Abstract
Type 3 effector proteins secreted via the bacterial type 3 secretion system (T3SS) are not only virulence factors of pathogenic bacteria, but also influence symbiotic interactions between nitrogen-fixing nodule bacteria (rhizobia) and leguminous host plants. In this study, we characterized NopM (nodulation outer protein M) of Rhizobium sp. strain NGR234, which shows sequence similarities with novel E3 ubiquitin ligase (NEL) domain effectors from the human pathogens Shigella flexneri and Salomonella enterica. NopM expressed in Escherichia coli, but not the non-functional mutant protein NopM-C338A, showed E3 ubiquitin ligase activity in vitro. In vivo, NopM, but not inactive NopM-C338A, promoted nodulation of the host plant Lablab purpureus by NGR234. When NopM was expressed in yeast, it inhibited mating pheromone signaling, a mitogen-activated protein (MAP) kinase pathway. When expressed in the plant Nicotiana benthamiana, NopM inhibited one part of the plant's defense response, as shown by a reduced production of reactive oxygen species (ROS) in response to the flagellin peptide flg22, whereas it stimulated another part, namely the induction of defense genes. In summary, our data indicate the potential for NopM as a functional NEL domain E3 ubiquitin ligase. Our findings that NopM dampened the flg22-induced ROS burst in N. benthamiana but promoted defense gene induction are consistent with the concept that pattern-triggered immunity is split in two separate signaling branches, one leading to ROS production and the other to defense gene induction.
Author Summary
Many Gram-negative bacterial pathogens possess type 3 secretion systems, which deliver effector proteins into eukaryotic host cells through needle-like structures. Effectors manipulate the host cell and many of them suppress host defense responses. Interestingly, certain symbiotic strains of rhizobia also possess such secretion systems. Rhizobia infect legume roots and induce root nodules, where the bacteria convert atmospheric nitrogen into ammonia. Here, we characterize the effector NopM of Rhizobium sp. strain NGR234. We demonstrate that NopM possesses E3 ubiquitin ligase activity, indicating that NopM can “tag" proteins with ubiquitin, and thus target them for proteasome-dependent degradation. Using a mutant approach, we demonstrate that enzymatically active NopM promotes establishment of symbiosis with Lablab purpureus, the host plant from which NGR234 was originally isolated. We further examine effects of NopM when directly expressed in eukaryotic cells and show that NopM interferes with specific signaling pathways. NopM expressed in the model plant Nicotiana benthamiana dampened generation of reactive oxygen species (ROS), which are formed in response to the bacterial flagellin peptide flg22. We suggest that NopM promotes nodule initiation by reducing the levels of harmful ROS during the infection process.
Introduction
Type 3 effector proteins of pathogenic Gram-negative bacteria are transported into eukaryotic host cells through the bacterial type 3 secretion system (T3SS), which forms a needle-like pilus [1]–[3]. Various effectors from phytopathogenic bacteria act as virulence factors by suppressing activation of plant defense genes, i.e. they inhibit innate immunity triggered by highly conserved ubiquitous microbial elicitors (microbe-associated molecular patterns – MAMPs) such as flagellin, also called pattern-triggered immunity. On the other hand, plants can also possess resistance (R) proteins that mediate defense (effector-triggered immunity) by directly or indirectly recognizing specific type 3 effectors (avirulence factors). Hence, type 3 effectors of pathogenic bacteria can positively or negatively affect pathogenicity [1]–[3].
Interestingly, certain rhizobia also use type 3 effectors during symbiosis with host legumes [4], [5]. Rhizobia are nitrogen-fixing bacteria which establish a specific mutualistic endosymbiosis with legumes and certain species of the genus Parasponia. As a result of rhizobial infection, roots of host plants develop nodules, in which the bacteria differentiate into bacteroids. For the host's benefit, atmospheric nitrogen is then reduced to ammonia by the bacterial nitrogenase enzyme. During nodule formation, various signal molecules are exchanged between the two partners [6], [7]. Flavonoids released by host plants into the rhizosphere interact with rhizobial transcriptional regulators (NodD proteins). As a result, symbiotic genes involved in synthesis of bacterial nodulation signals (Nod factors) are activated. In certain rhizobial strains, such as Rhizobium sp. strain NGR234 [8], NodD-flavonoid interactions also result in stimulated expression of ttsI. This gene encodes a transcriptional activator, which controls expression of genes that have a conserved cis-element in their promoters, named tts-box. In NGR234 and a number of other strains, genes encoding a bacterial type 3 secretion system (T3SS) and corresponding type 3 effectors are regulated by TtsI [9], [10]. Mutant analysis revealed that type 3 effectors of NGR234 can play a role during symbiosis. Depending on the host plant, positive, negative or no effects on symbiosis have been reported [11]–[16].
One approach to study the function of bacterial effectors is to express them singly in eukaryotic cells. The type 3 effector proteins NopL and NopT (nodulation outer proteins L and T) of strain NGR234 have been characterized in this way. When expressed in tobacco and Lotus japonicus, NopL suppressed expression of defense genes [17]. NopL was multiply phosphorylated within eukaryotic cells and interfered with mitogen-activated protein (MAP) kinase signaling in yeast and tobacco cells [16]. Indeed, nodules of certain bean cultivars colonized by NGR234 mutated in nopL rapidly developed necrotic areas, indicating a lack of suppression of defense [16]. The protease NopT, another type 3 effector of NGR234 belonging to the YopT-AvrPphB effector family, influenced nodulation of host plants either positively or negatively [14], [15]. Accordingly, when transiently expressed in tobacco plants, proteolytically active NopT elicited a rapid hypersensitive reaction, suggesting that NopT action induced an R-protein mediated defense response in this non-host plant [14]. Similarly, resistance (R) genes (Rj2 and Rfg1) of certain soybean cultivars are involved in host-specific nodulation and prevented establishment of symbiosis with specific strains in a T3SS-dependent manner [18].
The leucine-rich repeat (LRR) protein NopM (nodulation outer protein M) was first identified in Sinorhizobium fredii HH103 using a proteomic approach, in which secreted proteins from a T3SS-deficient mutant were compared to proteins from wild-type bacteria [19]. Homologous sequences exist in various rhizobial strains, namely Rhizobium sp. strain NGR234 (nopM formerly y4fR), Bradyrhizobium japonicum USDA110 (blr1904 and blr1676) and B. elkanii USDA 61 (nopM). During the course of the present study, T3SS-dependent secretion of NopM has been reported for strain NGR234 and a nopM deletion mutant induced fewer nodules on the host Lablab purpureus compared to the parent strain [15]. The nopM promoter activity depended on TtsI, which is predicted to bind to the conserved tts box (TB1) in the promoter region of nopM [10]. Based on sequence comparisons, rhizobial NopM proteins are predicted type 3 effectors belonging to the IpaH effector family with representatives in Shigella flexneri (such as IpaH9.8 and IpaH1.4) and Salomonella enterica (SspH1, SspH2, SlrP) [20]–[24]. The NopM sequence is also related to the YopM effector of Yersinia pestis [25]. Sequence similarities, albeit less related, also exist for non-characterized effectors from other bacteria, including the phytopathogens Pseudomonas syringae and Ralstonia solanacearum (e.g. HpX29 of R. solanacearum strain RS1000) [26].
IpaH family effectors are E3 ubiquitin ligases with a NEL (novel E3 ligase) domain. Enzymatic activity has been demonstrated for effectors from S. flexneri (such as IpaH9.8 and IpaH1.4) and S. enterica (SspH1, SspH2, SlrP) [20]–[24]. E3 ubiquitin ligases mediate transfer of ubiquitin from an E2 ubiquitin conjugating enzyme to a given target protein in eukaryotic cells, which is thereby marked for degradation. Ubiquitin-mediated proteasome-dependent protein degradation is conserved in eukaryotic cells. Ubiquitination itself requires three enzymatic components. First, an ubiquitin-activating enzyme (E1) forms a thioester bond between a catalytic cysteine and the carboxy terminal glycine residue of ubiquitin. The ubiquitin is then transferred to an ubiquitin-conjugating enzyme (E2). Finally, an E3 ubiquitin ligase facilitates the covalent conjugation of ubiquitin from an ubiquitin-loaded E2 to one or more lysine residues in a given protein substrate [27].
Bacterial E3 ubiquitin ligases delivered into host cells mimic the activities of host E3 ubiquitin ligases and ubiquitinate specific target proteins. For example, IpaH9.8 of S. enterica blocks the innate immune system of human cells by interfering with the nuclear factor κB (NF-κB) pathway. IpaH9.8 interacts with NEMO (NFκB essential modifier or IKKγ; an essential component of the multi-protein IKK (IκB kinase) complex) and the ubiquitin-binding adaptor protein ABIN-1. As a result, NEMO is polyubiquitinated and NF-κB activation is suppressed [28]. The SlrP effector of S. enterica targets thioredoxin and ERdj3, an endoplasmic reticulum luminal chaperone [29].
In this study, we characterize NopM of Rhizobium sp. strain NGR234. We demonstrate that NopM possesses E3 ubiquitin ligase activity. Our mutant analysis reveals that NopM acts as an E3 ubiquitin ligase during symbiosis with the host L. purpureus. NopM activity also inhibits mating pheromone signaling when expressed in yeast, and MAMP-triggered generation of reactive oxygen species (ROS) when expressed in Nicotiana benthamiana plants, while stimulating expression of MAMP-induced defense genes at the same time. We discuss our results in the light of the role of NopM in symbiosis.
Results
NopM possesses E3 ubiquitin ligase activity
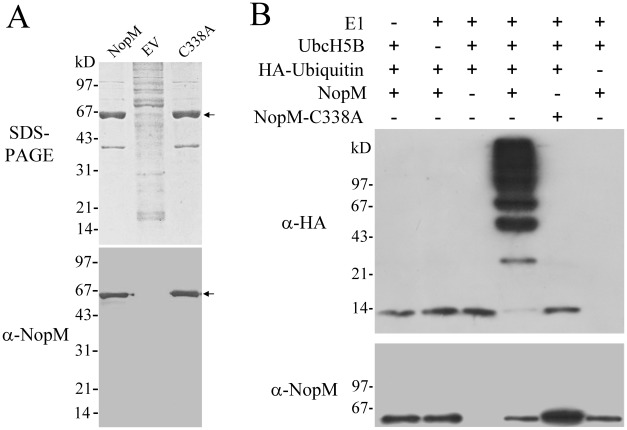
The coding region of nopM was cloned into pET28b resulting in plasmid pET-nopM. A second plasmid, pET-nopM(C338A), was constructed in which the cysteine residue 338 of NopM was replaced by alanine. Residue C338 in the C-terminal NEL domain is predicted to be an essential catalytic residue required for the ubiquitin transfer [20]–[22]. Escherichia coli BL21 (DE3) harboring the constructed plasmids were induced with IPTG and extracted proteins were purified using nickel–nitrilotriacetic acid affinity chromatography. When analyzed by SDS-PAGE, a strong band with an apparent molecular mass of about 65 kD was detected, which corresponded to His-tagged NopM and NopM-C338A, respectively (calculated molecular weight of NopM ≈60.5 kD). This band was not seen, when proteins from E. coli BL21 (DE3) harboring the empty vector pET28b were purified in a similar way. Immunoblot analysis revealed that corresponding anti-NopM antibodies recognized His-tagged NopM and NopM-C338A proteins (Figure 1A).
Figure 1. NopM possesses E3 ubiquitin ligase activity.
(A) Purification and immunoblot analysis of His-tagged NopM and point-mutated His-tagged NopM-C338A (marked by an arrow). Proteins purified by nickel–nitrilotriacetic acid affinity chromatography were obtained from E. coli BL21 (DE3) harboring pET-nopM (lane NopM) and pET-nopM(C338A) (lane C338A), respectively. BL21 (DE3) with the empty vector pET28b was used as a control (lane EV). The SDS-PAGE gel (0.5 µg loaded proteins) was stained with Coomassie Brilliant Blue R-250 and the corresponding immunoblot with the rabbit serum raised against NopM was developed with 3, 3′-diamino-benzidine. (B) In vitro ubiquitination reactions with indicated purified proteins followed by immunoblot analysis with chemiluminescence reagents using anti-HA and anti-NopM antibodies. Ubiquitination reactions were performed in the presence or absence of HA-tagged ubiquitin, E1, UbcH5B, His-tagged NopM and His-tagged NopM-C338A for 1 h at 37°C.
His-tagged NopM and NopM-C338A purified from E. coli cells were then tested using an in vitro E3 ubiquitin ligase assay using HA-tagged ubiquitin as a substrate. After incubation, reaction mixtures were separated by SDS-PAGE and corresponding immunoblots were performed with anti-HA or anti-NopM antibodies. As shown in Figure 1B, when ubiquitination reactions were performed with His-NopM, anti-HA antibodies recognized a ladder of ubiquitinated proteins in the range of 24 to >200 kD. The size of proteins detected by the anti-HA antibodies (27 kD, 45 kD and 63 kD bands) were multiples of the size of HA-ubiquitin (9 kD), indicating formation of polyubiquitination chains. In contrast, reactions with NopM-C338A did not result in a ladder of polyubiquitinated proteins. Anti-NopM antibodies recognized a 65-kD protein band corresponding to His-tagged NopM and NopM-C338A, respectively. No additional bands of higher molecular weight were observed, indicating that NopM itself was not autoubiquitinated (Figure 1B). Taken together, these findings show that NopM is an E3 ubiquitin ligase and that the mutant protein NopM-C338A lacks this enzyme activity.
Only enzymatically active NopM promotes nodulation of Lablab purpureus
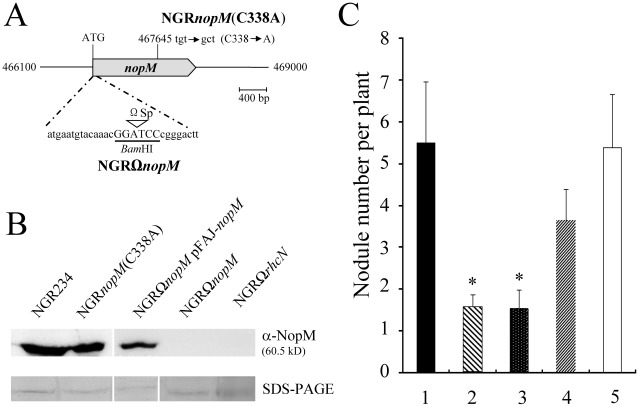
Two mutant derivatives of Rhizobium sp. NGR234 were constructed to examine the function of NopM during symbiosis. A nopM knock-out mutant, called NGRΩnopM, was generated, which contained an Ω spectinomycin interposon close to the ATG start codon. A point mutant, called NGRnopM(C338A), was constructed by using a corresponding DNA sequence encoding NopM-C338A (Figure 2A). Proteins from apigenin-induced culture supernatants were concentrated and used for immunoblots with the anti-NopM antibodies. NopM and NopM-C338A (ca. 60-kD bands) were detected in culture supernatants from the parent strain NGR234 and the NGRnopM(C338A) mutant, respectively. As expected, no bands were seen for the knock-out mutant NGRΩnopM. Strain NGRΩrhcN, a mutant lacking a functional T3SS served as a negative control [11]. NGRΩnopM carrying the plasmid pFAJ-nopM (containing nopM including its promoter sequence) secreted NopM, indicating complementation by this plasmid (Figure 2B).
Figure 2. Characterization of the mutant strains NGRΩnopM and NGRnopM(C338A).
(A) Schematic representation of the knock-out mutant NGRΩnopM (with an insertion of a 2-kb spectinomycin Ω interposon (Ωsp) at the constructed BamHI restriction site) and of the point mutant NGRnopM(C338A) (cysteine 338 mutated to alanine; additional Aor51HI restriction site). (B) Immunoblot analysis of NopM of Rhizobium sp. NGR234 and indicated mutant derivatives. Immunoblots were performed with secreted proteins and the anti-NopM antibodies. As a loading control, secreted proteins in the molecular weight range of NopM were visualized on a parallel SDS-PAGE gel by silver staining (C) Symbiotic phenotype of Rhizobium sp. NGR234 (column 1), NGRΩnopM (column 2), NGRnopM(C338A), (column 3), NGRΩnopM carrying pFAJ-nopM (column 4) and NGRnopM(C338A) carrying pFAJ-nopM (column 5) on the host plant L. purpureus (cv. Chaojibiandou). In total, 70 plants (14 plants per strain) were inoculated. The numbers of nodules formed per plant were determined 35 days post inoculation. Data indicate means ± SE. Asterisks indicate significant reduced nodule formation as compared to the parent strain NGR234 (Kruskal-Wallis rank sum test; P<0.01).
Nodulation phenotypes of the examined strains differed when the legume L. purpureus was inoculated. Figure 2C shows the results for a representative nodulation experiment. The parent strain NGR234 induced about 5–6 nodules per plant under the tested growth conditions. In contrast, NGRΩnopM induced significantly fewer nodules (1–2 nodules per plant), indicating that NopM was required for optimal nodulation of this host plant. Plants inoculated with NGRnopM(C338A) showed a similar reduction in nodulation, indicating that the C338 residue is essential for the nodule-promoting effect of NopM. The symbiotic phenotype of NGRΩnopM and NGRnopM(C338A) on L. purpureus could be complemented when plasmid pFAJ-nopM was introduced into these mutants. The nodule number was significantly increased and reached values comparable to those of the parent strain NGR234 (Figure 2C).
Nodulation tests were also performed with Phaseolus vulgaris (cv. Yudou No 1). Optimal nodulation of this plant with NGR234 required NopT, another type 3 effector of NGR234 [14]. Nodulation data with either NGRΩnopM or NGRnopM(C338A) were similar to those obtained from the parent strain NGR234, however. Similarly, nodulation tests with the constructed mutants showed no obvious differences for Flemingia congesta (data not shown), although nodulation of this host plant is improved by a functional T3SS [12].
Taken together, the mutant analysis revealed that the symbiotic phenotype of the constructed nopM mutants depended on the tested host legume and that the positive effect of NopM on L. purpureus nodulation likely depended on its E3 ubiquitin ligase activity.
Effects of NopM in yeast cells
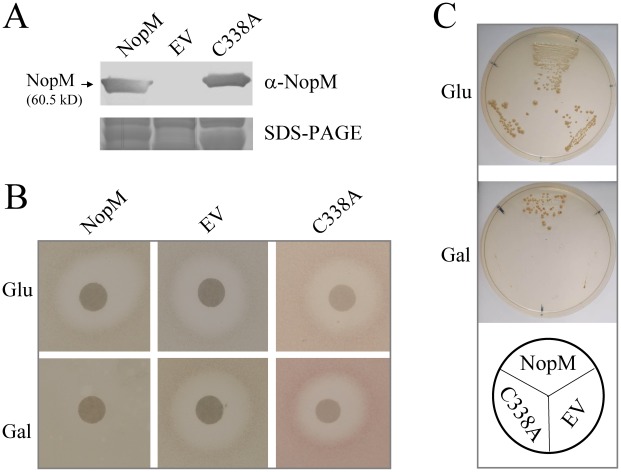
When expressed in yeast, IpaH9.8 of S. flexneri blocked mating pheromone (α-factor) response signaling, a specific MAP kinase pathway [20]. We used the same type of assay to study the effect of NopM when expressed in yeast. The α-factor is perceived by a G protein-coupled receptor and activation of the signal cascade results in arrest of the cell cycle and transcription of mating genes. Accordingly, application of α-factor to the center of an agar plate of strain W303-1A (MATa) results in a typical halo of growth inhibition [30]. The coding sequence of nopM and the point-mutated sequence encoding NopM-C338A were cloned into the expression vector pESC-leu, which has a galactose-inducible promoter (GAL1). W303-1A cells carrying the resulting plasmids (pESC-nopM and pESC-nopM(C338A), respectively) expressed NopM and NopM-C338A on galactose plates: An immunoblot with anti-NopM antibodies exhibited a band corresponding to the predicted size of NopM (60.5 kD), which was absent in cells transformed with the empty vector pESC-leu (Figure 3A). Upon exposure to α-factor, yeast cells expressing nopM under the GAL1 promoter failed to form a halo, indicating that NopM interfered with the mating pheromone signaling pathway. Using the same assay, NopM-C338A did not inhibit mating pheromone response signaling (Figure 3B).
Figure 3. NopM inhibits mating pheromone signaling in yeast.
(A) Immunoblot analysis of NopM and NopM-C338A isolated from yeast strain W303-1A carrying pESC-nopM (lane NopM) and pESC-nopM(C338A) (lane C338A), respectively. Proteins from W303-1A containing the empty vector pESC-leu (lane EV) were analyzed as a control. Extracted proteins (0.5 µg) were probed using the anti-NopM antibodies. A parallel SDS-PAGE gel was stained with Coomassie Brilliant Blue R-250 (B) Halo assay with glucose (Glu) or galactose (Gal) containing plates of yeast strain W303-1A harboring the plasmids pESC-nopM, the empty vector pESC-leu or pESC-nopM(C338A), respectively. A disk impregnated with 10 µg of α-factor was added to the center of the plate. (C) Growth of strain SY2227 (STE4 expression on SD/galactose plates) transformed with pESC-nopM (NopM expression on SD/galactose plates), pESC-nopM(C338A) (NopM-C338A expression on galactose plates) or the empty vector pESC-leu. Yeast cells were spread on SD medium plates containing either 2% (w/v) glucose (Glu) or 2% (w/v) galactose (Gal) without leucine.
A similar growth inhibition assay was performed with yeast strain SY2227, which expresses the G protein β-subunit STE4 when the fungus is grown on galactose-containing media. Overproduction of STE4 activates the mating pheromone signaling pathway and therefore causes cell growth arrest in the absence of α-factor [31]. Figure 3C shows the growth phenotype of this strain transformed with pESC-nopM, pESC-nopM(C338A) or the empty vector pESC-leu. Yeast transformed with pESC-nopM showed normal growth on SD/galactose plates. In contrast, cells transformed with pESC-nopM(C338A) or pESC-leu poorly grew on SD/galactose (Figure 3C). Hence, NopM, but not NopM-C338A, inhibited STE4-induced mating pheromone signaling.
Effects of NopM in Nicotiana benthamiana
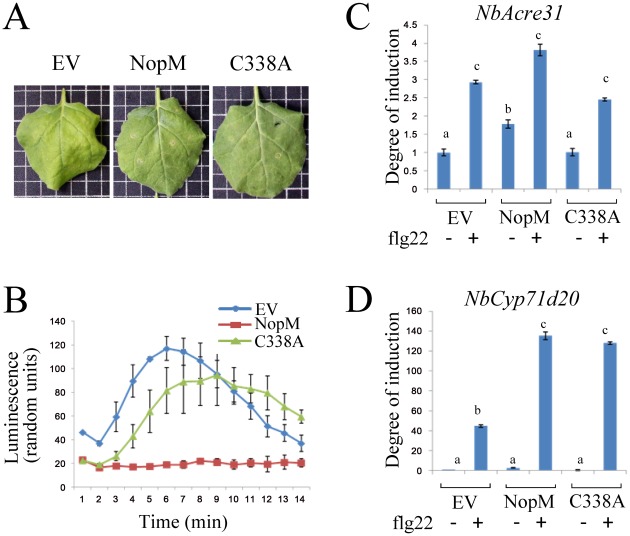
To investigate effects of NopM within plant cells, NopM and NopM-C338A were transiently expressed in N. benthamiana. DNA encoding NopM or NopM-C338A was cloned into the binary vector pCAMBIA-T, which contains a 35S cauliflower mosaic virus 35S promoter. Agrobacterium tumefaciens cells carrying these vectors were then used for infiltration of N. benthamiana leaves. Immunoblot analysis with anti-NopM antibodies revealed the presence of NopM and NopM-C338A proteins in transformed tissue (Figure S1 in Text S1, panel A). Leaves expressing NopM (2 days after infiltration) showed no hypersensitive reaction (Figure 4A). Trypan blue based cell death staining of leaves (5 days after infiltration) showed that neither NopM nor NopM-C338A caused visible changes as compared to leaf tissue transformed with the empty vector (Figure S1 in Text S1, panel B). The P. syringae pv. tomato DC3000 effector HopQ1, which is known to induce a hypersensitive reaction in N. benthamiana [32], was used as a positive control. As expected, the HopQ1 expressing tissue was necrotic and strongly stained by trypan blue.
Figure 4. Effects of NopM and NopM-C338A on N. benthamina cells.
Leaves were infiltrated with A. tumefaciens strain GV3101 carrying pCAMBIA-nopM, pCAMBIA-nopM(C338A) or the empty vector pCAMBIA-T. (A) Leaves were photographed two days post infiltration. (B) ROS production in transformed leaf disks. Two days post infiltration, leaf disks were treated with 1 µM of the elicitor flg22. (C) Levels of NbAcre31 transcripts. Two days after infiltration, leaf disks were treated with 1 µM flg22 in BSA/NaCl solution. Control leaves were mock treated with BSA/NaCl solution in the absence of flg22. RNA was isolated from tissue harvested 30 min later. qRT-PCR was performed with three biological and three technical replicates. Data (means ± SE) indicate the degree of induction as compared to leaves infiltrated with A. tumefaciens carrying the empty vector. Different letters above columns indicate statistically different transcript levels (D) Levels of NbCyp71d20 transcripts. Expression data were obtained as for NbAcre31.
Transient generation of reactive oxygen species (ROS) induced by MAMPs is a rapid signaling response, which depends on Rboh enzymes (respiratory burst oxidase homologs) and is activated by calcium-dependent protein kinases (CDPKs) [33], [34]. When challenged with flg22, a conserved, 22-amino acid motif of the bacterial MAMP flagellin [1], N. benthamiana leaf disks respond with a ROS burst, which can be measured with luminol and horseradish peroxidase [35]. An example for such an experiment is shown in Figure 4B. Interestingly, the flg22-induced ROS burst was nearly completely abolished in leaf disks expressing NopM (statistical analysis of all data from 4 independent time series; significant differences as compared to controls transformed with the empty vector; one-way ANOVA, P = 0.002). In disks expressing NopM-C338A, however, the ROS burst in response to flg22 was similar to empty vector controls (p = 0.24). Accordingly, differences between disks expressing NopM and NopM-C338A were significant (one-way ANOVA, P = 0.008), suggesting that ROS suppression depends on the ubiquitin E3 ligase activity of NopM (Figure 4B).
Transcript levels of the flg22-responsive defense genes NbAcre31 (encoding a putative calcium-binding protein) and NbCyp71d20 (encoding a putative cytochrome P450) are upregulated in response to flg22 [36]. Quantitative reverse transcription (qRT)-PCR was used to examine the effect of NopM on expression of these genes (Figure 4C and D). In the absence of flg22, NopM expression resulted in slightly elevated transcript levels of NbAcre31 (one-way ANOVA, p = 0.03), but not of NbCyp71d20 (p = 0.08). In contrast, cells expressing NopM-C338A neither showed increased transcript levels of NbAcre31 nor of NbCyp71d20. As expected, leaf tissue challenged with flg22 showed stimulated expression, particularly for NbCyp71d20. When compared to empty vector controls, effects of the flg22 treatment on NbCyp71d20 activation were significantly stronger in either NopM or NopM-C338A expressing tissues (Figure 4D). Thus, NopM promoted flg22-induced NbCyp71d20 expression independently of its E3 ubiquitin ligase activity.
The flg22-induced expression of NbAcre31 and NbCyp71d20 depends on the MAP kinase SIPK (salicylic acid-induced protein kinase) [37]. The amounts of active MAP kinases in N. benthamiana were visualized on immunoblots with anti-p42/44-phospho-ERK antibodies, which recognize activated SIPK and WIPK (wound-induced protein kinase). Expression of NopM in N. benthamiana did not result in MAP kinase activation. A treatment of leaves with flg22 for 15 min caused MAP kinase activation in control plants transformed with the empty vector as well as in NopM or NopM-C338A expressing plants, indicating that NopM did not block flg22-induced MAP kinase signaling (Figure S1 in Text S1, panel C). In the presence of NopM and flg22, activation of SIPK might be slightly stronger, but a more quantitative approach would be required to reveal small changes in MAP kinase activation.
Discussion
We show in this study that the LRR protein NopM of the rhizobial symbiont NGR234 is an E3 ubiquitin ligase belonging to the IpaH effector family. Effectors of this family are also known as NEL (novel E3 ligase) domain effectors. These enzymes are structurally unrelated to other bacterial E3 ubiquitin ligases, which have a HECT or RING/U-box domain [38]. The NEL domain in the C-terminal region of NopM was functional in NopM, whereas the NopM-C338 mutant protein was inactive. The C338 residue of NopM likely acts as a nucleophile, forming a thioester bond with ubiquitin. Enzymatic activities of NEL domain effectors have been only reported for the human pathogens S. flexneri and S. enterica [20]–[24]. Thus, NopM represents a first studied example for a NEL domain effector delivered into plant cells.
Inoculation tests with the constructed NGR234 mutants, NGRΩnopM and NGRnopM(C338A), showed reduced nodulation on L. purpureus, indicating the importance of the C338 residue during establishment of symbiosis. We suggest that NopM functions as an E3 ubiquitin ligase during the infection process and that ubiquitination of one or more host proteins helps to promote nodulation on L. purpureus. In some other host plants, however, effects of NopM on nodulation were either not observed or even negative ([15] and this study). It is tempting to speculate that NopM function reflects an evolutionary adaptation to protein substrates of specific hosts. Indeed, NGR234 has been originally isolated from L. purpureus [39] whose nodulation is promoted by NopM. Negative effects of NopM on nodulation in other legumes are potentially related to specific R-proteins of the host plant as shown for T3SS-dependent nodulation of certain soybean cultivars [18].
Similar to many Gram-negative pathogenic bacteria, rhizobial T3SSs are believed to deliver type 3 effector proteins into host cells. Translocation of rhizobial type 3 effectors into legume cells has been questioned [40]. However, recent evidence has provided strong support, based on transfer of adenylate cyclase fused to rhizobial effectors [41], [42]. Our findings indicate that NopM, but not NopM-C338A, promoted nodule formation in L. purpureus. In fact, delivery into host cells is a prerequisite for bacterial E3 ubiquitin ligases, as they function in combination with ubiquitin and E1/E2 enzymes, which are present only in the eukaryotic cell.
Yeast is a model to investigate effects of type 3 effector proteins in eukaryotic cells [43]. The IpaH9.8 effector of S. flexneri blocked the mating pheromone response signaling in yeast and ubiquitinated Ste7, the mitogen-activated protein kinase kinase of this pathway [20], [21]. Our data point to a similar activity of NopM in yeast and these findings prompted us to investigate whether NopM can interfere with MAP kinase signaling in plants. Interestingly, N. benthamiana plants expressing NopM did not show suppression of flg22-induced MAP kinase signaling. Instead, the flg22-associated ROS burst was nearly completely abolished in plants expressing NopM. These findings are consistent with the concept that early flagellin signaling is split in two separate signaling branches, one leading to MAP kinase activation and the other to calcium-dependent protein kinase (CDPK) mediated ROS production [34], [37].
Suppression of flg22-triggered ROS in N. benthamiana depended on the C338 residue of NopM, suggesting ubiquitination of a MAMP signaling component. The E3 ubiquitin ligase activity of NopM is reminiscent of the virulence function of AvrPtoB, a type 3 effector of P. syringae with a functional E3 ubiquitin ligase RING/U-box E3 domain [44]. In Arabidopsis thaliana, AvrPtoB targets proteins such as the flagellin receptor complex FLS2-BAK1 [45] and the chitin receptor kinase CERK1 [46]. In contrast to ROS suppression, expression of either NopM or NopM-C338A in N. benthamiana promoted flg22-triggered accumulation of NbCyp71d20 transcripts. Hence, ubiquitination activity of NopM was not essential to induce this effect. We suggest that an interaction between NopM and a N. benthamiana protein is sufficient to cause a partial deregulation of immune signaling in flg22-challenged tissue.
Taken together, we provide genetic and biochemical evidence that NopM is a type 3 effector with a functional NEL domain. Inoculation tests with the constructed point mutant NGRnopM(C338A) suggest that the E3 ubiquitin ligase activity of NopM is required for optimal nodulation of the host plant L. purpureus. When expressed in N. benthamiana, NopM suppresses the flg22-elicited ROS burst, suggesting that NopM blocks ROS-associated defense responses. Future work is required to test whether NopM can also suppress ROS formation in legume roots. Indeed, ROS generation could be detrimental during the rhizobial infection process [5], [47] and it is tempting to speculate that NopM keeps ROS generation in L. pupurpureus infection threads below a harmful threshold level.
Materials and Methods
Strains, plasmids and primers
Bacterial strains and plasmids used for this study are listed in Table S1 of Text S1. Plasmids were constructed according to standard methods and corresponding PCR primers are listed in Table S2 of Text S1.
Expression of NopM and NopM-C338A in E. coli
The sequence encoding NopM of Rhizobium (Sinorhizobium fredii) sp. NGR234 (accession number AAB91674) was cloned into the pET28b vector, resulting in plasmid pET-nopM. PCR-based site-directed mutagenesis was used to mutate the cysteine 338 (TGT codon) of NopM into alanine (GCT codon) and the resulting plasmid was named pET-nopM(C338A). The plasmids were then transformed into E. coli BL21 (DE3) cells. The His-tagged NopM and NopM-C338A proteins were purified from isopropyl-β-D-thiogalactopyranoside (IPTG) induced cultures by nickel affinity chromatography with Ni-NTA resin beads (Qiagen, Hilden, Germany). For immunization of a New Zealand rabbit, Ni-NTA purified His-tagged NopM was separated by SDS-PAGE and gel bands containing NopM were cut from the gel.
Ubiquitination test
Purified ubiquitin-activating enzyme from human (E1), UbcH5B (E2), and HA-ubiquitin were purchased from Boston Biochem (Cambridge, MA, USA). His-tagged NopM and NopM-C338A from E. coli cultures grown at 27°C for 12 h were purified by nickel affinity chromatography according to the manufacturer's recommendations under non-denaturing conditions (Qiagen, Germany). Ubiquitination assays were performed in a 40-µl volume containing the reaction buffer (25 mM Tris HCl (pH 7.5), 50 mM NaCl, 5 mM ATP, 10 mM MgCl2, 0.1 mM DTT), 2 µg HA-ubiquitin, 0.5 µg of E1, and 2 µg of UbcH5B in the presence or absence of 1 µg of His-tagged NopM, or NopM-C338A, respectively. Reactions were incubated at 37°C for 1 h and stopped by addition of an equal volume of Laemmli sample buffer (62.5 mM Tris HCl (pH 6.8), 10% (v/v) glycerol, 2% (w/v) SDS, 0.005% (w/v) bromophenol blue) containing 100 mM DTT. Reaction mixtures were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with specific antibodies (anti-NopM antibodies at 1∶10 000 dilution; anti-HA antibodies from (Abcam, England) at 1∶ 4 000 dilution). Immunoblots were developed with enhanced chemiluminescence reagents (GE Healthcare).
Construction of NGRΩnopM and NGRnopM(C338A)
For construction of the mutant NGRΩnopM, a 2.5-kp fragment containing nopM was cloned into pBluescript II KS(+), generating pSK-nopM2500. PCR-based site-directed mutagenesis was used to generate a BamHI restriction site close to the ATG codon of nopM. A spectinomycin-resistant (Spr) Ω interposon was excised from pHP45 [48] with BamHI and ligated into the BamHI site, generating pSK-nopMΩ. The construct was then cloned into the suicide vector pJQ200SK [49]. The resulting plasmid (pJQ-nopMΩ) was mobilized from E. coli DH5α into Rhizobium sp. NGR234 by triparental mating using the pRK2013 helper plasmid [50]. Gene replacement was forced by selecting for the resistance of the Ω interposon marker (Spr) and for growth on 5% (w/v) sucrose. The obtained mutant NGRΩnopM was confirmed by Southern blot analysis using the DIG DNA labeling and detection kit as specified by the supplier (Roche, Basel, Switzerland).
For construction of NGRnopM(C338A), plasmid pSK-nopM2500 was mutated by a PCR-based site-directed mutagenesis approach, thereby creating the restriction site Aor51HI. The insert of this plasmid (named pSK-nopM(C338A)) with DNA encoding NopM-C338A was then cloned into the suicide vector pJQ200SK, resulting in plasmid pJQ-nopM(C337A). After conjugation, Rhizobium sp. NGR234 bacteria were first cultivated on agar plates containing gentamycin and rifampin and then on plates containing rifampin and 5% (w/v) sucrose. Genomic DNA from candidate colonies served as a template for a PCR (primers 11 and 12; Table S2 in Text S1). The amplicon from the mutant NGRnopM(C338A) was completely cleaved by Aor51HI into two smaller fragments.
For complementation analysis of the constructed mutants, a 2081-bp fragment containing the coding region and promoter sequence of nopM was cloned into pFAJ1702 [51]. The obtained plasmid (pFAJ-nopM) was then mobilized into Rhizobium sp. NGR234 and selection was performed on agar plates containing tetracycline.
Isolation of secreted proteins
Secreted proteins from culture supernatants from Rhizobium sp. strains NGR234 (parent strain), NGRΩnopM (this study), NGRnopM(C338A) (this study), NGRΩnopM carrying pFAJ-nopM (this study) and NGRΩrhcN [11] were isolated according to a previously described procedure [12], [52]. Briefly, cultures (RMS medium) were induced with 1 µM apigenin and cultivated at 27°C on a rotary shaker for 40 h. Proteins from culture supernatants were precipitated by addition of TCA (10%, w/v) and incubation over night at 4°C. After centrifugation (10 000× g, 4°C, 30 min), precipitates were washed twice with 5 ml of cold 80% acetone and resuspended in 100 µl of rehydration buffer (8 M urea, 2% w/v CHAPS, 0.01% w/v bromophenol blue). Secreted proteins (corresponding to 100 ml of cell culture) were subjected to immunoblot analysis with antiserum against NopM (1∶5 000 dilution) followed by staining with chemiluminescence reagents (Thermo Scientific, Waltham, MA USA).
Nodulation tests
Nodulation tests were performed in plastic jars using the host plants Lablab purpureus cv. Chaojibiandou, Phaseolus vulgaris cv. Yudou No 1, and Flemingia congesta. Seeds were surface sterilized and germinated on agar plates, and plantlets were transferred to 300-mL plastic jar units linked with a cotton wick (a mixture of vermiculite and expanded clay in the upper vessel; nitrogen-free nutrient solution in the lower vessel). Plants (1 plant per jar) were inoculated with 109 bacteria (strain NGR234 and mutant derivatives; see Table S1 in Text S1). Plants were cultivated at 26±2°C in a temperature-controlled greenhouse. The nodulation test results were statistically analyzed with the Kruskal-Wallis rank sum test, which is suitable for unequal replications. A P-value of ≤0.01 was considered as significant. All data are presented as means ± SE (standard error).
Expression of NopM and NopM-C338A in yeast and halo assay with α-factor
Standard media and techniques were used for transformation, maintenance, and growth of Saccharomyces cerevisiae [53]. Strains (haploid strain W303-1A (MATa) strain SY2227) and constructed plasmids encoding NopM or NopM-C338A are listed in Table S1 of Text S1. For immunoblot analysis, yeast cells were cultured at 30°C in liquid SD/-Leu medium (Clontech) supplemented with 2% galactose. Membranes were incubated with anti-NopM antibodies at a 1∶5 000 dilution and blots were developed with 3, 3′-diamino-benzidine (Boster, Wuhan, China). The halo assay with the mating pheromone was performed by placing a filter disk impregnated with 8 µg of the mating pheromone α-factor (Sigma-Aldrich; dissolved in 8 µl H2O) to the center of each agar plate. The plates were sealed, incubated at 27°C for 1 week and then photographed.
Expression of NopM and NopM-C338A in N. benthamiana
Plasmids (pCAMBIA-nopM, pCAMBIA-nopM(C338A), the empty vector pCAMBIA-T and pGWB417-HopQ1-myc; see Table S1 in Text S1) were transformed into chemically competent Agrobacterium tumefaciens strain GV3101 by heat shock. Leaves from 4-week old Nicotiana benthamiana plants were infiltrated with bacteria (OD600 = 0.5) re-suspended in infiltration buffer (10 mM MgCl2, 10 mM MES pH 5.6). Expression of NopM was detected by immune blot analysis with anti-NopM antibodies at a 1∶1000 dilution. Blots were developed with CDPstar reagents (New England Biolabs). Staining of N. benthamiana leaves was performed with trypan blue as described previously [54].
Measurement of reactive oxygen species (ROS) generation
Leaf discs (0.38 cm2) were floated on water overnight and ROS released by the leaf tissue were measured using a chemiluminescent assay [35]. The water was replaced with 500 µl of an aqueous solution containing 20 µM luminol (Sigma-Aldrich) and 1 µg of horseradish peroxidase (Fluka, Buchs, Switzerland). ROS was elicited with 1 µM flg22 peptide (QRLSTGSRINSAKDDAAGLQIA) in all experiments. Mock treatments without flg22 were performed with the BSA/NaCl solution (1% w/v BSA, 1% w/v mM NaCl) used to solubilize flg22. Luminescence was measured over a time period of 30 min using a luminometer (MicroLumat LB96P; EG&G Berthold). Data from 12 leaf disks derived from 4 independent infiltrations were statistically analyzed by one-way ANOVA considering P≤0.05 as significantly different.
MAP kinase activation assay
Two days post A. tumefaciens transformation, N. benthamiana leaves transiently expressing NopM, NopM-C338A and empty vector controls were infiltrated with 1 µM flg22 peptide or mock-treated with BSA/NaCl for 15 min. Leaf discs (50 mg) were then frozen in liquid nitrogen and proteins were extracted in 100 µl extraction buffer (50 mM Tris-HCl pH 7.5, 150 nM NaCl, protease inhibitor cocktail from Sigma-Aldrich) for 30 min at 4°C. Subsequently 100 µl Lämmli loading buffer (2×) was added to each sample. Samples were subjected to immunoblot analysis using the anti-p42/44-phospho-ERK antibody (Sigma-Aldrich). Blots were developed using CDP-star technology (NEB).
Quantitative reverse transcription (qRT)-PCR
One day post infiltration, N. benthamiana leaf discs expressing NopM, NopM-C338A or the empty vector (EV) control were collected, and then floated overnight in water. Leaf discs were subsequently treated with 1 µM flg22 or mock-treated with BSA/NaCl solution for 30 min and then frozen in liquid nitrogen. Total RNA was extracted using the NucleoSpin RNA Plant extraction kit (Machery-Nagel). The absence of genomic DNA was checked by PCR amplification of the housekeeping NbEF1α gene by using 1 µg of RNA (NbEF1α amplification crosses an exon/intron boundary). For analysis of gene expression, first-strand cDNA was synthesized from 1 µg of RNA using AMV reverse transcriptase (Promega) and an oligo (dT) primer (Microsynth), according to the manufacturer's instructions. For quantitative PCR, 5 µl of a 1/100 µl dilution of cDNA were combined with SYBR master mix. PCRs were performed in triplicates with the 7500 Real Time PCR system (Applied Biosystems). Data were collected and analyzed with the respective ABI analyzing program. The NbEF1α RNA was analyzed as an internal control and used to normalize the values for transcript abundance. All samples were related to the empty vector (EV) control. Primers for the genes NbCyp71D20, NbAcre31 and NbEF1α are listed in Table S2 of Text S1. Data derived from three biological repeats were statistically analyzed by ANOVA (one-way ANOVA) considering P≤0.05 as significantly different.
Accession number of NopM
NopM of Rhizobium (Sinorhizobium fredii) strain NGR234: AAB91674
Supporting Information
Text S1 contains Table S1 (Strains and plasmids used in this study), Table S2 (Primers used in this study) and Figure S1 (Analysis of N. benthamiana expressing NopM and NopM-C338A by immunoblot analysis, trypan blue based cell death staining and MAP kinase activation in response to flg22). Text S1 also contains references cited in Table S1 and an explanatory legend to Figure S1.
(PDF)
Acknowledgments
We thank Yong-Jun Lu (Sun Yat-sen University, Guangzhou, China) for yeast strain W303-1A, George F. Sprague (University of Oregon, Eugene, OR USA) for yeast strain SY2227, William Broughton (University of Geneva, Switzerland) for NGRΩrhcN and Jan Michiels (Katholieke Universiteit Leuven, Belgium) for pFAJ1702. Cheng Jin and Ye Tian (Sun Yat-sen University) are acknowledged for construction of pJQ-nopM(C338A) and pCAMBIA-T, respectively.
Footnotes
The authors have declared that no competing interests exist.
This study was supported by the National Basic Research Program of China (973 program, No. 2010CB126501), by the National Natural Science Foundation of China (grant 31070219), by the Guangdong Key Laboratory of Plant Resources (grant plant01k19) and by the Science Foundation of the State Key Laboratory of Biocontrol (grants SKLBC09B05 and SKLBC201123). DRH and TB acknowledge funding by the Swiss National Science Foundation and the Systems-X initiative. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boller T, He SY. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hann DR, Gimenez–Ibanez S, Rathjen JP. Bacterial virulence effectors and their activities. Curr Opin Plant Biol. 2010;13:388–393. doi: 10.1016/j.pbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Fauvart M, Michiels J. Rhizobial secreted proteins as determinants of host specificity in the rhizobium–legume symbiosis. FEMS Microbiol Lett. 2008;285:1–9. doi: 10.1111/j.1574-6968.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- 5.Saeki K. Rhizobial measures to evade host defense strategies and endogenous threats to persistent symbiotic nitrogen fixation: a focus on two legume-rhizobium model systems. Cell Mol Life Sci. 2011;68:1327–1339. doi: 10.1007/s00018-011-0650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper JE. Early interactions between legumes and rhizobia: disclosing complexity in a molecular dialogue. J Appl Microbiol. 2007;103:1355–1365. doi: 10.1111/j.1365-2672.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- 8.Schmeisser C, Liesegang H, Krysciak D, Bakkou N, Le Quéré A, et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol. 2009;75:4035–4045. doi: 10.1128/AEM.00515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehner S, Schober G, Wenzel M, Lang K, Göttfert M. Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol Plant Microbe Interact. 2008;21:1087–1093. doi: 10.1094/MPMI-21-8-1087. [DOI] [PubMed] [Google Scholar]
- 10.Wassem R, Kobayashi H, Kambara K, Le Quéré A, Walker GC, et al. TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol Microbiol. 2008;68:736–748. doi: 10.1111/j.1365-2958.2008.06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 12.Marie C, Deakin WJ, Viprey V, Kopciñska J, Golinowski W, et al. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol Plant Microbe Interact. 2003;16:743–751. doi: 10.1094/MPMI.2003.16.9.743. [DOI] [PubMed] [Google Scholar]
- 13.Skorpil P, Saad M, Boukli M, Kobayashi NM, Ares-Orpel H, et al. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol Microbiol. 2005;57:1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 14.Dai WJ, Zeng Y, Xie ZP, Staehelin C. Symbiosis-promoting and deleterious effects of NopT, a novel type 3 effector of Rhizobium sp. strain NGR234. J Bacteriol. 2008;190:5101–5110. doi: 10.1128/JB.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kambara K, Ardissone S, Kobayashi H, Saad MM, Schumpp O, et al. Rhizobia utilize pathogen-like effector proteins during symbiosis. Mol Microbiol. 2009;71:92–106. doi: 10.1111/j.1365-2958.2008.06507.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Chen XJ, Lu HB, Xie ZP, Staehelin C. Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation and interference with mitogen activated protein kinase signaling. J Biol Chem. 2011;286:32178–32187. doi: 10.1074/jbc.M111.265942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartsev AV, Deakin WJ, Boukli NM, McAlvin CB, Stacey G, et al. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 2004;134:871–879. doi: 10.1104/pp.103.031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang SM, Tang F, Gao MQ, Krishnan HB, Zhu HY. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc Natl Acad Sci U S A. 2010;107:18735–18740. doi: 10.1073/pnas.1011957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues JA, López-Baena FJ, Ollero FJ, Vinardell JM, Espuny MR, et al. NopM and NopD are rhizobial nodulation outer proteins: identification using LC-MALDI and LC-ESI with a monolithic capillary column. J Proteome Res. 2007;6:1029–1037. doi: 10.1021/pr060519f. [DOI] [PubMed] [Google Scholar]
- 20.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Singer, AU, Rohde JR, Lam R, Skarina T, Kagan O, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Li H, Hu L, Wang J, Zhou Y, et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 23.Bernal-Bayard J, Ramos-Morales F. Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem. 2009;284:27587–27595. doi: 10.1074/jbc.M109.010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quezada CM, Hicks SW, Galan JE, Stebbins CE. A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci U S A. 2009;106:4864–4869. doi: 10.1073/pnas.0811058106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soundararajan V, Patel N, Subramanian V, Sasisekharan V, Sasisekharan R. The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun. 2011;17:548–557. doi: 10.1177/1753425910377099. [DOI] [PubMed] [Google Scholar]
- 26.Mukaihara T, Tamura N. Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrpB-regulated gene products. Microbiology. 2009;155:2235–2244. doi: 10.1099/mic.0.027763-0. [DOI] [PubMed] [Google Scholar]
- 27.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 28.Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NFκB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal-Bayard J, Cardenal-Munoz E, Ramos-Morales F. The Salmonella type III secretion effector, Salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J Biol Chem. 2010;285:16360–16368. doi: 10.1074/jbc.M110.100669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman GA, Garrison TR, Dohlman HG. Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 2002;344:617–631. doi: 10.1016/s0076-6879(02)44744-1. [DOI] [PubMed] [Google Scholar]
- 31.Cole GM, Stone DE, Reed SI. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol Cell Biol. 1990;10:510–517. doi: 10.1128/mcb.10.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, et al. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, et al. Calcium–dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 2007;19:1065–1080. doi: 10.1105/tpc.106.048884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keppler LD, Baker CJ, Atkinson MM. Active oxygen production during a bacterial-induced hypersensitive reaction in tobacco suspension cells. Phytopathology. 1989;79:974–978. [Google Scholar]
- 36.Heese A, Hann DR, Gimenez–Ibanez S, Jones AME, He K, et al. The receptor–like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Rathjen JP. Hierarchy and roles of pathogen–associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 2011;156:687–699. doi: 10.1104/pp.110.171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hicks SW, Galán JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinick MJ. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J Appl Microbiol. 1980;49:39–53. [Google Scholar]
- 40.Büttner D, Bonas U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol. 2006;9:193–200. doi: 10.1016/j.mib.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Schechter LM, Guenther J, Olcay EA, Jang S, Krishnan HB. Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl Environ Microbiol. 2010;76:3758–3761. doi: 10.1128/AEM.03122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wenzel M, Friedrich L, Göttfert M, Zehner S. The type III-secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol Plant Microbe Interact. 2010;23:124–129. doi: 10.1094/MPMI-23-1-0124. [DOI] [PubMed] [Google Scholar]
- 43.Siggers KA, Lesser CF. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB. Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A. 2006;103:2851–2856. doi: 10.1073/pnas.0507892103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shan L, He P, Li J, Heese A, Peck SC, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gimenez–Ibanez S, Hann DR, Ntoukakls V, Petutschnig E, Lipka V, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 47.Tavares F, Santos CL, Sellstedt A. Reactive oxygen species in legume and actinorhizal nitrogen-fixing symbioses: the microsymbiont's responses to an unfriendly reception. Physiol Plant. 2007;130:344–356. [Google Scholar]
- 48.Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 49.Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 50.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dombrecht B, Vanderleyden J, Michiels J. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol Plant Microbe Interact. 2001;14:426–430. doi: 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- 52.Hempel J, Zehner S, Göttfert M, Patschkowski T. Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J Biotechnol. 2009;140:51–58. doi: 10.1016/j.jbiotec.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Guthrie C. mRNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 54.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1 contains Table S1 (Strains and plasmids used in this study), Table S2 (Primers used in this study) and Figure S1 (Analysis of N. benthamiana expressing NopM and NopM-C338A by immunoblot analysis, trypan blue based cell death staining and MAP kinase activation in response to flg22). Text S1 also contains references cited in Table S1 and an explanatory legend to Figure S1.
(PDF)