.01 | 0.99 | 0.65 |
MAF = minor allele frequency (HapMap); OA = osteoarthritis; OR = odds ratio; CI = confidence interval;
association with hand- and/or hip- and/or knee_OA;
hip- and/or knee-OA;
hand- and/or knee-OA;
random effects model
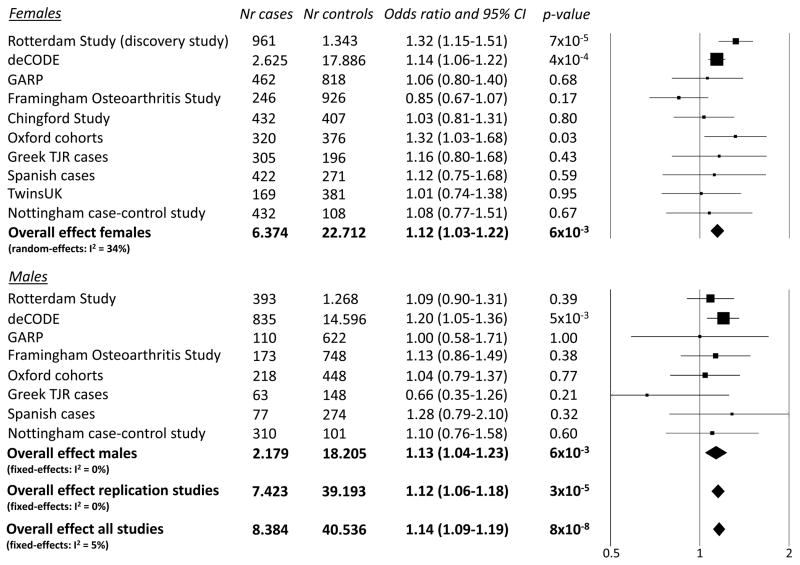
In Figure 1 the forest plot for the association result of the C-allele of rs3815148 with knee- and/or hand-OA is given. The estimated effect sizes were similar for knee-OA alone (OR 1.16) and hand-OA alone (OR 1.14). Interestingly, this SNP was also associated with progression of knee-OA in the Rotterdam Study (OR 1.31, 95%CI: 1.01–1.69) and the Chingford Study (OR 1.26, 95%CI: 0.73–2.17) (Figure 2). The meta-analysis for progression of knee-OA combining the results of the Rotterdam Study and the Chingford Study showed an OR of 1.30 per C-allele (95%CI: 1.03–1.64, p=0.03). We performed a sensitivity analysis in which the Rotterdam Study as discovery study was excluded from the meta-analysis of the SNP rs3815148 and observed that the association remained highly significant (OR 1.12, 95%CI: 1.06–1.18, p=3×10−5).
Figure 1.
Forest plot for the association of the rs3815148 SNP (GPR22 locus) with hand- and/or knee-osteoarthritis (allelic model).
Figure 2.
Forest plot for the association of the rs3815148 SNP (GPR22 locus) with knee OA progression (allelic model).
Functional analysis for the rs3815148 SNP
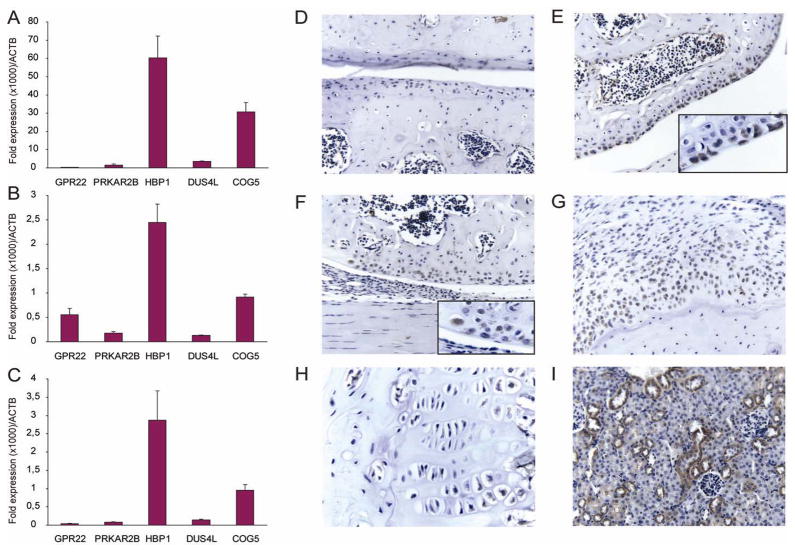
Although this SNP is annotated in the COG5 gene, there is a very large linkage disequilibrium (LD) block of over 500 kb in which this SNP resides with >400 SNPs which are highly correlated (Figure 3).18 The functional variant is likely to be one of the variants in LD with rs3815148 and could be located in any of the five genes annotated within this region, component of oligomeric golgi complex 5 (COG5), HMG-box transcription factor 1 (HBP1), dihydrouridine synthase 4-like (DUS4L), protein kinase, cAMP-dependent, regulatory, type II, beta (PRKAR2B) or G protein-coupled receptor 22 (GPR22). Therefore, we performed in silico and in vitro functional analysis to try to identify the gene underlying this association. There are no human mutations reported in the COG5, HBP1, DUS4L, PRAKAR2B and GPR22 genes resulting in clinical phenotypes. Expression databases show that all DUS4L, COG5 and HBP1 are ubiquitously expressed, with PRAKR2B especially in the immune system and the brain and GPR22 mainly in heart and the brain (http://www.genecards.org/). Furthermore, DUS4L, COG5 and HBP1 are expressed in cartilage according to the integrated cartilage gene database (http://bioinfo.hku.hk/iCartiGD/main), whilst GPR22 and PRKAR2B are not present in this database. For the GPR22 gene and the PRKAR2B gene, animal models are described in the literature, but skeletal and joint tissues were not studied in depth. It is known that knock-out mice of the PRKAR2B gene have diminished white adipose tissue19 and show decreased physical activity,20 while GPR22 gene knock-out mice develop heart failure in response to induced stress.21 Also, we tested whether rs3815148 or linked SNPs affected expression of any of the 5 genes in lymphoblast cell lines (n=400)22 and/or in the cortex of the brain (n=193)23 using publically available databases. In the expression database of the brain, only the DUS4L and COG5 genes were represented and no clear significant associations were observed between SNPs in the region surrounding the rs3815148 SNP and expression levels of those 2 genes. In the lymphoblast cell lines, there was a significant association between rs3757713 (D′=1.0, r2=0.95 with rs3815148) and expression levels of GPR22 (p=4×10−12, Figure 4). We also studied mRNA expression levels of the 5 genes of interest in cells derived from several tissues of the joint. Expression analyses by quantitative PCR in cultured synovial fibroblasts and primary chondrocyte cultures showed detectable transcripts of all 5 genes with COG5 and HBP1 being most abundantly expressed, while DUS4L, PRKAR2B and GPR22 are very low in all cell types (Figure 5A–C). As GPR22 showed the only remarkable association with altered expression in the lymphoblast cell lines, we studied the presence of GPR22 protein in knee joints from normal and osteoarthritic mice. Immunohistochemistry did not show GPR22 positive cells in normal mouse articular cartilage or synovium. However, GPR22 positive chondrocytes were found in the upper layers of the articular cartilage of mouse knee joints that were challenged by in vivo papain treatment or in the presence of interleukin-1 driven inflammation. GRP22 positive chondrocyte-like cells were also found in osteophytes in instability induced osteoarthritis (Figure 5D–I).
Figure 3.
Regional LD plot for the chromosome 7 locus (rs3815148 SNP, GPR22). r2 measures are based on the HapMap CEU samples. Each diamond represents one SNP. The brightness of each diamond is proportional to the r2 values for that SNP, the lower the r2, the lighter the colour of the diamond.
 r2 ≥ 0.8
r2 ≥ 0.8
 0.5 ≤ r2 < 0.8
0.5 ≤ r2 < 0.8
 0.2 ≤ r2 < 0.5
0.2 ≤ r2 < 0.5
 r2 <0.2
r2 <0.2
Analysis of SNPs shown to be associated with OA from the literature
The SNPs rs225014 and rs12885300 situated in the DIO2 gene, rs4140564 of the PTGS2 gene and rs7639618 of the DVWA gene were tested for association with hip-, knee- and hand OA in the Rotterdam Study. Results for these SNPs and for the GDF5 gene (rs6088813) which was also selected for replication in this GWAS, are shown in Table 4. The T-allele of SNP rs12885300 (DIO2 gene) had a trend towards a protective effect for hip OA (OR 0.84, 95%CI 0.68–1.03). The T-allele of SNP rs7639618 (DVWA gene) was associated with an increased risk of hand OA (OR 1.18, 95%CI 1.02–1.38). The C-allele of rs6088813 (GDF5) was associated with a decreased risk of hand OA (OR 0.82, 95%CI 0.73–0.92) and a non-significant trend in the same direction for knee OA (OR 0.79, 95%CI 0.89–1.01). No other statistically significant associations were observed.
Table 4.
Association results of SNPs in the DIO2, PTGS2, DVWA and GDF5 genes identified in previous genome-wide (linkage) studies
| rs-number | gene | minor allele | Paper of first report | OA-phenotype in first report | Association tested in males/females | Hip OA | Knee OA | Hand OA |
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) p-value | ||||||||
| rs225014 | DIO2 | C | Meulenbelt et al.3 | hip | females | 1.1 (0.9–1.4) 0.28 | 1.0 (0.8–1.1) 0.53 | 1.1 (0.9–1.2) 0.44 |
| rs12885300 | DIO2 | T | Meulenbelt et al.3 | hip | females | 0.8 (0.7–1.0) 0.09 | 1.1 (0.9–1.3) 0.34 | 0.9 (0.8–1.0) 0.13 |
| rs4140564 | PTGS2 | C | Valdes et al.4 | knee | females | 1.0 (0.6–1.5) 0.81 | 1.2 (0.9–1.6) 0.31 | 0.9 (0.6–1.2) 0.45 |
| rs7639618 | DVWA | T | Miyamoto et al.5 | knee, hip | females/males | 0.9 (0.7–1.1) 0.25 | 1.1 (0.9–1.2) 0.53 | 1.2 (1.0–1.4) 0.03 |
| rs6088813 | GDF5 | C | Miyamoto et al.28 | knee, hip | females/males | 1.0 (0.9–1.2) 0.77 | 0.9 (0.8–1.0) 0.08 | 0.8 (0.7–0.9) 7×10−4 |
OA = osteoarthritis; OR = odds ratio; CI = confidence interval
Discussion
In this paper we present a GWAS of OA to find DNA sequence variation associated with OA at multiple joint sites and with progression of knee-OA.
The C-allele of the SNP rs3815148 showed an increased risk of 14% for knee- and hand-OA and was also associated with progression of knee OA (increased risk of 30% per allele). This DNA variant is located in intron 12 of the COG5 gene, while there is high linkage disequilibrium in the surrounding region (Figure 4). In silico and in vitro functional experiments showed that the T-allele of the rs3757713 SNP (D′=1, r2=0.95 with rs3815148) was associated with expression levels of GPR22 in lymfoblasts (p-value 4×10−12) and immunohistochemistry experiments showed a presence of the GPR22 protein in cartilage and osteophytes in OA-induced mouse models, whilst this protein was absent in normal cartilage. In our opinion, this suggests that the GPR22 gene might be the causal gene in the associations found. GPR22−/− mice were only tested for heart-specific outcomes,21 it would be interesting to study their skeletal phenotype as well. To our knowledge, ligands that may bind to GPR22 have not been identified yet, which makes this receptor a so called “orphan receptor”. Since the GPR22 gene encodes a G-protein-coupled receptor, this is potentially an interesting drug target. While we tend to focus on a specific role in joint tissues, it could also be that this gene is involved in locomotor function/disability since it is expressed in regions of the brain which are involved in locomotor function (frontal cortex and basal ganglia).24 This may lead to, for example, a different gait pattern or joint loading which could lead to OA. Although we present results to suggest GPR22 as the gene underlying the genetic association, the understanding of the precise mechanism remains unknown and it is possible that one or more of the other genes in the same LD block are important for OA.
In this study, we focused on a spectrum of OA phenotypes for several reasons. Because multiple joint-sites can be affected by OA, we searched for loci that showed association with more than one OA phenotype. This method is likely to identify more general susceptibility genes for OA rather than site-specific aspects. In addition, in our discovery study we had limited power to identify joint-specific alleles, such as the GPR22 SNP, to be modestly associated with OA. We realize that our broad phenotype approach can introduce heterogeneity, although there was no overlap in cases and controls in the combined phenotype analyses, and that we could have increased the chances of missing true OA susceptibility alleles that are site-specific. Yet, the consistent and robust pattern of association we observe for the GPR22 locus together with the expression data provides sufficient evidence to implicate this locus in OA etiology. However, the expression patterns observed did not clarify the exact underlying mechanism and therefore more experiments are warranted. While we observed several other potentially interesting loci, these did not reach genome-wide significance and so larger GWAS sample sizes and meta-analysis will be useful to identify further OA susceptibility loci, including those that might act at a joint-site specific level.
To date, genome-wide linkage and association studies have shown that there are DNA variants in 4 genes found to be consistently associated with OA.2–5,25 These are the rs225014 and rs12885300 SNPs of the DIO2 gene, rs4140564 of the PTGS2 gene, rs7639618 of the DVWA gene and rs143383 (rs6088813 proxy SNP with D′=1.0 and r2=0.93) of the GDF5 gene. In Table 4, the association results of those 5 SNPs are shown for the Rotterdam Study based on the GWAS dataset. The 2 SNPs found to be associated with hip-OA in females in the DIO2 gene were discovered through a linkage study.3 Those 2 SNPs were not associated with hip-OA in the Rotterdam Study although the rs12885300 SNP showed a trend towards a decreased risk in female carriers of the T-allele which is the same direction as described in the paper of Meulenbelt et al.3 The SNP in the PTGS2 gene was not associated with knee-OA in females of the Rotterdam Study. In the original papers by Valdes et al.4 and Meulenbelt et al.3 an association with a more severe OA phenotype (total joint replacement or a K/L grade >=3) was described, which could be an explanation for the lack of replication in the Rotterdam Study using the less severe, radiographic knee-OA phenotype (K/L >=2). In addition, the DVWA SNP was not associated with knee-OA in our population. An increased risk for hand-OA was observed for the T-allele of rs7639618 SNP (DVWA), however this effect is in the opposite direction compared to the association observed in the GWAS for knee-OA by Miyamoto et al. 5 This SNP was also not associated with knee-OA in other Caucasian studies,6,7 which could indicate that this SNP has different effects in different ethnic populations. The GDF5 gene, identified with our search strategy showed an association with knee- and/or hand-OA with an OR of 0.91 and 95%CI: 0.84–0.98 in the meta-analysis. This was previously also shown by Miyamoto et al.[25],25 Chapman et al.[2]2 Vaes et al.26 and in a large meta-analysis (n=5085 cases and 8135 controls) performed within the TREAT-OA consortium27 for knee-OA. Although the GDF5 locus does not reach genome-wide significance, it is considered a true and consistent association in the OA field.
In conclusion, we have identified a novel locus involved in susceptibility for prevalence and progression of OA, SNP rs3815148 close to the GPR22 gene, in the largest association analysis in the genetics of OA to date with 14,938 OA cases and approximately 39,000 controls.
Supplementary Material
Acknowledgments
The generation and management of GWAS genotype data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) (project nr. 050-060-810), and funding from the European Commission framework 7 programme TREAT-OA (grant 200800).
The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam.
We thank Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Scientific support for this project was also provided through the Tufts Clinical and Translational Science Institute (Tufts CTSI) under funding from the National Institute of Health/National Center for Research Resources (UL1 RR025752). Points of view or opinions in this paper are those of the authors and do not necessarily represent the official position or policies of the Tufts CTSI.
Rik Lories is the recipient of a post-doctoral fellowship from the Flanders Research Foundation (FWO Vlaanderen).
TwinsUK acknowledges financial support from the Wellcome Trust, the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London and Arthritis Research Campaign.
deCODE also wishes to thank Gudmar Thorleifsson, Thorvaldur Ingvarsson and Helgi Jonsson for their valuable contribution to this work.
The Leiden University Medical Centre, the Dutch Arthritis Association and Pfizer Inc., Groton, CT, USA support the GARP study. The genotypic work was supported by the Netherlands Organization of Scientific Research (MW 904-61-095, 911-03-016, 917 66344 and 911-03-012), Leiden University Medical Centre and the Centre of Medical System Biology in the framework of the Netherlands Genomics Initiative (NGI).
The Framingham Osteoarthritis Study would like to acknowledge NIH AR47785 and AG18393 which funded the Framingham Osteoarthritis Study.
The Study of Osteoporotic Fractures (SOF) and the Osteoporotic Fractures in Men Study (Mr OS) are supported by National Institutes of Health funding. The National Institute on Aging (NIA) provides support under the following grant numbers: AG05407, AR35582, AG05394, AR35584, AR35583, R01 AG005407, R01 AG027576-22, 2 R01 AG005394-22A1, AR048841, and 2 R01 AG027574-22A1.
The Chingford Study is funded by the arc and acknowledges the support of the staff and volunteers-particularly Alan Hakim, Vicky Thompson, Maxine Daniels, as well as staff who have read x-rays including David Hunter, Geraldine Hassett, Nigel Arden and Deborah Hart.
Footnotes
Conflict of Interest
All authors report no financial conflict of interest that might influence the results or interpretation of this manuscript.
References
- 1.Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2004;12(Suppl A):S39–44. doi: 10.1016/j.joca.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Chapman K, Takahashi A, Meulenbelt I, Watson C, Rodriguez-Lopez J, Egli R, Tsezou A, Malizos KN, Kloppenburg M, Shi D, et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′ UTR of GDF5 with osteoarthritis susceptibility. Human molecular genetics. 2008;17:1497–1504. doi: 10.1093/hmg/ddn038. [DOI] [PubMed] [Google Scholar]
- 3.Meulenbelt I, Min JL, Bos S, Riyazi N, Houwing-Duistermaat JJ, van der Wijk HJ, Kroon HM, Nakajima M, Ikegawa S, Uitterlinden AG, et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Human molecular genetics. 2008;17:1867–1875. doi: 10.1093/hmg/ddn082. [DOI] [PubMed] [Google Scholar]
- 4.Valdes AM, Loughlin J, Timms KM, van Meurs JJ, Southam L, Wilson SG, Doherty S, Lories RJ, Luyten FP, Gutin A, et al. Genome-wide association scan identifies a prostaglandin-endoperoxide synthase 2 variant involved in risk of knee osteoarthritis. Am J Hum Genet. 2008;82:1231–1240. doi: 10.1016/j.ajhg.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyamoto Y, Shi D, Nakajima M, Ozaki K, Sudo A, Kotani A, Uchida A, Tanaka T, Fukui N, Tsunoda T, et al. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nature genetics. 2008;40:994–998. doi: 10.1038/ng.176. [DOI] [PubMed] [Google Scholar]
- 6.Meulenbelt I, Chapman K, Dieguez-Gonzalez R, Shi D, Tsezou A, Dai J, Malizos KN, Kloppenburg M, Carr A, Nakajima M, et al. Large Replication Study and Meta-Analyses of Dvwa as an Osteoarthritis Susceptibility Locus in European and Asian Populations. Human molecular genetics. 2009 Jan 30; doi: 10.1093/hmg/ddp053. [DOI] [PubMed] [Google Scholar]
- 7.Valdes AM, Spector TD, Doherty S, Wheeler M, Hart DJ, Doherty M. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Annals of the rheumatic diseases. 2008 Dec 3; doi: 10.1136/ard.2008.102236. [DOI] [PubMed] [Google Scholar]
- 8.Altman RD, Fries JF, Bloch DA, Carstens J, Cooke TD, Genant H, Gofton P, Groth H, McShane DJ, Murphy WA, et al. Radiographic assessment of progression in osteoarthritis. Arthritis Rheum. 1987;30:1214–1225. doi: 10.1002/art.1780301103. [DOI] [PubMed] [Google Scholar]
- 9.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nature genetics. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 13.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 14.Lories RJ, Derese I, De Bari C, Luyten FP. In vitro growth rate of fibroblast-like synovial cells is reduced by methotrexate treatment. Annals of the rheumatic diseases. 2003;62:568–571. doi: 10.1136/ard.62.6.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dell’Accio F, De Bari C, Luyten FP. Molecular markers predictive of the capacity of expanded human articular chondrocytes to form stable cartilage in vivo. Arthritis Rheum. 2001;44:1608–1619. doi: 10.1002/1529-0131(200107)44:7<1608::AID-ART284>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 16.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis and cartilage/OARS, Osteoarthritis Research Society. 2007;15:1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Lories RJ, Peeters J, Bakker A, Tylzanowski P, Derese I, Schrooten J, Thomas JT, Luyten FP. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum. 2007;56:4095–4103. doi: 10.1002/art.23137. [DOI] [PubMed] [Google Scholar]
- 18.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummings DE, Brandon EP, Planas JV, Motamed K, Idzerda RL, McKnight GS. Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature. 1996;382:622–626. doi: 10.1038/382622a0. [DOI] [PubMed] [Google Scholar]
- 20.Newhall KJ, Cummings DE, Nolan MA, McKnight GS. Deletion of the RIIbeta-subunit of protein kinase A decreases body weight and increases energy expenditure in the obese, leptin-deficient ob/ob mouse. Mol Endocrinol. 2005;19:982–991. doi: 10.1210/me.2004-0343. [DOI] [PubMed] [Google Scholar]
- 21.Adams JW, Wang J, Davis JR, Liaw C, Gaidarov I, Gatlin J, Dalton ND, Gu Y, Ross J, Jr, Behan D, et al. Myocardial expression, signaling, and function of GPR22: a protective role for an orphan G protein-coupled receptor. Am J Physiol Heart Circ Physiol. 2008;295:H509–521. doi: 10.1152/ajpheart.00368.2008. [DOI] [PubMed] [Google Scholar]
- 22.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nature genetics. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 23.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. A survey of genetic human cortical gene expression. Nature genetics. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 24.O’Dowd BF, Nguyen T, Jung BP, Marchese A, Cheng R, Heng HH, Kolakowski LF, Jr, Lynch KR, George SR. Cloning and chromosomal mapping of four putative novel human G-protein-coupled receptor genes. Gene. 1997;187:75–81. doi: 10.1016/s0378-1119(96)00722-6. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nature genetics. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 26.Vaes RB, Rivadeneira F, Kerkhof JM, Hofman A, Pols HA, Uitterlinden AG, van Meurs JB. Genetic variation in the GDF5 region is associated with osteoarthritis, height, hip axis length and fracture risk: the Rotterdam study. Annals of the rheumatic diseases. 2008 Nov 24; doi: 10.1136/ard.2008.099655. [DOI] [PubMed] [Google Scholar]
- 27.Evangelou Eea. Large-scale analysis of association between GDF5 (rs143383) and FRZB (rs7775 and rs288326) variants and hip, knee and hand osteoarthritis. Arthritis Rheum. 2009 doi: 10.1002/art.24524. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.