Abstract
Background
The discovery of genetic associations is an important factor in the understanding of human illness to derive disease pathways. Identifying multiple interacting genetic mutations associated with disease remains challenging in studying the etiology of complex diseases. And although recently new single nucleotide polymorphisms (SNPs) at genes implicated in immune response, cholesterol/lipid metabolism, and cell membrane processes have been confirmed by genome-wide association studies (GWAS) to be associated with late-onset Alzheimer's disease (LOAD), a percentage of AD heritability continues to be unexplained. We try to find other genetic variants that may influence LOAD risk utilizing data mining methods.
Methods
Two different approaches were devised to select SNPs associated with LOAD in a publicly available GWAS data set consisting of three cohorts. In both approaches, single-locus analysis (logistic regression) was conducted to filter the data with a less conservative p-value than the Bonferroni threshold; this resulted in a subset of SNPs used next in multi-locus analysis (random forest (RF)). In the second approach, we took into account prior biological knowledge, and performed sample stratification and linkage disequilibrium (LD) in addition to logistic regression analysis to preselect loci to input into the RF classifier construction step.
Results
The first approach gave 199 SNPs mostly associated with genes in calcium signaling, cell adhesion, endocytosis, immune response, and synaptic function. These SNPs together with APOE and GAB2 SNPs formed a predictive subset for LOAD status with an average error of 9.8% using 10-fold cross validation (CV) in RF modeling. Nineteen variants in LD with ST5, TRPC1, ATG10, ANO3, NDUFA12, and NISCH respectively, genes linked directly or indirectly with neurobiology, were identified with the second approach. These variants were part of a model that included APOE and GAB2 SNPs to predict LOAD risk which produced a 10-fold CV average error of 17.5% in the classification modeling.
Conclusions
With the two proposed approaches, we identified a large subset of SNPs in genes mostly clustered around specific pathways/functions and a smaller set of SNPs, within or in proximity to five genes not previously reported, that may be relevant for the prediction/understanding of AD.
Keywords: Late-Onset Alzheimer's Disease, GWAS, SNPs, Random Forest
Background
It is predicted the number of people who suffer from Alzheimer's disease (AD) will increase from 5 million to 13.4 million in the United States of America and will be 115.4 million worldwide by 2050 [1,2]. There is currently no treatment to stop or reverse the progress of this disease. This neurodegenerative disorder is believed to be caused by an inability to clear β-amyloid (increasing all its forms: monomer, oligomer, insoluble fibrils, and plaques) from the Central Nervous System provoking neuronal impairment and cell death, and by tangled tau formation when cells are dying [3]. Genetic variation is an important contributor to the risk for this disease, estimated to be up to seventy-nine percent in the late-onset AD (LOAD) more frequent form of the disease [4]. A few genes have been confirmed by independent studies to be implicated with LOAD, summarized below.
Alzheimer's can be divided into early-onset AD (EOAD) and LOAD. There are thus far three established genes involved in EOAD and follow autosomal dominant inheritance APP (β-amyloid precursor protein), PSEN1 and PSEN2 (presenilin-dependent γ-secretase activity cuts amyloid precursor proteins into β-amyloid peptides) [5,6]. Another well established genetic risk factor is APOE (it encodes a lipoprotein that may interact with accumulated β-amyloid); it manifests in the more common LOAD and its inheritance does not follow Mendelian principles [7,8]. APOE has three common alleles, ε4, ε3, and ε2, and each of these variants of the gene are determined by two single nucleotide polymorphisms (SNPs). In European populations, ε4ε4 homozygotes are the most likely to develop disease, followed by ε3ε4 heterozygotes and ε3ε3 homozygotes, with ε2 heterozygotes having the least risk [8,9]. However, a person who has one or two copies of ε4 may never develop AD, while another who does not carry the ε4 alleles may [8].
APOE genotypes could be useful in combination with other genetic variations to predict disease risk since the scientific literature suggests the existence of additional genetic factors associated with LOAD. In the past two years, at least eight genes mapped to the immune system, cholesterol metabolism, and cell membrane processes have been confirmed by independent genome-wide association studies (GWAS) to be implicated with LOAD (See AlzGene database [10]). The genetic factors are CLU (it encodes apoliprotein J and may have a similar function as to that of APOE), PICALM (it encodes a protein involved in intracellular traffic of neurotransmitters between proteins and lipids), CR1 (it encodes the main receptor of complement C3b protein thought to be involved in β-amyloid clearance through phagocytosis) [5,11,12], BIN1 (it is involved in synaptic vesicle endocytocis) [5,13,14]; moreover, recently two separate studies conducted by Hollingworth P., et al and Naj, A.C. et al identified MS4A6A/MS4A4E (these encode cell membrane proteins), CD2AP (encodes a protein involved in endocytocis), EPHA1 (it produces a membrane bound protein involved in cell and axon guidance and synaptic function; additionally, it is involved in cell morphology, motility, and inflammation), and provided further support for CD33 (it is involved in cell-cell interaction and function regulation of cells in the immune system and also mediates endocytocis through a process independent from clathrin) [14,15]. Different SNPs in CD33 were previously identified by Bertram, L. et al [14-16]. CLU, PICALM, CR1 and BIN1 were confirmed by Naj, A.C. et al and BIN1 and CR1 were confirmed by Hollingworth P., et al as LOAD susceptibility loci [14,15]. In the study by Naj, A.C. et al, the genetic effect for the most salient SNPs at each locus had estimated population attributable fractions (PAF) of 2.72% - 5.97%; nonetheless, the authors caution that the true PAF might be different [15]. These newly confirmed genes could be mapped to pathways related to the innate and adaptive immune response - (CLU, CR1, CD33, EPHA1) [14,17], cell membrane processes including endocytocis (PICALM, BIN1, CD33, CD2AP) [14], and cholesterol/lipid metabolism (CLU) [14,17].
A few years ago, another gene that was shown to have an increased associated risk with LOAD was GAB2 although with inconsistent reproducibility by independent GWAS [18,19]. GAB2 protein may be involved in protection from the formation of insoluble tau deposits known as neurofibrillary tangles (NFTs) [9] and may participate in the production of β-amyloid [20]. Reiman, Eric. M. et al utilized stratification and linkage disequilibrium (LD) analysis and found six SNPs, part of a common haplotype block covering the GAB2 gene, to have a strong interaction with APOE in three groups of APOE ε4 carriers [9].
APOE by itself, or in combination with GAB2, remains to some extend a weak predictor for the risk of developing AD [8]. We used a published GWAS data set from Reiman and colleagues [9] to analyze it for AD risk determination in new loci by different models in APOE ε4 positive and negative samples.
One of the challenges trying to identify multiple interacting genetic mutations associated with disease in studying the etiology of complex diseases arises from the fact that there are millions of genome-wide variants, many of them untyped in the study samples of GWAS, and the number of possible combinations encountered in "interaction analysis" grows exponentially with the number of variants. As a result, it is computationally prohibitive to perform a comprehensive test for interaction analysis between four or more factors and disease. Heuristic approaches must be developed to analyze these data, that leverage and combined statistical and data mining methods [21].
We devised two informatics approaches to identify new genetic biomarkers. The first approach utilizes statistical and data mining methods. The second approach also leverages prior biological knowledge to refine the analysis. In both approaches, multi-locus (classifier building) analysis is done with a reduced number of variants that first passed, for instance, a single-locus (logistic regression) threshold.
Results and discussion
Approach I: Choice of SNPs without prior biological knowledge for model building
Step 1
In order to cast a wide net to filter the data and take into account the correlation among some of the SNPs due to LD, the association analysis was run with a p-value = 1E-3. SNPs from APOE and GAB2 were excluded from the analysis, since these are already known to be associated with AD in this data set [9]. This gave 199 SNPs with p-values ≤ 1E-3 and 1 < ORs < 5. Table 1 lists the top seven scoring SNPs; the complete list of all 199 SNPs is found in additional file 1, table SA.
Table 1.
Logistic regression top scoring SNPs, approach I
| Gene Symbol dbSNP RS ID | Distance to Gene | Unadj. p-value | FDR_BH p-value | OR (95% CI) | Pathway/Disease/function |
|---|---|---|---|---|---|
| NISCH rs6784615 | intron | 7.16E-07 | 4.47E-02 | 2.21 (1.61-3.02) | Interaction with PAK4 for reduction of LIMK1 phosphorylation; neuronal migration and axon/dendrite outgrowth [22,23]. |
| RABEP1 rs4356530 | upstream 27742 | 8.59E-07 | 4.47E-02 | 2.21 (1.61-3.02) | Endocytosis [24,32]. |
| THEMIS rs9398855 | intron | 2.25E-06 | 8.78E-02 | 2.13 (1.56-2.92) | Immnune response [25-27]. |
|
NDUFA12 rs249153 |
downstream 40719 | 4.13E-06 | 1.29E-01 | 1.62 (1.32-2.00) | AD, Parkinson's, Hungtinton's, oxidative phosphorylation [28-32]. |
| MUC21 rs2517509 | downstream 72544 | 4.93E-06 | 1.39E-01 | 3.14 (1.92-5.12) | Prevention of cell-cell interaction of Integrins [55]. |
| TUSC1 rs10115381 | upstream 328744 | 5.33E-06 | 1.39E-01 | 2.08 (1.52-2.85) | Shwachman-Diamond syndrome [56]. |
| CTNNA3 rs10996618 | downstream 187123 | 1.19E-05 | 2.03E-01 | 2.03 (1.48-2.78) | AD (4 studies), Adherens junction, Inmune response [10,32]. |
FDR_BH = Benjamini & Hochberg (1995) step-up False Discovery Rate control.
Step 2
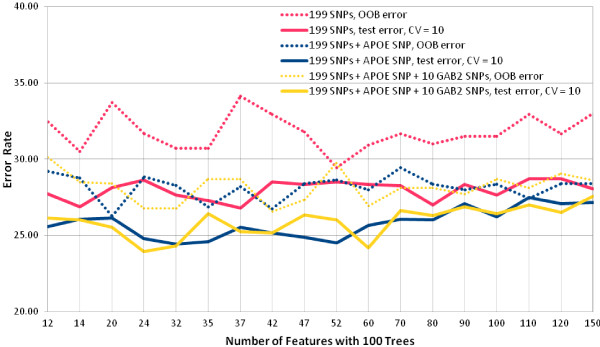
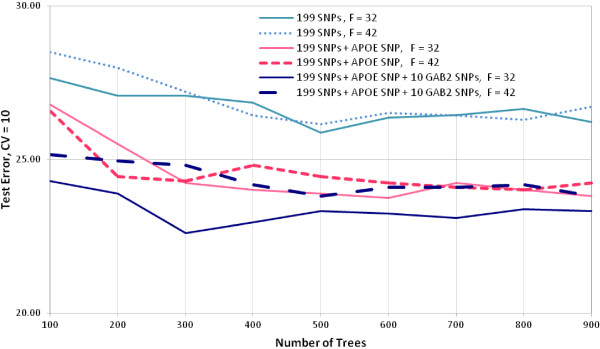
After univariate association analysis, the Random Forest (RF) classifier performance assessment was done with the 199 SNPs data. With 100 trees, Figure 1 shows the test and out of bag (OOB) errors for different number of features (SNPs). The figure suggests that increasing the number of attributes above 70 actually leads to a gradual increase in test error rate, 10-fold cross validation (CV), for the 199 SNPs and APOE SNP and the 199 SNPs, APOE SNP and GAB2 SNPs sets. OOB error rate (estimated class ŷi is determined from models where row i is out-of- bag) is not a good estimation of test error in all instances here; however, as features are added to the forest the OOB error becomes a better estimator of the test error for these two data sets. Figure 2 shows the classifier tuning; the additional induced randomness on the selection of number of attributes for choosing the splits seems to have worked, giving a modest improvement with average 10-fold CV error rates in the range of 23-27%.
Figure 1.
RF performance assessment, different number of features and number of trees fixed at 100; approach I.
Figure 2.
RF tuning, best number of attributes at different number of trees; approach I. F = number of features.
In order to further improve the classifiers, a supervised instance resample filter was applied to the data. The original case-control distribution in the data is 61% cases and 39% controls. After the data is filtered the distribution of the data becomes 52% cases and 48% controls. A big reduction in misclassification was obtained by first resampling the data to make its distribution more balanced followed by RF. The results at 100, 300, and 600 trees and various numbers of attributes are listed in Table 2. When APOE and GAB2 SNPs alone are used, the average classification error rate is 33.3%. This error rate is reduced when the 199 SNPs are used for classification. Random forests built with eighteen SNPs from the 199 SNPs give an average 10-fold CV error of 11.7%, and when the 199 SNPs are added to the bag containing either an APOE SNP or an APOE SNP and GAB2 SNPs the average 10-fold CV error is reduced to between 9.3% and 9.5% for 11 and 18 attributes respectively.
Table 2.
RF modeling, filtered data, approach I
| Data | RF | 10-Fold | CV | % | Error | |
|---|---|---|---|---|---|---|
| (APOE ε4+ & ε4- samples) | Number of Trees | F = 7 | F = 11 | F = 18 | F = 32 | F = 42 |
| APOE & 10 GAB2 SNPs | ||||||
| 100 | 33.3 | 33.2 | ||||
| 300 | 33.3 | 33.2 | ||||
| 600 | 33.4 | 33.5 | ||||
| 199 SNPs | ||||||
| 100 | 11.9 | 12.3 | 12.5 | 12.3 | ||
| 300 | 11.3 | 11.3 | 12.1 | 11.6 | ||
| 600 | 10.6 | 11.6 | 11.2 | 11.1 | ||
| APOE & 199 SNPs | ||||||
| 100 | 10.9 | 9.9 | 10.4 | 10.3 | ||
| 300 | 8.9 | 9.6 | 9.5 | 10.3 | ||
| 600 | 9.0 | 9.1 | 10.1 | 9.9 | ||
| APOE, 10 GAB2, & 199 SNPs | ||||||
| 100 | 9.9 | 10.1 | 9.9 | 11.2 | ||
| 300 | 8.9 | 9.1 | 9.5 | 10.3 | ||
| 600 | 9.1 | 9.4 | 9.9 | 10.3 | ||
F = number of features.
A manual compilation of pathway, disease association, or biological function information reveals some of the 199 SNPs are associated with genes involved in calcium signalling, cell adhesion, endocytosis, and immune response in addition to synaptic function. This information was added to both Table 1 and the additional file 1, table SA. Some of the genes linked to the 199 SNPs appear among genes previously identified in GWAS as posted in the AlzGene database [10]. Furthermore, some of the top 199 SNPs are novel SNPs part of or in proximity (may be acting as flags) to genes that may be engaged in a cascade of events leading to AD. Some of these genes and their relevance to AD are discussed below.
For instance, NISCH codes for a cytosolic protein Nischarin which negatively affects cell migration by forming inhibitory complexes with PAK family kinases among other proteins [22]. PAK4 suppression decreases the phosphorylation of LIMK1, key for axon/dendrite outgrowth and neuronal migration [23]. Another gene, RABEP1 codes for rabaptin-1 which interacts with Gap-43. One of the main roles of Gap-43 is adjustment of neurotransmitter release, endocytosis, and long-term potentiation and its expression and function is altered in AD [24]. Recently identified THEMIS produces a protein also known as GAB2 associated protein (Gasp) which plays a crucial regulatory role in positive selection during thymocyte development [25-27]. Post-positive selection, thymocytes differentiate into CD4 or CD8 single-positive (SP) thymocytes as determined by their restriction to MHC class II and I respectively. SP CD4 and CD8 in time leave the thymus for other organs and form part of the adaptive immune system. It is thought Gasp may function through a new molecular pathway downstream of T-cell receptor (TCR) signalling [27]. Further studies will be needed to establish the role this gene may play in the etiology of AD.
Changes of the expression of mitochondrial genes such as NDUFA12, part of complex I, may alter the oxidative metabolism in AD [28,29]. Complex I initiates electron transfer by oxidizing NADH and transferring the electrons to coenzyme Q while pumping protons across the mitochondrial membrane creating an electro-chemical proton gradient [29]. The high rate of oxygen consumption needed for normal function, polyunsaturated fatty acid and transition metal ion composition, and limited antioxidant defence mechanisms renders neurons vulnerable to oxidative damage [29,30]. Energy decline and mitochondrial dysfunction are a major, early event in AD. Complex I deficiency decreases energy production by oxidative phosphorylation this in turn increases reactive oxygen species (ROS) which often causes structural and functional cell membrane changes setting off a vicious cycle that ends in apoptosis.
Research by Rhein et al found tau induces mitochondrial dysfunction and increases levels of ROS and together with β-amyloid synergistically alters complex I function and energy balance with aging in AD [31]. Tau has specific sensitivity of complex I oxidative phosphorylation system. Furthermore, β-amyloid directly interacts with mitochondria via the translocase of the outer membrane (TOM) system. Additionally, maternal family history of AD links maternal inheritance of mitochondria to predisposition to AD and glucose hypometabolism [31]. NDUFA12 is already listed as part of the AD pathway [32].
Approach II: Choice of SNPs with prior biological knowledge for model building
Step 1
Table 3 highlights 19 SNPs with the smallest p-values from the logistic regression and stratified analysis, in strong LD, and within six genes potentially relevant to AD. The LD patterns and gene overlays for the SNPs are shown in the supplementary information (additional file 2, figure S1, additional file 3, figure S2, additional file 4, figure S3, additional file 5, figure S4, additional file 6, figure S5, and additional file 7, figure S6).
Table 3.
CMH top scoring SNPs in LD, approach II.
| Gene (Chr.) dbSNP RS ID | Physical Position | Distance to Gene | Minor Allele (MAF) | p-value from χ2 | OR (95% CI) |
|---|---|---|---|---|---|
| APOE ε4- SAMPLES | |||||
| ST5 (11) | |||||
| rs4910068 | 8830651 | intron | C (0.25) | 9.47E-05 | 1.57 (1.25-1.98) |
| rs10743089 | 8744744 | intron | A (0.33) | 2.12E-05 | 1.59 (1.28-1.97) |
| APOE ε4+ SAMPLES | |||||
| TRPC1 (3) | |||||
| rs4259003 | 144006245 | intron | A (0.21) | 6.82E-05 | 2.36 (1.53-3.64) |
| rs9784320 | 144024724 | intron | C (0.25) | 9.68E-04 | 1.87 (1.28-2.71) |
| rs2033912 | 143999057 | intron | T (0.22) | 1.71E-03 | 1.86 (1.26-2.75) |
| rs891159 | 81526843 | intron | C (0.24) | 2.40E-05 | 2.34 (1.56-3.49) |
| ATG10 (5) | |||||
| rs1485587 | 81362798 | intron | G (0.48) | 8.11E-05 | 1.82 (1.35-2.45) |
| rs4703879 | 81589571 | intron | A (0.24) | 1.01E-03 | 1.86 (1.28-2.71) |
| ANO3 (11) | |||||
| rs1389421 | 25747721 | upstream 561825 | G (0.45) | 3.39E-06 | 2.07 (1.52-2.83) |
| rs10834774 | 25715397 | upstream 594149 | C (0.20) | 4.01E-03 | 1.86 (1.21-2.85) |
| rs11028909 | 25729021 | upstream 580525 | G (0.20) | 4.13E-03 | 1.84 (1.21-2.81) |
| NDUFA12 (12) | |||||
| rs249153 | 93848520 | downstream 40719 | C (0.19) | 3.19E-06 | 1.63 (1.33-2.00) |
| rs249154 | 93848687 | downstream 40552 | C (0.18) | 3.22E-05 | 1.54 (1.25-1.89) |
| APOE ε4+ and ε4- SAMPLES | |||||
| NISCH (3) | |||||
| rs6784615 | 52481466 | intron | C (0.09) | 6.63E-07 | 2.13 (1.57-2.88) |
| rs9855470 | 52468315 | intron | A (0.06) | 4.86E-05 | 2.11 (1.46-3.05) |
| rs6445486 | 52481531 | intron | A (0.06) | 2.77E-04 | 1.90 (1.34-2.71) |
| rs10865972 | 52466487 | intron | C (0.06) | 3.38E-04 | 1.87 (1.32-2.65) |
| rs4687619 | 52493826 | intron | T (0.06) | 3.54E-04 | 1.93 (1.34-2.79) |
| rs6810027 | 52499614 | intron | C (0.05) | 5.80E-04 | 1.89 (1.31-2.73) |
All p-values are uncorrected.
Four genes, distinct from the ones already identified in approach I, are discussed next. ST5, suppression of tumorigenicity 5, encodes three proteins. One of the proteins, p126, is an activator of mitogen-activated protein kinase MAPK1 also known as ERK2 [33]. ERK1 and ERK2 are some of several proline-directed kinases that have been shown to phosphorylate tau protein [34]. Tau binds and stabilizes microtubules in cells and in neurons; intracellular transport occurs in axons through microtubules [35]. Hyperphosphorylation reduces tau binding to microtubules and may increase neurofibrillary tangles (NFTs) in cell bodies and dendrites of neurons [36]. There is direct correlation between NFTs and memory decline in AD patients [36]; however, it remains to be seen how important of a role ERK2 plays in the hyperphosphorylation of tau. Another gene, TRPC1 codes for a TRP cation channel protein expressed in the neurons of the hippocampus and cortex among other regions of the brain. TRPC1 is activated by either G receptor proteins or intracellular Ca2+ depletion [37]. Strübing et al discovered that TRPC1 channels uniquely adjust neuronal function independently of synapse processes [37]. In addition, they demonstrated that TRPC1 can form heteromeric channels i.e. TRPC1/TRPC5. TRPC5 is expressed in the hippocampus; TRPC1/TRPC5 is activated by Gq-coupled receptors and not by Ca2+ depletion and its regulation is not neurotransmitter specific [37]. Calcium signalling necessary for axonal regeneration in the adult CNS and for growth cone response of spinal neurons in Xenopus to myelin-associated glycoprotein is mediated by TRPC1 channels [38]. Calcium disequilibrium has been observed to lead to neuronal injury and apoptosis [39]. Key modulators of calcium homeostasis such as presenilins and CALHM1 have been associated with EOAD [39]; however, there have not been prior studies on TRPC1 in AD patients.
ATG10 is an E2-like ligase protein involved in two ubiquitin-like modifications essential for autophagosome formation [40]. Autophagy is an intracellular degradation mechanism responsible for clearance of misfolded proteins, pathogens, and organelles (organelles such as functionally disabled mitochondria in aging) [41]. Double-membrane autophagosomes enclose cytoplasmic proteins and later degrade them by fusing with lysosomes. Autophagy initiation enhances the clearance of tau and offers a cytoprotective role. Overactive or dysfunctional autophagy may promote neuronal cell death in disease states contributing to the pathology of multiple neurodegenerative disorders [42]. Blocking autophagosome formation by knockout of either ATG5 or ATG7 genes causes ubiquitinated protein aggregates and eventual neurodegeneration, demonstrating that autophagy is both constitutive and essential for neuronal functioning [42].
A genome-wide screen study by Lipinski et al showed that ROS are common mediators upstream of the activation of the type III PI3 kinase (critical protein in autophagy initiation) in response to β-amyloid peptide. On the other hand, lysosomal blockage also caused by β-amyloid is independent of ROS. Furthermore, they proved that autophagy is transcriptionally down-regulated during normal aging in the human brain in contrast to the autophagy up-regulation observed in later stages of AD human brains. In addition, AD drugs they tested have inhibitory effects on autophagy, decreasing input into the lysosomal system; they hypothesized this may ameliorate cellular stress in AD [43].
A fourth gene, ANO3 encodes anoctamin 3, rs1389421 is in a 49 kb LD region upstream of ANO3 as seen in additional file 5, figure S4. The anoctamin family of ten highly hydrophobic membrane proteins is also known as TMEM16 [44]. Some anoctamins function as Ca2+ activated Cl- channels (CaCCs) in the retinal photoreceptor synaptic terminals and the olfactory sensory neurons; others participate in tumor progression [44,45]. Some studies indicate that olfactory neurogenesis disruption is linked to AD [46]. ANO2, 3, and 4 are mostly expressed in neuronal tissues. ANO3 and ANO4 mRNA are equally expressed in spinal cord, brain stem, cerebellum, and eye; however, it is not known how ano3 and ano4 function thus far [44,47]. Ist2p, ANO in S. cerevisiae, is translated locally at the peripheral endoplasmic reticulum (ER) and may be inserted into the plasma membrane by the fusion of peripheral ER with the plasma membrane. If these anion selective proteins in mammals are transported by a similar novel mechanism, it is believed they might have effects in protein synthesis in axons and dendrites [45].
Step 2
The 19 SNPs in LD with six genes that resulted from the analysis in step 1 above were used next in RF. Table 4 shows that when combining the 19 SNPs with the APOE SNP the average 10-fold CV error rate is reduced to 20.5%. The 10-fold CV error rate is reduced to 16.9% when the data set is the 19 SNPs, APOE SNP, and 10 GAB2 SNPs, and using 600 trees and 11 features for tree building. The higher 10-fold CV error rate obtained in approach II as compared to that of approach I may not be due to LD. As Meng, Y.A. et al explain, in RF if a SNP is near the root of a tree in the forest and a second SNP in LD with the first SNP is close to the leaf of the same tree, the permutation of the first SNP value will not increment the prediction error of the tree because the second SNP can be a substitute for the first SNP. However, the prediction error might be still somewhat increased [48].
Table 4.
RF modeling, filtered data, approach II.
| Data | RF | 10-Fold | Cross | Validation | % Error |
|---|---|---|---|---|---|
| (APOE ε4+ and ε4- samples) | Number of Trees | F = 6 | F = 7 | F = 11 | F = 18 |
| APOE & 10 GAB2 SNPs | |||||
| 100 | 33.3 | 33.2 | |||
| 150 | 33.4 | 33.5 | |||
| 200 | 33.4 | 33.2 | |||
| 250 | 33.2 | 33.2 | |||
| 300 | 33.3 | 33.2 | |||
| 600 | 33.4 | 33.5 | |||
| 19 SNPs | |||||
| 100 | 26.9 | 27.5 | 27.9 | ||
| 150 | 26.7 | 27.0 | 27.5 | ||
| 200 | 26.8 | 27.0 | 27.4 | ||
| 250 | 27.1 | 27.3 | 27.6 | ||
| 300 | 27.2 | 27.1 | 27.6 | ||
| 600 | 27.1 | 27.4 | 27.4 | ||
| APOE & 19 SNPs | |||||
| 100 | 20.3 | 20.8 | 20.7 | ||
| 150 | 19.9 | 20.6 | 20.8 | ||
| 200 | 20.1 | 20.7 | 20.9 | ||
| 250 | 20.3 | 20.6 | 20.6 | ||
| 300 | 20.1 | 20.6 | 20.6 | ||
| 600 | 19.8 | 20.4 | 20.7 | ||
| APOE, 10 GAB2, & 19 SNPs | |||||
| 100 | 17.6 | 17.6 | 18.2 | ||
| 150 | 17.2 | 17.0 | 17.6 | ||
| 200 | 17.2 | 17.2 | 18.1 | ||
| 250 | 17.3 | 17.1 | 17.9 | ||
| 300 | 17.2 | 17.3 | 17.9 | ||
| 600 | 17.3 | 16.9 | 17.6 | ||
F = number of features
Conclusions
It is believed that LOAD is a complex disease caused by the interaction of multiple genetic and environmental factors. In the past two years, at least eight genes have been confirmed to be associated with LOAD. These are common risk variants of moderate to small effects the same as APOE. The new variants functionality could be mapped to the immune response, cholesterol metabolism, and cell membrane processes pathways [14]. However, a percentage of AD heritability is still missing. The purpose of this study was to explore new associations between multiple SNPs and AD by data mining approaches. We analyzed a published AD GWAS data set by a couple of two-step approaches that first filtered the data with a low threshold to obtain a data subset used in a second step for multi-locus analysis. In one approach statistical and data mining techniques were implemented, and in the other approach biological domain knowledge and LD analysis were done prior to the multi-locus analysis. A 10-fold CV was done with the multi-locus analysis which helped remove bias from the reported error rate. Previously AD associated SNPs [7,9] were removed from the data to avoid obscuring other possible significant variants. There is overlap between the SNPs identified with both approaches; some of the genes associated with the SNPs used to build the classifiers have not been reported before as currently listed in the AlzGene database [10].
The model built for approach I confirmed, that APOE and GAB2 genotypes alone can produce a moderate determinant of LOAD status [9], being able to discriminate between cases and controls with about 33% error rate (10 fold CV). By adding close to 200 other genome-wide SNPs that had a relatively high score of association with LOAD from the GWAS data set, the error rate of the model was greatly reduced, from 33% to about 10%. While many of the 200 SNPs were in the vicinity of genes that could potentially be involved in AD pathways, some of them were not.
The model built for approach II leveraged biological domain knowledge to select a small number of SNPs from genes that had relevance to LOAD. This model used only 19 to 30 SNPs (Table 4), while the model in approach I used one order of magnitude more, about 200 SNPs (Table 2). The model in approach II was less successful in lowering the LOAD classification error rate - to only about 17%, vs. the 10% that the model in approach I did. Approach II; however, with its limited number of biologically-relevant SNPs, would be much easier to test, as opposed to the model in approach I, which included 200 genome-wide SNPs. A model employing dozens of SNPs might be harder to test, but it could be that dozens of genetic variants linked to many different pathways could be involved in the etiology of AD as is the case for other complex diseases [19], and as it is beginning to emerge from the GWAS outcomes from the past two years.
In order to improve the results from the analysis of this data, a (joint) meta-analysis could be done with another AD data set conducted on the same platform. The combined data sets would give more statistical power for gene-gene interaction effects and make possible fine mapping of variants with larger effect sizes. The functionality of the selected SNPs here could be further assessed by mapping the variants to genes that interact with or are in the same pathways as those already implicated in AD, and querying of genomic annotations of SNPs representing variation in microRNA target sites.
The two approaches described here are only a starting point that can be further refined to better understand the possible causes of LOAD. Similar approaches - that combine high throughput genomics techniques, statistical and data mining analysis, and leverage biological domain knowledge - can be applied to study other complex diseases that have a strong genetic component.
Methods
Data
A published AD GWAS data set was obtained from the Translational Genomics Research Institute [9]. The data includes results on 312,316 SNPs that passed quality control checks across the genome genotyped with the Mapping 500K Array set from Affymetrix on 1411 LOAD cases and controls from a discovery group and two replicate groups. Each of the three groups is divided into two sub-groups of APOE ε4 carriers and ε4 non-carriers. In addition to genotypes, the original data includes phenotypes such as gender, age of disease onset, and age at death. The analysis presented here focuses on genotype interactions and excludes these phenotypes from the analysis.
The data can be described as 312,316 nominal predictors along with a two class response variable y. The response variable is unevenly distributed; it is 61% cases and 39% controls. Also, the data has missing values. Furthermore, in order to avoid false-positive results due to population stratification, the data is from a Caucasian population of European ancestry; the samples were obtained from the United States and from the Netherlands.
The data was originally used to identify a novel interaction between LOAD and two genes, APOE and GAB2 [9]. In this analysis, known GAB2 and APOE SNPs are first excluded and then re-added in the model building phase. In the next section, we describe the approaches employed and briefly explain the RF algorithm.
Analytical approaches for Alzheimer's disease association analysis
In order to identify new genetic variants that increase disease risk, we implement some of the latest algorithm versions for disease association, LD, and data mining with the most recent genetic variant annotation files. A two step analysis, to reduce multiple genetic interactions to be tested, is implemented by two approaches: one statistically driven and a second incorporating sample stratification and biological knowledge. In the first step of both approaches, SNPs are filtered at a less stringent threshold for disease association. The multiple testing threshold correction, Bonferroni, assumes there are M independent tests (αp = αe /M where αp is the point-wise error and αe is the experimental error); however, the independence assumption fails in genetic association studies since there is correlation among some of the SNPs due to LD. Thus, we use a threshold of p-value = 1E-3 and take into account positive ORs (a positive OR means the minor allele increases disease risk relative to the major allele) for the genome-wide screening step. Furthermore, for the first step of approach II, the data is also filtered by the p-values and ORs from chi-square tests, and by significant LD values of selected SNPs within or close (~5 Kb) to neurobiological relevant genes. For both two step methods, known GAB2 and APOE SNPs (originally published with this data set [9]) are first removed so they do not obscure the finding of other statistically significant SNPs and then they are re-added in the RF building phase.
Random forest (or random forests) is an ensemble classifier that consists of many decision trees and outputs the class that is the mode (most frequent outcome) of the class's output by individual trees [49]. Ensemble methods use multiple models to obtain better predictive performance than could be obtained from any of the constituent models. For example, if individual classifiers would have an error rate of ε = 0.35, an ensemble of twenty-five independent base classifiers will make a wrong prediction at a smaller rate of 0.06 by the formula [50].
RF is a special case of bagging algorithm which is simple to train and tune. Bagging, or bootstrap aggregating, is a parallel ensemble method that induces additional randomness by allowing bag size to be chosen [51]. For each classifier in the ensemble, a sample is drawn uniformly and with replacement from the original training data set. If the training data has more rows expressing cases than expressing controls, the randomness causes more frequent cases rows in the bag than control rows. This results in cases rows getting classified much better than the control rows. The aim is to have both classes classified in a way to lead to overall low error rate. RF, as a classifier, induces additional randomness in the selection of features from a subset for deciding the splits at the nodes in each tree and if the skewed features in the data are de-selected one may improve the model predictions or vice versa. The size of the subset is decided by first taking the square root of the total number of attributes in the data set or by the log2 of the total number of attributes + 1. The additional randomness in RF helps to reduce variance (correlation among attributes) and maintain bias. It is standard to let the trees grow deep and not to prune them since the average of trees that are put together is taken to reduce the variance.
In the second step for both approaches, RF algorithms are optimized to build stable classifiers with new SNPs, the most significant APOE SNP (rs4420638) identified by Coon et al [7], and 10 GAB2 SNPs (Table 1[9]) for AD prognosis.
Approach I: Choice of SNPs without prior biological knowledge for model building
Step 1
Logistic regression with a less stringent p-value than the Bonferroni cut-off of 0.05/312,316 = 1.55E-7 is performed using PLINK v 1.07 [52] on all samples. A filter is set at a p-value = 1E-3 and the association analysis is run with removal of APOE and GAB2 SNPs. In addition, SNPs with p-values ≤ 1E-3 and 1 < ORs < 5 are selected. The raw genotype data corresponding to each of the selected SNPs is extracted by running Perl scripts and PLINK code. And to identify the genes corresponding to these SNPs, annotation files updated by the chip manufacturer (Affymetrix) with the Human Genome v 19 are queried.
Step 2
After pre-formatting the data subset, the data mining analysis is run using WEKA v 3-6-6 [53]. All the analysis in this study involves a cross validation of 10 folds without pruning the trees. To start the RF model building, the number of SNPs to include in the model is calculated by taking the square root of the total number of SNPs. Four data sets are used to build the RF classifiers, and all of the sets have y (disease) in the last column as class attribute. The first data set includes the APOE SNP and 10 GAB2 SNPs, the second set consists of the SNPs from step 1, the SNPs from step 1 plus the APOE SNP form the third set, and the fourth set comprises the SNPs from step 1, the APOE SNP, and 10 GAB2 SNPs. In order to assess the performance of each of the classifiers, the number of trees is held constant at 100 and the number of features (SNPs) is varied. Then, the classifiers are tuned by holding constant the various numbers of attributes, which gave the smallest test and OOB error rates (for the three data sets) in the performance step, and by changing the number of trees.
A supervised instance resample filter is applied to each data set. This produces a random subsample of each data set using sampling with replacement. The filter is set to bias the class distribution towards a uniform distribution; the original case-control distribution in the data is 61% cases and 39% controls.
After classification modelling, a manual compilation of epistasis information relevant to AD on the 199 SNPs is done.
Approach II: Choice of SNPs with prior biological knowledge for model building
Step 1
For the second approach, prior biological knowledge is used to supplement statistical analysis in selecting SNPs from genes that are more likely to play a role in AD. SNPs from APOE and GAB2 are excluded; then, the data is filtered by p-values ≤ 1E-3 and 1 < ORs < 5 from logistic regression and the Cochran-Mantel-Haenszel (CMH) test. For CMH, the stratification (three groups) is done based on APOE ε4 carrier status using PLINK.
A list of twenty-three SNPs selected from logistic regression and CMH based on their p-values and ORs, and situated in the vicinity of genes potentially relevant to AD is uploaded into Haploview v 4.2 [54] in order to find their linkage to other SNPs within a 300 kb region. The default settings are kept; the "Download HapMap info Track", release 22 version 2 and release 21 with panel CEU (Caucasian European), and the "Solid Spine" method to detect strong LD are utilized [54].
Step 2
The RF is implemented at various fixed number of attributes and trees with four different data sets. The APOE SNP and 10 GAB2 SNPs are a first set, the SNPs in LD from step 1 are a second set, and the SNPs from step 1 together with the APOE SNP are a third set. The SNPs from step 1 added to the APOE SNP and GAB2 SNPs make a fourth set. The classification building is performed in the same manner as for approach I - step 2.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the experiments: VD. Analyzed the data and wrote the paper: NB. Made major edits: VD. Both authors read and approved the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table SA. Complete list of 199 SNPs from logistic regression approach I with corresponding pathway, disease or biological function information which may be pertinent to AD.
Figure S1. LD display for SNPs across the 300 kb region surrounding the ST5 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb pairs interval surrounding the ST5 locus. Bottom: zoomed LD SNP region. The SNPs identified, rs4910068 and rs10743089, were found to be in significant LD with a D' value of 0.83. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S2. LD display for SNPs across the 300 kb region surrounding the TRPC1 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the TRPC1 locus. Bottom two: zoomed LD SNP region. The SNPs identified, rs4259003, rs9784320, and rs2033912, were found to be in significant LD; rs4259003 and rs9784320, rs4259003 and rs2033912, and rs9784320 and rs2033912 with D' values of 1.0. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S3. LD display for SNPs across the 300 kb region surrounding the ATG10 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the ATG10 locus. Bottom three: zoomed LD SNP region. The SNPs identified, rs891159, rs1485587, and rs4703879, were found to be in significant LD; rs891159 and rs1485587, rs891159 and rs4703879, and rs1485587 and rs4703879 with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S4. LD display for SNPs across the 300 kb region surrounding the ANO3 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the ANO3 locus. Bottom two: zoomed LD SNP region. The SNPs identified, rs1389421, rs10834774, and rs11028909, were found to be in significant LD; rs1389421 and rs10834774, rs1389421 and rs10834774, and rs11028909 and rs10834774 with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S5. LD display for SNPs across the 300 kb region surrounding the NDUFA12 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb pairs interval surrounding the NDUFA12 locus. Bottom: zoomed LD SNP region. The SNPs identified, rs249153 and rs249154, were found to be in significant LD with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S6. LD display for SNPs across the 300 kb region surrounding the NISCH locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the NISCH locus. Bottom: zoomed LD SNP region. The SNPs identified, rs6784615, rs9855470, rs6445486, rs10865972, rs4687619, and rs6810027, were found to be in significant LD; rs6784615 and rs9855470, rs6784615 and rs6445486, rs6784615 and rs10865972, rs6784615 and rs4687619, rs6784615 and rs6810027, rs9855470 and rs6445486, rs9855470 and rs10865972, rs9855470 and rs4687619, rs9855470 and rs6810027, rs6445486 and rs10865972, rs6445486 and rs4687619, rs6445486 and rs6810027, rs10865972 and rs4687619, rs10865972 and rs6810027, and rs4687619 and rs6810027 with D' values of 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Contributor Information
Natalia Briones, Email: natalia.briones@asu.edu.
Valentin Dinu, Email: valentin.dinu@asu.edu.
References
- Park A. Alzheimer's Unlocked. (cover story) Time. 2010;176(17):53. [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Jones L, Owen MJ, Williams J. Alzheimer's disease genetics: current knowledge and future challenges. Int J Geriatr Psychiatry. 2010. [DOI] [PubMed]
- Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010;330(6012):1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120(4):545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry. 2007;68(4):613–618. doi: 10.4088/JCP.v68n0419. [DOI] [PubMed] [Google Scholar]
- Avramopoulos D. Genetics of Alzheimer's disease: recent advances. Genome Med. 2009;1(3):34. doi: 10.1186/gm34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54(5):713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O. European Alzheimer's Disease Initiative Investigators; de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67(8):961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI. et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D. et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT. et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83(5):623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, Pahwa JS, Jones N, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S. et al. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer's disease. PLoS One. 2010;5(11):e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68(2):270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Bertram L. Alzheimer's Genetics in the GWAS Era: A Continuing Story of 'Replications and Refutations'. Curr Neurol Neurosci Rep. 2011;11(3):246–253. doi: 10.1007/s11910-011-0193-z. [DOI] [PubMed] [Google Scholar]
- Nizzari M, Venezia V, Repetto E, Caorsi V, Magrassi R, Gagliani MC, Carlo P, Florio T, Schettini G, Tacchetti C, Russo T, Diaspro A, Russo C. Amyloid precursor protein and Presenilin1 interact with the adaptor GRB2 and modulate ERK 1,2 signaling. J Biol Chem. 2007;282(18):13833–13844. doi: 10.1074/jbc.M610146200. [DOI] [PubMed] [Google Scholar]
- Dinu V, Zhao H, Miller PL. Integrating domain knowledge with statistical and data mining methods for high-density genomic SNP disease association analysis. J Biomed Inform. 2007;40(6):750–760. doi: 10.1016/j.jbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Alahari SK, Reddig PJ, Juliano RL. The integrin-binding protein Nischarin regulates cell migration by inhibiting PAK. EMBO J. 2004;23(14):2777–2788. doi: 10.1038/sj.emboj.7600291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Soundararajan M, Kumar R, Knapp S. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008;33(8):394–403. doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Chakravarthy B, Rashid A, Brown L, Tessier L, Kelly J, Menard M. Association of Gap-43 (neuromodulin) with microtubule-associated protein MAP-2 in neuronal cells. Biochem Biophys Res Commun. 2008;371(4):679–683. doi: 10.1016/j.bbrc.2008.04.119. [DOI] [PubMed] [Google Scholar]
- Fu G, Vallee S, Rybakin V, McGuire MV, Ampudia J, Brockmeyer C, Salek M, Fallen PR, Hoerter JA, Munshi A, Huang YH, Hu J, Fox HS, Sauer K, Acuto O, Gascoigne NR. Themis controls thymocyte selection through regulation of T cell antigen receptor-mediated signaling. Nat Immunol. 2009;10(8):848–856. doi: 10.1038/ni.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick MS, Oda H, Hayakawa K, Sato Y, Eshima K, Kirikae T, Iemura S, Shirai M, Abe T, Natsume T, Sasazuki T, Suzuki H. Gasp, a Grb2-associating protein, is critical for positive selection of thymocytes. Proc Natl Acad Sci USA. 2009;106(38):16345–16350. doi: 10.1073/pnas.0908593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrecque N, Baldwin T, Lesage S. Molecular and genetic parameters defining T-cell clonal selection. Immunol Cell Biol. 2010. [DOI] [PubMed]
- Mancuso M, Calsolaro V, Orsucci D, Carlesi C, Choub A, Piazza S, Siciliano G. Mitochondria, cognitive impairment, and Alzheimer's disease. Int J Alzheimers Dis. 2009;2009:951548. doi: 10.4061/2009/951548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T. The energy-transducing NADH: quinone oxidoreductase, complex I. Mol Aspects Med. 2002;23(5):345–368. doi: 10.1016/S0098-2997(02)00011-0. [DOI] [PubMed] [Google Scholar]
- Cardoso SM, Proenca MT, Santos S, Santana I, Oliveira CR. Cytochrome c oxidase is decreased in Alzheimer's disease platelets. Neurobiol Aging. 2004;25(1):105–110. doi: 10.1016/S0197-4580(03)00033-2. [DOI] [PubMed] [Google Scholar]
- Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Drose S, Brandt U, Savaskan E, Czech C, Gotz J, Eckert A. Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer's disease mice. Proc Natl Acad Sci USA. 2009;106(47):20057–20062. doi: 10.1073/pnas.0905529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEGG PATHWAY Database. http://www.genome.jp/kegg/pathway.html
- Majidi M, Hubbs AE, Lichy JH. Activation of extracellular signal-regulated kinase 2 by a novel Abl-binding protein, ST5. J Biol Chem. 1998;273(26):16608–16614. doi: 10.1074/jbc.273.26.16608. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buee L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19(20):3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33(32):9511–9522. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- Brunden KR, Trojanowski JQ, Lee VM. Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat Rev Drug Discov. 2009;8(10):783–793. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubing C, Krapivinsky G, Krapivinsky L, Clapham DE. TRPC1 and TRPC5 form a novel cation channel in mammalian brain. Neuron. 2001;29(3):645–655. doi: 10.1016/S0896-6273(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Shim S, Goh EL, Ge S, Sailor K, Yuan JP, Roderick HL, Bootman MD, Worley PF, Song H, Ming GL. XTRPC1-dependent chemotropic guidance of neuronal growth cones. Nat Neurosci. 2005;8(6):730–735. doi: 10.1038/nn1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Dreses-Werringloer U, Vingtdeux V. Calcium signaling in neurodegeneration. Mol Neurodegener. 2009;4:20. doi: 10.1186/1750-1326-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21(7):691–698. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28(27):6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci USA. 2010;107(32):14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic VM, Brockmann M, Stohr H, Weber BH, Strauss O. Evolution and functional divergence of the anoctamin family of membrane proteins. BMC Evol Biol. 2010;10:319. doi: 10.1186/1471-2148-10-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell HC, Yu K, Xiao Q, Chien LT, Qu Z. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J Physiol. 2009;587(Pt 10):2127–2139. doi: 10.1113/jphysiol.2008.163709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Faull RL, Eriksson PS. The effect of neurodegenerative diseases on the subventricular zone. Nat Rev Neurosci. 2007;8(9):712–723. doi: 10.1038/nrn2216. [DOI] [PubMed] [Google Scholar]
- Schreiber R, Uliyakina I, Kongsuphol P, Warth R, Mirza M, Martins JR, Kunzelmann K. Expression and function of epithelial anoctamins. J Biol Chem. 2010;285(10):7838–7845. doi: 10.1074/jbc.M109.065367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YA, Yu Y, Cupples LA, Farrer LA, Lunetta KL. Performance of random forest when SNPs are in linkage disequilibrium. BMC Bioinformatics. 2009;10:78. doi: 10.1186/1471-2105-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Mach Learning. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Tan P, Steinbach Michael, Kumar V. Introduction to data mining. Boston: Pearson Addison Wesley; 2006. [Google Scholar]
- Breiman L. Bagging predictors. Mach Learning. 1996;24(2):123–140. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software. ACM SIGKDD Explorations Newsletter. 2009;11(1):10. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Yi Y, Kamata-Sakurai M, Denda-Nagai K, Itoh T, Okada K, Ishii-Schrade K, Iguchi A, Sugiura D, Irimura T. Mucin 21/epiglycanin modulates cell adhesion. J Biol Chem. 2010;285(28):21233–21240. doi: 10.1074/jbc.M109.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujkijyanont P, Beyene J, Wei K, Khan F, Dror Y. Leukaemia-related gene expression in bone marrow cells from patients with the preleukaemic disorder Shwachman-Diamond syndrome. Br J Haematol. 2007;137(6):537–544. doi: 10.1111/j.1365-2141.2007.06608.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SA. Complete list of 199 SNPs from logistic regression approach I with corresponding pathway, disease or biological function information which may be pertinent to AD.
Figure S1. LD display for SNPs across the 300 kb region surrounding the ST5 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb pairs interval surrounding the ST5 locus. Bottom: zoomed LD SNP region. The SNPs identified, rs4910068 and rs10743089, were found to be in significant LD with a D' value of 0.83. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S2. LD display for SNPs across the 300 kb region surrounding the TRPC1 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the TRPC1 locus. Bottom two: zoomed LD SNP region. The SNPs identified, rs4259003, rs9784320, and rs2033912, were found to be in significant LD; rs4259003 and rs9784320, rs4259003 and rs2033912, and rs9784320 and rs2033912 with D' values of 1.0. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S3. LD display for SNPs across the 300 kb region surrounding the ATG10 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the ATG10 locus. Bottom three: zoomed LD SNP region. The SNPs identified, rs891159, rs1485587, and rs4703879, were found to be in significant LD; rs891159 and rs1485587, rs891159 and rs4703879, and rs1485587 and rs4703879 with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S4. LD display for SNPs across the 300 kb region surrounding the ANO3 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the ANO3 locus. Bottom two: zoomed LD SNP region. The SNPs identified, rs1389421, rs10834774, and rs11028909, were found to be in significant LD; rs1389421 and rs10834774, rs1389421 and rs10834774, and rs11028909 and rs10834774 with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S5. LD display for SNPs across the 300 kb region surrounding the NDUFA12 locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb pairs interval surrounding the NDUFA12 locus. Bottom: zoomed LD SNP region. The SNPs identified, rs249153 and rs249154, were found to be in significant LD with D' = 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.
Figure S6. LD display for SNPs across the 300 kb region surrounding the NISCH locus. Top: Entrez gene track overlaid with Hapmap genotyped SNPs across a 300 kb region surrounding the NISCH locus. Bottom: zoomed LD SNP region. The SNPs identified, rs6784615, rs9855470, rs6445486, rs10865972, rs4687619, and rs6810027, were found to be in significant LD; rs6784615 and rs9855470, rs6784615 and rs6445486, rs6784615 and rs10865972, rs6784615 and rs4687619, rs6784615 and rs6810027, rs9855470 and rs6445486, rs9855470 and rs10865972, rs9855470 and rs4687619, rs9855470 and rs6810027, rs6445486 and rs10865972, rs6445486 and rs4687619, rs6445486 and rs6810027, rs10865972 and rs4687619, rs10865972 and rs6810027, and rs4687619 and rs6810027 with D' values of 1. Standard color scheme for Haploview: D' < 1 and LOD < 2 are white, D' = 1 and LOD < 2 are blue, D' < 1 and LOD ≥ 2 are shades of pink/red, D' = 1 and LOD ≥ 2 are bright red. LOD = log of the odds.