Abstract
Background
Molecular and epidemiological evidence demonstrate that altered gene expression and single nucleotide polymorphisms in the apoptotic pathway are linked to many cancers. Yet, few studies emphasize the interaction of variant apoptotic genes and their joint modifying effects on prostate cancer (PCA) outcomes. An exhaustive assessment of all the possible two-, three- and four-way gene-gene interactions is computationally burdensome. This statistical conundrum stems from the prohibitive amount of data needed to account for multiple hypothesis testing.
Methods
To address this issue, we systematically prioritized and evaluated individual effects and complex interactions among 172 apoptotic SNPs in relation to PCA risk and aggressive disease (i.e., Gleason score ≥ 7 and tumor stages III/IV). Single and joint modifying effects on PCA outcomes among European-American men were analyzed using statistical epistasis networks coupled with multi-factor dimensionality reduction (SEN-guided MDR). The case-control study design included 1,175 incident PCA cases and 1,111 controls from the prostate, lung, colo-rectal, and ovarian (PLCO) cancer screening trial. Moreover, a subset analysis of PCA cases consisted of 688 aggressive and 488 non-aggressive PCA cases. SNP profiles were obtained using the NCI Cancer Genetic Markers of Susceptibility (CGEMS) data portal. Main effects were assessed using logistic regression (LR) models. Prior to modeling interactions, SEN was used to pre-process our genetic data. SEN used network science to reduce our analysis from > 36 million to < 13,000 SNP interactions. Interactions were visualized, evaluated, and validated using entropy-based MDR. All parametric and non-parametric models were adjusted for age, family history of PCA, and multiple hypothesis testing.
Results
Following LR modeling, eleven and thirteen sequence variants were associated with PCA risk and aggressive disease, respectively. However, none of these markers remained significant after we adjusted for multiple comparisons. Nevertheless, we detected a modest synergistic interaction between AKT3 rs2125230-PRKCQ rs571715 and disease aggressiveness using SEN-guided MDR (p = 0.011).
Conclusions
In summary, entropy-based SEN-guided MDR facilitated the logical prioritization and evaluation of apoptotic SNPs in relation to aggressive PCA. The suggestive interaction between AKT3-PRKCQ and aggressive PCA requires further validation using independent observational studies.
Keywords: Prostate cancer, Apoptosis, Single nucleotide polymorphisms, Gene-gene interactions, Multifactor dimensionality reduction (MDR), Statistical epistasis networks (SEN)
Background
Prostate cancer (PCA) is the most frequently diagnosed cancer and the 2nd leading cause of cancer-related deaths among men in the United States [1]. The American Cancer Society estimates that 26-29% of all new cancer cases and cancer-related deaths are attributed to PCA cancer. Well established PCA risk factors include older age, black race, and family history of PCA. However, other potential contributors of this disease may include lifestyle and genetic factors as well as imbalances within important biological pathways.
Apoptosis or programmed cell death is one such biological process that moderates cell differentiation, proliferation, death, whole body homeostasis and tumorigenesis [2-4]. This process is controlled by cell death (e.g. BAD, CASP, BIK) and cell survival proteins (e.g. BCL2, NFκB, AKT3) that induce or block apoptosis, respectively, as summarized in Table 1[2,3,5]. Decreased apoptotic cell death and increased cell proliferation may lead to clonal expansion and tumor growth [2]. Failure to undergo apoptosis permits survival of transformed cells, leading to subsequent genetic alteration, genomic instability, and ultimately a more invasive cancer phenotype [3]. Imbalances in apoptosis-related genes may play an important role in PCA susceptibility as well as disease progression. For example, several independent studies have shown that overexpression of cell survival indicators (e.g., BCL-2, CARD8, IKBKE, PRKCQ, and PIK3CB, AKT3) or down-regulation of cell death markers (e.g., BCL2L14) are associated with more aggressive phenotypes, higher Gleason grade, increased tumor progression, and poor PCA prognosis [6-19].
Table 1.
Selected genes involved in the regulation of apoptosis
| Gene | Function* | |
|---|---|---|
| Pro- & Anti- apoptotic | Tumor Protein 53 (TP53) | Transcriptionally regulates target genes that induce cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism in response to cellular stresses. |
|
Tumor Necrosis Factor (TNF) |
Binds and functions through its receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR to regulate cell proliferation, differentiation, and apoptosis. |
|
| PRKCQ | Protein kinase C family member; substrate for Caspase-3; phosphorylates BAD and required to activate NFκB (via CARD11 phosphorylation) and AP-1. | |
| Pro-apoptotic | FAS &FAS Ligand (FASL) | Death domain-containing receptor, binding of FASL to FAS allows the formation of a death-inducing signaling complex. |
| CASPASE (CASP) | Gene family involved in the execution of apoptosis. There are 2 classes of caspases, which include: initiators (e.g., CASP2, CASP8, CASP9, and CASP10) and effectors (e.g., CASP3, CASP6, CASP7). Initiator caspases activate pro-forms of effector caspases, enabling effectors to trigger the apoptosis process. |
|
|
CARD8 (Caspase recruitment domain family, member 8) |
CARD family protein; involved in various pathways which regulate caspases or NFκB; isoforms interact with caspases to signal apoptosis. |
|
| BCL2-associated X (BAX) | Forms a heterodimer with BCL2 and functions as an apoptotic activator involved mitochondrial release of cytochrome c. |
|
|
BCL2-antagonist/killer 1 (BAK1) |
Induces apoptosis by increasing cytochrome c release; interacts with the TP53 after exposure to cell stress. |
|
|
BCL2-associated agonist of cell death (BAD) |
Forms heterodimers with BCLXL and BCL2 to reverse their death repressor activity. | |
| BCL2-like 10 (BCL2L10) | Interacts with BCL2 proteins (e.g., BCL2, BCL2L1/BCLXL, and BAX). |
|
| BCL2-like 11 (BCL2L11) | (aka BIM); Interacts with other members of the BCL2 protein family (e.g., BCL2, BCL2L1/BCLXL, and MCL1) to act as an apoptosis activator. |
|
| BCL2-like 14 (BCL2L14) | Apoptosis facilitator; interacts with BCL2 family members; p53- target gene. |
|
|
BH3 interacting domain death agonist (BID) |
Induced by CASP8; CASP8 cleaves the protein encoded by this gene, and the COOH-terminal part translocates to mitochondria, which triggers cytochrome c release. |
|
|
BCL2-interacting killer (BIK) |
Interacts with survival-promoting proteins to enhance programmed cell death. |
|
|
BCL2/adenovirus E1B 19 kDa interacting protein 3-like (BNIP3L) |
(aka NIX); BCL2/adenovirus E1B 19 kd-interacting protein (BNIP) gene that may function simultaneously with BNIP3 and play a role in tumor suppression. |
|
| PRKCD | Translocates into nucleus during apoptosis. Nuclear PRKCD regulates initiation of cytosolic apoptosis machinery, and subsequent caspase activation and DNA fragmentation. |
|
| Anti-apoptotic | AKT3 | Phosphorylate and inactivate BAD. Activates NFκB via IκB kinase regulation. Regulates cell signals in response to insulin and growth factors. |
| B-cell CL/lymphoma 2 (BCL2) | Blocks the release of pro-apoptotic cytochrome c and caspase activation. |
|
| NFκB | Inhibit caspases 3, 6, 7 stimulation via IAP (inhibitor of apoptosis) activation. |
|
| PIK3CB | Interacts with growth factor receptors; activates AKT3; target for PRKC. |
|
| RAF1 | Inhibits BIM and BAD activation via ERK1/2 stimulation. | |
| BCL2-related protein A1 (BCL2A1) | Reduces cytochrome c release from mitochondria and blocks caspase activation. |
|
| Baculoviral IAP repeat-containing 2 (BIRC2) | (aka CIAP1); Inhibits apoptosis by binding to tumor necrosis factor receptor-associated factors TRAF1 and TRAF2. |
|
| PRKCE | Blocks mitochondrial-dependent caspase activation; phosphorylates and activates RAF-1; phosphorylates and inactivates BAD; activates AKT via DNA-dependent protein kinase (DNA-PK). |
|
| IKBKE | Induces BCL-2 expression via NFκB signaling and interaction. | |
*Gene functions based on NCBI and/or selected publications (as indicated in the manuscript)
There is mounting epidemiological evidence that genetic alterations in apoptosis-related genes play an important role in tumorigenesis. Apoptosis-associated sequence variants, when considered individually, may minimally influence the risk of developing numerous cancers, such as multiple myeloma, squamous cell carcinoma, chronic lymphocytic leukemia, non-Hodgkin lymphoma, colorectal, ovarian, breast, pancreatic, and non-small cell lung [20-37]. However, the impact of individual apoptosis-related single nucleotide polymorphisms (SNPs) and their interactions on PCA outcomes remains understudied.
Genome wide association studies (GWAS), involving the evaluation of millions of SNPs within various biological pathways, has resulted in the detection of numerous PCA susceptibility loci [38]. However, most GWAS and PCA epidemiology studies place emphasis on individual SNP effects. Consequently, researchers often ignore the fact that complex diseases such as PCA are governed by complex gene-gene and gene-environment interactions within distinct biological pathways. Consequently, we sought to evaluate millions of interactions among apoptosis SNPs and their joint modifying impact on PCA risk and disease progression.
This report focuses on the impact of complex interactions among 172 apoptosis-related SNPs on PCA among 2,286 European-American male participants of the Cancer Genetic Markers of Susceptibility Study (CGEMS). The exhaustive assessment of all possible two-, three-, and four-way interactions among 172 SNPs, totaling more than 36 million SNP combinations is computationally burdensome. The statistical challenge stems from the prohibitive amount of data needed for multiple hypothesis testing. To address this issue, for the first time, we use statistical epistasis networks (SEN) guided multi-factor dimensionality reduction (MDR) to efficiently pre-process our genetic data, prior to modeling higher order interactions. As previously reported by Hu and co-workers (2011), SEN uses network science to generate a large connected cluster of pairwise interactions embedded within a genetic dataset [39]. The resulting network, consisting of a subset of high susceptibility SNPs, is used to guide SNP interactions using MDR. MDR is a rigorous statistical tool designed, in part, to evaluate main effects and complex interactions in relation to discrete outcomes [40-48]. This report may serve as a foundation for researchers interested in a state-of the art bioinformatics technique, namely SEN-guided MDR, which facilitates the logical prioritization of SNPs for gene-gene interaction analyses in relation to PCA susceptibility and prognosis.
Results
Cancer Genetic Markers of Susceptibility (CGEMS) Population Description
CGEMS study participants consisted of middle aged non-Hispanic Caucasian men, ranging in age from 55 to 81. Relative to control subjects, men diagnosed with PCA were more likely to have a family history of the disease (11.4% versus 6.3%), PSA levels ≥ 4 ng/ml (48.5% versus 6.5%), and abnormal DRE exams (60.7% versus 46.2%) [data not shown]. Men with aggressive PCA had both a Gleason score ≥ 7 and tumor stage III/IV. Although non-aggressive PCA cases primarily had a Gleason score < 7 and tumor stage I/II, a small percentage (1.8%) of them had a Gleason score > 6 and a tumor stage I/II. Among cases, there were no significant differences in family history of disease or PSA levels between men diagnosed comparing men with aggressive and non-aggressive disease (data not shown). The vast majority of subjects with aggressive PCA had higher Gleason scores than subjects with non-aggressive PCA, although 9 of the 488 non-aggressive cases (1.08%) had a Gleason score greater than 6.
Single SNP Effects
We examined 172 apoptosis-related sequence variants in relation to PCA risk and disease aggressiveness, as shown in Tables 2 and 3. Age and family history of PCA did not modify the relationship between the sequence variants and the risk of developing PCA or aggressive disease. Inheritance of one or more TNFRSF10B rs1001793 or TNFSF10 rs4894559 A alleles was nominally associated with a 1.23-1.38 fold increase in PCA risk (p ≤ 0.039), as shown in Table 2. Whereas, eight apoptosis-related SNPs were associated with a moderate 18-50% reduction in disease susceptibility [BIK rs4988366 AG & AG + GG; BNIP3L rs10503786 TT & TC + TT; CASP9 rs1052571 TT; IKBKE rs1539243 TC & TC + TT; PRKCE rs608139 CC; PRKCE rs935673 GG & AG + GG; TNFRSF1A rs4149576 AG, GG, & AG + GG; and TNFRSF1A rs4149577 TC & TC + CC] (p ≤ 0.043). However, after applying Bonferonni correction, none of these findings remained statistically significant under the per-allele, dominant and recessive genetic models.
Table 2.
Association of apoptosis SNPs with prostate cancer
| Marker (Alleles and position) |
Allele | Cases N (%) | Controls N (%) | OR (95%CI) | Adj OR (95%CI)* | p value |
Bonf. p value |
p trend |
Permut. p value |
|---|---|---|---|---|---|---|---|---|---|
| BIK | AA | 944 (81.2) | 855 (77.0) | 1.00 (reference) | 1.00 (reference) | 0.048 | 1.000 | 0.017 | 1.000 |
| rs4988366 | AG | 208 (17.9) | 244 (22.0) | 0.77 (0.63-0.95) | 0.78 (0.63-0.96) | 0.014 | |||
| A41830193G | GG | 12 (1.0) | 14 (1.3) | 0.78 (0.36-1.69) | 0.79 (0.36-1.71) | 0.523 | |||
| AG + GG | 220 (18.9) | 258 (23.2) | 0.78 (0.63-0.95) | 0.78 (0.64-0.96) | 0.014 | 1.000 | 0.831 | ||
| BNIP3L | CC | 556 (47.8) | 477 (42.9) | 1.00 (reference) | 1.00 (reference) | 0.041 | 1.000 | 0.012 | 0.998 |
| rs10503786 | TC | 480 (41.3) | 489 (44.0) | 0.84 (0.71-1.00) | 0.85 (0.71-1.01) | 0.055 | |||
| C26325853T | TT | 103 (8.9) | 122 (11.0) | 0.72 (0.54-0.97) | 0.73 (0.55-0.98) | 0.029 | |||
| TC + TT | 583 (50.1) | 611 (55.0) | 0.82 (0.70-0.97) | 0.82 (0.70-0.97) | 0.020 | 1.000 | 0.928 | ||
| CASP9 | CC | 320 (27.5) | 348 (31.3) | 1.00 (reference) | 1.00 (reference) | 0.075 | 1.000 | 0.023 | 1.000 |
| rs1052571 | TC | 589 (50.6) | 553 (49.8) | 0.88 (0.71-1.10) | 0.88 (0.71-1.09) | 0.265 | |||
| C15595919T | TT | 253 (21.8) | 210 (18.9) | 0.76 (0.60-0.97) | 0.75 (0.59-0.95) | 0.026 | |||
| TC + CC | 842 (72.4) | 763 (68.7) | 0.84 (0.68-1.03) | 0.83 (0.68-1.02) | 0.090 | 1.000 | 0.998 | ||
| IKBKE | CC | 837 (72.0) | 740 (66.6) | 1.00 (reference) | 1.00 (reference) | 0.017 | 1.000 | 0.005 | 0.903 |
| rs1539243 | TC | 291 (25.0) | 325 (29.3) | 0.79 (0.66-0.95) | 0.78 (0.65-0.95) | 0.014 | |||
| C203036182T | TT | 36 (3.1) | 47 (4.2) | 0.68 (0.43-1.06) | 0.68 (0.43-1.06) | 0.086 | |||
| TC + TT | 327 (28.1) | 372 (33.5) | 0.78 (0.65-0.93) | 0.77 (0.64-0.92) | 0.006 | 0.943 | 0.522 | ||
| PRKCE | TT | 841 (72.3) | 788 (70.9) | 1.00 (reference) | 1.00 (reference) | 0.030 | 1.000 | 0.139 | 0.987 |
| rs608139 | TC | 304 (26.1) | 288 (25.9) | 0.99 (0.82-1.19) | 0.98 (0.51-1.19) | 0.909 | |||
| T45789207C | CC | 18 (1.5) | 36 (3.2) | 0.47 (0.26-0.83) | 0.50 (0.28-0.88) | 0.010 | |||
| TC + CC | 322 (27.7) | 324 (29.2) | 0.93 (0.78-1.12) | 0.93 (0.78-1.12) | 0.435 | 1.000 | 1.000 | ||
| PRKCE | AA | 556 (47.8) | 482 (43.4) | 1.00 (reference) | 1.00 (reference) | 0.045 | 1.000 | 0.013 | 1.000 |
| rs935673 | AG | 503 (43.3) | 503 (45.3) | 0.87 (0.73-1.03) | 0.87 (0.73-1.04) | 0.107 | |||
| A46034008G | GG | 105 (9.0) | 128 (11.5) | 0.71 (0.53-0.95) | 0.71 (0.53-0.95) | 0.019 | |||
| AG + GG | 608 (52.3) | 631 (56.8) | 0.84 (0.71-0.99) | 0.84 (0.71-0.99) | 0.034 | 1.000 | 0.991 | ||
| TNFRSF10B | GG | 547 (47.0) | 537 (48.3) | 1.00 (reference) | 1.00 (reference) | 0.055 | 1.000 | 0.253 | 1.000 |
| rs1001793 | AG | 448 (38.5) | 458 (41.2) | 0.96 (0.80-1.15) | 0.96 (0.80-1.14) | 0.653 | |||
| G22956894A | AA | 131 (11.3) | 93 (8.4) | 1.38 (1.03-1.85) | 1.38 (1.03-1.85) | 0.029 | |||
| AG + AA | 579 (49.8) | 551 (49.6) | 1.01 (0.86-1.19) | 1.03 (0.87-1.22) | 0.892 | 1.000 | 1.000 | ||
| TNFRSF1A | AA | 375 (32.2) | 376 (33.8) | 1.00 (reference) | 1.00 (reference) | 0.079 | 1.000 | 0.070 | 1.000 |
| rs4149576 | AG | 545 (46.9) | 545 (79.1) | 0.78 (0.62-0.98) | 0.79 (0.63-0.99) | 0.034 | |||
| A6319376G | GG | 234 (20.1) | 183 (16.5) | 0.78 (0.61-0.99) | 0.79 (0.62-1.01) | 0.043 | |||
| AG + GG | 779 (67.0) | 728 (65.5) | 0.78 (0.63-0.97) | 0.79 (0.63-0.98) | 0.024 | 1.000 | 0.854 | ||
| TNFRSF1A | TT | 326 (28.0) | 270 (24.3) | 1.00 (reference) | 1.00 (reference) | 0.087 | 1.000 | 0.205 | 1.000 |
| rs4149577 | TC | 571 (49.1) | 591 (5302) | 0.80 (0.66-0.98) | 0.80 (0.66-0.98) | 0.027 | |||
| T6317783C | CC | 265 (22.8) | 251 (22.6) | 0.87 (0.69-1.11) | 0.88 (0.70-1.12) | 0.266 | |||
| TC + CC | 836 (71.9) | 842 (75.8) | 0.82 (0.68-0.99) | 0.83 (0.68-1.00) | 0.038 | 1.000 | 0.141 | ||
| TNFSF10 | GG | 630 (54.2) | 668 (60.1) | 1.00 (reference) | 1.00 (reference) | 0.026 | 1.000 | 0.014 | 0.977 |
| rs4894559 | AG | 456 (39.2) | 380 (34.2) | 1.27 (1.07-1.51) | 1.27 (1.06-1.51) | 0.007 | |||
| G173716071A | AA | 76 (6.5) | 65 (5.9) | 1.24 (0.88-1.78) | 1.23 (0.87-1.74) | 0.227 | |||
| AG + AA | 532 (45.7) | 445 (40.1) | 1.26 (1.06-1.49) | 1.26 (1.07-1.49) | 0.007 | 1.000 | 0.594 | ||
Abbreviations: Bonf., Bonferroni; Permut., Permutation; *adjusted for age and family history of PCA
Table 3.
Association of apoptosis SNPs with aggressive prostate cancer
| Marker (Alleles and position) |
Allele | Cases N (%) | Controls N (%) | OR (95%CI) | Adj OR (95%CI)* | p value |
Bonf. p value |
p trend |
Permut. p value |
|---|---|---|---|---|---|---|---|---|---|
| AKT3 | CC | 295 (42.9) | 244 (50.0) | 1.00 (reference) | 1.00 (reference) | 0.065 | 1.000 | 0.028 | 1.000 |
| rs10157763 | TC | 307 (44.7) | 192 (39.3) | 1.31 (1.02-1.67) | 1.33 (1.04-1.71) | 0.027 | |||
| C240321082T | TT | 75 (10.9) | 45 (9.2) | 1.21 (0.82-1.78) | 1.39 (0.92-2.09) | 0.344 | |||
| TC + TT | 382 (55.6) | 237 (48.6) | 1.32 (1.04-1.66) | 1.34 (1.06-1.70) | 0.020 | 1.000 | 0.943 | ||
| AKT3 | GG | 438 (63.8) | 345 (70.7) | 1.00 (reference) | 1.00 (reference) | 0.005 | 0.853 | 0.038 | 0.493 |
| rs10927067 | AG | 227 (33.0) | 119 (24.4) | 1.50 (1.15-1.95) | 1.51 (1.16-1.96) | 0.002 | |||
| G240249285A | AA | 20 (2.9) | 19 (3.9) | 0.83 (0.44-1.58) | 0.82 (0.43-1.57) | 0.568 | |||
| AG + AA | 247 (36.0) | 138 (28.3) | 1.43 (1.12-1.84) | 1.41 (1.10-1.82) | 0.005 | 0.977 | 0.511 | ||
| AKT3 | TT | 483 (70.3) | 380 (77.9) | 1.00 (reference) | 1.00 (reference) | 0.002 | 0.356 | 0.023 | 0.250 |
| rs12031994 | TC | 192 (27.9) | 94 (19.3) | 1.61 (1.21-2.13) | 1.61 (1.22-2.14) | 0.009 | |||
| T240243350C | CC | 13 (1.9) | 14 (2.9) | 0.73 (0.34-1.57) | 0.73 (0.34-1.57) | 0.422 | |||
| TC + CC | 205 (29.8) | 108 (22.1) | 1.51 (1.15-1.98) | 1.50 (0.15-1.96) | 0.003 | 0.548 | 0.372 | ||
| AKT3 | GG | 436 (63.5) | 341 (69.9) | 1.00 (reference) | 1.00 (reference) | 0.030 | 1.000 | 0.068 | 0.984 |
| rs2125230 | AG | 231 (33.6) | 129 (26.4) | 1.40 (1.09-1.81) | 1.40 (1.08-1.82) | 0.010 | |||
| G240211889A | AA | 21 (3.1) | 18 (3.7) | 0.91 (0.48-1.74) | 0.91 (0.48-1.74) | 0.781 | |||
| AG + AA | 252 (36.7) | 147 (30.1) | 1.35 (1.06-1.73) | 1.34 (1.05-1.72) | 0.017 | 1.000 | 0.938 | ||
| AKT3 | GG | 296 (43.1) | 238 (48.8) | 1.00 (reference) | 1.00 (reference) | 0.083 | 1.000 | 0.027 | 1.000 |
| rs2125231 | AG | 314 (45.7) | 203 (41.6) | 1.24 (0.97-1.59) | 1.25 (0.98-1.60) | 0.082 | |||
| G240088340A | AA | 75 (10.9) | 42 (8.6) | 1.43 (0.95-2.17) | 1.44 (0.95-2.18) | 0.087 | |||
| AG + AA | 389 (56.6) | 245 (50.2) | 1.29 (1.02-1.63) | 1.28 (1.01-1.62) | 0.031 | 1.000 | 0.992 | ||
| AKT3 | CC | 295 (42.9) | 243 (49.8) | 1.00 (reference) | 1.00 (reference) | 0.060 | 1.000 | 0.022 | 1.000 |
| rs2345994 | TC | 312 (45.4) | 195 (40.0) | 1.32 (1.03-1.69) | 1.32 (1.03-1.70) | 0.028 | |||
| C240258316T | TT | 78 (11.4) | 45 (9.2) | 1.43 (0.95-2.14) | 1.44 (0.95-2.16) | 0.084 | |||
| TC + TT | 390 (56.8) | 240 (49.2) | 1.32 (1.05-1.67) | 1.35 (1.06-1.70) | 0.018 | 1.000 | 0.946 | ||
| AKT3 | CC | 435 (63.3) | 348 (71.3) | 1.00 (reference) | 1.00 (reference) | 0.009 | 1.000 | 0.029 | 0.714 |
| rs4132509 | AC | 227 (33.0) | 121 (24.8) | 1.50 (1.15-1.95) | 1.51 (1.16-1.96) | 0.002 | |||
| C240269125A | AA | 18 (2.6) | 16 (3.3) | 0.90 (0.45-1.79) | 0.88 (0.44-1.75) | 0.764 | |||
| AC + AA | 245 (35.7) | 137 (28.1) | 1.43 (1.11-1.84) | 1.44 (1.11-1.85) | 0.005 | 1.000 | 0.581 | ||
| BCL2L14 | GG | 436 (63.5) | 343 (70.3) | 1.00 (reference) | 1.00 (reference) | 0.041 | 1.000 | 0.025 | 0.998 |
| rs885720 | AG | 231 (33.6) | 131 (26.8) | 1.39 (1.07-1.79) | 1.39 (1.08-1.80) | 0.012 | |||
| G12139366A | AA | 21 (3.1) | 14 (2.9) | 1.18 (0.59-2.25) | 1.20 (0.60-2.39) | 0.639 | |||
| AG + AA | 252 (36.7) | 145 (29.7) | 1.37 (1.07-1.76) | 1.37 (1.07-1.76) | 0.013 | 1.000 | 0.843 | ||
| CARD8 | CC | 462 (67.2) | 362 (74.2) | 1.00 (reference) | 1.00 (reference) | 0.016 | 1.000 | 0.005 | 0.882 |
| rs10405717 | TC | 202 (29.4) | 111 (22.7) | 1.45 (1.11-1.90) | 1.45 (1.11-1.90) | 0.007 | |||
| C53443620T | TT | 21 (3.1) | 10 (2.0) | 1.70 (0.79-3.68) | 1.70 (0.79-3.68) | 0.173 | |||
| TC + TT | 223 (32.5) | 121 (24.8) | 1.45 (1.12-1.89) | 1.47 (1.13-1.91) | 0.005 | 0.754 | 0.459 | ||
| IKBKE | TT | 251 (36.5) | 209 (42.8) | 1.00 (reference) | 1.00 (reference) | 0.008 | 1.000 | 0.003 | 0.682 |
| rs11578093 | TG | 332 (48.3) | 233 (47.7) | 1.19 (0.93-1.52) | 1.17 (0.91-1.50) | 0.177 | |||
| T203059049G | GG | 102 (6.0) | 46 (9.4) | 1.85 (1.25-2.74) | 1.81 (1.22-2.69) | 0.002 | |||
| TG + GG | 434 (63.2) | 279 (57.2) | 1.31 (1.03-1.66) | 1.27 (1.00-1.62) | 0.027 | 1.000 | 0.982 | ||
| IKBKE | CC | 502 (73.1) | 341 (69.9) | 1.00 (reference) | 1.00 (reference) | 0.048 | 1.000 | 0.076 | 1.000 |
| rs1539243 | TC | 172 (25.0) | 125 (25.6) | 0.93 (0.71-1.22) | 0.95 (0.73-1.24) | 0.622 | |||
| C203036182T | TT | 14 (2.0) | 22 (4.5) | 0.43 (0.22-0.86) | 0.44 (0.22-0.88) | 0.016 | |||
| TC + TT | 186 (27.1) | 147 (30.1) | 0.86 (0.66-1.11) | 0.88 (0.68-1.13) | 0.244 | 1.000 | 1.000 | ||
| PIK3CB | TT | 215 (31.3) | 131 (26.8) | 1.00 (reference) | 1.00 (reference) | 0.060 | 1.000 | 0.021 | 1.000 |
| rs500687 | TC | 334 (48.6) | 234 (48.0) | 0.87 (0.66-1.14) | 0.87 (0.66-1.14) | 0.318 | |||
| T139942901C | CC | 134 (19.5) | 121 (24.8) | 0.67 (0.49-0.94) | 0.66 (0.48-0.92) | 0.019 | |||
| TC + CC | 468 (68.1) | 355 (72.7) | 0.81 (0.62-1.04) | 0.80 (0.62-1.03) | 0.099 | 1.000 | 1.000 | ||
| RAF1 | TT | 508 (73.9) | 377 (77.3) | 1.00 (reference) | 1.00 (reference) | 0.010 | 1.000 | 0.473 | 0.764 |
| rs13060691 | TG | 167 (24.3) | 90 (18.4) | 1.38 (1.03-1.84) | 1.38 (1.03-1.84) | 0.030 | |||
| T12653013G | GG | 10 (1.5) | 16 (3.3) | 0.44 (0.20-0.97) | 0.45 (0.20-1.01) | 0.060 | |||
| TG + GG | 177 (25.8) | 106 (21.7) | 1.24 (0.94-1.63) | 1.24 (0.94-1.63) | 0.123 | 1.000 | 1.000 | ||
Abbreviations: Bonf., Bonferroni correction; Permut., Permutation; *adjusted for age and family history of PCA (PCA)
In terms of disease progression, eleven apoptosis SNPs were modestly associated with a 1.28-1.81 fold increase in the risk of developing aggressive PCA [AKT3 rs10157763 TC & TC + TT; AKT3 rs10927067 AG & AG + AA; AKT3 rs12031994 TC & TC + CC;AKT3 rs2125230 AG & AG + AA; AKT3 rs2125231 AG + AA; AKT3 rs2345994 TC & TC + CC; AKT3 rs4132509 AC & AC + AA; BCL2L14 rs885720 AG & AG + AA; CARD8 rs10405717 TC & TC + TT; IKBKE rs11578093 GG & TG + GG; and RAF1 rs13060691 TG] (p ≤ 0.031), as summarized in Table 3. In addition, possession of IKBKE rs1539243 TT and PIK3CB rs500687 CC genotypes were moderately linked with a 44-56% reduction in aggressive PCA susceptibility (p ≤ 0.019). However, none of these loci remained significant after adjusting for multiple comparison bias.
Combined Gene Effects
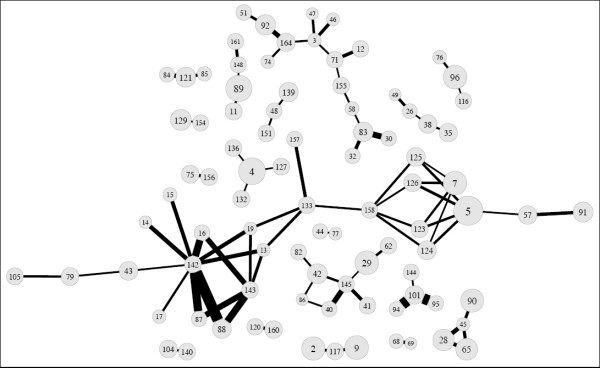
We investigated the ability of gene-gene interactions to predict PCA outcomes using statistical epistasis networks (SEN) guided multifactor dimensionality reduction (MDR), information gain (IG) measure, and hierarchical interactions graphs (hIG). SEN was used to generate a network to visualize the genetic architecture of PCA outcomes, prior to an exhaustive search of SNP interactions. This approach uses information theory to build an epistasis network from the strongest pairwise SNP interactions. A network, consisting of vertices (i.e., SNPs) joined by edges (i.e., SNP pairs), enabled us to limit our MDR analysis to a subset of the most informative SNPs in relation to PCA outcomes. We were unable to build a statistically significant epistasis network for PCA susceptibility. However, the network for aggressive PCA was topographically significant (p < 0.05), given the results from 1000 permutated data sets. This network consisted of 91 vertices (i.e., individual SNP effects), 80 edges (i.e., pairwise interactions), and 18 total components (i.e., "sub-networks" of vertices and edges). The largest connected component of the aggressive PCA SEN diagram involved 24 vertices and 34 edges, as shown in Figure 1 andAdditional file 1. Consequently, MDR analysis was limited to < 13,000 interactions among 24 SNPs, instead of more than 36 million interactions for 172 SNPs. SEN-guided MDR revealed complex interactions between AKT3 rs12031994-PRKCQ rs571715 as well as AKT3 rs12031994-BID rs366542-PRKCQ rs571715 that were significantly associated with disease aggressiveness (p ≤ 0.009). However, the network and entropy graphs indicated these findings were mainly driven by AKT3 rs12031994. This is evident by the marker's large vertex size (i.e., large main effect) in the SEN graph (Figure 1) and a lack of a strong pairwise interaction between AKT3 rs12031994-PRKCQ rs571715 in the entropy graph. The AKT3-PRKCQ interaction resulted in joint mutual information (I) of only 0.54% relative to the mutual information (I) of 0.87% for AKT3 alone. Therefore, prior to repeating MDR, we removed the AKT3rs12031994 marker. This resulted in the detection of a modest synergistic interaction between AKT3 rs2125230-PRKCQ rs571715 (p = 0.011), as shown in Table 4. Based on MDR, this model was the best two-factor model. It was selected 9 out of 10 times by MDR based on a cross-validation consistency (CVC) score of 90% and predictive accuracy of 56.3%. Furthermore, we observed a higher mutual information for the combined effect of AKT3 rs2125230-PRKCQ rs571715 (I = 0.66%), in contrast to AKT3 rs2125230 alone (I = 0.45%) or PRKCQrs571715 alone (I = 0.21%), as revealed in Figure 2.
Figure 1.
Visualization of the interaction among apoptosis-related sequence variants using Statistical Epistasis Network Modeling (SEN)-guided MDR. The Statistical Epistasis Network (SEN) was generated using a pairwise interaction strength cut-off or 1%. Overall, the model has 91 vertices (single SNPs), 80 edges (pairwise SNPs), and 18 components ("sub-networks" of vertices and edges). The largest connected component, shown in the center and spanning the entire length of this model, has 24 vertices and 34 edges. Each number in Figure 1 corresponds with a specific apoptosis-related SNP analyzed in the current study, as summarized in Additional file 2.
Table 4.
Multi-dimensionality reduction models for apoptosis-related markers in relation to aggressive prostate cancer (PCA)
| Best Model | Cross Validation Consistency (CVC)* |
Average Testing Accuracy (ATA)* |
Permutation Testing p value* |
|---|---|---|---|
|
One Factor AKT3 rs2125230 |
9/10 | 0.512 | 0.282 |
|
Two Factor AKT3 rs2125230 PRKCQ rs571715 |
9/10 | 0.563 | 0.011 |
|
Three Factor AKT3 rs2125230 BID rs366542 PRKCQ rs571715 |
6/10 | 0.529 | 0.127 |
|
Four Factor AKT3 rs2125230 BCL2L14 rs2448050 BID rs366542 TNFRSF1A rs4149576 |
2/10 | 0.517 | 0.321 |
*Adjusted for age and family history of prostate cancer (PCA)
Figure 2.

Interaction Entropy model. This graphical model, describes the percent entropy that is explained by each apoptosis-related SNP or pairwise combination within our study population. Positive percent entropy indicates information gain (IG) or synergy; whereas, negative percent indicates redundancy or lack of information gain (IG). Schematic coloration used in the visualization tools represents a continuum from synergy (i.e. non-additive) to redundancy. The colors range from red representing a high degree of synergy (positive information gain (IG)), orange a lesser degree, and gold representing independence and a midway point between synergy and redundancy. On the other hand, green and blue represent redundancy, which is not apparent in the current analysis.
To confirm the validity of the SEN results, MDR analysis was also conducted on all 172 SNPs (Figure 1 andAdditional file 1) as well as 148 loci that were not included in the main component of the network. As expected, we did not detect any significant two-, three-, or four- way interaction models (p ≥ 0.184).
Discussion
Previous genome-wide association studies (GWAS) have identified linkages about 40 PCA loci, including between PCA genetic alterations detected in the 8q24 region, β-microseminoprotein (MSMB), and allele -8 of the microsatellite DG8S737 [38]. These studies have limited their scope to individual SNPs across the entire genome. Such assessments tend to ignore the genetic architecture of PCA that potentially involves complex interactions along key regulatory pathways. For the first time, the current study evaluates complex interactions among 172 apoptosis-related SNPs in relation to PCA risk and disease aggressiveness among 2,286 European-American men using SEN-guided MDR. Specifically, SEN was used to build a topographically significant aggressive PCA epistasis network, prior to evaluating complex interactions. This inferred epistasis network consisted of 24 SNPs and 34 pairwise interactions, and reduced MDR analysis from > 36 million to < 13,000 SNP interactions. Consequently, we observed a non-linear and modest interaction between AKT3 rs2125230-PRKCQ rs571715 in relation to aggressive PCA. This state-of-the-art bioinformatics technique facilitates the logical prioritization of SNPs for gene-gene interaction analyses in relation to complex diseases.
Unfortunately, there are no published reports on the functional consequence of these two intronic SNPs in AKT3 and PRKCQ in relation to mRNA stability/expression protein expression/structure/function or PCA outcomes. However, we speculate that the AKT3 rs2125230 and PRKCQ rs571715 sequence variants, with minor allele frequencies ranging from 14.4-22.9 among men of European descent, may alter transcription regulation, leading to increased mRNA expression. Increased mRNA/protein expression AKT3 rs2125230 and PRKCQ rs571715 may cause: decreased apoptosis, an escape of transformed cells from programmed cell death, increased accumulation of genetic alterations, genomic instability, and ultimately an invasive PCA phenotype. Thus, in vitro and in vivo assays using (short hairpin RNAs) shRNAs or small interfering RNAs (siRNAs) are needed to elucidate the impact of AKT3-PRKCQ genetic alterations on protein expression, apoptosis capacity, and prostate tumorigenesis.
The impact of a non-linear interaction along the AKT3 rs2125230-PRKCQ rs571715 axis in relation to aggressive PCA may be attributed to markers involved in the apoptosis signaling pathway. Overexpression of PRKCQ and AKT3 are associated with invasive cancer phenotypes [6-19]. In fact, AKT3 is responds to insulin and growth factors, transduces signals including cell death, and is upregulated in androgen-independent PCA cell lines [49]. PRKCQ is also associated with apoptosis. In particular PRKCQ, a protein kinase C (PRKC) family member, promotes cell survival by inactivating BAD (BCL2-associated agonist of cell death), which subsequently results in NFκB activation.
Although AKT3 and PRKCQ are involved in pro-survival pathways, their interaction is not fully understood. However, their interactions with other related protein kinases may offer biological clues on the mechanism by which AKT3 and PRKCQ synergistically influence aggressive PCA. For example, PRKCQ interacts with another AKT family member, AKT1, to activate NFκB [50]. If the AKT3-PRKCQ axis has a similar function as other protein kinases, namely AKT1 and PRKCQ, then these pro-survival markers may synergistically activate NFκB. As a result, activated NFκB may enable the tumor to escape programmed cell death and progress toward an aggressive PCA phenotype.
Numerous observational studies evaluated the impact of one or more apoptosis-related SNPs in relation to cancer outcomes [20-37]. Among 15 case-control studies, less than 10% of the sequence variants evaluated in the current study were significantly associated with various tumors, including of the colon, rectal, ovarian, breast, pancreatic, and non-small cell lung cancers [20-37]. In particular, modest cancer risk estimates were observed among 8 apoptosis-related SNPs detected in CASP3, CASP8, CASP9, TP53, NFKB2, and NFKBIA. However, some of these studies were limited by a small sample size, small number of analyzed SNPs, or failure to consider the impact of multiple SNPs on disease susceptibility. In the current study, there were 24 SNPs detected in 10 apoptosis-related genes [i.e., AKT3, BIK, BNIP3L, CARD8, CASP9, IKBKE, PRKCE, TNFSF10, and TNFRSF10 (B, D)]. These apoptosis-related SNPs, after adjusting for confounders, were modestly associated with PCA risk and/or aggressive disease. Yet, these findings lost statistical significance after adjusting for multiple hypothesis testing. Main effects were not observed for AKT3 rs2125230 and PRKCQ rs571715 in relation to PCA risk or disease progression in our study set. Wang and co-workers (2009) evaluated interactions among 5 apoptosis-related SNPs, including death receptor 4 (DR4), and pack-years of smoking in relation to bladder cancer using entropy-based MDR [51]. MDR analysis revealed a significant additive interaction between DR4 -397 G > T and smoking on bladder cancer. Unfortunately, this study analyzed a relatively small number of sequence variants in the DR4 apoptosis-related gene; hence, making it difficult to compare study findings. In a post-hoc analysis, we did not observe any interactions among the selected apoptosis-related markers and sources of reactive oxygen species, antioxidants, and anti-inflammatory agents, including cigarette smoking, dietary supplements, aspirin, ibuprofen, and meat-derived carcinogens (data not shown).
We considered the strengths, limitations and future directions of the current study. Although SEN-guided MDR only identified a nominally significant network for disease aggressiveness in the current study, this approach overcomes the computational challenge of detecting all possible two-, three- and four-way SNP combinations involved in PCA progression. SEN, in the current study, was used to prioritize > 10 million possible interactions by focusing on a "sub-network" of informative SNPs in relation to aggressive PCA. Reduction of our genetic data set to the most informative markers improved the feasibility to detect interactions that may have otherwise remained undetected. Unfortunately, the genome-wide association studies (GWAS) database used for the current study did not include apoptosis-related sequence variants (e.g., TNF-308 rs1800629, TNFSF10 rs1131532, BCL2 -936 rs2279115) previously reported in published cancer epidemiology studies [52-54]. Future studies in our laboratory will focus on high-throughput targeted sequencing to evaluate the impact of novel and commonly reported sequence variants on PCA susceptibility and disease prognosis. In light of recent GWAS reports, it is tempting to assume that extremely large case-control study sets, involving tens of thousands subjects are required to evaluate millions of SNP interactions in relation to PCA outcomes. However, the current study had adequate statistical power to evaluate individual and SNP combination effects in relation to prostate cancer. In particular, MDR has 80% statistical power to evaluate all possible two-, three-, and four-way gene-gene interactions with as low as 200 cases and 200 controls [55]. MDR remains effective even in the presence of 5% genotyping errors and/or 5% missing data [55]. It is anticipated that 5% of the about 13,000 possible interactions among 24 apoptosis-related SNPs will result in approximately 650 significant relationships due to chance alone. However, MDR coupled with permutation testing adjust for multiple comparison bias. Given the low prediction accuracy affiliated with the interaction between AKT3 and PRKCQ, our study findings require replication within independent study sets. However, recent simulation studies demonstrate that even modest disparities in genotype allele frequencies among study participants of independent study sets may interfere with the capacity to replicate complex interactions [56]. Thus, to ensure reproducibility within future studies, we plan to select study sets with similar genetic architecture (i.e., ancestry identification markers and SNP genotypes) as CGEMS PCA case-control subjects.
Conclusion
In summary, we have identified a marginal interaction between two apoptosis-related SNPs linked to PCA disease aggressiveness using SEN-guided MDR. Future molecular and pre-clinical studies may help to clarify the functionality of these genetic variants and their role in PCA disease progression. Our state-of-the art bioinformatics approach will enable researchers to pre-process millions of SNP interactions using GWAS or cancer consortia genotype data. In order to replicate findings within independent study sets, researchers will have to ensure that comparative sub-populations have the same genetic architecture and ancestry backgrounds. Therefore, the application of SEN-guided MDR may ultimately help researchers identify and validate gene-gene and/or gene-environment interactions to serve as effective cancer prognostication tools.
Methods
Study population
The Prostate, Lung, Colon, and Ovarian (PLCO) Cancer Screening Trial is a randomized, well-designed, and multi-center investigation sponsored and coordinated by the National Cancer Institute (NCI) [57,58]. Between 1993 and 2001, the PLCO Trial recruited study participants aged 55 to 81 years old to evaluate the effect of screening on disease specific mortality, relative to standard care. Men randomized to the PCA arm of the trial, received annual PSA and DRE exams. For the current study, 2,286 European-American men were included in our study if they had GWAS data available through the Cancer Genetic Markers of Susceptibility (CGEMS) data portal. (http://cgems.cancer.gov/) [38]. The CGEMS study population consists of nationally available genetic data for nearly 500,000 sequence variants. Incident PCA cases (488 non-aggressive and 687 aggressive) were identified through various sources including: screening exams; reports from patients, physicians, or relatives; or linkage with the National Death Index or state cancer registries. Incident PCA cases were pathologically confirmed with aggressive (Gleason score ≥ 7 and tumor stage III/IV) or non-aggressive [Gleason score (< 7) and tumor stage I/II] disease. Controls (n = 1,111) were matched to cases identified based on age, time since initial screening, and year of blood draw using incidence density sampling. All participants signed informed consent documents approved by both the NCI and local institutional review boards. Access to clinical and background data collected through examinations and questionnaires was approved for use by the PLCO.
Gene selection
A panel of 73 apoptosis-associated genes was selected from published cancer epidemiology or molecular biology studies as well as pathway databases and tools, including Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg), Gene Ontology (GO), BioCarta (http://www.biocarta.com), ProteinLounge (http://www.proteinlounge.com), and Ingenuity (http://www.ingenuity.com) [59-63]. We searched PubMed for articles using the following keywords: [(prostate OR prostatic) AND (cancer OR neoplasms) AND apoptosis (gene variants OR single nucleotide polymorphisms OR targets) AND epidemiology]. Pathway tools, such as KEGG, BioCarta, ProteinLounge, and Ingenuity were used to visualize gene-gene and protein-protein interactions essential to regulating apoptosis [59-61,63]. The Entrez Gene database of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) was used to ensure that selected targets were involved in the apoptotic pathway [64].
Criteria for SNP selection and data management
Prior to uploading our initial list (i.e., 73 apoptosis genes) into the CGEMS data portal, we secured the HUGO gene name equivalents for the targets of interest using NCBI Entrez Gene. SNP profile data was available for more than 1197 SNPs [38]. However, upon further investigation, many of these SNPs were eliminated if they were: not located within 2.5 kb of the 5' or 3' end of the gene; or detected within a gene that was not related to apoptosis. Among SNPs that were associated with apoptosis, we focused on gene regions located in coding, promoter, or "near gene" regions. "Near gene" regions were defined as 2.5 kb up- and downstream of the 5' or 3' ends of the selected genes, respectively.
We excluded any sequence variants that had a minor allele frequency (MAF) < 5% as reported in the NCBI Entrez SNP [64]. In addition, one SNP was removed since its genotype frequency distribution among controls deviated substantially from the Hardy-Weinberg Equilibrium, p ≤ 0.005 (n = 1). Following data cleanup, 172 apoptotic SNPs were analyzed among 2,286 men of European descent (687 aggressive cases, 488 non-aggressive cases, and 1,111 controls).
Predicted function of selected SNPs
The SNPinfo (http://snpinfo.niehs.nih.gov/) webserver enabled us to annotate and/or predict the functional consequence of selected apoptosis variants, as summarized in Additional file 2[65]. This server consists of several pipelines to predict whether alternative alleles of a SNP may alter one or more of the following: transcriptional regulation via transcription factor binding site (TFBS) activity; premature termination of amino-acid sequence (stop codons); the splicing pattern or efficiency at mRNA splice sites, exonic splicing enhancers (ESE) or silencers (ESS); protein structures or properties by changing single amino acids (i.e., non-synonymous SNPs); or mRNA transcription or protein translation by altering microRNA (miRNA) binding sites.
Statistical analysis for single gene effects
Univariate and multivariate analyses were used to evaluate apoptosis associated SNPs among men of European descent in relation to PCA outcomes. To assess whether possession of one or more apoptotic alleles influence the risk of developing PCA, we tested for significant differences in the distribution of homozygous major, heterozygous, or homozygous minor genotypes between cases and controls using the chi-square test of heterogeneity. A case-only analysis was used to examine the relationship between apoptosis-related alleles and aggressive PCA. We evaluated differences in the distribution and inheritance of apoptosis-related genes comparing men with aggressive disease (Gleason score ≥ 7 and tumor stage > 2) to those with non-aggressive disease (i.e., other PCA cases that did not have both Gleason score ≥ 7 and tumor stage > 2). The associations between PCA outcomes and selected polymorphic genes, expressed as odds ratios (ORs) and corresponding 95% confidence intervals (CIs), were estimated using unconditional multivariate logistic regression (LR) models adjusted for age and family history of PCA. LR analyses for genetic variants and PCA outcomes were conducted using the major or common genotype as the referent category. All chi-square test and LR analyses were conducted using SAS 9.2 (SAS Institute Inc., Cary, NC) and SVS software (Golden Helix, Inc., Bozeman, MT). Statistical significance was assessed using a p-value < 0.05. Adjustments for multiple comparisons were made using Bonferroni correction and permutation testing.
Statistical power
We performed calculations to determine the statistical power of our sample to detect significant relationships between apoptosis-related polymorphisms and PCA development. The expected risk estimates of our study were determined by specifying values for a number of parameters, including: an average minor allele frequency of at least 26.0%, NCI's estimate of PCA disease prevalence (19%); statistical power (80%); sample size (1,175 PCA cases and 1,111 disease-free individuals or 687 aggressive PCA cases and 488 non-aggressive cases). We assumed the outcome was in complete linkage disequilibrium with an apoptosis-predisposing variant (r2 = 1.0). Based on the aforementioned parameters, we have > 80% power to detect genetic markers with odds ratios (ORs) of ≥ 1.4 (or 0.71 for protective effects) for PCA risk and ≥ 1.6 (or 0.62 for protective effects) for disease aggressiveness, assuming a codominant model with 1 degree of freedom (df). Power calculations were performed using Power for Genetic Association Version 2 Software [66].
Analysis of gene interactions using multi-factor dimensionality reduction (MDR)
To evaluate the single- and joint- modifying effects of 172 candidate apoptosis-related SNPs within a large dataset, such as CGEMS, is computationally challenging [67,68]. In order to overcome this problem, open-source and freely available MDR 2.0 software (htpp://http://www.epistasis.org) was used to analyze interactions among apoptotic sequence variants in relation to PCA outcomes [69]. MDR is a data reduction approach, designed to detect and characterize high-order interactions in case-control studies. With MDR, information from various genetic loci is categorized into high-risk or low-risk groups of disease. The resulting one-dimensional multi-locus genotype variable is then evaluated for its ability to classify and predict a disease outcome through cross-validation and permutation testing. MDR uses a 10-fold cross validation to estimate the testing accuracy of a model by leaving out 1/10th of the data as an independent test set. The model is developed on 9/10th of the data and then evaluated on the remaining test set. This process is repeated for each 1/10th of the data and the resulting prediction accuracies are averaged.
In the current study, the model with the greatest cross validation consistency (i.e., CVC ≥ 8/10) and highest prediction accuracy [i.e., Average Testing Accuracy (ATA)] was selected as the best predictor of disease outcome. Accuracy is a function of the percentage of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) as defined as (TP + TN)/(TP + TN + FP + FN). ATAs are averaged across all 10 pieces of the data, in order to provide an estimate of the predictive ability of the loci in relation to the outcome of interest. We used cross-validation consistency (CVC) to measure the degree to which the same best MDR model was selected across the 10 divisions of the data. Models with a CVC of ≥ 8/10 using a 10-fold cross-validation were considered more carefully. If the MDR model met the CVC criteria, we selected models that had the highest ATAs. The combination of CVC and permutation testing were used to control for multiple hypothesis testing. Permutation testing results were statistically significant at the ≤ 0.05 level.
The current version of MDR used in this study enabled the incorporation and adjustment of multiple covariates [70]. To remove the covariate effects [i.e., age-group and family history of PCA (yes or no)], we integrated two sampling methods (i.e., over- and under-sampling). This approach is computationally efficient, since it allows for the adjustment of multiple covariates without significantly increasing computational burden.
Visualization of interaction models using interaction entropy algorithms, hierarchical graphs and statistical epistasis network (SEN)
The interaction entropy algorithm, based on information theory, is a method to verify, visualize, and interpret combination effects identified by parametric (e.g., LR) and non-parametric (e.g., MDR) statistical test[43,71-74]. Jakulin and Brakto (2003) have developed a metric to gauge whether the gain in information (i.e., information gain) about a class variable (i.e., ability to predict disease status) from a combination of two variables provides more information than each variable considered independently [72,73]. In the current study, measures of interaction were used to build interaction entropy graphs to visualize and interpret interactions among selected apoptosis-related markers in relation to PCA risk and disease progression. Individual and all possible pairwise loci were assigned a mutual information (I) percentage score in relation to disease risk or aggressiveness; whereby typical scores for genetic loci are < 5%. Pairwise SNP combinations were deemed important if the joint mutual information [I = (SNP1, SNP2; disease status)] was greater than the total mutual information of each individual locus considered separately [I (SNP1; disease status) + I (SNP2; disease status)]. Interactions were further visualized using an interaction entropy graph, which uses a color-coding scheme to interpret interactions. Strong interacting factors, coded either red or orange, indicated high and medium levels of synergistic effects on outcomes, respectively. Weak interacting factors, coded either blue or green, denoted high or modest levels of redundancy between markers, respectively. Gold depicted independence and a midway point between synergy and redundancy. Design of interaction entropy graphs was accomplished using the Orange software [75].
An important limitation of any gene-gene interaction analysis method is the combinatorial nature of the problem. Exhaustive analysis of all possible two-, three-, four-way combinations of SNPs creates a computational burden and significant multiple testing problems. We addressed this issue by pre-processing the data using statistical epistasis network (SEN) modeling, which has been described elsewhere [39]. Briefly, this tool uses information theory, as previously described, to develop a network based on the amount of mutual information from SNP1, SNP2, and a class variable (i.e., disease status). Thus, it describes the pairwise interactive effect on a discrete outcome. An epistasis network, or graph, consists of a set of vertices (individual SNPs) and edges (pairwise interactions) that attach the vertices. In the current study, the statistical epistasis networks (SEN) were built by incrementally adding edges if their strengths were greater than a given threshold. Each vertex and each edge connecting two vertices were assigned a weight commensurate with the strength of main and interactive effects, respectively. The weight of a vertex is depicted by its size; whereby, a larger size is indicative of a higher main effect. Similarly, the thickness of the line between SNP pairs is directly proportional to the strength of their interaction.
We used SEN to visually summarize the strongest individual SNP effects and SNP-pairwise interactions. The topology of the graph, consisting of SNPs and their pairwise interactions, was used to guide MDR modeling. This resulted in a reduction in the total number of SNP combinations. For instance, we reduced the gene-gene interaction analysis from 172 to 24 SNPs that resided within the major connected component with the largest number of vertices (i.e., main effects). The derived major component plot was validated by comparing: (1) the number of vertices and edges among the various connected "sub-networks"; and (2) the size of the major component plot of real data versus randomized data that underwent 1000 permutations. The latter compared data using a pairwise interaction strength cut-off of 1% and significance level of 0.05. To ensure that no significant interactions were identified, non-informative SNPs outside the major component plot were also analyzed by MDR. In essence, SEN-guided MDR facilitated logical prioritization of SNPs for gene-gene interaction analysis in relation to PCA outcomes.
Abbreviations
PCA: (PCA); SNP: (single nucleotide polymorphism); MDR: (multifactor dimensionality reduction); SEN: (Statistical Epistasis Network).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NAL: conceptualized the project, conducted the statistical analysis and initial MDR modeling, interpreted study results, and composed the initial manuscript draft. ENR, SY, LRK: Intellectually contributed toward the introduction and/or discussion. TH: performed statistical epistasis network modeling. JZ and JR: conducted MDR modeling and/or SEN-guided MDR analysis. GNB & LRK: co-supervised data-management and statistical/bioinformatics analyses. LRK, DWH, GNB, JHM, KSK: served as mentors in the study design and implementation as well as data interpretation. All authors contributed to draft reviewing, editing, and approving the final manuscript draft.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1. Corresponding Gene and dbSNP IDs for apoptosis-related sequence variants depicted in the Statistical Epistasis Network Modeling Graph, presented in Figure 1.
Table S2. Functional consequence of selected apoptosis-related Polymorphisms.
Contributor Information
Nicole A Lavender, Email: nlavender@tgen.org.
Erica N Rogers, Email: enroge01@louisville.edu.
Susan Yeyeodu, Email: syeyeodu@nccu.edu.
James Rudd, Email: jrudd@nccu.edu.
Ting Hu, Email: ting.hu@dartmouth.edu.
Jie Zhang, Email: j0Zhang@louisville.edu.
Guy N Brock, Email: g0broc01@lousiville.edu.
Kevin S Kimbro, Email: kkimbro@nccu.edu.
Jason H Moore, Email: Jason.H.Moore@dartmouth.edu.
David W Hein, Email: dwhein01@louisville.edu.
La Creis R Kidd, Email: lrkidd01@louisville.edu.
Acknowledgements
The authors thank CGEMS and PLCO projects for making their data available for use in this study. This work was partially supported by the James Graham Brown Cancer Center (JGBCC) Pilot Project Initiative Grant to LRK, and the JGBCC Bucks for Brains "Our Highest Potential" in Cancer Research Endowment to LRK, National Cancer Institute/National Institute of Health grants (R03 CA128028, 3R01 CA034627-19S) to LRK, and the National Institute of Health/National Institute of Environmental Health Sciences grants P30-ES014443 to GNB, National Institute of Environmental Health Sciences training grant T32 ES011564 to DWH, and P20-MD000175 NIH NCMHD to KSK.
References
- American Cancer S. Cancer Facts and Figures 2012. Atlanta, Georgia: American Cancer Society; 2012. [Google Scholar]
- Tapia-Vieyra JV, Mas-Oliva J. Apoptosis and cell death channels in prostate cancer. ArchMedRes. 2001;32(3):175–185. doi: 10.1016/s0188-4409(01)00274-0. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Carcinogenesis and apoptosis: paradigms and paradoxes. Carcinogenesis. 2006;27(10):1939–1945. doi: 10.1093/carcin/bgl035. [DOI] [PubMed] [Google Scholar]
- Limoli CL, Hartmann A, Shephard L, Yang CR, Boothman DA, Bartholomew J, Morgan WF. Apoptosis, reproductive failure, and oxidative stress in Chinese hamster ovary cells with compromised genomic integrity. Cancer Res. 1998;58(16):3712–3718. [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Furuya Y, Krajewski S, Epstein JI, Reed JC, Isaacs JT. Expression of bcl-2 and the progression of human and rodent prostatic cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 1996;2(2):389–398. [PubMed] [Google Scholar]
- Colombel M, Symmans F, Gil S, O'Toole KM, Chopin D, Benson M, Olsson CA, Korsmeyer S, Buttyan R. Detection of the apoptosis-suppressing oncoprotein bc1-2 in hormone-refractory human prostate cancers. Am J Pathol. 1993;143(2):390–400. [PMC free article] [PubMed] [Google Scholar]
- McDonnell TJ, Troncoso P, Brisbay SM, Logothetis C, Chung LW, Hsieh JT, Tu SM, Campbell ML. Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res. 1992;52(24):6940–6944. [PubMed] [Google Scholar]
- Fleischmann A, Huland H, Mirlacher M, Wilczak W, Simon R, Erbersdobler A, Sauter G, Schlomm T. Prognostic relevance of Bcl-2 overexpression in surgically treated prostate cancer is not caused by increased copy number or translocation of the gene. The Prostate. 2011. in press doi: 10.1002/pros.21504. [DOI] [PubMed]
- Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14(19):2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Robinson M, King P, Hasan T, Charlton R, Martin J, Carr TW, Neal DE. p53 expression and clinical outcome in prostate cancer. Br J Urol. 1993;72(5 Pt 2):778–781. doi: 10.1111/j.1464-410x.1993.tb16267.x. [DOI] [PubMed] [Google Scholar]
- Shurbaji MS, Kalbfleisch JH, Thurmond TS. Immunohistochemical detection of p53 protein as a prognostic indicator in prostate cancer. Hum Pathol. 1995;26(1):106–109. doi: 10.1016/0046-8177(95)90122-1. [DOI] [PubMed] [Google Scholar]
- Stackhouse GB, Sesterhenn IA, Bauer JJ, Mostofi FK, Connelly RR, Srivastava SK, Moul JW. p53 and bcl-2 immunohistochemistry in pretreatment prostate needle biopsies to predict recurrence of prostate cancer after radical prostatectomy. J Urol. 1999;162(6):2040–2045. doi: 10.1016/S0022-5347(05)68095-0. [DOI] [PubMed] [Google Scholar]
- Pickard MR, Edwards SE, Cooper CS, Williams GT. Apoptosis regulators Fau and Bcl-G are down-regulated in prostate cancer. Prostate. 2010;70(14):1513–1523. doi: 10.1002/pros.21186. [DOI] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL. et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Guo JP, Shu SK, He L, Lee YC, Kruk PA, Grenman S, Nicosia SV, Mor G, Schell MJ, Coppola D. et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am J Pathol. 2009;175(1):324–333. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KM, Kalifa S, Das JR, Bhatti T, Gay M, Williams D, Taliferro-Smith L, De Marzo AM. The role of PI 3-kinase p110beta in AKT signally, cell survival, and proliferation in human prostate cancer cells. Prostate. 2010;70(7):755–764. doi: 10.1002/pros.21108. [DOI] [PubMed] [Google Scholar]
- Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. The role of IKK in constitutive activation of NF-kappaB transcription factor in prostate carcinoma cells. J Cell Sci. 2002;115(Pt 1):141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM. et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454(7205):776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricks-Santi L, Mason T, Apprey V, Ahaghotu C, McLauchlin A, Josey D, Bonney G, Dunston GM. p53 Pro72Arg polymorphism and prostate cancer in men of African descent. Prostate. 2010;70(16):1739–1745. doi: 10.1002/pros.21209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res. 2011;728(1-2):67–79. doi: 10.1016/j.mrrev.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EB, Jeon HS, Yoo SS, Choi YY, Kang HG, Cho S, Cha SI, Choi JE, Park TI, Lee BH. et al. Polymorphisms in apoptosis-related genes and survival of patients with early-stage non-small-cell lung cancer. Ann Surg Oncol. 2010;17(10):2608–2618. doi: 10.1245/s10434-010-1082-4. [DOI] [PubMed] [Google Scholar]
- Bye H, Prescott NJ, Matejcic M, Rose E, Lewis CM, Parker MI, Mathew CG. Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis. 2011;32(12):1855–1861. doi: 10.1093/carcin/bgr211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L. et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J National Cancer Inst. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC. et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20(16):3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Wang X, McWilliams RR, Bamlet WR, de Andrade M, Petersen GM. Association of breast cancer susceptibility variants with risk of pancreatic cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(11):3044–3048. doi: 10.1158/1055-9965.EPI-09-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergentanis TN, Economopoulos KP. Association of two CASP8 polymorphisms with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;120(1):229–234. doi: 10.1007/s10549-009-0471-5. [DOI] [PubMed] [Google Scholar]
- Enjuanes A, Benavente Y, Bosch F, Martin-Guerrero I, Colomer D, Perez-Alvarez S, Reina O, Ardanaz MT, Jares P, Garcia-Orad A. et al. Genetic variants in apoptosis and immunoregulation-related genes are associated with risk of chronic lymphocytic leukemia. Cancer Res. 2008;68(24):10178–10186. doi: 10.1158/0008-5472.CAN-08-2221. [DOI] [PubMed] [Google Scholar]
- Sigurdson AJ, Bhatti P, Doody MM, Hauptmann M, Bowen L, Simon SL, Weinstock RM, Linet MS, Rosenstein M, Stovall M. et al. Polymorphisms in apoptosis- and proliferation-related genes, ionizing radiation exposure, and risk of breast cancer among U.S. Radiologic Technologists. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16(10):2000–2007. doi: 10.1158/1055-9965.EPI-07-0282. [DOI] [PubMed] [Google Scholar]
- Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S. et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- Lan Q, Zheng T, Chanock S, Zhang Y, Shen M, Wang SS, Berndt SI, Zahm SH, Holford TR, Leaderer B. et al. Genetic variants in caspase genes and susceptibility to non-Hodgkin lymphoma. Carcinogenesis. 2007;28(4):823–827. doi: 10.1093/carcin/bgl196. [DOI] [PubMed] [Google Scholar]
- Lou Y, Fang CQ, Li JH. A study on the expression of CASP9 gene and its polymorphism distribution in non-small cell lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24(1):59–62. [PubMed] [Google Scholar]
- Consortium BCA. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J National Cancer Inst. 2006;98(19):1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- Du J, Huo J, Shi J, Yuan Z, Zhang C, Fu W, Jiang H, Yi Q, Hou J. Polymorphisms of nuclear factor-kappaB family genes are associated with development of multiple myeloma and treatment outcome in patients receiving bortezomib-based regimens. Haematologica. 2011;96(5):729–737. doi: 10.3324/haematol.2010.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Pfeifer D, He LJ, Qiao F, Zhang Z, Arbman G, Wang ZL, Jia CR, Carstensen J, Sun XF. Association of NFKBIA polymorphism with colorectal cancer risk and prognosis in Swedish and Chinese populations. Scand J Gastroenterol. 2007;42(3):345–350. doi: 10.1080/00365520600880856. [DOI] [PubMed] [Google Scholar]
- Schildkraut JM, Iversen ES, Wilson MA, Clyde MA, Moorman PG, Palmieri RT, Whitaker R, Bentley RC, Marks JR, Berchuck A. Association between DNA damage response and repair genes and risk of invasive serous ovarian cancer. PLoS One. 2010;5(4):e10061. doi: 10.1371/journal.pone.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Trentham-Dietz A, Garcia-Closas M, Newcomb PA, Titus-Ernstoff L, Hampton JM, Chanock SJ, Haines JL, Egan KM. Genetic variation in TP53 and risk of breast cancer in a population-based case control study. Carcinogenesis. 2007;28(8):1680–1686. doi: 10.1093/carcin/bgm097. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N. et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39(5):645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- Hu T, Sinnott-Armstrong NA, Kiralis JW, Andrew AS, Karagas MR, Moore JH. Characterizing genetic interactions in human disease association studies using statistical epistasis networks. BMC Bioinforma. 2011;12:364. doi: 10.1186/1471-2105-12-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duell EJ, Bracci PM, Moore JH, Burk RD, Kelsey KT, Holly EA. Detecting pathway-based gene-gene and gene-environment interactions in pancreatic cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17(6):1470–1479. doi: 10.1158/1055-9965.EPI-07-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason RA, Morlock EV, Karagas MR, Kelsey KT, Marsit CJ, Schned AR, Andrew AS. EGFR pathway polymorphisms and bladder cancer susceptibility and prognosis. Carcinogenesis. 2009;30(7):1155–1160. doi: 10.1093/carcin/bgp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Karagas MR, Nelson HH, Guarrera S, Polidoro S, Gamberini S, Sacerdote C, Moore JH, Kelsey KT, Demidenko E. et al. DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. HumHered. 2008;65(2):105–118. doi: 10.1159/000108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27(5):1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- Lavender NA, Benford ML, VanCleave TT, Brock GN, Kittles RA, Moore JH, Hein DW, Kidd LC. Examination of polymorphic glutathione S-transferase (GST) genes, tobacco smoking and prostate cancer risk among men of African descent: a case-control study. BMC Cancer. 2009;9:397. doi: 10.1186/1471-2407-9-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender NA, Komolafe OO, Benford M, Brock G, Moore JH, Vancleave TT, States JC, Kittles RA, Kidd LC. No association between variant DNA repair genes and prostate cancer risk among men of African descent. Prostate. 2010;70(2):113–119. doi: 10.1002/pros.21048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Kamat AM, Huang M, Grossman HB, Dinney CP, Lerner SP, Wu X, Gu J. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis. 2007;28(10):2160–2165. doi: 10.1093/carcin/bgm167. [DOI] [PubMed] [Google Scholar]
- Benford ML, VanCleave TT, Lavender NA, Kittles RA, Kidd LR. 8q24 sequence variants in relation to prostate cancer risk among men of African descent: a case-control study. BMC Cancer. 2010;10:334. doi: 10.1186/1471-2407-10-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCleave TT, Moore JH, Benford ML, Brock GN, Kalbfleisch T, Baumgartner RN, Lillard JW Jr, Kittles RA, Kidd LC. Interaction among variant vascular endothelial growth factor (VEGF) and its receptor in relation to prostate cancer risk. Prostate. 2010;70(4):341–352. doi: 10.1002/pros.21067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani K, Thompson DA, Barthel A, Sakaue H, Liu W, Weigel RJ, Roth RA. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J Biol Chem. 1999;274(31):21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- Bauer B, Krumbock N, Fresser F, Hochholdinger F, Spitaler M, Simm A, Uberall F, Schraven B, Baier G. Complex formation and cooperation of protein kinase C theta and Akt1/protein kinase B alpha in the NF-kappa B transactivation cascade in Jurkat T cells. J Biol Chem. 2001;276(34):31627–31634. doi: 10.1074/jbc.M103098200. [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng G, Zhang Z, Fu G. Genetic variants in the death receptor 4 gene contribute to susceptibility to bladder cancer. Mutat Res. 2009;661(1-2):85–92. doi: 10.1016/j.mrfmmm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Nam RK, Zhang WW, Jewett MA, Trachtenberg J, Klotz LH, Emami M, Sugar L, Sweet J, Toi A, Narod SA. The use of genetic markers to determine risk for prostate cancer at prostate biopsy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11(23):8391–8397. doi: 10.1158/1078-0432.CCR-05-1226. [DOI] [PubMed] [Google Scholar]
- Jung JH, Chae YS, Moon JH, Kang BW, Kim JG, Sohn SK, Park JY, Lee MH, Park HY. TNF superfamily gene polymorphism as prognostic factor in early breast cancer. J Cancer Res Clin Oncol. 2010;136(5):685–694. doi: 10.1007/s00432-009-0707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd LR, Coulibaly A, Templeton TM, Chen W, Long LO, Mason T, Bonilla C, Akereyeni F, Freeman V, Isaacs W. et al. Germline BCL-2 sequence variants and inherited predisposition to prostate cancer. Prostate Cancer ProstaticDis. 2006;9(3):284–292. doi: 10.1038/sj.pcan.4500884. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genetic epidemiology. 2003;24(2):150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4(6):e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohagan JK, Prorok PC, Hayes RB, Kramer BS. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Control ClinTrials. 2000;21(6 Suppl):251S–272S. doi: 10.1016/s0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- Hayes RB, Sigurdson A, Moore L, Peters U, Huang WY, Pinsky P, Reding D, Gelmann EP, Rothman N, Pfeiffer RM. et al. Methods for etiologic and early marker investigations in the PLCO trial. MutatRes. 2005;592(1-2):147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008. pp. D480–D484. [DOI] [PMC free article] [PubMed]
- Kanehisa M, Goto S, Hattori M, oki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006. pp. D354–D357. [DOI] [PMC free article] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenuity Systems. 2008.
- Biocarta LLC. 2000.
- National Center for Biotechnology Information. 2011.
- Xu J, Kibel AS, Hu JJ, Turner AR, Pruett K, Zheng SL, Sun J, Isaacs SD, Wiley KE, Kim ST. et al. Prostate cancer risk associated loci in African Americans. Cancer EpidemiolBiomarkers Prev. 2009;18(7):2145–2149. doi: 10.1158/1055-9965.EPI-09-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Kiralis J, Moore JH. Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions. BioData mining. 2009;2(1):5. doi: 10.1186/1756-0381-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26(4):445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241(2):252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Gui J, Andrew AS, Andrews P, Nelson HM, Kelsey KT, Karagas MR, Moore JH. A simple and computationally efficient sampling approach to covariate adjustment for multifactor dimensionality reduction analysis of epistasis. Hum Hered. 2010;70(3):219–225. doi: 10.1159/000319175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill WL. Multivariate information transmission. Psychometrika vol. 1954;19:97–116. [Google Scholar]
- Jakulin A, Bratko I. Analyzing attribute interations. Lecture Notes in Artificial Intelligence. 2003;2838:229. [Google Scholar]
- Jakulin A, Bratko I, Smrke D, Demsar J, Supan B. Attribute interactions in medical data analysis. Lecture Notes in Artificial Intelligence. 2003;2780:229. [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. JTheorBiol. 2006;241(2):252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Mramor M, Leban G, Demsar J, Zupan B. Visualization-based cancer microarray data classification analysis. Bioinformatics. 2007;23(16):2147–2154. doi: 10.1093/bioinformatics/btm312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Corresponding Gene and dbSNP IDs for apoptosis-related sequence variants depicted in the Statistical Epistasis Network Modeling Graph, presented in Figure 1.
Table S2. Functional consequence of selected apoptosis-related Polymorphisms.