Abstract
The cell biological principles that govern innate immune responses in Drosophila are unknown. Here, we report that Toll signaling in flies was dictated by the subcellular localization of the adaptor protein dMyD88. dMyD88 was located at the plasma membrane by a process dependent on a C-terminal phosphoinositide-binding domain. In vivo analysis revealed that lipid binding by dMyD88 was necessary for its antimicrobial and developmental functions, as well as for the recruitment of the downstream cytosolic adaptor Tube to the cell surface. These data are reminiscent of the interactions between the mammalian Toll adaptors MyD88 and TIRAP with one major exception. In the mammalian system, MyD88 is the cytosolic adaptor that depends on the phosphoinositide-binding protein TIRAP for its recruitment to the cell surface. We therefore propose that dMyD88 is the functional homologue of TIRAP, and that both proteins function as sorting adaptors to recruit downstream signaling adaptors to activated receptors.
Introduction
An emerging theme in the study of innate immune signal transduction is the importance of subcellular localization for protein function. Observations in support of this claim were first made through the study of the Toll-like Receptors (TLRs), which can be found at the plasma membrane as well as on various endosomal organelles (Barton and Kagan, 2009). Recently, this principle has been extended to the cytosolic signaling proteins that act downstream from TLRs (Kagan and Medzhitov, 2006; Kagan et al., 2008; Rowe et al., 2006). It is at this level that the first mechanistic insight has come to explain the link between subcellular localization and protein function. For example, TLR4 activates two signaling pathways that are dictated by the localization of the TIR domain-containing adaptor pairs, TIRAP-MyD88 and TRAM-TRIF (Barton and Kagan, 2009). Signaling induced by the TIRAP-MyD88 pair occurs from the cell surface and mediates the expression of inflammatory cytokines by a process dependent on NF-κB and AP-1 (Horng et al., 2002; Kagan and Medzhitov, 2006; Yamamoto et al., 2002). TIRAP contains a lipid-binding domain that permits its localization to phosphatidylinositol 4,5-bisphosphate (PIP2) rich regions of the plasma membrane (Kagan and Medzhitov, 2006). By binding these PIP2-rich regions of the cell surface, TIRAP is positioned to recruit, or “sort,” MyD88 to this location in order to promote signaling. While MyD88 interacts directly with the downstream enzyme IRAK4 to induce signal transduction, TIRAP cannot. Thus, TIRAP is considered a sorting adaptor, whose primary function is to deliver the signaling adaptor MyD88 to plasma membrane-localized TLRs (Barton and Kagan, 2009). Similar cell biological rules can be applied to the other TIR-containing adaptor pair, TRAM-TRIF. This adaptor pair mediates the expression of Type I Interferons (IFN) from endosomes following microbe-induced endocytosis of TLR4 by a process dependent on IRF3 (Doyle et al., 2002; Kagan et al., 2008; Tanimura et al., 2008). TRAM contains a bipartite localization motif consisting of a phosphoinositide-binding domain and a myristoylation motif, which permits its delivery to the plasma membrane and endosomes (Kagan et al., 2008; Rowe et al., 2006). It is from this latter location that TRAM engages activated TLR4 and induces the recruitment of TRIF (Kagan et al., 2008; Tanimura et al., 2008). Like MyD88, TRIF is a bona fide signaling adaptor in that it interacts directly with downstream signaling enzymes, such as the E3 ubiquitin ligases TRAF3 and TRAF6 (Hacker et al., 2006; Sato et al., 2003). TRAM cannot directly bind to these factors. Thus, like TIRAP, TRAM is considered a sorting adaptor, whose function is to recruit TRIF to endosome-localized TLR4 to promote signal transduction.
While the sorting-signaling adaptor paradigm has emerged as an important regulatory process in mammalian innate immunity, its significance in lower eukaryotes is less clear. The reason for this confusion is that no known homologs of the sorting adaptors exist in lower eukaryotes. For example, Drosophila melanogaster has long been the model organism of choice to study evolutionary aspects of innate immunity (Hoffmann, 2003), but neither TIRAP nor TRAM have been identified in flies. Despite the lack of identified sorting adaptors in flies, Drosophila Toll, like most mammalian TLRs, requires two adaptor molecules to link it to the dowstream kinases of the IRAK/Pelle family (Sun et al., 2002; Sun et al., 2004). Since Drosophila Toll requires two adaptor proteins, we considered the possibility that one of these might serve a sorting adaptor function. The receptor proximal adaptor shows substantial sequence homology to the mammalian MyD88 protein, and thus, was named dMyD88 (or dmMyD88) (Horng and Medzhitov, 2001; Tauszig-Delamasure et al., 2002). The other adaptor, Tube, has no known mammalian homolog. Interestingly, biochemical analysis indicates that the interactions that the conserved adaptor MyD88 undergoes during signal transduction differ between mammals and flies. For example, in mammals, MyD88 binds directly to IRAK family members (Lin et al., 2010; Motshwene et al., 2009), but in flies, dMyD88 cannot bind directly to the IRAK homologue Pelle (Sun et al., 2002). Rather, Tube serves this function with dMyD88 only binding indirectly to Pelle (Sun et al., 2002).
Because the known sorting adaptors are not found in lower eukaryotes, the question remains as to whether or not the subcellular positioning of adaptors is only important for Toll signaling in mammals. Considering that sorting adaptors bind indirectly to signaling enzymes in mammals, as well as the fact that the known adaptor protein Tube lies between dMyD88 and Pelle (Sun et al., 2002), we considered the possibility that MyD88 may have different functions in mammals and flies. We hypothesized that unlike its mammalian namesake, dMyD88 may function analogously to TIRAP by serving the role of a sorting adaptor in Drosophila Toll signaling. Herein, we show that dMyD88 contains a C-terminal phosphoinositide-binding motif that directs it to the plasma membrane. Moreover, we demonstrate the functional importance of this localization domain in vivo and establish that signaling is at least initiated from the plasma membrane by dMyD88’s ability to recruit downstream signaling components to the cell surface. We propose that the use of sorting-signaling adaptor pairs in Toll signaling predates mammals, and that the cell biological principles that govern innate signaling pathways represent a fundamental rule throughout evolution.
Results
dMyD88 is Localized to the Plasma Membrane
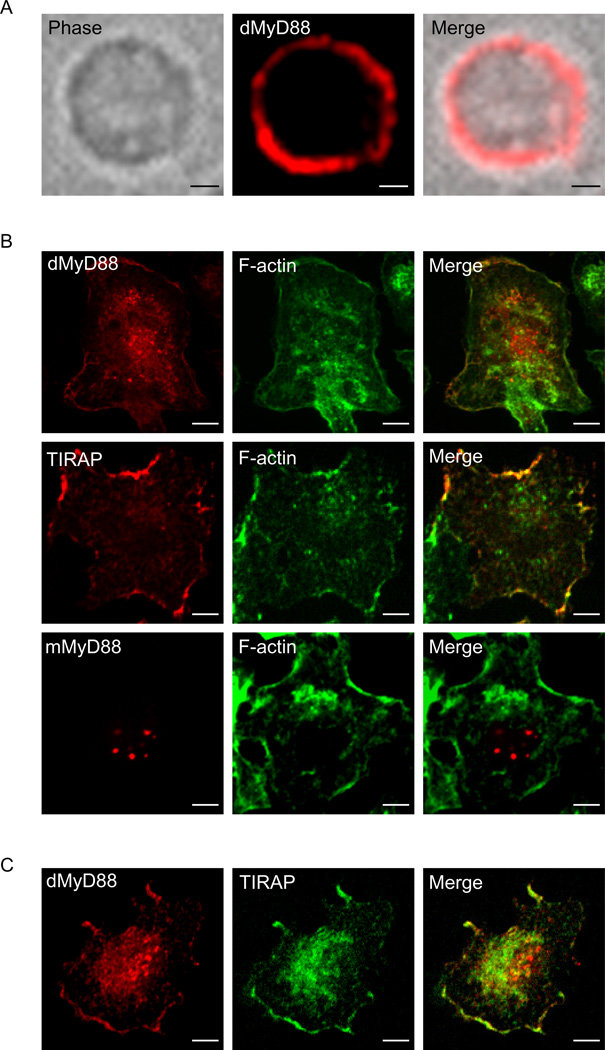
A defining feature of mammalian sorting adaptors is their ability to localize to regions of the cell that contain TLRs even before signaling has been initiated (Barton and Kagan, 2009; Kagan and Medzhitov, 2006; Kagan et al., 2008). This function is thought to facilitate rapid detection of activated receptors and promote the recruitment of downstream adaptors to these sites. dMyD88 has been found at the plasma membrane in Drosophila embryos (Sun et al., 2004), but whether this localization was simply the result of developmental Toll signaling in vivo is unknown. To address the intrinsic pre-signaling localization of dMyD88, we examined the subcellular distribution of this adaptor in Drosophila S2 cells. In fly cells, dMyD88 was localized to the plasma membrane (Figure 1a and Figure S1). Similarly, dMyD88 was found to be enriched at plasma membrane ruffles in primary mouse macrophages (Figure 1b), suggesting that dMyD88 does not require any fly-specific factors for its plasma membrane recruitment. Similar results were obtained when examining the localization of TIRAP (Figure 1b), whereas mouse MyD88 (mMyD88) was found in foci scattered throughout the cell (Figure 1b). The fact that dMyD88 localization appeared similar to that of TIRAP prompted us to ask if these two proteins were recruited to similar regions of the plasma membrane. To address this question, we examined the distribution of dMyD88 and TIRAP within the same cell. dMyD88 and TIRAP were both concentrated at overlapping regions of membrane ruffles in wild type (WT) mouse macrophages (Figure 1c), raising the possibility that these adaptors utilize a similar mechanism of protein localization. Collectively, these data indicate that dMyD88 resides at the plasma membrane, and that recruitment of dMyD88 to the cell surface does not require a fly specific factor.
Figure 1. dMyD88 localizes to the plasma membrane.
(a) S2 cells were transfected with FLAG-dMyD88 and stained with FLAG antibodies. (b) Fluorescence micrographs of primary mouse macrophages transfected with dMyD88-YFP, FLAG-TIRAP, or FLAG-mMyD88. Cells were co-stained with FLAG antibodies and Alexa Fluor-647 phalloidin in order to visualize F-actin. As evidenced by the co-staining with F-actin in the merged images, dMyD88 and TIRAP both localize to membrane ruffles. (c) Fluorescence micrographs of primary mouse macrophages expressing TIRAP-GFP and FLAG-dMyD88. Note that both TIRAP and dMyD88 are concentrated in overlapping regions of membrane ruffles. All images are representative of at least three independent experiments where over 500 cells were examined per condition and >95% of the cells displayed similar staining. Scale bars represent 5µm. See also Figure S1.
dMyD88 is a Phosphoinositide-Interacting Protein
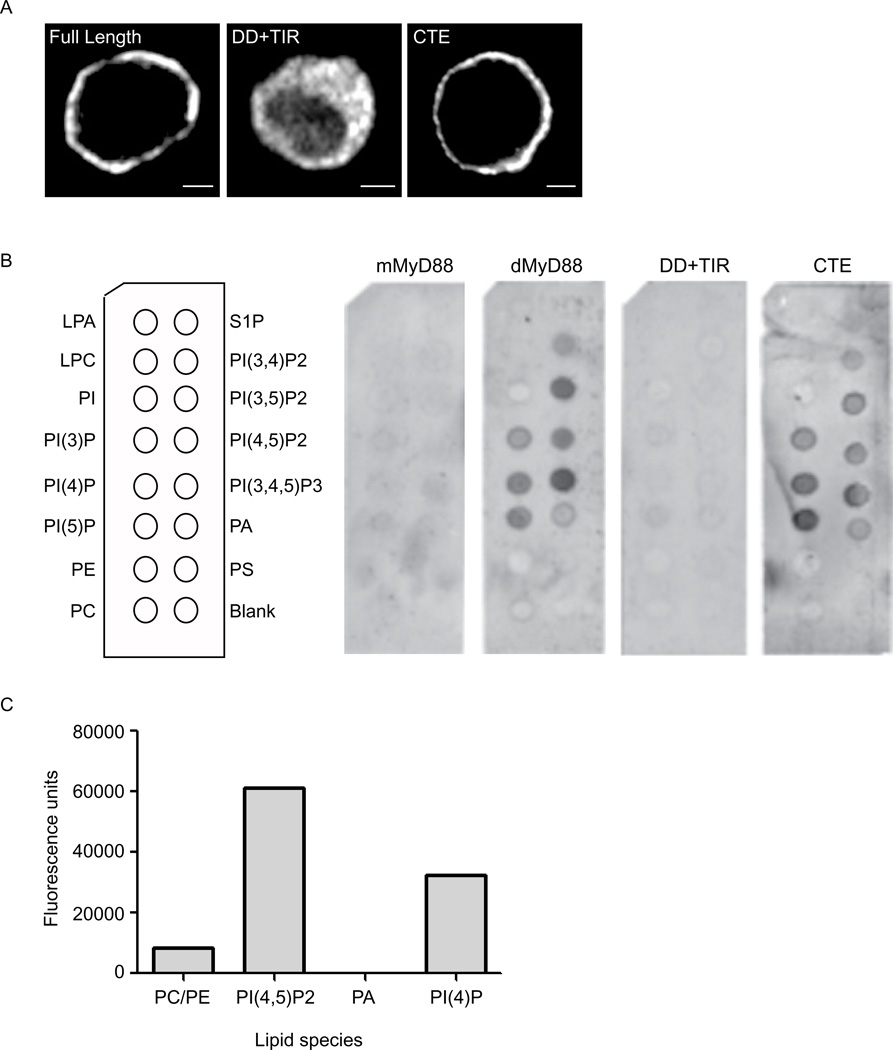
Sequence homology had originally identified dMyD88 as the Drosophila homolog of mammalian MyD88 (Horng and Medzhitov, 2001; Tauszig-Delamasure et al., 2002). Both proteins contain an N-terminal death domain and a TIR domain. However, unlike its mammalian counterpart, dMyD88 contains a C-terminal extension (CTE) that does not have similarity to any known protein domains. We hypothesized that this CTE was responsible for dMyD88 localization to the plasma membrane. To test this hypothesis, deletion mutants were generated, and the resulting proteins were examined for their ability to phenocopy the localization of full length dMyD88. Consistent with our hypothesis, the CTE was sufficient to phenocopy full length dMyD88 localization (Figure 2a). In contrast, dMyD88 mutants lacking the C-terminal domain (DD+TIR) were cytosolic (Figure 2a). These results provide evidence that membrane localization of dMyD88 occurs independently of its signaling domains. Thus, dMyD88 exhibits a property common to other sorting adaptors, as both TIRAP and TRAM use TIR-independent means of membrane localization (Kagan and Medzhitov, 2006; Kagan et al., 2008; Rowe et al., 2006).
Figure 2. dMyD88 localization is dictated by a C-terminal phosphoinositide binding domain.
(a) Fluorescence micrographs of S2 cells transfected with either full length dMyD88 (FL) or various dMyD88 deletion mutants. The N-terminal construct (DD+TIR) contains amino acids 1–386, whereas amino acids 375–537 comprise the CTE construct. Scale bars represent 5µm. (b) PIP strips of various lipids (shown in left panel) were incubated with GST-tagged full length dMyD88, mouse MyD88 (mMyD88), and the dMyD88 deletion mutants described in (a). dMyD88 and its CTE interact with all PIs tested, whereas mMyD88 and the N-terminus (DD+TIR) do not. (c) Quantitative analysis of dMyD88’s ability to bind liposomes containing 18% of the indicated phosphoinositide. Shown is a representative experiment (n=6) demonstrating that dMyD88 interacts preferentially to PIP2 when compared to other phospholipids. See also Figure S2.
Examination of the localization motif of dMyD88 revealed that this region had a higher isoelectric point than the rest of the protein (10.16 and 6.71, respectively). The localization domains of TIRAP and TRAM also possess this characteristic, which is a feature found in domains that bind phospholipids (McLaughlin et al., 2002). Since both TIRAP and TRAM are phosphoinositide-interacting proteins (Kagan and Medzhitov, 2006; Kagan et al., 2008), we performed in vitro protein-lipid interaction assays to assess the ability of dMyD88 to bind to membrane lipids. More specifically, we used PIP strips and PIP arrays, which are commercially available nitrocellulose membranes that contain a variety of lipid species (Figure 2b and Figure S2). Glutathione S-transferase (GST) tagged dMyD88, but not mouse MyD88, bound detectably to all the negatively-charged phosphoinositides (Figure 2b). This interaction with phosphoinositides was specific, as dMyD88 did not interact with unphosphorylated phosphatidylinositol or any of the general membrane lipid species examined, such as phosphatidylethanolamine, phosphatidylcholine, or phosphatidylserine (Figure 2b). Consistent with our hypothesis that the C-terminal localization domain is responsible for this interaction, the CTE of dMyD88 bound the same phosphoinositides as the full length protein (Figure 2b). In contrast, the N-terminal portion of dMyD88 (DD + TIR) was defective for lipid binding in vitro (Figure 2b). As an independent approach, we examined the ability of GST-dMyD88 to bind liposomes containing either PI(4)P, PI(4,5)P2 or PA. GST-dMyD88 interacted preferentially with liposomes containing PI(4,5)P2 and to a lesser extent liposomes containing PI(4)P (Figure 2c). Similar results were obtained when the CTE was used in this pulldown assay (data not shown). Taken together, these data indicate that dMyD88 is a phosphoinositide-interacting protein and that the CTE, which is responsible for its localization, is also responsible for this interaction.
The C-terminal Phosphoinositide Binding Domain of dMyD88 is Necessary to Control Infections in Flies
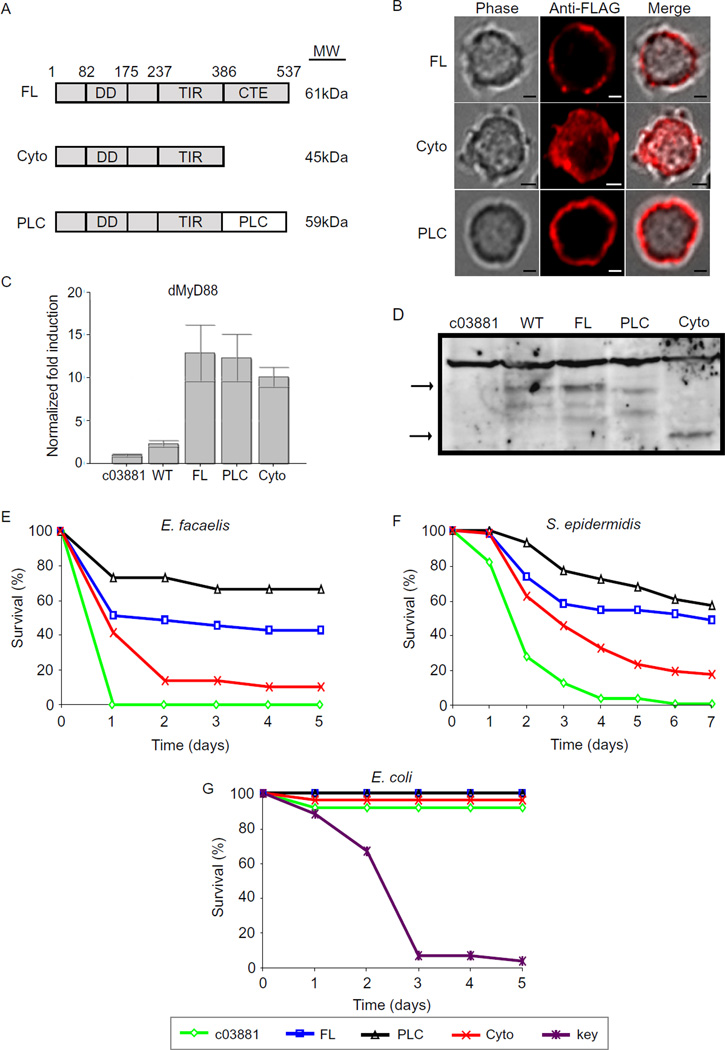
It is very common for phosphoinositide-binding proteins to exhibit promiscuous binding in vitro, but a single lipid usually mediates protein localization in vivo (Lemmon, 2003). To determine which lipid species is likely mediating dMyD88 localization within cells, we generated a series of chimeric alleles where the natural lipid binding domain was replaced with domains that interact with single phosphoinositides. We focused specifically on lipids that are found to some extent at the plasma membrane as this is the natural site of dMyD88 residence. In S2 cells, chimeric dMyD88 proteins that bind specifically to PI(4)P through the attachment of the pleckstrin homology (PH) domain of FAPP1 did not phenocopy the localization of the WT allele (Figure S3a). In addition, neither the PI(3,4,5)P3 specific domains (PH domain of GRP1) nor the PI(3)P-specific domain (PX domain of Gp91phox) exhibited a localization similar to WT dMyD88 (Figure S3a). In contrast, the dMyD88 chimera containing a PI(4,5)P2 (PIP2) specific PH domain from PLCδ1 (dMyD88-PLC) phenocopied the localization of the WT protein (Figure 3 and Figure S3a). The PLCδ1 PH domain was chosen for our studies because it is one of the only known domains that interact specifically with PIP2 in vitro and in vivo (Lemmon, 2003). This specificity is due it its unusually high affinity for this lipid. Thus, the fact that dMyD88-PLC phenocopied the localization of WT dMyd88 made PIP2 a likely candidate for the in vivo regulator of dMyD88 localization.
Figure 3. Phosphoinositide binding by dMyD88 is necessary for immune defenses in flies.
(a) Schematic representation of the dMyD88 constructs used to generate transgenic flies. The molecular weights (MW) of the resulting proteins are indicated. (b) Fluorescence micrographs of S2 cells expressing the indicated dMyD88 constructs. The localization of full length dMyD88 (FL) is phenocopied by the PIP2-specific chimera (dMyD88-PLC). Scale bars represent 5µm. (c,d) Lysates from dMyD88 homozygous mutant (c03881), wild type dMyD88 heterozygote (WT), and dMyD88 homozygous mutant flies complemented with the transgenes described in (a) were analyzed either by immunoblotting with a dMyD88 antibody (obtained from S. Wassermann (Sun et al., 2002) or by performing RT-PCR using dMyD88 Taqman probes. No significant difference in RNA or protein amounts was observed between the various transgenic flies. (e, f, g) Male flies of the indicated genotype were infected with E. facaelis (e), S. epidermidis (f), or E. coli (g). Each infection was done with at least 25 flies for each genotype, and the surviving flies were counted daily. Log-rank analysis demonstrated a statistically significant difference between survival of dMyD88-FL flies and dMyD88 mutants (p=<0.0001) for both E. facaelis and S. epidermidis infections, but not following E. coli infection (p=0.1986). Survival of dMyD88-FL flies and dMyD88-Cyto flies was statistically significant following treatment with E. facaelis (p=0.0091) and S. epidermidis (p= 0.0020), but not following E. coli infection (p=0.2673). The difference between survival of dMyD88-FL flies and dMyD88-PLC was not significant for any of the above infections. See also Figure S3.
To determine if PIP2 binding by dMyD88 is sufficient for its signaling functions, we reconstituted dMyD88 mutant flies with the dMyD88-PLC allele. We also reconstituted dMyD88 mutant flies with the full length dMyD88 allele (dMyD88-FL) or an allele lacking its localization domain (dMy88-Cyto). The collection of these transgenic lines permits the side-by-side examination of the functional consequence of altering the affinity of dMyD88 for phosphoinositides, in particular PIP2. All transgenic fly lines produced comparable amounts of dMy88 RNA and their respective dMyD88 proteins (Figure 3c & Figure 3d), and the alleles used to generate these lines displayed the expected subcellular distributions in S2 cells (Figure 3b).
Because dMyD88 mutant flies are highly susceptible to infection by Gram-positive bacteria (Kambris et al., 2003; Tauszig-Delamasure et al., 2002), the transgenic lines were tested for their ability to rescue this immune phenotype. None of the parental lines showed a difference in survival following E. facaelis infection, suggesting that any variation between the dMyD88 mutant flies complemented with the dMyD88 transgenes reflect the expression of the various dMyD88 alleles and not the insertion of the transgenes (Figure S3b). As expected, infection by either E. faecalis or S. epidermidis caused rapid and complete lethality of dMyD88 mutant flies, and this phenotype was complemented by the expression of full length dMyD88 (Figure 3e & 3f). In contrast, flies expressing the cytosolic allele (dMy88-Cyto) displayed reduced resistance to infection (Figure 3e & 3f). These results indicate that plasma membrane localization is necessary for the immune functions of dMyD88. Interestingly, dMyD88-PLC flies survived even better than their dMyD88-FL counterparts (Figure 3e & 3f), suggesting that PIP2 binding by dMyD88 is sufficient for the immune functions of this adaptor. The possibility that the PLCδ1 PH domain has any intrinsic signaling functions was ruled out by assaying its ability to rescue Toll signaling in dMyD88-deficient S2 cells. Unlike the cells expressing dMyD88-FL and dMyD88-PLC, no significant Drosomycin expression was observed in cells expressing the PLCδ1 PH domain alone (Figure S3c). To confirm that non-Toll immune pathways were unaffected by the dMyD88 transgenes, infections were performed with the Gram-negative bacteria E.coli, which is eradicated by the Imd pathway (Leulier et al., 2000). None of the transgenic lines were sensitive to infection by E. coli (Figure 3g), whereas the Imd pathway mutant Kenny succumbed to infection by this bacteria.
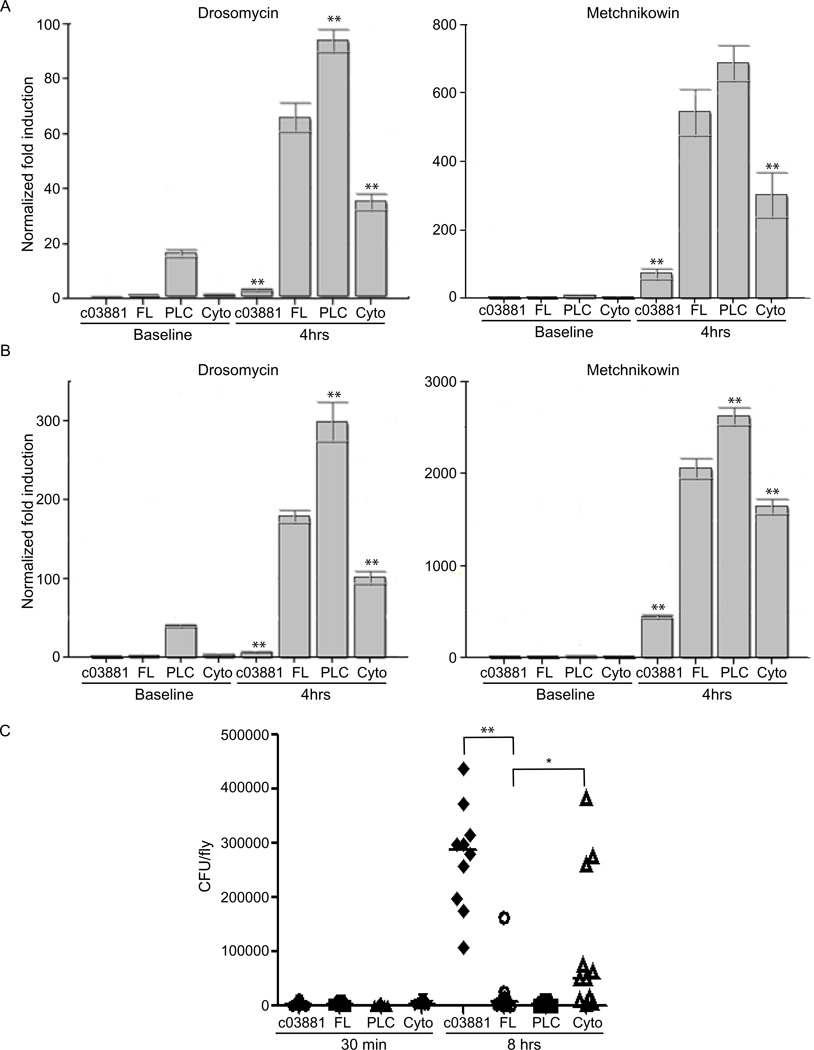
To confirm that the observed survival defect in dMyD88-Cyto flies resulted from reduced Toll signaling, we measured the expression of antimicrobial peptides (AMP) known to be induced following activation of the Toll pathway (De Gregorio et al., 2002). Consistent with a defect in Toll signaling, dMyD88 mutant flies expressing dMyD88-Cyto displayed decreased expression of both Drosomycin (Drs) and Metchnikowen (Mtk) in response to infection with either E. facaelis or S. epidermidis (Figure 4a and 4b). However, induction of Diptericin (Dpt), an AMP induced by the Imd pathway, was not affected (Figure S4). Interestingly, dMyD88-PLC displayed higher expression of Toll-activated AMPs both at baseline and following infection. This observation likely explains the heightened resistance to bacterial infection exhibited by flies expressing this transgene.
Figure 4. Phosphoinositide binding by dMyD88 is necessary for AMP expression and restricting bacterial load.
(a, b) dMyD88 homozygous mutant flies complemented with dMyD88-FL, dMyD88-PLC, and dMyD88-Cyto were challenged by septic injury with E. facaelis (a) or S. epidermidis (b). Total RNA extracts from 10 flies of each genotype were analyzed for Drosomyocin and Metchnikowin induction by quantitative RT-PCR both at baseline and 4 hours after infection. (c) Individual male flies of the indicated genotype were injected with E. facaelis and homogenized 0.5 or 8 hours after infection. Serial dilutions of the homogenized flies were plated on kanamycin-containing agar plates to determine bacterial loads. Due to large variances, the data in (c) was log transformed for statistical analysis. *p<0.05, **p<0.01, Student’s t-test. See also Figure S4.
We hypothesized that the differences in survival after infections may result from an inability to kill and clear the bacteria. To address this possibility, male flies were challenged with E. facaelis, and individual flies were homogenized to measure their bacterial loads. Consistent with the survival defect, dMyD88-Cyto flies were unable to control the infection, as a higher bacterial load was observed in these mutant flies when compared to flies that readily control the infection (FL or PLC counterparts) (Figure 4c). However, as compared to flies that display dMyd88 at the plasma membrane (FL or PLC), the bacterial load in dMyD88-Cyto flies varied greatly from fly to fly with some flies having bacterial loads as high as the null mutants and others having bacterial loads as low as dMyD88-FL flies. These results suggest that a diffusion mediated process of dMyD88 recruitment results in an unreliable antimicrobial response, which likely explains the decreased ability of dMyD88-Cyto flies to survive bacterial infections. Collectively, these data indicate that membrane localization is necessary for maximal Toll signaling in Drosophila, and that the C-terminal phosphoinositide binding of dMyD88 is both necessary and sufficient for dMyD88 localization and antimicrobial functions.
The C-terminal Phosphoinositide Binding Domain of dMyD88 is Necessary for Toll Signaling in Development
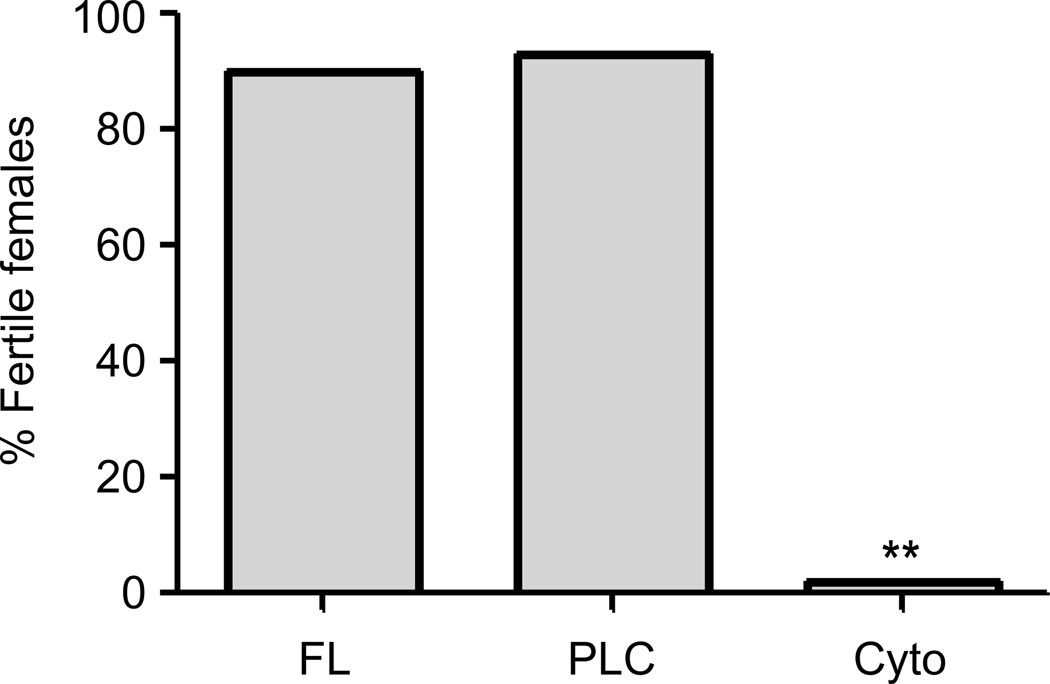
In addition to its role in fly immunity, dMyD88 is maternally required for the establishment of the dorsoventral axis during Drosophila embryogenesis (Charatsi et al., 2003; Kambris et al., 2002). This need for maternal deposition of the dMyD88 transcript renders homozygous dMyD88 mutant females sterile (Charatsi et al., 2003; Kambris et al., 2002). Thus, to test if PIP2 binding by dMyD88 is necessary for development, homozygous dMyD88 mutant flies expressing the various trangenes were tested for their ability to rescue the sterility phenotype observed in dMyD88 homozygous females. Consistent with our finding that plasma membrane localization is necessary for the function of dMyD88, females expressing the cytosolic allele (dMyD88-Cyto) failed to produce viable progeny (Figure 5). In contrast, both the full length and the PLC alleles restored female fertility (Figure 5). Taken together, these data indicate that plasma membrane localization of dMyD88 is necessary for its developmental function.
Figure 5. The phosphoinositide binding domain of dMyD88 is necessary for female fertility.
dMyD88 homozygous mutant flies complemented with dMyD88-FL, dMyD88-PLC, and dMyD88-Cyto using an ovary-specific driver (V32-Gal4) were put to lay and assayed for the their ability to make viable progeny. The sterility of dMyD88 homozygous females is complemented by dMyD88-FL and dMyD88-PLC, but not dMyD88-Cyto. In all cases, at least 50 individual females were scored. **p<0.01, Fisher’s exact test.
dMyD88 Functions to Recruit Tube to the Plasma Membrane
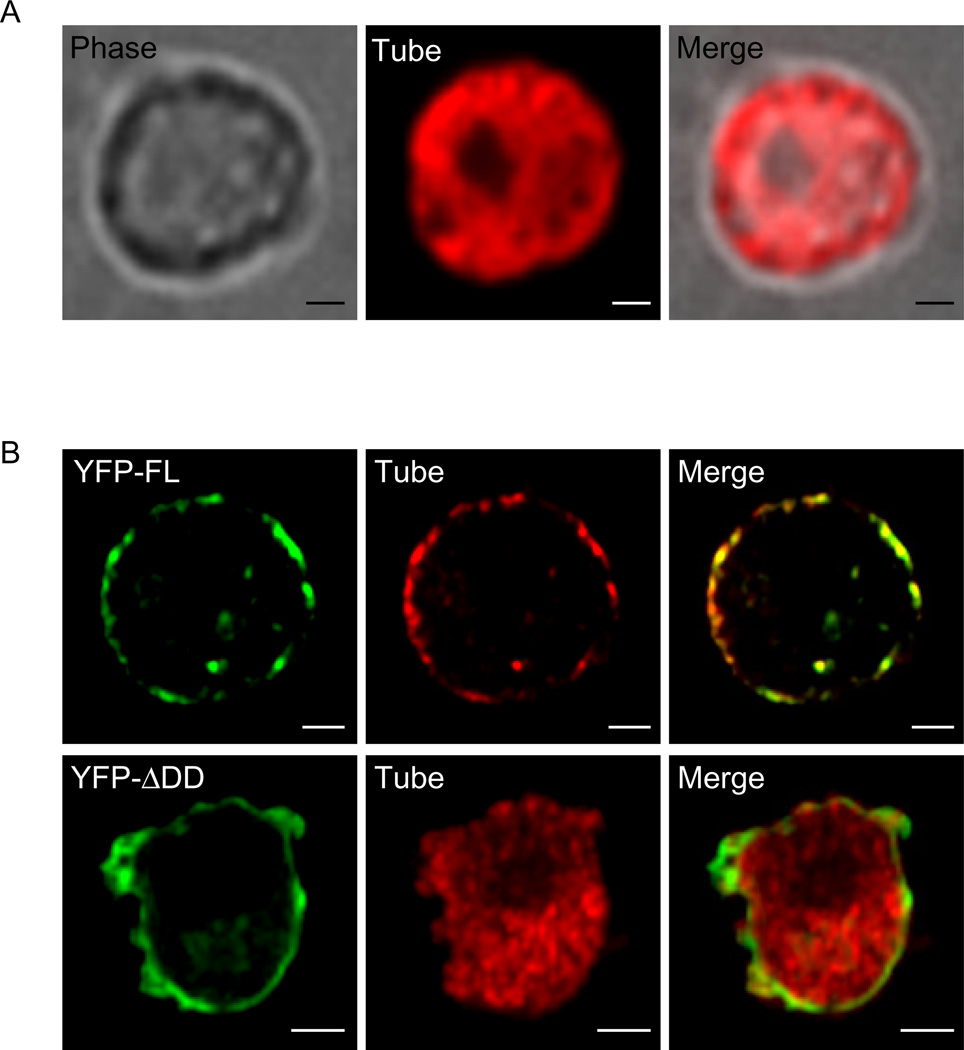
Until recently, it was assumed that Toll signaling occurred at the plasma membrane. Our data fully support this idea and suggest that PIP2-rich regions of the plasma membrane are the preferred site of Toll signaling. However, recent reports claim that Toll signaling occurs from an endocytic compartment (Huang et al., 2010; Lund et al., 2010). We reasoned that if dMyD88 is indeed functioning as a sorting adaptor, then it must recruit a signaling adaptor to the site of signaling. Given the genetic and biochemical data placing the adaptor Tube downstream of dMyD88 (Sun et al., 2002; Sun et al., 2004), we hypothesized that Tube would serve the role of the signaling adaptor. To address this prediction, we first needed to know where in the cell Tube resides. Expressing Tube in S2 cells revealed that Tube is found to be evenly distributed throughout the cytosol (Figure 6a). Since Tube localization differed from dMyD88, we next asked if dMyD88 was capable of recruiting Tube to the plasma membrane in a signaling domain dependent manner. To address this question, we co-expressed Tube with full length dMyD88 or a dMyD88 mutant lacking the death domain. Under both conditions, dMyD88 localized to the plasma membrane, further validating that its localization is independent of its signaling domains. Tube, on the other hand, was no longer observed to be cytosolic when co-expressed with full length dMyD88. Rather, Tube co-localized with full length dMyD88 at the plasma membrane (Figure 6b). This co-localization was not observed when Tube was expressed with the dMyD88 mutant lacking the death domain, indicating that a signaling domain is necessary for the observed cell surface recruitment (Figure 6b). Collectively, our results, along with previous in vivo data showing dMyD88-dependent Tube localization at the plasma membrane (Sun et al., 2004), suggest that signaling is at least initiated at the plasma membrane and that the subcellular localization of dMyD88 prepositions this adaptor to recruit downstream signaling proteins.
Figure 6. dMyD88 recruits Tube to the plasma membrane.
(a) S2 cells were transfected with FLAG-Tube and stained with FLAG antibodies. (b) S2 cells were co-transfected with either full length dMyD88 (YFP-FL) or a dMyD88 mutant lacking the death domain (YFP-ΔDD) and Tube. The dMyD88 vectors were under the control of the metallothionein promoter; thus, to induce expression of these constructs, 500µM CuSO4 was added to the cells 6 hours after transfection. Forty-eight hours after CuSO4 induction, Tube is redistributed to the cell surface with full length dMyD88, but not the ΔDD mutant. All images are representative of at least three independent experiments where over 500 cells were examined per condition and >95% of the cells displayed similar staining. Scale bars represent 5µm.
The C-terminal Extension is Conserved Throughout Invertebrate Evolution
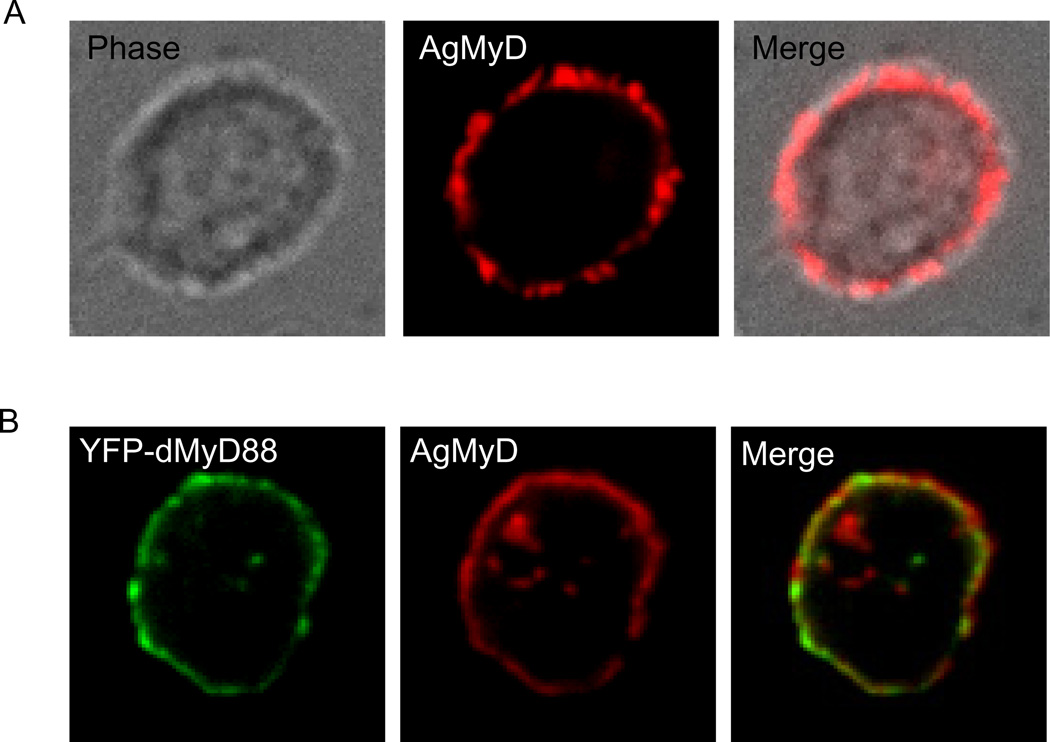
To determine if the properties of dMyD88 represent a common theme in the invertebrate lineage, we analyzed the MyD88 gene from other insect species for the presence of a CTE. All insect species analyzed had a CTE connected to the TIR domain of their MyD88 gene (Figure S5). Interestingly, none of these invertebrate species had a TIRAP homolog (Figure S5). In contrast to what we observed in the insect lineage, among the vertebrate species that we analyzed, none had a MyD88 allele with additional sequence following the TIR domain, but all had a TIRAP homolog (Figure S5). The strong correlation between the presence of the CTE and the absence of a TIRAP homolog supports our hypothesis that this localization domain endows MyD88 with sorting adaptor function. To further test this hypothesis, we cloned the MyD88 gene from a clinically relevant insect species and determined its localization. Specifically, we expressed the MyD88 homolog from Anopheles gambiae (AgMyD) in S2 cells and found that AgMyD localizes to the plasma membrane (Figure 7a). Since AgMyD localized to the cell surface, we were prompted to ask if its distribution overlapped with dMyD88 localization. When co-expressed within the same cell, AgMyD co-localized with dMyD88 at the plasma membrane (Figure 7b). These results suggest that localization of MyD88 to the plasma membrane is a fundamental principle of insect Toll signaling.
Figure 7. The presence of a C-terminal extension is conserved within the insect lineage.
(a) S2 cells were transfected with FLAG-AgMyD and stained with FLAG antibodies. (b) Fluorescence micrographs of S2 cells expressing YFP-dMyD88 and FLAG-AgMyD. All images are representative of at least three independent experiments where over 500 cells were examined per condition and >95% of the cells displayed similar staining. Scale bars represent 5µm. See also Figure S5.
Discussion
Research performed on the innate immune responses of Drosophila has undoubtedly contributed to our current knowledge of the mammalian TLR network. However, in an interesting twist of events, we now have a more advanced understanding of the cell biology of TLR signaling in mammals than the ancestral Toll pathway in flies. For example, it has become increasingly clear that the mammalian TLRs and their adaptors depend on basic cell biological trafficking factors for their subcellular localization and signaling functions. Whether the cell biological “rules” that govern TLR signaling in mammals applies to lower eukaryotes is unknown. A potential explanation for this gap in our knowledge is that the known mammalian adaptors that exhibit interesting cell biological properties, such as the sorting adaptors TIRAP and TRAM, are not found in flies. Although there are no obvious homologs for these sorting adaptors, the receptor proximal complex found in flies displays notable symmetry to the receptor proximal complex found in mammals. For example, both the mammalian and fly pathways rely on two adaptor proteins to bridge the activated receptor to downstream kinases. Based on this comparison, we considered the idea that the fly adaptors serve equivalent roles to the mammalian sorting-signaling adaptor pair. In particular, we hypothesized that dMyD88 functions as a sorting adaptor. In all cell biological assays performed, dMyD88 does not behave as its mammalian namesake, but rather phenocopies the sorting adaptor TIRAP. Like TIRAP, dMyD88 is localized to the plasma membrane at steady state in the absence of Toll signaling, and this plasma membrane localization is mediated by a C-terminal phosphoinositide-binding domain. Moreover, just as TIRAP can recruit MyD88 to the plasma membrane by a process dependent on its signaling domain (Kagan and Medzhitov, 2006), dMyD88 can recruit the downstream adaptor Tube to the plasma membrane by a similar process. Taken together, our cell biological and genetic analyses suggest that dMyD88 is functionally a sorting adaptor.
Although the idea that protein localization is important for adaptor function has attracted much attention, the importance of this localization in preventing infections in vivo has remained unclear. Our findings that transgenic flies containing dMyD88-Cyto exhibit defects in AMP expression, control of bacterial replication, and ultimately survival demonstrate the fundamental importance of sorting adaptor localization for immune defense in vivo. Moreover, our additional finding that dMyD88-PLC can complement the immune defects of dMyD88 mutant flies suggests that the only activity needed within the C-terminal localization domain is the ability to bind PIP2. Thus, we propose that PIP2 in the primary mediator of dMyD88 localization in flies.
Because PIP2 is enriched at the plasma membrane, our suggestion that this lipid is the primary mediator of dMyD88 localization implies that Toll signaling occurs at the cell surface. Recently, however, this idea has been contested by reports indicating that endocytosis is necessary for the proper activation of the Toll pathway (Huang et al., 2010; Lund et al., 2010). Based on its comparison to mammalian TLRs that activate signal transduction after receptor internalization, the discovery that endocytosis is necessary for the Drosophila Toll pathway suggests an evolutionarily conserved role for endocytosis in Toll signaling. Although this parallel is intriguing, there are important differences between these two pathways that indicate that endocytosis does not play similar roles in flies and mammals. One notable difference is that the mammalian TLRs require endocytosis to activate a second signaling pathway that is induced uniquely from endosomes (Kagan et al., 2008; Tanimura et al., 2008). For example, in the best characterized mammalian system, TLR4 first signals from the plasma membrane to activate NF-κB, and then later switches to the IRF3 activating pathway that is induced from endosomes. The Drosophila Toll pathway, on the other hand, is not known to induce distinct signaling pathways. Rather, flies activate a single dMyD88-Tube dependent pathway that results in AMP expression (Tauszig-Delamasure et al., 2002). While endocytosis may be involved in this Drosophila pathway, our results suggest that Toll signaling is at least initiated at the plasma membrane. We provide several lines of evidence to support this idea. First, dMyD88 recruits Tube to the plasma membrane. This recruitment only occurs in a signal dependent manner, as dMyD88 mutants lacking the death domain are incapable of delivering Tube to the cell surface. Secondly, dMyD88-PLC rescues the immune phenotype observed in dMyD88 mutants. As stated above, the PH domain of PLCδ1 binds to PIP2, which is highly enriched at the cell surface and virtually absent from endosomes. In fact, during endocytosis, PIP2 concentrations fall so drastically (Botelho et al., 2000) that it is unlikely that dMyD88-PLC would be present at very high quantities on early endosomes, and thus, unlikely that Toll signaling would be initiated from this internal cellular location in these flies. Furthermore, the fact that dMyD88-PLC signals better even at baseline provides further credence to the hypothesis that signaling must occur at the plasma membrane, as this result implies that dMyD88 localization to the cell surface is a rate-limiting step in the Toll signaling pathway. Collectively, our data suggest that the initial site of Toll signaling is from the plasma membrane, but subsequent endocytic events may extend the duration of the signaling response.
It is intriguing, though, that dMyD88-PLC does signal better than the WT adaptor. As described above, the PH domain from PLCδ1 is an excellent tool to study the sufficiency of PIP2 binding by dMyD88, as it is one of the only known domains that binds PIP2 with high affinity in vitro and in vivo (Lemmon, 2003). If an interaction domain works better to protect against infection, then why did nature choose to use the weaker PIP2 binding domain for dMyD88? One hypothesis is that the weaker PIP2 interacting domain may be evolutionarily advantageous for Toll signaling in flies. For example, it is possible that endocytosis is required for Toll signaling, and that the adaptor complex must be released from the membrane either for downregulation or continuation of the signal. It is also possible that the weaker PIP2 binding domain evolved to avoid constitutively high amounts of AMP production, which have been shown to be deleterious to the animal. For example, chronic activation of the Imd pathway leads to altered commensal populations and reduced lifespan (Libert et al., 2006; Ryu et al., 2008). Although Toll signaling has not be implicated in the regulation of commensal bacteria, it is possible that such strong baseline signaling as observed with dMyD88-PLC may affect the fly’s interaction with its commensal microbiota, and thus, may lead to death by allowing noxious bacterial species to dominate the gut.
In addition to its function in innate immunity, Toll signaling is important for development in Drosophila. The protein weckle is the only cytosolic factor thus far identified that is selectively required for Toll signaling in development, and it has been proposed to function by recruiting dMyD88 to the plasma membrane to engage Toll (Chen et al., 2006). Our data demonstrating that dMyd88 must encode a plasma membrane localization motif for its signaling functions in development suggest that any interactions between weckle and dMyD88 are not sufficient for plasma membrane recruitment and signaling. It is therefore possible that weckle acts at a different stage of Toll signaling, perhaps downstream of the sorting-signaling adaptors dMyD88 and Tube.
Although this study has focused on the role that dMyD88 plays in Toll signaling, our results also highlight the functional significance of Tube. Based on the sorting-signaling adaptor paradigm established in mammalian cells, our data indicate that Tube is the functional equivalent of mammalian MyD88. Classifying Tube as a signaling adaptor strengthens the analogy to the mammalian system, but it raises the question about why there is a need for these sorting-signaling pairs in flies. In the mammalian network, not all TLRs use the same adaptors (Akira and Takeda, 2004). For example, many plasma membrane-localized TLRs utilize both TIRAP and MyD88, whereas only MyD88 is needed for signaling by TLRs found on endosomes. The fact that MyD88 is needed in both subcellular compartments suggests that sorting adaptors, like TIRAP, evolved to recruit MyD88 to the proper signaling location. Based on this evolutionary perspective, we speculate that the need for dMyD88 evolved because Tube functions in additional locations within the cell. Eight other Tolls have been identified in flies (Tauszig et al., 2000), but few (if any) play a role in immunity. It is possible that the other Tolls are needed for various immune responses not yet characterized, and that these immune responses occur in a different cellular location than at the plasma membrane. Thus, based on the analogy to the mammalian system, dMyD88 would have evolved to bring the signaling adaptor Tube to the plasma membrane, whereas other sorting adaptors may be required to recruit Tube to other signaling locations. Further characterization of other Tolls and their dependence on the known adaptor molecules is needed to address this possibility.
Methods
Cell culture, transfections, and immunofluorescence
S2 cells were maintained at 25°C in Schneider’s Drosophila medium (GIBCO) supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were transfected with 1µg of expression vector using Fugene 6 according to the manufacturer’s protocol (Roche). Forty-eight hours after transfection, cells were washed in serum free media and then incubated on alcian blue coverslips for 30 minutes prior to fixation. Cells were fixed at 25°C in 2% paraformaldehyde for 20 minutes, made permeable for 10 minutes using 1X phosphate buffered saline (PBS) + 0.1% Triton-X, and then incubated with block buffer (2% goat serum, 50mM ammonium chloride in PBS) for 30 minutes. Samples were then incubated with the appropriate antibodies diluted in block buffer. Antibody binding was detected using AlexaFluor secondary antibodies from Molecular Probes and visualized with a spinning-disk confocal microscope (Zeiss Axiovert 200M). Images were captured and analyzed using Slidebook 5.0 software (Intelligent Imaging Innovations). Primary bone marrow-derived macrophages from wild type (C57B6) mice were prepared as previously described (Kagan and Roy, 2002). Macrophages were transfected by nucleofection (AMAXA) with the mouse macrophage tranfection reagent. Following transfection, the cells were plated on coverslips and incubated at 37°C for 4 hours prior to fixing. Staining of macrophages was performed as described above for S2 cells, except that AlexaFluor 647-phalloidin (Molecular Probes) was included with the secondary antibody to identify F-actin.
Protein purification and lipid binding assays
GST-fusion proteins were purified from BL21 E. coli using glutathione Sepharose 4B (Amersham) according to the manufacturer's instructions. Purity of each GST preparation was confirmed by SDS-PAGE and Coommasie staining. PIP strips and PIP arrays were performed as previously described (Kagan and Medzhitov, 2006). Quantitative lipid pulldown assays were performed as previously described (Kagan and Medzhitov, 2006) except that 10ug of each GST protein were mixed with 50ug of liposomes containing 18% of the indicated phosphoinositide. Fluorescence recovered by beads coated with GST-mMyD88 was subtracted from each sample to account for nonspecific binding of liposomes to the beads.
Fly strains and crosses
All flies were maintained at 25°C on a standard cornmeal medium. The mutant strains used in this study (PBc03881 and key) have been previously described (Kambris et al., 2003; Rutschmann et al., 2000). Transgenic flies were generated by P-element transformation of w1118 embryos using standard procedures (Rainbow Transgenic Flies, Inc.). Constructs used to generate transgenic flies were created by cloning dMyD88-FL (nucleotides 1–1614) and dMyD88-Cyto (nucleotides 1–1158) into pUAST (Brand and Perrimon, 1993). For the dMyD88-PLC chimera, the N-terminal sequence (nucleotides 1–1158) was fused to the PH domain of PLCδ1 by overlap extension PCR and then cloned into pUAST. Each construct was crossed into the dMyD88 homozygous mutant background, along with the hs-Gal4 driver for all infection assays. The genotype of these flies was dMyD88c03881, hsGal4/dMyD88c03881, UAS-dMyD88 transgene (where the transgene is either dMyD88-FL or dMyD88-PLC or dMyD88-Cyto). Because of the known leakiness of the heat-shock promoter (Kambris et al., 2003; Maurange and Paro, 2002), no heat shock was necessary in order to express the transgenes. The V32-Gal4 driver (Maxton-Kuchenmeister 1999) was used to obtain high expression of maternal Gal4 in the sterility assays. At least two independent lines were tested in every experiment for each dMy88 transgene. Homozygous dMyD88 mutant flies (dMyD88c03881/dMyD88c03881) and wild type dMyD88 heterozygotes (dMyD88c03881/CyO) were used as controls.
Infection experiments
Septic injury was performed by pricking 2–4 day old male flies with a tungsten needle previously dipped in a pellet from an overnight culture of E. facaelis (ATCC number 10100), S.epidermidis (gift from P. Watnik), or E. coli (XL1 Blue). Following bacterial infection, flies were immediately transferred to 29°C. Flies that died in the first 3 hours were discarded. Flies were scored daily for survival experiments or RNA was extracted from the infected flies at the indicated times using RNA-Bee (Fisher). RT-PCR was carried out with a Bio-Rad iQ5 real time cycler using Taqman probes as directed by the manufacturer.
Determination of CFU in flies
Flies were challenged with an overnight culture of E. facaelis as described above and incubated at 29°C for 8 hours. Individual flies were homogenized in 100ul of PBS, diluted serially, and spread onto Brain Heart Infusion agar plates supplemented with 25ug/mL of kanamycin. It should be noted that no kanamycin resistant bacteria grew out of lysates from non-infected flies.
Plasmids
pJL1-FLAG-dMyD88, pCMV2-FLAG-dMyD88, pJL1-FLAG-Tube, pCMV2-FLAG-TIRAP, pCMV2-FLAG-mMyD88, and TIRAP-GFP were described previously (Horng and Medzhitov, 2001; Kagan and Medzhitov, 2006). The pCMV2-FLAG-dMyD88 vector was used as a template to clone dMyD88 into pEYFP-C1 (Clonetech) and pGEX4T1 (Amersham). The sequences coding for the N-terminal (DD+TIR) construct (nucleotides 1–1158) and the CTE vector (nucleotides 1125–1614) were amplified by PCR using pCMV2-FLAG-dMyD88 as a template and then cloned into either pJL1-FLAG vector (Horng and Medzhitov, 2001) or pGEX4T1. Metallothionine vectors encoding YFP-FL or YFP-ΔDD (nucleotides 709–1614) were generated by subcloning the YFP-tagged dMyD88 cDNAs from pEYFP-C1 into pJL252 (Horng and Medzhitov, 2001). The AgMyD gene was cloned into the pJL1-FLAG vector by amplifying the AgMyD from Anopheles gambiae cDNA using the following forward and reverse primers (F: ATGTTGGATAATCCCGTAAAACC; R: TTACACAGCGCTAGCTAGTTTTC).
Supplementary Material
Acknowledgements
We would like to thank R. Medzhitov, J.L. Imler, C. Kocks, S. Wassermann, M. Muskavitch, and P. Watnick for providing reagents used in this study. We would particularly like to thank P. Watnick for the generous use of the fly facility and fly equipment in her lab. We thank Z. Wang for helpful conversations and assistance with infection assays, as well as the members of the Kagan lab for insightful discussions. This work is supported by grants 5T32HD007466 (L.R.M), R00AI072955 (JCK) and the Harvard Digestive Disease Center P30 DK34854 (JCK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, York JD, Meyer T, Grinstein S. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151:1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Charatsi I, Luschnig S, Bartoszewski S, Nusslein-Volhard C, Moussian B. Krapfen/dMyd88 is required for the establishment of dorsoventral pattern in the Drosophila embryo. Mech Dev. 2003;120:219–226. doi: 10.1016/s0925-4773(02)00410-0. [DOI] [PubMed] [Google Scholar]
- Chen LY, Wang JC, Hyvert Y, Lin HP, Perrimon N, Imler JL, Hsu JC. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr Biol. 2006;16:1183–1193. doi: 10.1016/j.cub.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Hacker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc Natl Acad Sci U S A. 2001;98:12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HR, Chen ZJ, Kunes S, Chang GD, Maniatis T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc Natl Acad Sci U S A. 2010;107:8322–8327. doi: 10.1073/pnas.1004031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Bilak H, D'Alessandro R, Belvin M, Imler JL, Capovilla M. DmMyD88 controls dorsoventral patterning of the Drosophila embryo. EMBO Rep. 2003;4:64–69. doi: 10.1038/sj.embor.embor714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Hoffmann JA, Imler JL, Capovilla M. Tissue and stage-specific expression of the Tolls in Drosophila embryos. Gene Expr Patterns. 2002;2:311–317. doi: 10.1016/s1567-133x(02)00020-0. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Chao Y, Chu X, Pletcher SD. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell. 2006;5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signaling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund VK, DeLotto Y, DeLotto R. Endocytosis is required for Toll signaling and shaping of the Dorsal/NF-kappaB morphogen gradient during Drosophila embryogenesis. Proc Natl Acad Sci U S A. 2010;107:18028–18033. doi: 10.1073/pnas.1009157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C, Paro R. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002;16:2672–2683. doi: 10.1101/gad.242702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, Yamamoto M, Akira S, O'Neill LA, Fitzgerald KA, Golenbock DT. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci U S A. 2006;103:6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A. 2002;99:12871–12876. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Towb P, Chiem DN, Foster BA, Wasserman SA. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. Embo J. 2004;23:100–110. doi: 10.1038/sj.emboj.7600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N, Saitoh S, Matsumoto F, Akashi-Takamura S, Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368:94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci U S A. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–97. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, Hoshino K, Takeuchi O, Kobayashi M, Fujita T, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.