Abstract
Microglia are cells from non-neuronal lineages that reside in the central nervous system. In zebrafish, early macrophages migrate from the yolk sac to the brain and retina at 26–30 hours post fertilization (hpf) and transform into microglia at 55–60 hpf. The migration of macrophages into the central nervous system requires signaling by macrophage colony stimulating factor-1 receptor (csf-1r), which is encoded by the gene fms. In this study, we show that the targeted knockdown of csf-1r with morpholino oligonucleotides delays migration of macrophages from the yolk sac to the retina, and this delay in macrophage migration results in microphthalmia, delay in cell cycle withdrawal among retinal progenitors and the absence of neuronal differentiation. When embryos were allowed to survive beyond the time when morpholino-dependent translation inhibition is lost, microglia re-occupy the retina and neuronal differentiation partially recovers. Our data demonstrate that microglia are required for normal retinal growth and neurogenesis. This study provides new insight into the neurogenic role of microglia during retinal development in zebrafish.
Keywords: microglia, retinal development, neurogenesis
Introduction
Microglia are cells from non-neuronal lineages that reside in the CNS. It is generally accepted that microglia are phagocytic cells involved in establishing host defense mechanisms against invading agents and the clearance of endogenous cell debris in the CNS[1,2]. Depending upon the nature of the injury, microglia may either be neuroprotective or neurotoxic or both [3]. The function of microglia in the retina is similar to that elsewhere in the CNS.
In zebrafish, early macrophages differentiate in the yolk sac; some then join the blood circulation, while others migrate to the head of the embryo. Macrophages that colonize the retina and forebrain migrate to the cephalic mesenchyme and then secondarily invade the nervous system. These cells first enter the retina between 26 and 30 hpf. The migration of macrophages from the yolk sac to the brain and retina requires signaling by macrophage colony stimulating factor-1 receptor (csf-1r). The fms gene, which codes for csf-1r, is expressed in all macrophage lineage cells and is crucial for their colonization of embryonic tissues [4, 5]. Recently, a concept has emerged that in both the normal and injured brain, microglia function to regulate neurogenesis [6, 7]. Array studies performed in our lab identified progranulin-a (pgrn-a, GenBank: NM_001001949) as a gene that was strongly upregulated during the proliferative phase of photoreceptor regeneration in zebrafish [8]. Furthermore, this gene was expressed exclusively by microglia in the CNS and in the macrophage lineage cells outside of the CNS. However, there is still a lack of knowledge about the function of microglia during retinal development.
In this study, embryonic zebrafish were used to experimentally investigate the function of microglia during retinal neurogenesis. Following the targeted knockdown of csf-1r, the migration of macrophage to the retina and brain is delayed. This results in microphthalmia and a delay in the differentiation of retinal neurons. Labeling mitotic cells with BrdU revealed that retinal progenitors have an extended cell cycle period and a reduced rate of cell cycle exit. Escape from the inhibition of csf-1r knockdown results in the partial recovery of neuronal differentiation. Collectively, these data indicate that microglia function as essential regulators of neurogenesis in the embryonic and larval retina of zebrafish.
Materials and methods
Experimental Animals
AB strain zebrafish were used in this study. The animals were maintained in aquaria at 28.5°C with a 10/14-hour dark/light cycle. Embryos were collected after natural spawns, housed at 28.5°C, and staged by hours post fertilization (hpf). Protocols for all procedures using animals were approved by the University Committee for Use and Care of Animals at the University of Michigan and the Institutional Animal Care Committee at Nankai University and conform to National Institutes of Health guidelines.
Morpholino oligonucleotides and microinjections
Morpholino oligonucleotides, either complementary to the translation start site of zebrafish fms mRNA sequence (GenBank: AF240639) or containing a 5-bp mismatch (control morpholinos), were synthesized by Gene Tools, LLC (Philomath, OR). The sequence of the morpholino oligonucleotides is 5’- AAGAGCGCGAAGAACATCTCAGAGC-3’ (antisense start codon were underlined), and the sequence of the mismatch oligos is 5’-AAcAcCGCcAAcAACATCTCAcAGC-3’ (mismatch nucleotides were underlined). Morpholinos were diluted in 1× Danieau buffer [9] at 1 ng/µl. Embryos were injected with 4 ng morpholino oligonucleotides at the 1- to 4-cell stage. To suppress off-target p53-mediated apoptosis [10], we co-injected a zebrafish p53 oligo at 2 ng with the fms morpholino oligonucleotides. The antisense sequence for the p53 oligos is 5’- GCGCCATTGCTTTGCAAGAATTG-3’ (www.gene-tools.com).
Acridine Orange Labeling
The embryos were dechorionated at 24 hpf and placed in acridine orange solution (5 µg/ml in embryo medium) (Sigma) for 15 min followed by extensive washes in embryo medium. The embryos were anesthetized and viewed under a fluorescence dissecting microscope, and cells in the retina that were positively labeled with acridine orange were counted.
Western blot analysis
The specificity of the translation-blocking morpholinos was tested by Western blot analysis. Thirty to forty embryos were lysed in buffer with protease inhibitors (Complete Mini, Roche, Mannheim, Germany), and protein concentrations were quantified by using the BCA Protein Assay Kit (Complete Mini, Roche, Mannheim, Germany). The proteins were separated in a 12% SDS-PAGE gel and were transferred to a nitrocellulose membrane (Sigma-Aldrich, St. Louis, MO). The membrane was then blocked in 5% nonfat dry milk in PBS for 2 hours and incubated with the following antibodies: anti-csf-1r (1:500; Anaspec, Fermont, CA) and anti-GAPDH (1:1,000; Millipore, Billerica, MA). The blots were rinsed with PBS and incubated with peroxidase-conjugated goat anti-rabbit antibodies (1:10,000; Sigma-Aldrich, St. Louis, MO) for 1 hour. The bound antibody was visualized using the enhanced chemiluminescence assay (Kodak Chemiluminescence BioMax Film, Rochester, NY).
Immunohistochemistry
Embryos were fixed overnight in 4% paraformaldehyde, cryoprotected in 20% sucrose in 0.1M phosphate-buffered saline (pH 7.2), frozen in OCT (Sakura Finetek, Torrance, CA). Cryosections (10 µm) were mounted on glass slides for immunohistochemistry or in situ hybridization. Immunohistochemistry was performed using standard procedures. In brief, sections were rinsed in 0.1 M phosphate-buffered saline and 0.5% Triton X-100 (PBST), incubated with 20% normal sheep serum (NSS) in PBST, and incubated overnight at 4°C in primary antibodies diluted in 2% NSS-PBST. The primary antibodies and concentrations used were as follows: mouse monoclonals Zn12 (1:200), Zpr1 (1:200), and Zpr3 (1:200) all from the Zebrafish International Resource Center (ZIRC, University of Oregon, Eugene, OR) for labeling ganglion cells, cones, and rods, respectively. After washing with PBST, the sections were incubated for 1.5 hrs at room temperature in PBST containing 2% NSS and fluorescently labeled secondary antibodies (Molecular Probes, Eugene, OR), diluted at 1:500. The sections were counterstained with a 1:1,000 dilution of DAPI (4',6-diamidino-2-phenylindole) (Sigma) to label the nuclei. Following this step, sections were washed extensively in PBST, and sealed with mounting media and glass coverslips. Ten animals were processed at each time point.
BrdU labeling
Bromodeoxyuridine (BrdU; Sigma) was used to label mitotically active cells. The embryos were systemically labeled with BrdU at 72 hpf by soaking them for 20 minutes in 5 mM BrdU and 15% DMSO in 4°C E3 medium as described previously [11]. BrdU was detected with a monoclonal antibody (Becton Dickinson Immunocytochemistry Systems, San Jose, CA) diluted 1:100. The total cell population was labeled using DAPI (Sigma) diluted at 1:1,000 to label nuclei. Twenty retina sections from ten embryos in each group were counted to determine the ratio of BrdU positive cells to all nuclei.
In situ hybridization
All embryos were grown in 0.003% 1-phenyl-2-thiourea (PTU, Sigma) to block pigmentation and mediate visualization until 72 hpf. In situ hybridization was performed on whole mount embryos or cryosections using a standard protocol [12]. Briefly, sense and antisense riboprobes were synthesized from linearized plasmids, and digoxigenin (DIG)-labeled probes were generated by in vitro transcription using the DIG RNA labeling kit (Roche Diagnostics, Indianapolis, IN). Two probes were used in this study. Microglia were labeled using an mRNA probe for pgrn-a. The cDNA encoding pgrn-a was linearized with SmaI, and the riboprobes were synthesized with T3 polymerase. An mRNA probe for athnal-homologue5 (ath5) was used as a marker to explore whether the neurogenesis initiated on time. The cDNA encoding ath5 was linearized with ApaI, and the riboprobes were synthesized with SP6 polymerase. For whole mount in situ hybridization, probe with a concentration of 1 ng/µl was added to Eppendorf tubes and hybridized overnight at 55°C. For the in situ hybridization of the sections following prehybridization, 100 ng pgrn-a probe in 100 µl of hybridization buffer was pipetted onto each slide, coverslipped, and hybridized overnight at 55°C. Negative controls were riboprobes encoding the sense strand of the respective cDNAs, which failed to hybridize. The next day, the embryos or sections were washed, and digoxigenin was immunolabeled using an alkaline-phosphatase-conjugated antibody (Roche). NBT/BCIP (Roche) served as the enzymatic substrate on the third day.
Photography and Image Analysis
Images of the immunohistochemistry were photographed with a Leica TCS SP5 confocal microscope. Images of the in situ hybridization were photographed with a Nikon DMX1200 digital camera mounted on a Nikon Eclipse E800 epifluorescent compound microscope. The larvae for whole mount in situ hybridization of cryosections were observed with an Olympus SZX10 dissecting microscope. The resulting images were compiled in Adobe Photoshop CS2 (Adobe, San Jose, CA, USA) and resized and occasionally modified for contrast and brightness using the Image-Adjustments-Contrast-Brightness setting. All of the images within an experiment were manipulated in the same manner. Immunostaining and image analysis was conducted identically for each experimental group to minimize the possibility of introducing variability between samples. Image J 1.38X software (Wayne Rasband, NIH, http://rsb.info.nih.gov/ij/) was used to convert the fluorescent images of the BrdU to 8-bit grayscale prior to thresholding, positive staining and determining the positive area of each image. Excel was then used to calculate the percentile of the positive area on each slide. The values were averaged across similarly treated retinas, and statistical analysis was performed by ANOVA.
Results and Discussion
The csf-1r knockdown results in the delayed migration of macrophages into the retina and microphthalmia
In this study, we used an fms-targeted morpholino to inhibit the translation of the fms gene. Because changes in the cell cycle often result in the activation of apoptosis and increased cell death, we first examined whether cells undergo apoptosis in fms morphants at 24hpf. Generally, the overall death rate in the developing zebrafish retina is very low (1–2%). By the co-injection of fms and p53 morpholinos, we did not observe a significant increase in the levels of cell death in the fms morphant retina at 24 hpf by acridine orange labeling in either the retina or the whole body (figure not shown). This result suggested that cell death did not contribute to the retinal phenotype observed in fms morphants.
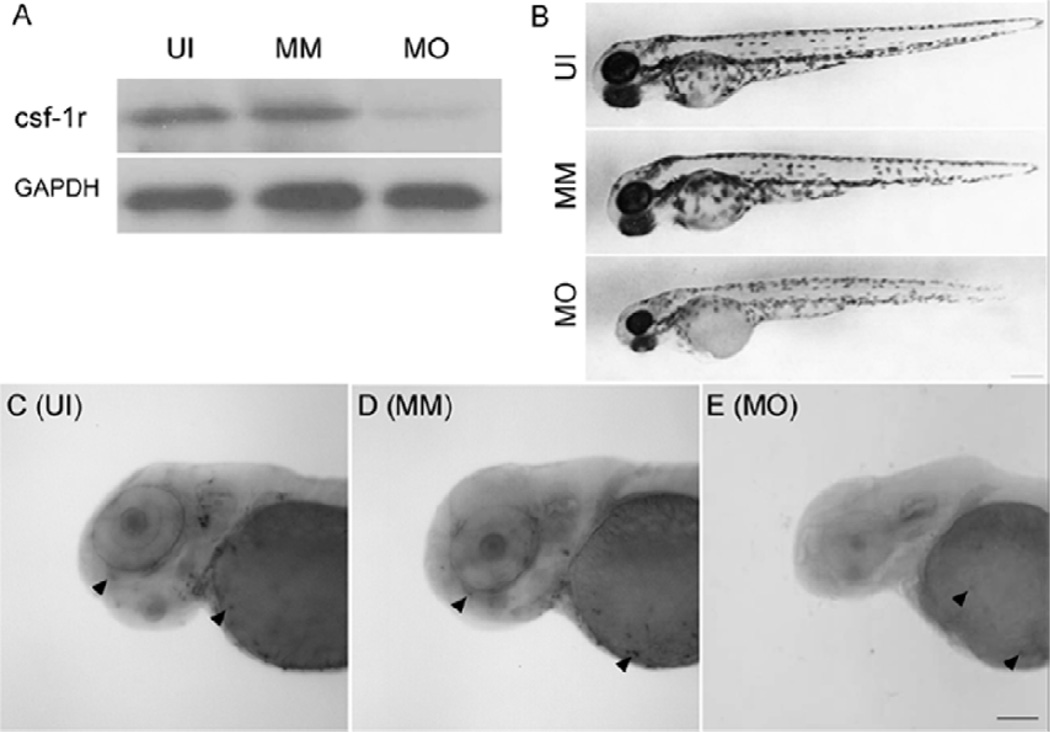
A western blot test showed injection of the fms MO resulted in specific suppression of csf-1r protein expression at 72 hpf in the fms morphants (Fig. 1A). The delayed migration of the macrophages from the yolk sac to the retina was verified by whole-mount in situ hybridization with the microglia-specific mRNA probe pgrn-a. In uninjected and mismatch control animals pgrn-a was expressed in microglia in the brain and retina at 72hpf (Fig. 1C, D). However, following knockdown of csf-1r, pgrn-a-expressing cells were present on the surface of the yolk sac, but there were no labeled cells in the brain or retina (Fig. 1E). These results indicate we were able to exclude microglia from the brain and retina until 72 hpf, thereby providing a model for evaluating the effects of microglia on the developing retina.
Figure 1. fms-targeted morpholino oligonucleotides inhibit the migration of microglia from the yolk sac to the retina and brain at 72 hpf.
Panel A is the result of a western blot with the csf-1r antibody at 72 hpf. Panel B illustrates the gross development of uninjected (UI), mismatch control (MM) and fms-morphant (MO) larvae. Fms morphant larvae are slightly smaller than are wild-type and mismatch controls, and have less pigmentation of the body at 72 hpf. However, the fms morphants have dramatically smaller eyes than do the other two groups. Panels C–E show a whole mount in situ hybridization with the riboprobe progranulin-a (pgrn-a), which is specific to microglia. For uninjected and mismatch control larvae, the microglia (arrowheads) were detected in the yolk sac, retina and brain at 72 hpf. Note the absence of microglia in the brain and retina in fms morphants (E) and the presence of microglia in the yolk sac (arrowheads). Scale bar: B, 250 µm; C–E, 100 µm.
Compared to uninjected and mismatch control embryos, two differences were found in the gross appearance of fms morphants at 72 hpf (Fig. 1B). The first difference was less pigmentation in the bodies of fms morphants. The function of the zebrafish fms gene was first investigated in the Danio. rerio pigment pattern mutant panther line [4]. fms is expressed and required by D. rerio xanthophore precursors for their ventralward migration and differentiation, and promotes the normal patterning of melanocyte death and migration during adult stripe formation [13]. Our result is consistent with this previous study in that when we knocked down the fms gene, the morphants had a lighter pigmentation. The second finding was that fms morphants had remarkably smaller eyes (Fig. 1B) than did uninjected and mismatch controls. However, the panther line did not show gross abnormalities in their eyes [4]. In panther larvae, a mean number of three macrophages penetrated the optic tectum and had differentiated into microglia at 72 hpf [5]. In our study, the fms morphants we identified had no microglia detected in the brain or retina by whole mount in-situ hybridization with a pgrn-a mRNA probe specific to microglia. Our results suggested that even a few microglia (i.e., 1–3) may be sufficient to promote normal development of the retina and eye.
The onset of neurogenesis and neuronal differentiation are delayed following knockdown of csf-1r
Neurogenesis in the zebrafish retina is initiated in a small, discrete patch adjacent to the optic stalk in the ventronasal retina and then follows an order from the inner section to the outer [14,15]. Subsequent cellular differentiation then propagates away from this patch as a wave throughout the neuroepithelium [16]. The first retinal progenitors to withdraw from the mitotic cycle at 28 hpf lie in a small cluster in the ganglion cell layer [17]. From 48hpf, the cells in the outer nuclear layer begin to withdraw from the mitotic cycle and lamination has spread across the retina [18]. Most major classes of retina cells, including photoreceptors, can be identified at 72 hpf [19].
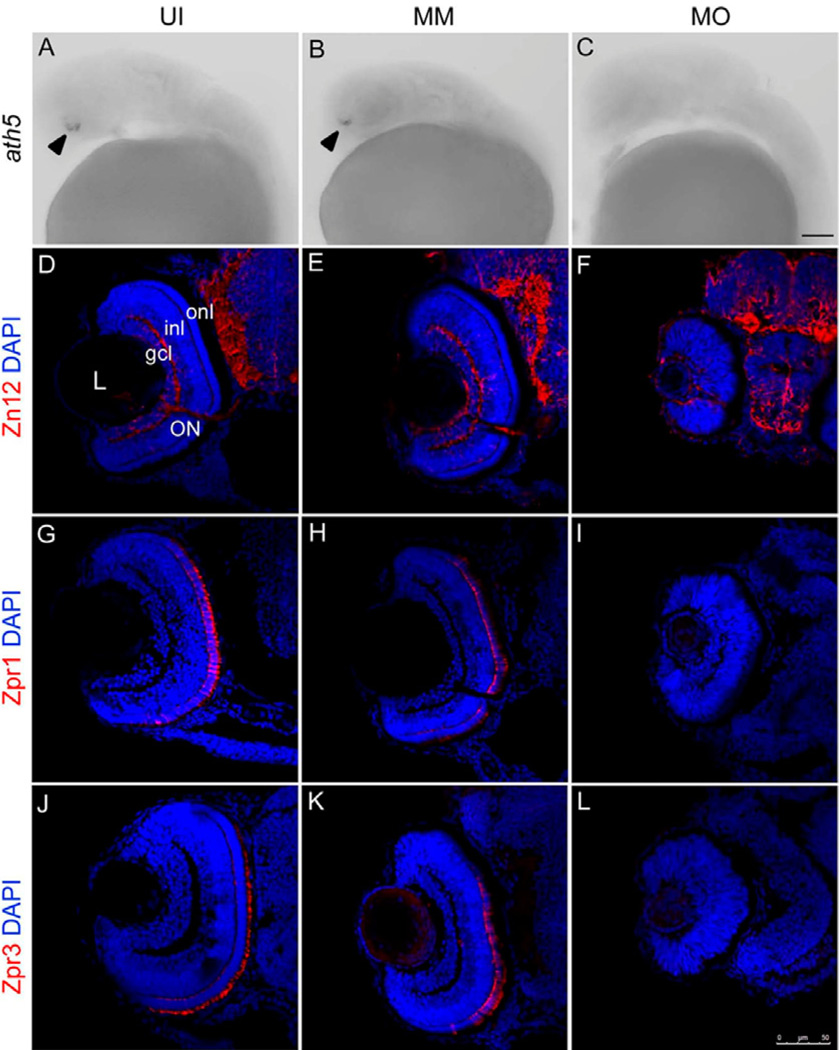
Athnal-homologue5 (ath5) is a bHLH transcription factor that is expressed in a wave-like pattern in cells immediately after their final mitosis, and is required for the specification of the first-born retinal neurons (retinal ganglion cells) [17, 20]. In this study, we used ath5 as a marker at 28 hpf to explore whether the differentiation of ganglion cells initiated on time. In situ hybridization revealed that in uninjected and mismatch control retinas at 28hpf, the expression of ath5 matches that described previously (see [14, 15]; Fig. 2A–B). In contrast, following knockdown of csf-1r, at 28hpf there was no ath5 mRNA expression (Fig. 2C). These data suggested that neuroepithelial cells were defective in the process of neurogenesis at the early time point, and the differentiation of ganglion cells was disrupted in the fms morphant retina.
Figure 2. fms knockdown delays neurogenesis initiation at 28 hpf and affects neuronal differentiation at 72 hpf.
Panels A–C show the in situ analysis of ath5 expression during retina development in the retina of uninjected (UI), mismatch control (MM) and fms morphants (MO) at 28 hpf. Note that the expression of ath5 was detected in the retina of uninjected and mismatch controls (arrowheads), but was absent in fms morphant retina. Panels D–L illustrate sections taken through the retinas at 72 hpf. Panels D–F illustrate Zn12 staining; panels G–I illustrate Zpr1 staining; panels J–K illustrate Zpr3 staining. Note that in the fms morphant retinas, only a handful of differentiated Zn12-positive cells were present at 72 hpf, possessed rudimentary optic nerves and had the appearance of an undifferentiated neuroepithelium. Neither Zpr1 nor Zpr3 was detected, while the retinas are well laminated and differentiated, showing strong expression of Zn12, Zpr1 and Zpr3 in the uninjected and mismatch control larvae at 72 hpf. L, lens; gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer; ON, optic nerve. Scale bar: A–C, 50µm; D–L, 75 µm.
Cell type-specific antibodies were used to evaluate neuronal differentiation at 72 hpf. The Zpr1 antibody labels a cell surface epitope on red/green-sensitive double cone photoreceptors in the zebrafish [21, 22]. The Zpr3 antibody labels an antigenic region on the rod opsin protein [23]. The Zn12 antibody labels a cell surface epitope on ganglion cells [15]. In uninjected and mismatch control retinas, the three nuclear layers were well-formed that included differentiated RGCs and photoreceptors (Fig. 2D, E, G, H, J, K). In contrast, following knockdown of csf-1r there was little evidence of cellular laminae, there were only few differentiated RGCs and a rudimentary optic nerve (Fig. 2F) and no presence of differentiated photoreceptors (Fig. 2I, L). These data show that the differentiation of all of the major cell types in the retina largely absent following knockdown of csf-1r. In the above results, we have demonstrated that the absence of microglia in the brain and retina resulted in a severe delay in the differentiation of all major cell types (ganglion cells, cones and rods) in the developing retina until 72 hpf.
The delayed migration of microglia causes defects in cell proliferation
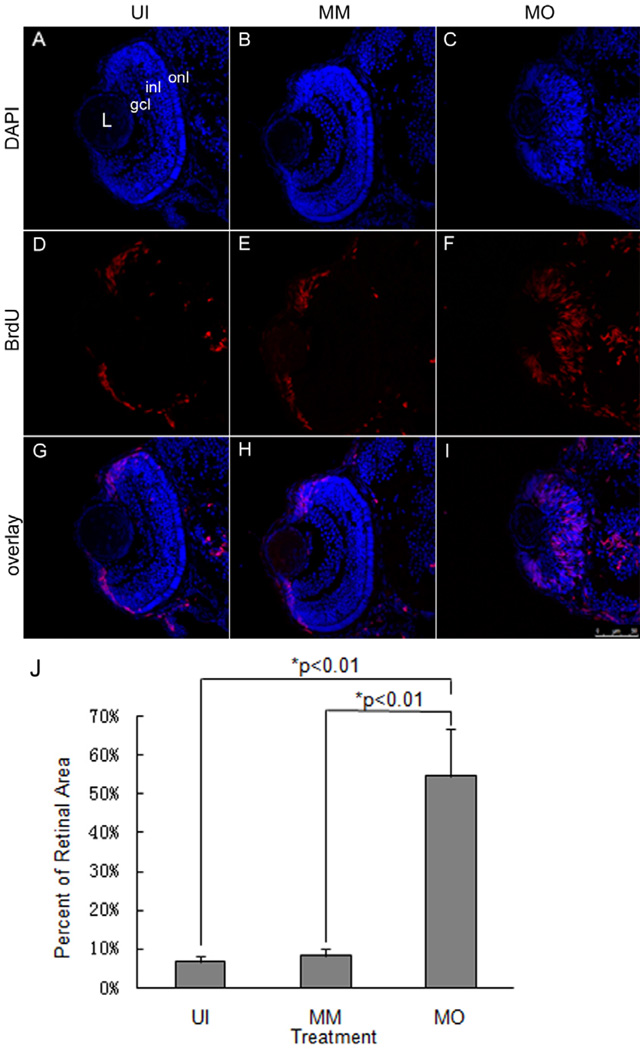
To understand the basis of the defect in retinal development in fms morphant embryos, we examined the cell cycle progression of retinal precursor cells in fms morphants by BrdU systemic labeling. This method labels all cells that are in the S-phase, and therefore, will mark proliferative progenitor cells [24]. In the uninjected and mismatch control embryos, retinal neuroepithelial cells are restricted to the ciliary marginal zone (CMZ) by 72 hpf, an area that remains proliferative for the life of the zebrafish. Labeling with BrdU showed that most of the cells of the central retina in uninjected and mismatch control larvae did not incorporate BrdU because they had become postmitotic, whereas cells located within the CMZ remained strongly labeled (Fig. 3D, E). In contrast, the majority of the cells in fms morphant retinas continued to incorporate BrdU at 72 hpf, including a large number of cells in the central retina (Fig. 3F). To quantify these findings, we scored the percent of the total retinal cells that were BrdU-positive and revealed that there were no statistically significant differences between the uninjected and mismatch controls, but there was a significantly greater proportion of the retina that contained proliferating cells in fms morphants (Fig. 3J). This result suggested that the absence of microglia in the brain and retina results in defects in the proliferation of retinal progenitor cells, whereby the cell cycle in the progenitor cells is prolonged or/and cell cycle exit is compromised. Microglia regulate the cell cycle and influence cell-type fate and differentiation pathways in the zebrafish retina. There are at least two possible roles for the microglia pathway in regulating the cell cycle of retina progenitor cells: it could be required for normal S-phase progression and/or it should be required for S-phase exit and G2/M progression [25]. Further experiments can be performed in fms morphant retinas to address this question, including checking the key regulators and genes for S/M transition or cell cycle analysis by flow cytometry.
Figure 3. The fms morphant retina remains as neuroepithelium at 72 hpf.
Systemic BrdU was used to label mitotically active cells in the retinas of uninjected (UI), mismatch controls (MM) and fms morphants (MO) at 72 hpf, respectively. Note the expanded distribution of BrdU-positive cells in the fms morphant retina. L, lens; gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer. Scale bar: 75 µm. Panel J illustrates the quantitative comparison of the proportion of the retina occupied by BrdU-positive cells (n=10 larvae/treatment group). There were statistically significant differences between the control and morphant retinas (p<0.01; ANOVA).
Retinal neurogenesis partially recovers in fms morphants
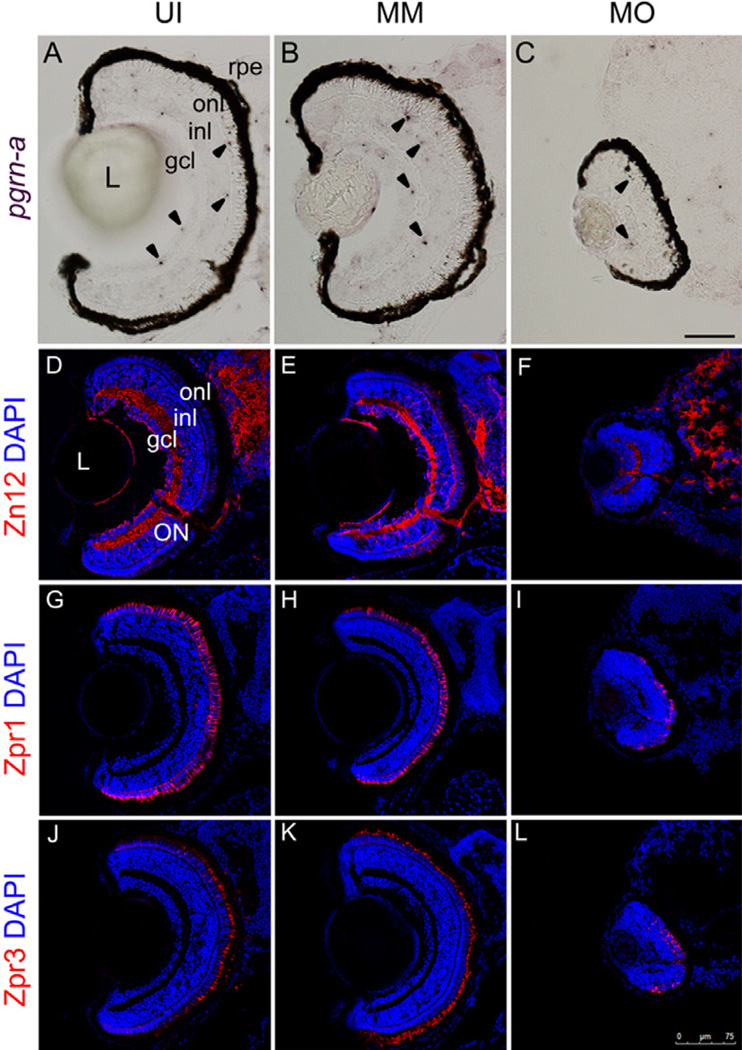
The effect of a morpholino knockdown is temporary, and the effect disappears after 5 days. Morphants from a single clutch were randomly selected and sacrificed at 7dpf. By performing in situ hybridization on sections using the microglia-specific mRNA probe pgrn-a, we observed that the microglia in both the brain and the retina in fms morphants at 7 dpf (Fig. 4C). These data suggest that the effect of this csf-1r morpholino was to temporarily delay the migration of the macrophages. We then checked the expression of Zn12, Zpr1 and Zpr3 in the uninjected, mismatch controls and fms morphants to evaluate the differentiation and development of the ganglion cells and photoreceptors at 7 dpf. Within the cohort examined at 72 hpf, there was little neuronal differentiation in the retina; in contrast, by 7 dpf, the retinas of the fms morphants were laminated, and the retinal neurons expressed the appropriate cell type-specific markers (Fig. 4F, I and L), although the eyes of the morphants continued to be smaller than those of the uninjected and mismatch controls.
Figure 4. Recovery of microglia migration into the retina and brain results in delayed cellular differentiation at 7 dpf.
Panels A–C illustrate expression patterns of progranulin-a (pgrn-a) by in situ hybridization in uninjected (UI), mismatch controls (MM) and fms morphants (MO) at 7 dpf. Note that the microglia (arrowheads) have already migrated and colonized into the brain and retina in fms morphants at 7 dpf, although the morphants continue to have smaller eyes. Larvae are allowed to survive to 7 dpf and are stained with cell type-specific antibodies: Zn12 for ganglion cells(D–F), Zpr1 for cones (G–I) and Zpr3 for rods(J–L). The retinas of the fms morphants expressed the appositional cell type-specific markers (F, I, L). L, lens; gcl, ganglion cell layer; inl, inner nuclear layer; onl, outer nuclear layer; rpe, retinal pigment epithelium; ON, optic nerve. Scale bar: A–C, 50µm; D–L, 75 µm.
Following the knockdown of fms, the retina progenitors fail to exit the cell cycle, and the retina persists as a proliferative neuroepithelium. However, escape from the inhibition of fms translation results in an incomplete recovery of the neuronal differentiation. The spatial patterns of gene expression in morphant retinas suggested that the neurogenesis was not stopped by the fms morpholino but was only delayed. The absence of microglia did not alter positional information, cell identities or cell fate decisions in the retina; rather, microglia were required for retinal progenitors to initiate the switch from proliferation to differentiation.
Overall, our data indicate that the cell cycle of retinal progenitor cells can be disrupted by microglia, although the competency of retinal progenitor cells is intrinsically determined. The further identification and testing of the molecular determinants of neuronal differentiation in the zebrafish retina should yield results that will have implications for understanding the regulation of microglia in the developing zebrafish retina.
Highlights.
The delayed migration of macrophages into the retina causes microphthalmia.
The absence of microglia affects onset of retinogenesis and neuro differentiation.
Microglia function as essential regulators of neurogenesis in the zebrafish retina.
Acknowledgements
The authors thank Laura Kakuk-Atkins and Dilip Pawar for technical assistance. This work was supported by National Institutes of Health Grants R01 EY07060 (PFH) and the Fundamental Research Funds for the Central Universities 65012211 (YL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 2.Hailer NP. Immunosuppression after traumatic or ischemic CNS damage: It is neuroprotective and illuminates the role of microglial cells. Prog. Neurobiol. 2008;84:211–233. doi: 10.1016/j.pneurobio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Langmann T T. Microglia activation in retinal degeneration. J. Leukocyte Biol. 2007;81:1345–1351. doi: 10.1189/jlb.0207114. [DOI] [PubMed] [Google Scholar]
- 4.Parichy DM, Ransom DG, Paw B, Zon LI, Jonhson SL. An rothologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish Danio rerio. Development. 2000;127:3031–3144. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 5.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 6.Cui Q, Yin Y, Benowitz LI. The role of macrophages in optic nerve regeneration. Neuroscience. 2009;158:1039–1048. doi: 10.1016/j.neuroscience.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick AD. Influence of microglia on retinal progenitor cell turnover and cell replacement. Eye. 2009;23:1939–1945. doi: 10.1038/eye.2008.380. [DOI] [PubMed] [Google Scholar]
- 8.Craig SEL, Calinescu AA, Hitchcock PF. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebrafish. J. Ocul. Biol. Dis. Inform. 2008;1:73–84. doi: 10.1007/s12177-008-9011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasevicius A, Ekker SC. Effective targeted gene 'knockdown' in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 10.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochocinska MJ, Hitchcock PF. Cellular expression of the bHLH transcription factor, neuroD, in the retina of the embryonic and larval zebrafish. J. Comp. Neurol. 2007;501:1–12. doi: 10.1002/cne.21150. [DOI] [PubMed] [Google Scholar]
- 12.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 13.Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130:817–833. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J. Comp. Neurol. 1999;404:515–536. [PubMed] [Google Scholar]
- 15.Hu M, Easter SS. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev. Biol. 1999;207:309–321. doi: 10.1006/dbio.1998.9031. [DOI] [PubMed] [Google Scholar]
- 16.Hitchchck PF, Raymond PA. The Teleost Retina as a Model for Developmental and Regeneration Biology. Zebrafish. 2004;1:257–271. doi: 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- 17.Pittman AJ, Law MY, Chien CB. Pathfinding in a large vertebrate axon tract: isotypic interactions guide retinotectal axons at multiple choice points. Development. 2008;135:2865–2871. doi: 10.1242/dev.025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Easter SS, Malicki JJ JJ. The zebrafish eye: developmental and genetic analysis. Results Probl. Cell. Differ. 2002;40:346–370. doi: 10.1007/978-3-540-46041-1_17. [DOI] [PubMed] [Google Scholar]
- 19.Bailey TJ, Fossum SL, Fimbel SM, Montgomery JE, Hyde DR. The inhibitor of phagocytosis, O-phospho-L-serine, suppresses Müller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Exp. Eye Res. 2010;91:601–612. doi: 10.1016/j.exer.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JA, Goldman D, Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J. Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin Z, Barthel LK, Raymond PA. Genetic evidence for shared mechanisms of epimorphic regeneration in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9310–9315. doi: 10.1073/pnas.0811186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson SM, Park L, Stenkamp DL. Retinal homeobox 1 is required for retinal neurogenesis and photoreceptor differentiation in embryonic zebrafish. Dev. Biol. 2009;328:24–39. doi: 10.1016/j.ydbio.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell C, Elsaeidi F, Goldman D. Injury-dependent Müller glia and ganglion cell reprogramming during tissue regeneration requires Apobec2a and Apobec2b. J. Neurosci. 2012;32:1096–1109. doi: 10.1523/JNEUROSCI.5603-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locker M, Agathocleous M, Amato MA, Parain K, Harris WA, Perron M. Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]