Abstract
Perturbations in the composition and assembly of extracellular matrices (ECMs) contribute to progression of numerous diseases, including cancers. Anchoring of laminins at the cell surface enables assembly and signaling of many ECMs, but the possible contributions of altered laminin anchoring to cancer progression remain undetermined. In this study, we investigated the prominence and origins of defective laminin anchoring in cancer cells and its association with cancer subtypes and clinical outcomes. We found loss of laminin anchoring to be widespread in cancer cells. Perturbation of laminin anchoring originated from several distinct defects which all led to dysfunctional glycosylation of the ECM receptor dystroglycan. In aggressive breast and brain cancers, defective laminin anchoring was often due to suppressed expression of the glycosyltransferase LARGE. Reduced expression of LARGE characterized a broad array of human tumors where it was associated with aggressive cancer subtypes and poor clinical outcomes. Notably, this defect robustly predicted poor survival in patients with brain cancers. Restoring LARGE expression repaired anchoring of exogenous and endogenous laminin and modulated cell proliferation and tumor growth. Together, our findings suggest that defects in laminin anchoring occur commonly in cancer cells, are characteristic of aggressive cancer subtypes, and are important drivers of disease progression.
Keywords: laminin, LARGE, dystroglycan, extracellular matrix, assembly
Introduction
The heterogeneity of cancers is evident by a growing list of genetic and epigenetic changes that perturb cell regulatory networks. Among these changes are regulatory defects that alter the interaction of cells with their microenvironment, leading to a loss of normal tissue architecture, invasion of surrounding tissues, and metastasis (1). Extracellular matrices (ECMs) are prominent and influential components of the cellular microenvironment in both normal and cancerous tissues, and changes in the expression, deposition, cross-linking, and degradation of ECM components have been clearly implicated in the progression of cancers (2–6).
Normal cell-ECM communications are modulated not only by the presence of ECM ligands and receptors, but also by the higher order assembly of ECM components into multimeric complexes. Many widely expressed ECM molecules, including fibronectins, collagens, fibrillins, and laminins are assembled into polymers. These assembly processes modulate the architecture of the matrices, and their biophysical and signaling properties, with direct consequences on cell function and tissue organization (7, 8). Therefore, changes in the ordered assembly of ECM molecules have the potential to strongly modulate cancer cell behavior.
Assembly of laminin isoforms into polymeric arrays across cell surfaces is believed to initiate the higher order organization of basement membranes (BMs), with resulting impact on BM integrity and signaling functions (7, 9). This assembly takes place via interactions of the laminin side arms, and is receptor-facilitated wherein anchoring of the laminin’s C-terminal domains to the cell surface initiates polymerization (10). Multiple laminin receptors have been implicated in this process of receptor-facilitated assembly including the integrins, dystroglycan and sulfatides (10–14). Cues from laminins modulate several cellular homeostatic functions in normal cells which are often found deregulated in carcinomas, including cell proliferation, adhesion, migration, differentiation, cell polarity, responsiveness to soluble factors and angiogenesis (15–18). In functionally normal epithelial cells, loss of laminin anchoring alone leads to loss of cell polarity signals and tissue-specific gene expression (13). Laminins have also been proposed to regulate resistance of cancer cells to therapeutic agents and apoptotic stimuli (19, 20). The effect of laminins on these many cell functions underscores the potential influence of deregulated laminin anchoring and assembly on cancer progression, from the growth and invasion of cancers to therapeutic responsiveness.
In this study, we investigate the prevalence of defective laminin anchoring, and the underlying causes, in cancer cells of distinct subtypes and tissue origins. We report that defective laminin anchoring is a prominent phenotype of cancer cells, caused by multiple distinct mechanisms. The most common of these defects is identified as reduced expression of the glycosyltransferase like-acetylglucosaminyltransferase (LARGE). Loss of LARGE expression is found to be a surrogate marker for the loss of laminin anchoring, and leads to enhanced cell proliferation and tumor growth. Reduced expression of LARGE is evident in diverse human cancers and associated with more aggressive tumor subtypes and decreased patient survival. These results reveal that inability to anchor laminin is a prevalent malignant phenotype, is associated with poor clinical outcomes, and is a potential target for the modulation of aggressive cancer cell behavior.
Materials and Methods
Molecular biology and viral transduction assays
Standard molecular biology methods were used (as detailed in supplementary information). Briefly, LARGE and β3-N-acetylglucosaminyltransferase-1 (β3GnT1), were cloned into the retroviral vector pBMN-IRES-PURO or adenoviral pShuttle vector (provided by Dr G. Inesi, CPMCRI, San Francisco) for generation of pAdeasy-LARGE which was transfected into HEK 293 cells. Viral particles were plaque-purified, amplified and further purified by CsCl density-gradient. Retroviral infections were achieved as previously described (13).
Generation and maintenance of cells in culture
All cancer cell lines used in this study were obtained from the American Type Culture Collection or from collections developed in the laboratories of Drs. Steve Ethier and Adi Gazdar, and have been carefully controlled for quality and identity as previously described (21). Primary normal human breast cells were obtained as a generous gift from Dr. Martha Strampfer (LBNL, Berkeley, California). The generation of immortalized floxed DG mouse mammary epithelial cells (Floxed-DG), knockout of DG (DG−/−) and re-expression of DG (DG+) or vector control has been previously described (13). The same method was used to generate the immortalized floxed-β1 integrin and β1 integrin knockout (β1−/−) mouse mammary epithelial cells.
Laminin assembly, immunofluorescence and biochemical assays
Laminin labeling, laminin assembly, immunofluorescence and biochemical assays were performed as previously described (13) and detailed in supplementary information. For surface labeling, samples were directly stained with primary antibody in growth media at 4 °C for 90 min prior to fixation and permeabilization.
Cell proliferation and cell cycle analysis
MTT Cell Proliferation Assay was used according to manufacturer’s protocol (ATCC). Percent increase in cells proliferation was calculated by the formula (dx-d1/d1) X 100. Data obtained from MTT assay was confirmed by counting cells seeded on 6 well plates.
Cell cycle assessment was performed by flow cytometry. Briefly, cells were labeled with 10 μM Bromodeoxyuridine (BrdU), fixed in 70% ethanol and stained with an anti-BrdU antibody, FITC-conjugated secondary, counterstained with propidium iodide (PI) and analyzed by FACScan. Treatments with laminin fragments E1 and E4 were done 24 hrs prior to labeling with BrdU at 40 μg/ml and 20 μg/ml, respectively.
Orthotopic xenografts
One million cells were injected into mammary fat pad of 6–8 weeks old female Balb/c nu/nu mice (Harlan). Tumor size was evaluated every other day and tumor volume was calculated using the formula Length X Width2/2 and graphed (7 mice per group).
Bioinformatic analysis
LARGE expression in primary tumor versus normal tissue was compared using data deposited on Oncomine™; website (www.oncomine.com). Normalized mean gene expression levels for LARGE (Affymetrix 215543_s_at) in 36 human glioblastoma samples were obtained from a previous study (42). Kaplan-Meier survival plots for 343 glioma samples were obtained from data deposited in the Repository of Molecular Brain Neoplasia Data (Rembrandt), National Cancer Institute (2005), accessed July 21, 2011 (http://rembrandt.nci.nih.gov).
Whole transcriptome shotgun sequencing (RNA-seq) was performed on the Illumina GAII system using standard protocols as previously described (27) and analysis was performed with the ALEXA-seq software package (22). An average of 74.8 million (76bp paired-end) reads passed quality control per sample. Subtype specific expression was determined by Wilcoxon signed-rank test. Genes were considered differentially expressed if they displayed fold-change greater than 2 and had a p-value less than 0.05 after multiple testing corrections by Benjamini-Hochberg method. The MD Anderson (GSE25066A) dataset of breast cancers was screened to verify subtype-specific expression of LARGE in tumors based on subtypes assigned by Hatzis and colleagues (23). Analysis of the TCGA dataset appears in the Supplementary Materials.
Results
Defects of laminin-111 anchoring are a prominent feature of human breast cancer cells
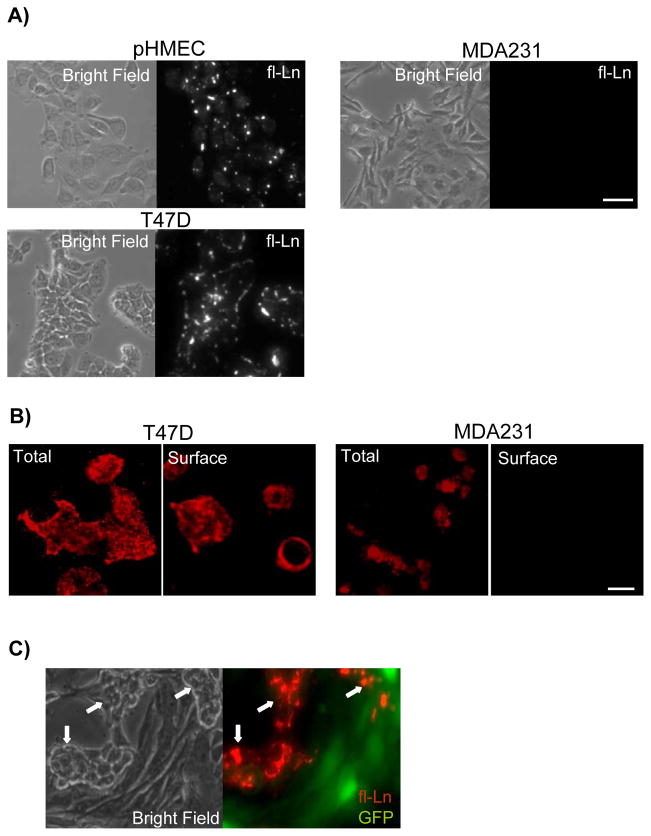
We assessed the capacity of cancer cells to anchor and assemble laminins at the cell surface, focusing first on breast cancer cells as a model system. To accomplish this we used a well established assay wherein cells incubated with exogenous laminin-111 are examined for accumulation of laminin on the cell surface (12, 13, 24, 25). In functionally normal mammary epithelial cell lines and primary cultures, fluorescently labeled laminin-111 (fl-Ln) accrued at the surface of living cells within minutes, beginning at discreet foci, and continued to accumulate over several hours (Figure 1A) (13, 24). However, while some breast cancer cells displayed normal anchoring and assembly of fl-Ln, others showed no detectable fl-Ln at the cell surface, even after 24 hours incubation (Figure 1A). The same defect was observed when assaying for assembly of endogenous laminin at the cell surface by immunofluorescence (Figure 1B). We then asked whether the variable capacity of cells to anchor laminin is reliant on inherent cell properties or on secreted factors transferable from neighboring cells, such as proteolytic enzymes or inhibitory peptides. Treatment of cells with the broad spectrum metalloproteinase inhibitor GM6001 did not restore laminin assembly at the cell surface (data not shown). Furthermore, in co-culture experiments in which MDA-MB-231 (MDA231) cells were mixed with T47D cells, fl-Ln anchoring was again observed uniquely on the T47D cells (Figure 1C). These data suggest that the ability to anchor laminin is cell autonomous and heterogeneous among cancer cells, likely arising from differences in laminin receptor functions.
Figure 1. Absence of laminin anchorage is a cell autonomous defect in breast cancer cells.
A) Normal primary human mammary epithelial cells (pHMECs) and breast cancer cell lines were treated with fl-Ln overnight and imaged by phase (left) and fluorescence (right) microscopy. B) Immunofluorescence staining of total and surface-bound endogenous laminin in breast cancer cell lines revealed laminin expression in all cells, but an absence of surface-bound laminin in MDA231 cells. C) T47D cells and GFP-expressing MDA231 cells were co-cultured, treated with fl-Ln overnight and imaged by phase (left) and fluorescence (right) microscopy. The morphologically distinct T47D cells (white arrows) retained the capacity for anchorage of fl-Ln (red, right) whereas the MDA231 cells (green, right) remained anchorage-deficient. (Bars = 25 μm).
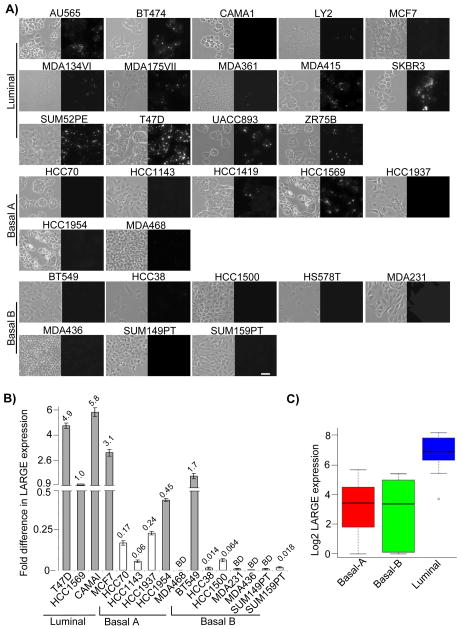
We determined the prevalence and origins of defective laminin anchoring among cancer cells by testing a large panel of human breast cancer cell lines for their ability to anchor fl-Ln. This particular panel of cancer cell lines has been developed as a model system displaying a molecular heterogeneity resembling that observed in human breast cancers (21). A key advantage of testing this panel of cancer cells is the large collection of gene expression and genomic data that has been assembled for these cell lines, which permits rapid in silico exploration of the molecular mechanisms underlying cellular phenotypes (21, 26). Less than 30% of the 29 cell lines tested exhibited clear fl-Ln binding, as revealed by the accumulated fluorescence signal at the cell surface (Figure 2A). In the other lines, the fluorescent signal was either entirely absent (15 out of 29) or it was detected in only a small subpopulation of cells (Figure 2A). The majority of cell lines displaying defects of laminin anchoring fell within the basal-like subtypes (summarized in the Table S1). Taken together, these data indicated that defects of laminin anchoring are prevalent in human breast cancer cells.
Figure 2. Absence of laminin anchorage and LARGE expression are common defects of breast cancer cell lines of distinct subtypes.
A) The indicated human breast cancer cell lines were treated with fl-Ln overnight and imaged by phase (left) and fluorescence (right) microscopy. Cell lines are grouped by subtype (labeled on the left) and shown in alphabetical order. (Bar = 25 μm). B) mRNA obtained from indicated cell lines were subjected to real time RT-PCR using specific primary for human LARGE and GAPDH. T47D and HCC1569 are shown as references. The fold difference in LARGE mRNA expression is indicated above each bar. White bars indicate cell lines designated to lack expression of LARGE mRNA and BD indicates signals that are below detection, as described in supplementary material and methods. C) The boxplot illustrates the gene-level expression of LARGE in 26 breast cancer cell line relative to the distribution of all gene expression values in breast cancer cell subtypes. Higher expression levels in luminal relative to basal-A/B (grouped) subtypes are statistically significantly (p=6.696e-05).
DG, not integrins, is required for anchoring of laminin-111 in mammary epithelial cells
Numerous laminin-binding receptors have been identified as potential mediators of laminin anchoring, including DG, integrins, and sulfatides (11–14, 25). We sought to identify changes in gene expression that could explain the loss of laminin assembly among these cell lines by interrogating the comprehensive gene expression data generated from this cell line panel by massively parallel RNA-sequencing (RNA seq) (27). None of the known laminin receptor genes exhibited a loss of gene expression that correlated with loss of laminin anchoring, including DG and the integrin subunits β1, β4,α1, α2, α3, α6 α7, α8, and α9 (Figures S1). Likewise, genes encoding enzymes essential for sulfatide synthesis exhibited generally low or undetectable expression among both luminal and basal-like carcinoma cells (Figure S1). Therefore, the defect of breast cancer cells to anchor laminin was not associated with variable expression of known laminin receptors.
Deletion of the DG gene from mammary epithelial cells and embryonic stem cells abolishes laminin-111 anchoring, and DG may perform this function in cooperation with β1 integrins (13, 24, 25). We directly tested the requirement for β1 integrin heterodimers in laminin-111 anchoring by eliminating their expression through deletion of the shared β1 integrin subunit by Cre-lox recombination in mammary epithelial cells established from β1 integrin fl/fl mice (Figure S2A). Ln anchoring in the β1 −/− cells was comparable to control cells, demonstrating that β1 integrin function is not essential for laminin-111 anchoring (Figure S2B). In contrast; deletion of DG completely abrogated laminin anchoring in normal mammary epithelial cells (Figure S2B).
Dysfunction of DG correlates with deficient laminin-111 anchoring in human breast cancer cells
The interaction of DG with laminins is reliant on O-linked carbohydrate modifications of the α-DG mucin domain, making altered glycosylation of DG a potential modulator of laminin anchoring (28). Defects of DG glycosylation cause loss of DG function in some muscular dystrophies (28), and similar defects have been observed in cancer cell lines (29–32). We assessed the glycosylation state of DG within the breast cancer cell line panel by performing western blot analysis using a well-characterized monoclonal antibody (IIH6) that binds to critical carbohydrate residues on α-DG that are required for binding to laminin. The vast majority of these cell lines (more than 70%) did not display any IIH6 immunoreactivity (Figure S3A). In contrast, the immunoblot analysis of β-DG subunit confirmed strong expression of the DG protein (Figure S3A). In addition, direct assessment of α-DG expression in some cells indicated that loss of IIH6 immunoreactivity was accompanied by a reduction in the mass of the α-DG subunit; consistent with α-DG hypoglycosylation (Figure S3B). There is a strong correlation between the IIH6 immunoreactivity and ability to anchor fl-Ln across the collection of human breast cancer cell lines tested (Table S1). All 15 cell lines deficient in laminin anchoring lacked IIH6 immunoreactivity. In aggregate, these results reveal that abnormal glycosylation of α-DG correlates strongly with the inability of human breast cancer cells to anchor laminin-111, and suggest that hypoglycosylation of DG is causative.
Deficient expression of LARGE is the predominant, but not exclusive, cause of loss of laminin anchoring in breast cancer cells
Genetic studies of congenital muscular dystrophies in humans have identified six genes that modulate DG glycosylation: POMT1, POMT2, POMGnT1, LARGE, FKTN and FKRP (28, 33). More recently, β3GNT1 has been implicated in DG glycosylation(30). RNA-seq expression data for the cell lines revealed minimal variation in mRNA expression of these genes (Figure S4A), with the sole exception of LARGE (Figure 2B and Figure S4B). LARGE expression levels were highly variable across the breast cancer cell lines and significantly higher in luminal lines compared to basal types (p=6.696e-05) (Figure 2C).
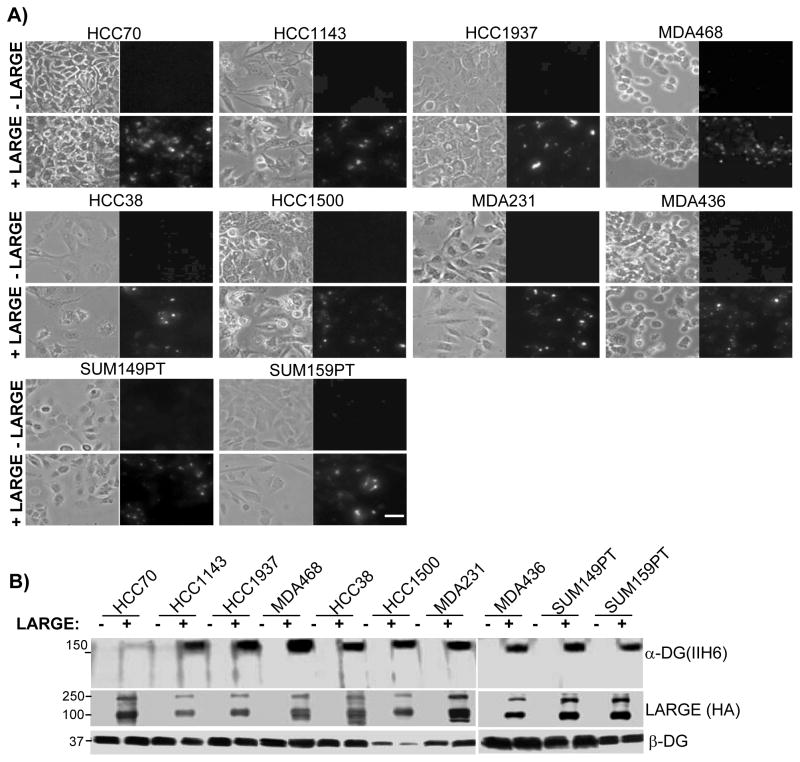
Cell lines exhibiting decreased expression of LARGE (indicated by white bars in Figure 2B) were unable to anchor laminin-111 and did not display IIH6 immunoreactivity. These data indicated that loss of laminin anchoring in these cells is caused by reduced expression of LARGE and dysfunctional glycosylation of DG. This hypothesis was directly tested in these cells by re-expression of LARGE. Restoring expression of LARGE by viral transduction repaired fl-Ln anchoring at the cell surface in all 10 cell lines that lack endogenous LARGE expression (Figure 3A). Consistently, in parallel with repairing laminin-binding, LARGE expression also restored IIH6 immunoreactivity (Figure 3B).
Figure 3. Expression of exogenous LARGE restores laminin anchorage and DG glycosylation in cells lacking LARGE expression.
A) The indicated cells infected with cDNA encoding the HA-tagged LARGE (+LARGE) or control vector (−LARGE) were treated with fl-Ln and imaged by phase (left) and fluorescence (right) microscopy. (Bars = 25 μm). B) Protein extracts from indicated cell line were immunoblotted using the indicated antibodies. Molecular weights are shown on the left.
Apart from loss of LARGE expression, at least two additional defects can cause the loss of laminin anchoring in breast cancer cells
Cell lines CAMAI, MCF7, HCC1954 and BT549 showed defects in laminin anchoring and DG glycosylation that were not attributed to lack of LARGE expression. Significantly, over-expression of LARGE restored anchoring of fl-Ln and endogenous laminin in most lines, but not in HCC1954, demonstrating additional heterogeneity within this subset of cells (Figure S5A and S5B). Likewise, over-expression of LARGE restored glycosylation of DG in all cell lines but HCC1954 (Figure S5C). In contrast, over-expression of β3GNT1 did not restore fl-Ln anchoring or DG glycosylation in any of the cell lines tested (Figure S5D and E).
We searched for alternative causes for loss of laminin anchoring in the CAMAI, MCF7, HCC1954 and BT549 lines by sequencing mRNA from genes implicated in DG glycosylation. Analysis of cDNA sequence did not reveal additional mutations, deletions or alternate splicing (Figures S4C). Thus, while these results do not specify the origin or number of additional defects, they do reveal that at least two defects other than reduced expression of LARGE can account for loss of laminin anchoring and DG glycosylation in cancer cells, a set that can be repaired by LARGE over-expression, and a set that cannot.
Suppression of LARGE expression enhanced cell proliferation and tumor growth
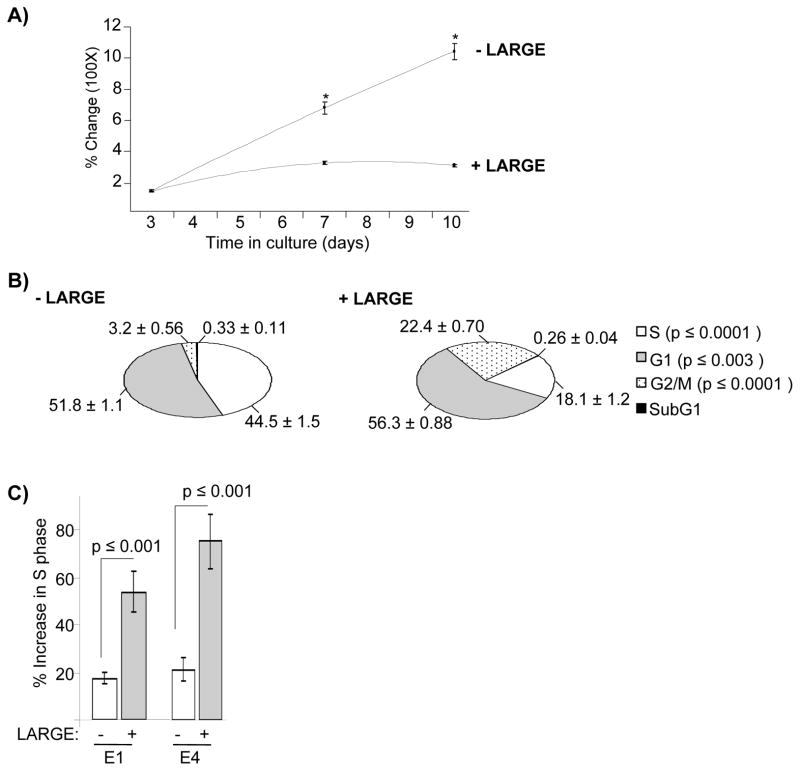
We observed that expression of LARGE resulted in slower proliferation in some cell lines. In particular, CAMAI cells exhibited markedly reduced cell proliferation upon stable expression of LARGE. Quantitative assessments indicated that LARGE-expressing cells were significantly less proliferative than cells expressing vector control and this difference was enhanced with time in culture (Figure 4A). Assessment of cell cycle progression indicated that expression of LARGE produced significant differences in phases of cell cycle, most noticeably by reducing the fraction of cells in S phase (Figure 4B). The presence of the assembly-blocking laminin fragments E1 and E4 significantly increased the percentage of LARGE-expressing cells in S phase relative to cells expressing the vector control (Figure 4C). These results indicate that loss of LARGE expression promotes cell proliferation by disabling laminin anchoring.
Figure 4. Restoration of laminin anchorage in breast cancer cells slows cell proliferation.
A) CAMAI cells expressing LARGE (+LARGE) or vector control (−LARGE) were maintained in culture and percent change in OD measurements obtained from MTT assays were plotted against time. Statistically significant differences are shown by * (p ≥ 0.0003) B) CAMAI cells were treated with BrdUrd, fixed and stained with an anti-BrdUrd antibody and propidium iodide and subjected to cell cycle analysis by FACS. The numbers on the pie chart indicated percentage (± standard errors) of cells in each phase of cell cycle. The insert on the right denotes cell cycle phases corresponding to the pie chart as well as the p values representing statistically significant differences between −LARGE and +LARGE. C) Percent change in S phase with (+) or without (−) treatment with laminin fragments E1 and E4 was calculated and plotted as histogram bars for CAMAI cells −LARGE (white bars) and +LARGE (gray bars). Standard errors and p values are shown.
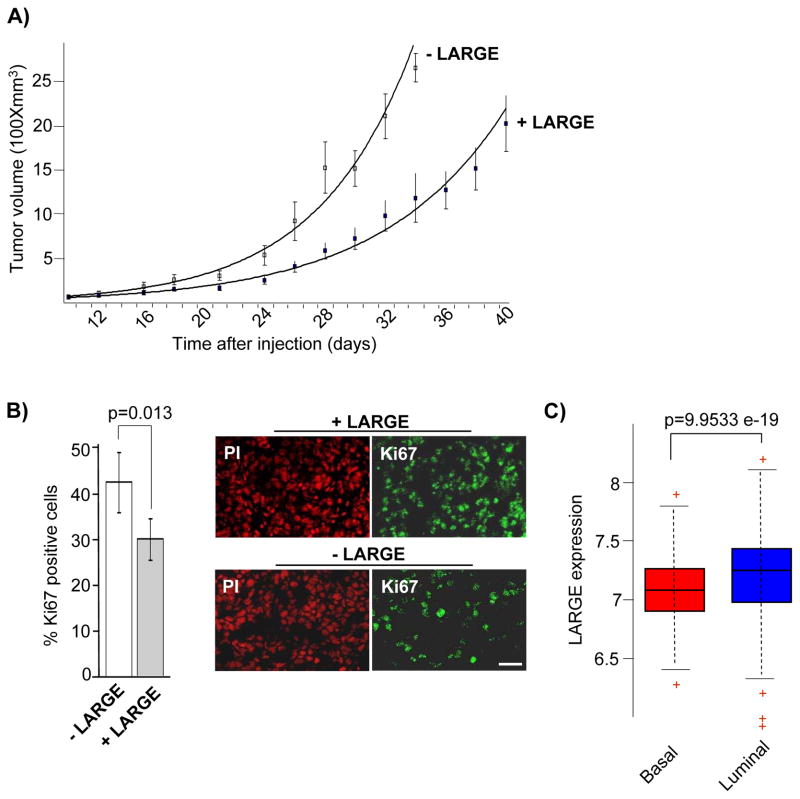
Next, we assessed the consequence of LARGE expression on the tumorigenic capacity of cancer cells in vivo using orthotopic xenograft models generated by MDA231 cells expressing vector control or LARGE. Both cell types generated palpable tumors in 100% of injected mice. However, tumors generated by cells expressing LARGE were significantly smaller and exhibited a decrease in cell proliferation (Figure 5A and B). In sum, reduced expression of LARGE enhanced cell proliferation and tumor growth in vivo.
Figure 5. Reduced expression of LARGE promotes tumorigenicity in vivo and is associated with aggressive human breast cancers.
A) Tumor volumes generated by orthotropic injection of MDA231 cells expressing either vector control (−LARGE) or LARGE (+LARGE) are plotted against time post-injection along with standard errors. Differences in tumor volumes formed by −LARGE and +LARGE cells are statistically significant (p ≤ ranging from 0.05 to 0.001) from day 18 onward. B) Tumors generated by MDA231 cells were sectioned, stained with a human specific anti-Ki67 antibody and propidium iodide, and imaged by confocal microscopy. Representative images are shown on the right. Two regions from of each tumor section was imaged to compare the percent of cells displaying Ki67 staining in tumors generated by vector control (−LARGE) or LARGE (+LARGE) expressing MDA231 cells. Standard errors and p values are shown. (Bar = 20 μm) C) The expression of LARGE mRNA was examined in microarray data obtained from a 508 sample tumor cohort (MD Anderson GSE25066A data set) previously categorized into different subtypes. Statistical significance (p=9.9533e-19) between the Basal and Luminal A and B (grouped) is shown.
Loss of LARGE expression is common in basal-like breast tumors and other human cancers
Analyses of breast cancer cell lines indicted that loss of LARGE expression is a surrogate measure for loss of laminin anchoring function and is more commonly observed in basal-like breast cancer cells. We examined the clinical relevance of these observations by analyzing two publically available datasets, each comprised of data from more than 500 patients, for LARGE expression in human breast cancers. In both datasets, reduced expression of LARGE mRNA was significantly associated with basal-like breast tumors (p=9.9533e-19 and p=2.29e-18, respectively) (Figure 5C and S6), a subtype which displays shortest overall and relapse free survival (34). These results corroborated the breast cancer cell line studies which showed reduced LARGE expression in the basal-like breast cancer cells. We then compared LARGE expression between normal and cancerous human tissues using the public datasets in Oncomine (Oncomine™ Compendia Bioscience, accessed July 2011) (Table S2). These analyses showed significantly decreased LARGE expression in infiltrating and superficial bladder carcinomas (35); various forms of colon adenocarcinomas (36); kidney clear cell adenocarcinoma (37); prostate adenocarcinomas (38) skin basal cell carcinoma (39) and a particularly strong decrease in brain cancers (40). Only squamous cell carcinomas showed significant overexpression of LARGE in primary cancers (41).
Loss of laminin anchoring is characteristic of aggressive subclasses of astrocytomas and poor prognosis
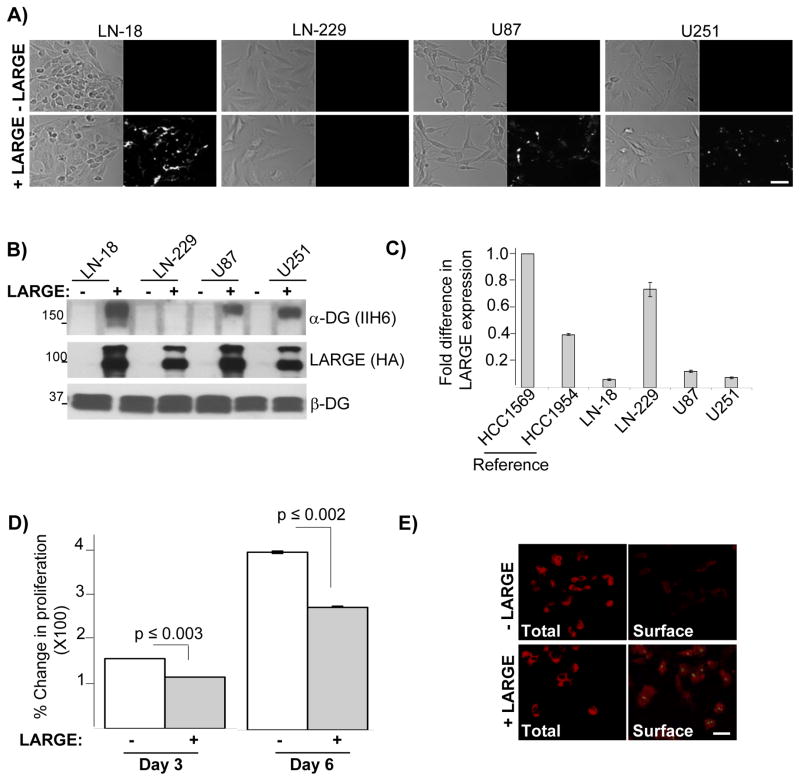
The highly significant reduction of LARGE expression in brain malignancies suggested that defects of laminin anchoring and assembly are functionally important in brain cancer cells, as they are in breast cancer cell lines. We tested this using cultured human glioblastoma cell lines. First, we confirmed that normal astrocytes displayed laminin anchoring and assembly, and that this capacity was lost in astrocytes obtained from mice lacking LARGE gene function (42) (Figure S7). In contrast, glioblastoma cell lines (LN-18, LN-229, U87 and U251) had deficiencies in laminin anchoring and DG glycosylation (Figure 6A and B) and, except for LN-229 cells, had reduced expression of LARGE (Figure 6C). Expression of exogenous LARGE in these cells restored laminin anchoring and DG glycosylation as observed in breast cancer cells - again with the exception of LN-229 (Figure 6A and B). Cell proliferation was also reduced, and surface accumulation of endogenous laminin increased, in LN-18 cells upon expression of LARGE (Figure 6D and E).
Figure 6. Defective laminin anchorage is a characteristic of glioblastoma cells arising primarily from lack of LARGE expression and conferring enhanced cell proliferation.
A) Indicated glioblastoma cells expressing LARGE (+LARGE) or vector control (−LARGE) were treated with fl-Ln and imaged by phase (left) and fluorescence (right) microscopy. B) Protein extracts from the cells described in A were subjected to immunoblot analysis with the indicated antibodies. C) Real-time RT-PCR analysis of LARGE expression from indicated glioblastoma cell lines were assessed and compared to two breast cancer cell lines. Expression levels are relative to LARGE expression in HCC1569 cells. D) LN-18 cells expressing vector control (white bars) or LARGE (gray bars) were maintained in culture and percent change in OD measurements obtained from MTT assays at indicated points were plotted along with standard errors. Statistically significant differences and the corresponding p are shown. E) Immunofluorescence of total (total) and surface-bound laminin (Surface) co-stained with LARGE in LN-18 cells expressing LARGE or vector control. Laminin and LARGE are shown in red and green, respectively. (Bars = 25 μm).
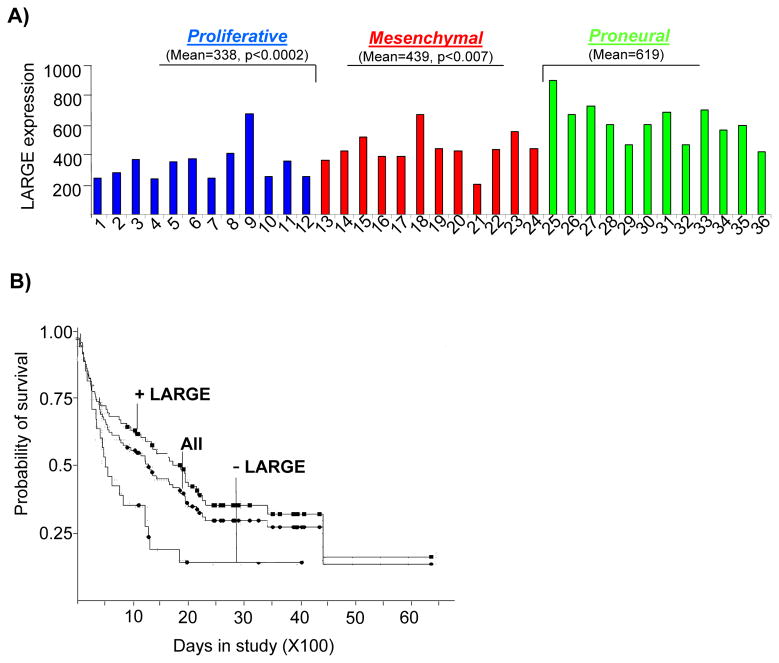
We investigated LARGE expression in a cohort of glioblastomas (GBMs) classified by aggressiveness and by expression profiling as proneural (PN), proliferative (Prolif), and mesenchymal (Mes) (43). Expression of LARGE was significantly lower in the two most aggressive subclasses, Mes and Prolif (Figure 7A). Patient outcome is markedly worse for Prolif and Mes compared to PN (median survival= 174.5 weeks for PN, 60.5 weeks for Prolif, and 65 weeks for Mes) (42). We next assessed the prognostic value of LARGE expression in independent cohorts of astrocytomas using the REMBANDT database (See Methods). As shown in Figure 7B, reduced expression of LARGE was strongly correlated with poor patient survival (p=7.9e-7).
Figure 7. Reduced LARGE expression is associated with aggressive gliomas and poor patient survival.
A) Normalized mean gene expression levels for LARGE in 36 human glioblastoma samples are displayed. The samples belong to three distinct molecular subclasses; proliferative (samples 1–12), mesenchymal (samples 13–25) and proneural (samples 26–37) which correlate strongly with patient survival. P values shown are relative to Proneural. B) Kaplan Meier survival plots for astrocytomas from the Rembrandt database are displayed with respect to LARGE expression levels. Survival data for all samples (All), samples with down regulated LARGE (− LARGE) and samples excluding LARGE down regulation (+LARGE) are shown. Log-rank test for − LARGE and + LARGE is p ≤ 0.037. Statistically significant association with LARGE expression and poor survival were also obtained in Kaplan Meier survival plots of all gliomas (not shown), p=7.9e-7.
Discussion
Our study shows that loss of laminin anchoring is a prominent phenotype of cancer cells of distinct pathophysiological and tissue origins. We reveal a clear association of this defect with aggressive cancer subtypes in breast cancers and glioblastomas, and demonstrate changes in cell proliferation and tumor growth as a result of this defect. Correspondingly, loss of laminin anchoring is also associated with poor clinical outcomes. Surprisingly, this defect is attributed to altered glycosylation of the laminin receptor DG, and not the expression of any known laminin receptors, including the integrins. Our assessment of a large panel of cancer cells provides a broad characterization of the molecular defects underlying the loss of laminin anchoring, pointing to at least three mechanistically distinct defects. This work introduces novel factors to the study of cancer progression, novel markers of cancer heterogeneity, and potentially novel therapeutic targets, with relevance to a very broad array of human cancers.
Laminin anchoring and assembly as a tumor suppressor
Several lines of evidence indicate that laminin anchoring acts as a tumor suppressor. We demonstrate that exogenous expression of LARGE in cancer cells slows cell proliferation by enabling assembly of laminins on the surface of cancer cells. Xenograft assays showed that restoration of LARGE expression moderates tumor growth in vivo. Work by Beltran et al. showed that suppression of LARGE expression enhances the invasiveness of one breast cancer cell line (31). Interactions of cells with ECM constituents, in particular laminins, contribute to tumor suppressive properties and play a dominant role in the normal morphogenesis and function of epithelia (17, 18). Importantly, the laminin anchoring activity of DG is a determinant of epithelial cell polarization (13), a characteristic of epithelial tissues which is lost in tumor cells and which exerts potent tumor suppressor properties (17, 46). Ultimately, in vivo assessment of tumor suppressive properties of laminin anchoring is necessary, including the analysis of transgenic models manipulating LARGE expression or function, but such models have not yet been developed. Mice completely lacking LARGE expression have been identified and serve as models of congenital muscular dystrophy, however loss of LARGE expression in these models results in muscle disease and death at ~16 weeks of age, precluding studies of cancer progression in this context (42).
Agents that elevate LARGE expression may prove therapeutic in a large number of cancers, however, the cause of LARGE suppression is not yet known and such agents have not yet been identified. The LARGE locus was first discovered as a tumor-specific deletion (47). However, examination of SNP and CGH data for the breast cancer cell lines did not reveal any evidence of deletions in the LARGE gene (data not shown). DNA methylation has previously been implicated in the loss of LARGE expression in one breast cancer cell line (MDA-MB-231 cells) (31). Broad measures of DNA methylation have been obtained for the panel of breast cancer cells, and included 2 probes within the LARGE promoter (48). Among the cell lines in this study exhibiting loss of LARGE expression, hypermethylation of the LARGE promoter was detected in only 2 lines (MDA-MB-231, and HCC1143 cells) (data not shown). Therefore, the frequent loss of LARGE expression in cancers is not uniquely a consequence of methylation changes at the LARGE promoter.
The findings reported here have numerous implications for the understanding and treatment of cancers. The decrease in cell proliferation and tumor growth associated with restoration of laminin anchoring suggests novel therapeutic opportunities focused on enhancement of LARGE expression or other mechanisms to restore laminin anchoring. Because these defects are more prevalent in aggressive cancer subtypes, novel therapeutic approaches targeting these pathways can address cancers that are currently most difficult to treat and have the fewest treatment options. The responsiveness of cancers cells to therapeutic manipulations is influenced by cell-ECM interactions (19, 20, 49), therefore, loss of laminin anchoring can potentially modulate response to therapies. Our results on cancer heterogeneity and subtype association suggest that assays for laminin assembly or LARGE expression might serve as prognostic biomarkers for diverse cancers, although useful antibodies for LARGE are currently lacking. Lastly, our study highlights the potential importance of ECM assembly processes in cancer heterogeneity and cancer cell behavior. A closer look at the assembly processes of other ECM molecules in cancer cells may expose more novel factors contributing to disease heterogeneity and progression.
Supplementary Material
Acknowledgments
We thank Heidi Feiler and Mark Lenburg for helpful discussion, Sean McAllister, Ryuichi Murase and Pouya Mohajer for help with xenograft assays, Peter Yurchenco for the generous gift of laminin fragments, Mark LaBarge and Martha Stampfer for primary human cultures, and Rajendra Pilakatta for help in adenovirus generation.
Grant Support
This work was supported by grants from the National Institutes of Health (CA109579) to J.L.M. and (CA058207) to J.W.G., the Department of Defense Breast Cancer Research Program (W81XWH-07-1-0416) to J.L.M., a Stand Up to Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409 to J.W.G., a Komen for the Cure Postdoctoral fellowship to A.A., a Belgian American Educational Foundation Postdoctoral fellowship to A.D., and a Canadian Institutes of Health Research Postdoctoral fellowship to O.L.G.
This work was supported by grants from the National Institutes of Health (CA109579) to J.L.M. and (CA058207) to J.W.G., the Department of Defense Breast Cancer Research Program (W81XWH-07-1-0416) to J.L.M., a Stand Up to Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409 to J.W.G.
Footnotes
No conflicts of interest exist among the authors.
References
- 1.Hu M, Polyak K. Microenvironmental regulation of cancer development. Curr Opin Genet Dev. 2008;18:27–34. doi: 10.1016/j.gde.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–15. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 3.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 6.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15:1277–94. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol. 2008;20:495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki T, Fassler R, Hohenester E. Laminin: the crux of basement membrane assembly. J Cell Biol. 2004;164:959–63. doi: 10.1083/jcb.200401058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colognato H, Winkelmann DA, Yurchenco PD. Laminin polymerization induces a receptor-cytoskeleton network. J Cell Biol. 1999;145:619–31. doi: 10.1083/jcb.145.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohikangas L, Gullberg D, Johansson S. Assembly of Laminin Polymers Is Dependent on beta1-Integrins. Exp Cell Res. 2001;265:135–44. doi: 10.1006/excr.2001.5170. [DOI] [PubMed] [Google Scholar]
- 12.Henry MD, Campbell KP. A role for dystroglycan in basement membrane assembly. Cell. 1998;95:859–70. doi: 10.1016/s0092-8674(00)81708-0. [DOI] [PubMed] [Google Scholar]
- 13.Weir ML, Oppizzi ML, Henry MD, Onishi A, Campbell KP, Bissell MJ, et al. Dystroglycan loss disrupts polarity and {beta}-casein induction in mammary epithelial cells by perturbing laminin anchoring. J Cell Sci. 2006;119:4047–58. doi: 10.1242/jcs.03103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Liquari P, McKee KK, Harrison D, Patel R, Lee S, et al. Laminin-sulfatide binding initiates basement membrane assembly and enables receptor signaling in Schwann cells and fibroblasts. J Cell Biol. 2005;169:179–89. doi: 10.1083/jcb.200501098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, et al. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12:197–207. doi: 10.1016/S1044-579X(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 17.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–46. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XH, Flores LM, Li Q, Zhou P, Xu F, Krop IE, et al. Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res. 2010;70:2256–63. doi: 10.1158/0008-5472.CAN-09-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, et al. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–16. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7:843–7. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 23.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonoudakis D, Singh M, Mohajer R, Mohajer P, Fata JE, Campbell KP, et al. Dystroglycan controls signaling of multiple hormones through modulation of STAT5 activity. J Cell Sci. 2010;123:3683–92. doi: 10.1242/jcs.070680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry MD, Satz JS, Brakebusch C, Costell M, Gustafsson E, Fassler R, et al. Distinct roles for dystroglycan, (β)1 integrin and perlecan in cell surface laminin organization. J Cell Sci. 2001;114:1137–44. doi: 10.1242/jcs.114.6.1137. [DOI] [PubMed] [Google Scholar]
- 26.Heiser LM, Wang NJ, Talcott CL, Laderoute KR, Knapp M, Guan Y, et al. Integrated analysis of breast cancer cell lines reveals unique signaling pathways. Genome Biol. 2009;10:R31. doi: 10.1186/gb-2009-10-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Barresi R, Campbell KP. Dystroglycan: from biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 29.Martin LT, Glass M, Dosunmu E, Martin PT. Altered expression of natively glycosylated alpha dystroglycan in pediatric solid tumors. Hum Pathol. 2007;38:1657–68. doi: 10.1016/j.humpath.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao X, Kobayashi M, Hatakeyama S, Angata K, Gullberg D, Nakayama J, et al. Tumor suppressor function of laminin-binding alpha-dystroglycan requires a distinct beta3-N-acetylglucosaminyltransferase. Proc Natl Acad Sci U S A. 2009;106:12109–14. doi: 10.1073/pnas.0904515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran-Valero de Bernabe D, Inamori KI, Yoshida-Moriguchi T, Weydert CJ, Harper HA, Willer T, et al. Loss of alpha-dystroglycan laminin binding in epithelium-derived cancers is caused by silencing of large. J Biol Chem. 2009;284:11279–84. doi: 10.1074/jbc.C900007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh J, Itahana Y, Knight-Krajewski S, Kanagawa M, Campbell KP, Bissell MJ, et al. Proteolytic enzymes and altered glycosylation modulate dystroglycan function in carcinoma cells. Cancer Res. 2004;64:6152–9. doi: 10.1158/0008-5472.CAN-04-1638. [DOI] [PubMed] [Google Scholar]
- 33.Hewitt JE. Abnormal glycosylation of dystroglycan in human genetic disease. Biochim Biophys Acta. 2009;1792:853–61. doi: 10.1016/j.bbadis.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–81. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 36.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–81. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–82. [PubMed] [Google Scholar]
- 39.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, et al. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64:55–63. doi: 10.1158/0008-5472.can-03-2144. [DOI] [PubMed] [Google Scholar]
- 42.Lee Y, Kameya S, Cox GA, Hsu J, Hicks W, Maddatu TP, et al. Ocular abnormalities in Large(myd) and Large(vls) mice, spontaneous models for muscle, eye, and brain diseases. Mol Cell Neurosci. 2005;30:160–72. doi: 10.1016/j.mcn.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 44.Godfrey C, Foley AR, Clement E, Muntoni F. Dystroglycanopathies: coming into focus. Curr Opin Genet Dev. 2011;21:278–85. doi: 10.1016/j.gde.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 45.van Reeuwijk J, Grewal PK, Salih MA, Beltran-Valero de Bernabe D, McLaughlan JM, Michielse CB, et al. Intragenic deletion in the LARGE gene causes Walker-Warburg syndrome. Hum Genet. 2007;121:685–90. doi: 10.1007/s00439-007-0362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang L, Muthuswamy SK. Polarity protein alterations in carcinoma: a focus on emerging roles for polarity regulators. Curr Opin Genet Dev. 20:41–50. doi: 10.1016/j.gde.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peyrard M, Seroussi E, Sandberg-Nordqvist AC, Xie YG, Han FY, Fransson I, et al. The human LARGE gene from 22q12.3-q13.1 is a new, distinct member of the glycosyltransferase gene family. Proc Natl Acad Sci U S A. 1999;96:598–603. doi: 10.1073/pnas.96.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fackler MJ, Umbricht C, Williams D, Argani P, Cruz LA, Merino VF, et al. Genome-Wide Methylation Analysis Identifies Genes Specific to Breast Cancer Hormone Receptor Status and Risk of Recurrence. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen OW, Ronnov JL, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–8. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.