Abstract
Progesterone receptor membrane component 1 (PGRMC1) is highly expressed in the granulosa and luteal cells of rodent and primate ovaries. Interestingly, its molecular weight as assessed by Western blot is dependent on its cellular localization with a ≈ 27 kDa form being detected in the cytoplasm and higher molecular weight forms being detected in the nucleus. The higher molecular weight forms of PGRMC1 are sumoylated suggesting that they are involved in regulating gene transcription, since sumoylation of nuclear proteins often is associated with regulation of transcriptional activity of the sumoylated protein.
In order to identify a set of candidate genes that are regulated by PGRMC1, a human granulosa/luteal cell line (hGL5 cells) was treated with PGRMC1 siRNA and changes in gene expression monitored by microarray analysis. The microarray analysis revealed that PGRMC1 generally functioned as a repressor of transcription, since depletion of PGRMC1 resulted in a disproportionate increase in the number of transcripts. Moreover, a pathway analysis implicated PGRMC1 in the regulation of apoptosis, which is consistent with PGRMC1’s known biological action. More importantly these results support the concept that PGRMC1 influences gene transcription. Additional studies reveal that progesterone (P4) acting through a PGRMC1-dependent mechanism suppresses the activity of the transcription factor, Tcf/Lef, thereby identifying one molecular pathway through which P4-PGRMC1 can regulate gene transcription and ultimately apoptosis.
Keywords: Progesterone, PGRMC1, Ovary, Gene Expression, Tcf/Lef Transcription Factor Activity, Sumoylation, Apoptosis
1. Introduction
With the identification of a receptor for P4 in the 1980s [1] insights into the mechanism through which P4 mediates its actions began to be revealed. This receptor, referred to as the progesterone receptor (PGR), localizes to the nucleus where it regulates gene transcription [1]. Although research has been predominately focused on the transcriptional actions of PGR, several groups have now shown that PGR can also localize to or near the plasma membrane where it interacts with signal transduction kinases such as Src and activates the MAP kinase cascade [2, 3].
To add further complexity to P4’s mechanism of action, two other putative progestin receptor families have been identified: the Progestin and AdipoQ family of Receptors (PAQR) discovered by Peter Thomas’ group [4, 5] and the Progesterone Receptor Membrane Component family [6] with the first member, PGRMC1, cloned by Martin Wehling’s group [7]. These two receptor families have been the subject of several reviews that have focused on their membrane-initiated or non-genomic mechanism of action [6–8]. Interestingly, PGRMC1 not only is detected within the plasma and endoplasmic reticular membranes but also within the nucleus [7, 9]. Surprisingly, Western blot analysis not only detects PGRMC1 as the predicted ≈ 27 kDa band but also as higher molecular weight bands [10]. Although it is generally assumed that the higher molecular weight forms are oligomers of the ≈ 27 kDa form [11], this concept has not be rigorously tested. Determining the nature of these higher molecular weight forms of PGRMC1 is important and could provide insights into PGRMC1’s action. In addition, the concept that PGRMC1 may regulate gene transcription is supported by the findings that P4’s anti-apoptotic action is dependent on de novo RNA and protein synthesis [12].
Therefore in the first section of this review, studies that shed light on the nature of these higher molecular weight forms of PGRMC1 will be presented. In the subsequent sections, studies that reveal a role for PGRMC1 in regulating gene transcription will be described. All of the studies presented in this review used cell lines derived from either human granulosa/luteal cells obtained from women undergoing in vitro fertilization (hGL5 cells) [13] or rat granulosa cells (SIGCs) [14]. To assess PGRMC1’s role in gene expression, cells were treated with PGRMC1 siRNA to deplete PGRMC1. Gene expression profiles were then assessed in PGRMC1-depleted hGL5 cells by microarray analysis, while PGRMC1-dependent transcription factor activity was determined using SIGCs.
2. Cellular Localization of PGRMC1
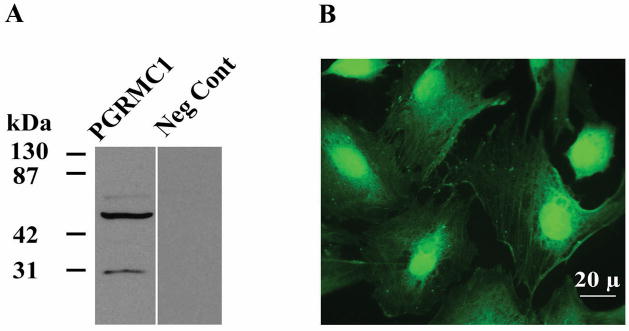
As indicated, Western blots from hGL5 cells (Figure 1A) detected PGRMC1 as a ≈ 27 kDa, ≈ 56 and ≈ 75 kDa bands. While the ≈ 27 kDa band is always detected, the presence of the higher molecular weight bands is often dependent on the amount of protein loaded on the gel. The detection of the higher molecular weight forms of PGRMC1 is consistent with Western blots that have been published for several of cell types [6, 10, 11, 15]. The vast majority of these Western blots used the “NT” antibody to PGRMC1 that was generated and made available by Martin Wehling’s group [10]. Presently there are several commercial antibodies available. Although we have not assessed all of them (Table 1), most detect PGRMC1 as a ≈ 27 kDa band as well as the higher molecular weight forms mimicking the Western blots obtained with Wehling’s “NT” antibody. Interestingly the antibody available from Proteintech group (12990-1AP) only works for human samples and only detects the ≈ 27 kDa form. The reasons for this remain to be determined.
Figure 1.
PGRMC1 expression in hGL5 cells which were derived from human granulosa/luteal cells obtained from women undergoing ovulation induction for infertility treatment. Panel A shows a Western blot in which PGRMC1 is detected as bands at ≈ 27, 56 and 75 kDa using the goat PGRMC1 antibody (1:150; Abcam Inc, Cambridge, MA). The immunocytochemical localization of PGRMC1 is shown in panel B and demonstrates that PGRMC1 is present at the membrane, throughout the cytoplasm and in the nucleus. Data taken from Peluso et al [26].
Table 1.
List of commercial antibody to PGRMC1
| Supplier | Cat. Number | Host | Comments |
|---|---|---|---|
| Sigma Chemical Co. | HPA002877 | rabbit polyclonal | Affinity purified |
| Sigma Chemical Co. | AV46752 | rabbit polyclonal | Affinity purified |
| Sigma Chemical Co. | SAB2101782 | rabbit polyclonal | Affinity purified |
| Proteintech | 12990-1-AP | rabbit polyclonal | Affinity purified |
| Abcam | ab88381 | rabbit polyclonal | |
| Abcfam | ab48012 | goat polyclonal | Sold by other companies * |
| Abcam | ab80941 | rabbit polyclonal | |
| Santa Cruz | sc-271275 | mouse monoclonal | |
| Santa Cruz | sc-133906 | rabbit polyclonal | |
| Santa Cruz | sc-98680 | rabbit polyclonal | |
| Santa Cruz | sc-82694 | goat polyclonal | |
| Santa Cruz | sc-135720 | mouse monoclonal |
Abnova, Abgen, Acris antibody, Antibodies-online, GmbH, Biorbyt, Everest Biotech, Genetex, Imgenex, Origene
Immunocytochemical studies detect PGRMC1 in the nucleus, cytoplasm and at segments of the plasma membrane (Figure 1B). Interestingly, analysis of Western blots that used lysate prepared from the cytoplasmic fraction of SIGCs only detected the lower molecular weight form of PGRMC1, while the higher molecular weight forms were predominately found within the nucleus [12].
It is generally believed that the higher molecular weight forms are oligomers of the ≈ 27 kDa form of PGRMC1 [7, 11]. However, we were unable to completely disrupt the putative oligomers with DTT, which is known to break the disulfide bonds that are thought to be responsible for dimerization of PGRMC1 molecules [7, 11]. If dimerization does not completely explain the presence of higher molecular weight forms of PGRMC1, what other mechanism could account for the higher molecular weight forms? An in silico analysis of PGRMC1 revealed three sumoylation sites (http://sumosp.biocuckoo.org/) and we have shown that PGRMC1 within the nucleus is sumoylated. This conclusion is based on the observations that the higher molecular weight forms of PGRMC1 are 1) reduced in the presence of a sumoylation inhibitor, Ginkgolic acid and 2) immunoprecipitated using a Sumo1 antibody (Lodde et al unpublished). Finally using an in situ proximity ligation assay, Sumo1 has been shown to be in direct contact with PGRMC1 within the nucleus (Figure 2) and the amount of sumoylated PGRMC1 is enhanced by progesterone treatment (Lodde et al unpublished). However, in addition to SUMO1, there are three other members of the SUMO family [16] and whether they are involved in sumoylation of PGRMC1 remains to be determined.
Figure 2.
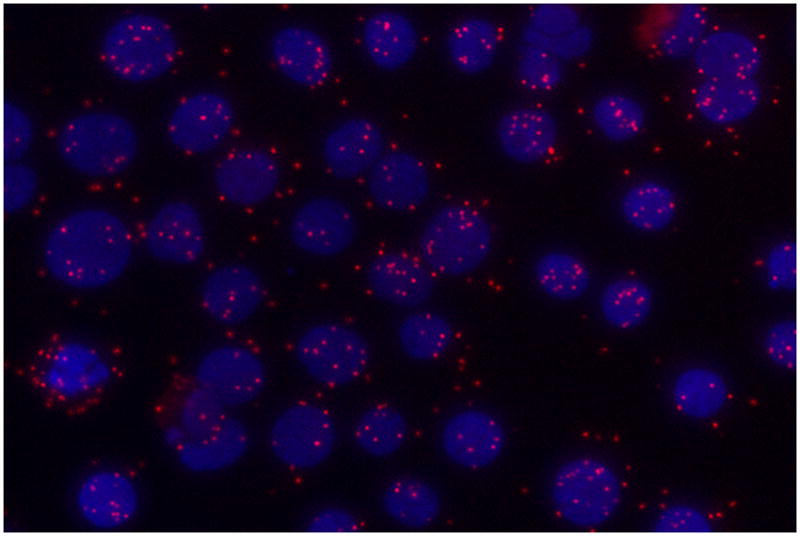
An image of an in situ proximity assay demonstrating that PGRMC1 and Sumo1 directly interact and are presumably covalently coupled. In this assay a rabbit antibody (Sigma Chemical Co., HPA002877) is used to detect PGRMC1 and a mouse antibody used to detect Sumo1(Invitrogen). Unlike standard co-localization protocols, each of the secondary antibodes used to detect each primary antibody was tagged with a complimentary oligonucleotide probe. Direct interaction between the two proteins is detected by the hybridization of the oligonucleotide probes, amplification and subsequent hybridization with the fluorescently labeled probe. A direct interaction between the two proteins is revealed by the presence of red fluorescent dots. The nuclei were stained with DAPI, which fluoresces blue. For details on this protocol see http://www.olink.com/products-services/duolink/situ-pla-technology.
These studies are the first to implicate sumoylation as a posttranslational modifier of PGRMC1 but considerably more work must be done to verify and to completely define the role that sumoylation plays in regulating PGRMC1’s action. Importantly, sumoylation of nuclear proteins is often associated with regulation of transcriptional activity of the sumoylated protein [16]. This further supports our hypothesis that nuclear PGRMC1 may function to transcriptional regulator.
3. PGRMC1 and P4-independent Gene Expression
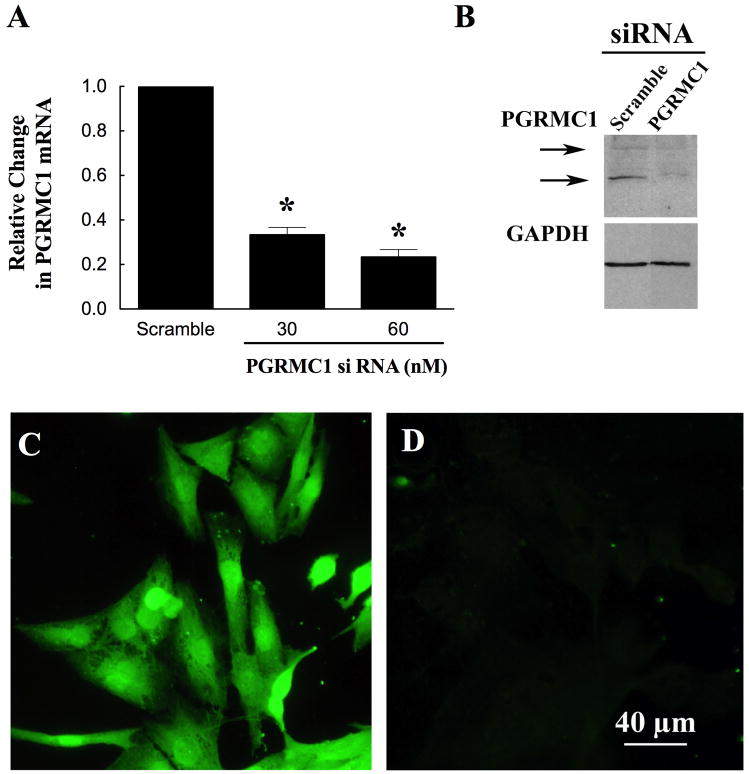
To begin to test this hypothesis, hGL5 cells were transfected with either PGRMC1 siRNA or a scrambled control siRNA. The transfections were done according the protocol described by Ambion using siPORT NeoFx as previously published [9]. After 3 days of culture with PGRMC1 siRNA, PGRMC1 levels were reduced by approximately 80% as assessed by real-time PCR (Figure 3A), Western blot analysis (Figure 3B), and immunocytochemistry (Compare Figure 3C with 3D). PGRMC1 siRNA treatment did not alter GAPDH levels (Figure 3B).
Figure 3.
The effect of PGRMC1 siRNA on the levels of PGRMC1 in hGL5 cells. hGL5 cells were cultured with either scramble (60 nM) or PGRMC1 siRNA (30 or 60 nM) for 3 days. PGRMC1 was assessed by real-time PCR (A), Western blot (B) or immunocytochemistry (C, D). Panel C show cells cultured with scramble, while cells treated with PGRMC1 siRNA are shown in panel D. Images shown in panels B, C and D were taken after treatment with 60 nM siRNA. Data from Peluso et al [26].
Having confirmed the effectiveness of PGRMC1 siRNA treatment, the effect of depleting PGRMC1 on the gene expression profile of hGL5 cells maintained in serum-supplement culture medium was assessed. For these micrarray studies, three independent samples of hGL5 cells were treated with PGRMC1 siRNA, while two samples of hGL5 cells were treated with scrambled control siRNAs. Briefly, RNA was isolated from each sample using the Qiagen RNeasy RNA isolation kit and the RNA samples assessed using the Human/Mouse-6 arrays. These array assays were processed at The University of Ct Hlth Ctr Genomics Core (http://genetics.uchc.edu/TG/Welcome.html).
The microarray data was analyzed using the Illumina BeadStudio program, using the GX gene expression module. The data was normalized using the average normalization method, and the mRNAs in the PGRMC1 siRNA treatment group compared with those of the scramble siRNA treatment group using the differential expression module and the Illumina custom algorithm. An mRNA was considered to be non-detectable if the probes had an overall detection value of less than 15. Average expression levels were then calculated.
As indicated the hGL5 RNA samples were analyzed on an Illumina Human/Mouse-6 chip that contained 48,701 mRNA probes. A specific and statistically significant signal was detected for 49% of the mRNA probes. A total of 2,042 mRNAs showed a 50% or greater increase in expression levels in the PGRMC1 siRNA treated cells relative to the scramble siRNA treatment group. In contrast, 1,775 probes showed at least a 50% decrease in expression in the PGRMC1 siRNA treated samples compared to the scramble siRNA treatment group. This suggests that PGRMC1 generally functioned as a repressor of transcription, since depletion of PGRMC1 resulted in a disproportionate increase in the number of transcripts (p < 0.0001 as assessed by Chi-Square with Yates correction).
To determine which biological pathways might be affected by the loss of PGRMC1, the DAVID bioinformatic resource at the National Institute of Allergy and Infectious Diseases was used. The genes that showed an increase or decrease in expression of 2-fold or greater were searched for statistically significant levels of over representation within Gene Ontology category: biological processes. This analysis revealed that genes involved in apoptosis were highly affected by depleting PGRMC1.
For example, depleting PGRMC1 increased mRNAs for caspases 3, 4 and 8 (Table 2). A comparison of Tables 2 and 3 indicated that most but not all of the changes in gene expression were directing the cell toward apoptosis. This would be consistent with PGRMC1’s role for maintaining the cells in a viable state and making them more resistant to inducers of apoptosis such as chemotherapeutic agents [9, 17].
Table 2.
Genes involved in apoptosis that increase at least 2 fold after PGRMC1 siRNA treatment of hGL5 cells
| Gene Name | Fold Change | Gene Description | Putative Effect on Apoptosis |
|---|---|---|---|
| NGFR | 3.89 | Nerve growth factor receptor | ↑ [27] |
| EAF2 | 2.86 | ELL associated factor 2 | ↑ [20] |
| CASP3 | 2.33 | Caspase 3, apoptosis-related cysteine peptidase | ↑ [28] |
| IHPK3 | 2.27 | Inositol hexaphosphate kinase 3 | ↑ [29] |
| CASP4 | 2.26 | Caspase 4, apoptosis-related cysteine peptidase, | ↑ [30] |
| TNFRSF9 | 2.17 | Tumor necrosis factor receptor superfamily, member 9 | ↑ [31] |
| CASP8 | 2.06 | Caspase 8 | ↑ [32] |
| YWHAH | 2.05 | 14-3-3 eta | ↓ [33] |
| NTN1 | 2.05 | Netrin 1 | ↓ [34] |
Table 3.
Genes involved in apoptosis that decrease by at least 2 fold after PGRMC1 siRNA treatment of hGL5 cells
| Gene Name | Fold Change | Gene Description | Putative Effect on Apoptosis |
|---|---|---|---|
| ITGB3BP | 2.73 | Integrin beta 3 binding protein (beta3-endonexin) | ↑ [35] |
| NME5 | 2.53 | Non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) | ↓ [36] |
| LTA | 2.24 | Lymphotoxin alpha (TNF superfamily, member 1; TNF-beta) | ↑ [37] |
| PPP2R1B | 2.23 | Protein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform, transcript variant 2 | ↑ [38] |
| ABL1 | 2.20 | v-abl Abelson murine leukemia viral oncogene homolog 1, transcript variant b | ↑ [39] |
| GADD45G | 2.04 | Growth arrest and DNA-damage-inducible, gamma | ↑ [40] |
Of the apoptosis-related genes, the increased expression of the caspases is the most interesting, since granulosa/luteal cells undergo apoptosis through a caspase-dependent mechanism [18]. Thus the ability of PGRMC1 to suppress the caspase expression could account in part for its ability to inhibit apoptosis.
Interesting, the PGRMC1-suppresssed genes include those that stimulate or inhibit mitosis. For example in the present microarray study, the expression of the mitosis-stimulating gene, Myb [19] was increased by 2.42 fold, while the anti-mitotic/pro-apoptotic gene, ELL associated factor 2 (Eaf2) [20] was increased by 2.86 fold. It is likely that ablation of PGRMC1 leads to a disregulation of the mitotic cascade resulting in “mitotic catastrophe” and ultimately apoptosis. It is possible then that PGRMC1 is somehow involved in controlling the timing of the precise chain of molecular events that regulate mitosis. These observations suggest that in granulosa cells PGRMC1’s anti-apoptotic action has two components: one that suppresses activators of apoptosis (i.e. caspase expression) and another that suppresses the expression of various cell cycle genes thereby preventing a “mitotic catastrophe”.
4. P4-PGRMC1 Regulated Gene Expression
Like hGL5 cells, PGRMC1 siRNA treatment of SIGCs increases basal expression of several apoptosis-related genes including caspase 4 and 8 [12]. Moreover in SIGCs, treatment with P4 induces the anti-apoptotic gene, Bcl2a1d and suppresses the pro-apoptotic gene, Bad with the expression of these genes being dependent on PGRMC1 [12].
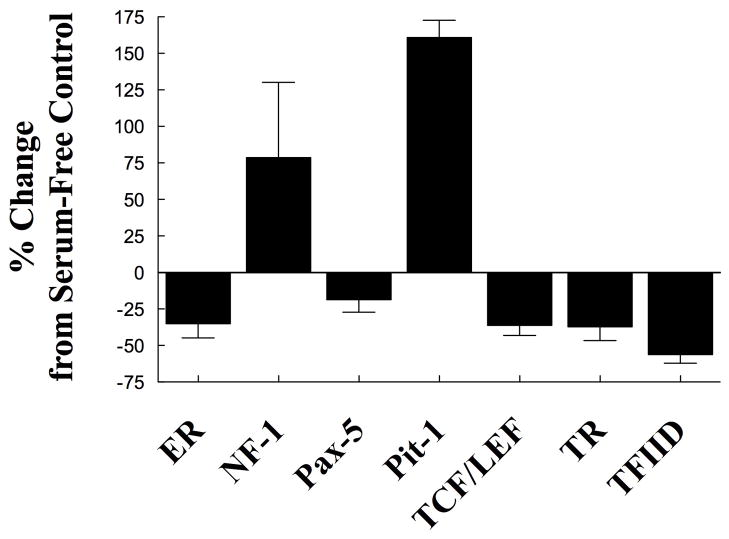
Although our studies indicate that PGRMC1 affects gene expression, they do not provide any insight into the mechanism through which P4 activates PGRMC1’s genomic action. To explore how P4-PGRMC1 interaction regulates gene expression, a study was conducted to determine which transcription factors are regulated by P4. This study revealed that P4 significantly increased the activity of NF-1 and Pit-1 and suppressed the activity of ER, Pax-5, TR, TFIID and Tcf/Lef (Figure 4). Further studies revealed that P4-induced suppression of Tcf/Lef activity was dependent on PGRMC1. Importantly, changes in Tcf/Lef activity influence the expression of genes that regulate many cellular functions including cell survival and cell proliferation [21]. Thus, P4’s ability to act through a PGRMC1 to suppress Tcf/Lef activity provides the first insights into how PGRMC1 regulates gene expression. In addition these findings support our concept that P4 activation of PGRMC1 prevents a “mitotic catastrophe” as part of its apoptotic mechanism of action.
Figure 4.
The effect of progesterone (1 μm) on the activity of various transcription factors. In this experiment SIGCs were incubated in serum-free medium for 6 h in the presence or absence of progesterone. The activity of these transcription factors was assessed using luciferase report assays provided by Signosis BioSingal Capture (Sunnyvale, CA). Values are expressed as a percentage change from serum-free control. Progesterone induced either a significant (p < 0.05) increase or decrease in activity.
5. Summary and Future Research
The data presented in this review begins to provide a basic framework that defines a mechanism through which P4-PGRMC1 interaction regulates gene expression and ultimately inhibits apoptosis [6, 17, 22, 23] and mitosis [24]. This mechanism is probably initiated in the cytoplasm where P4 stimulates the sumoylation of PGRMC1. Sumoylation not only increases its molecular weight but also is likely to be a required for PGRMC1’s transport into the nucleus. Transport into the nucleus is an essential component of this mechanism, since nuclear PGRMC1 is required for P4’s anti-apoptotic action [12].
Once in the nucleus, PGRMC1 regulates gene expression in P4-independent and P4-dependent manner. Some of the genes that are suppressed by PGRMC1 in the absence of P4 are involved in mediating the apoptotic pathway (i.e. caspases). A second set of PGRMC1-dependent genes that are regulated by P4 are involved in suppressing apoptosis and mitosis. That this component of PGRMC1’s action is P4-dependent is evidenced by P4’s inability to suppress Tcf/Lef activity in the absence of PGRMC1. Since Tcf/Lef activation suppresses genes that promote entry into the cell cycle [25], P4-PGRMC1 activation likely prevents an inappropriate entry into the cell cycle, thereby preventing a “mitotic catastrophe” and subsequent apoptosis.
Finally, it is important to appreciate that this is a skeletal framework for PGRMC1’s actions, which is presented in order to stimulate research that will eventually completely define the mechanism through which P4-PGRMC1 interaction preserves cell viability and regulates mitosis. Because this framework is incomplete, it is absolutely essential that each component be rigorously tested.
Highlights.
Progesterone can signal through progesterone receptor membrane component-1 (PGRMC1) to regulate the expression of various genes
The regulation of the expression of these genes ultimately inhibits apoptosis of granulosa cells.
This mechanism involves the ability of PGRMC1 to be sumoylated, to localize to the nucleus and to suppress Tcf/Lef activity.
Acknowledgments
The authors would like to thank Xiufang Liu for her excellent technical support of this work. These studies were supported by a grant from NICHD (HD RO1 52740). The authors would like to thank Dr. Robert Burghardt of Texas A&M University for providing the spontaneously immortalized granulosa cells and Dr. Bruce Carr (University of Texas Dallas) for providing the GL5 cells that were used in these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Lonard DM, O’Malley BW. A contemporary understanding of progesterone receptor function. Mech Ageing Dev. 2004;125:669–78. doi: 10.1016/j.mad.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Boonyaratanakornkit V, McGowan E, Sherman L, Mancini MA, Cheskis BJ, Edwards DP. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol Endocrinol. 2007;21:359–75. doi: 10.1210/me.2006-0337. [DOI] [PubMed] [Google Scholar]
- 3.Lange CA. Integration of progesterone receptor action with rapid signaling events in breast cancer models. J Steroid Biochem Mol Biol. 2008;108:203–12. doi: 10.1016/j.jsbmb.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Y, Bond J, Thomas P. Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc Natl Acad Sci U S A. 2003;100:2237–42. doi: 10.1073/pnas.0436133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Rice CD, Pang Y, Pace M, Thomas P. Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc Natl Acad Sci U S A. 2003;100:2231–6. doi: 10.1073/pnas.0336132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahill MA. Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Losel RM, Besong D, Peluso JJ, Wehling M. Progesterone receptor membrane component 1--many tasks for a versatile protein. Steroids. 2008;73:929–34. doi: 10.1016/j.steroids.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Thomas P, Pang Y, Zhu Y, Detweiler C, Doughty K. Multiple rapid progestin actions and progestin membrane receptor subtypes in fish. Steroids. 2004;69:567–73. doi: 10.1016/j.steroids.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Peluso JJ, Liu X, Saunders MM, Claffey KP, Phoenix K. Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J Clin Endocrinol Metab. 2008;93:1592–9. doi: 10.1210/jc.2007-2771. [DOI] [PubMed] [Google Scholar]
- 10.Meyer C, Schmid R, Schmieding K, Falkenstein E, Wehling M. Characterization of high affinity progesterone-binding membrane proteins by anti-peptide antiserum. Steroids. 1998;63:111–6. doi: 10.1016/s0039-128x(97)00143-8. [DOI] [PubMed] [Google Scholar]
- 11.Falkenstein E, Eisen C, Schmieding K, Krautkramer M, Stein C, Losel R, et al. Chemical modification and structural analysis of the progesterone membrane binding protein from porcine liver membranes. Mol Cell Biochem. 2001;218:71–9. doi: 10.1023/a:1007269507856. [DOI] [PubMed] [Google Scholar]
- 12.Peluso JJ, Liu X, Gawkowska A, Lodde V, Wu CA. Progesterone inhibits apoptosis in part by PGRMC1-regulated gene expression. Molecular and cellular endocrinology. 2010;320:153–61. doi: 10.1016/j.mce.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rainey WH, Sawetawan C, Shay JW, Michael MD, Mathis JM, Kutteh W, et al. Transformation of human granulosa cells with the E6 and E7 regions of human papillomavirus. J Clin Endocrinol Metab. 1994;78:705–10. doi: 10.1210/jcem.78.3.8126145. [DOI] [PubMed] [Google Scholar]
- 14.Stein LS, Stoica G, Tilley R, Burghardt RC. Rat ovarian granulosa cell culture: a model system for the study of cell-cell communication during multistep transformation. Cancer research. 1991;51:696–706. [PubMed] [Google Scholar]
- 15.Engmann L, Losel R, Wehling M, Peluso JJ. Progesterone regulation of human granulosa/luteal cell viability by an RU486-independent mechanism. J Clin Endocrinol Metab. 2006;91:4962–8. doi: 10.1210/jc.2006-1128. [DOI] [PubMed] [Google Scholar]
- 16.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 17.Crudden G, Chitti RE, Craven RJ. Hpr6 (heme-1 domain protein) regulates the susceptibility of cancer cells to chemotherapeutic drugs. J Pharmacol Exp Ther. 2006;316:448–55. doi: 10.1124/jpet.105.094631. [DOI] [PubMed] [Google Scholar]
- 18.Glamoclija V, Vilovic K, Saraga-Babic M, Baranovic A, Sapunar D. Apoptosis and active caspase-3 expression in human granulosa cells. Fertil Steril. 2005;83:426–31. doi: 10.1016/j.fertnstert.2004.06.075. [DOI] [PubMed] [Google Scholar]
- 19.Nakata Y, Shetzline S, Sakashita C, Kalota A, Rallapalli R, Rudnick SI, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol Cell Biol. 2007;27:2048–58. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn J, Xiao W, Jiang F, Simone F, Thirman MJ, Wang Z. Apoptosis induction and growth suppression by U19/Eaf2 is mediated through its ELL-binding domain. Prostate. 2007;67:146–53. doi: 10.1002/pros.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. Journal of cell science. 2007;120:385–93. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 22.Peluso JJ, Pappalardo A, Losel R, Wehling M. Progesterone membrane receptor component 1 expression in the immature rat ovary and its role in mediating progesterone’s antiapoptotic action. Endocrinology. 2006;147:3133–40. doi: 10.1210/en.2006-0114. [DOI] [PubMed] [Google Scholar]
- 23.Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone’s antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology. 2008;149:534–43. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodde V, Peluso JJ. A novel role for progesterone and progesterone receptor membrane component 1 in regulating spindle microtubule stability during rat and human ovarian cell mitosis. Biology of reproduction. 2011;84:715–22. doi: 10.1095/biolreprod.110.088385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sampson EM, Haque ZK, Ku MC, Tevosian SG, Albanese C, Pestell RG, et al. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. The EMBO journal. 2001;20:4500–11. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peluso JJ, Liu X, Gawkowska A, Johnston-MacAnanny E. Progesterone activates a progesterone receptor membrane component 1-dependent mechanism that promotes human granulosa/luteal cell survival but not progesterone secretion. The Journal of clinical endocrinology and metabolism. 2009;94:2644–9. doi: 10.1210/jc.2009-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabizadeh S, Bitler CM, Butcher LL, Bredesen DE. Expression of the low-affinity nerve growth factor receptor enhances beta-amyloid peptide toxicity. Proc Natl Acad Sci U S A. 1994;91:10703–6. doi: 10.1073/pnas.91.22.10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussein MR. Apoptosis in the ovary: molecular mechanisms. Hum Reprod Update. 2005;11:162–77. doi: 10.1093/humupd/dmi001. [DOI] [PubMed] [Google Scholar]
- 29.Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–40. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 30.Hitomi J, Katayama T, Eguchi Y, Kudo T, Taniguchi M, Koyama Y, et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Abeta-induced cell death. J Cell Biol. 2004;165:347–56. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma BY, Mikolajczak SA, Danesh A, Hosiawa KA, Cameron CM, Takaori-Kondo A, et al. The expression and the regulatory role of OX40 and 4-1BB heterodimer in activated human T cells. Blood. 2005;106:2002–10. doi: 10.1182/blood-2004-04-1622. [DOI] [PubMed] [Google Scholar]
- 32.Reed JC. Mechanisms of apoptosis. Am J Pathol. 2000;157:1415–30. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wakui H, Wright AP, Gustafsson J, Zilliacus J. Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3 eta protein. J Biol Chem. 1997;272:8153–6. doi: 10.1074/jbc.272.13.8153. [DOI] [PubMed] [Google Scholar]
- 34.Arakawa H. p53, apoptosis and axon-guidance molecules. Cell Death Differ. 2005;12:1057–65. doi: 10.1038/sj.cdd.4401601. [DOI] [PubMed] [Google Scholar]
- 35.Li D, Das S, Yamada T, Samuels HH. The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells. Mol Cell Biol. 2004;24:3838–48. doi: 10.1128/MCB.24.9.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang KC, Ok DW, Hong JC, Kim MO, Kim JH. Cloning, sequencing, and characterization of the murine nm23-M5 gene during mouse spermatogenesis and spermiogenesis. Biochem Biophys Res Commun. 2003;306:198–207. doi: 10.1016/s0006-291x(03)00916-1. [DOI] [PubMed] [Google Scholar]
- 37.Gray PW, Aggarwal BB, Benton CV, Bringman TS, Henzel WJ, Jarrett JA, et al. Cloning and expression of cDNA for human lymphotoxin, a lymphokine with tumour necrosis activity. Nature. 1984;312:721–4. doi: 10.1038/312721a0. [DOI] [PubMed] [Google Scholar]
- 38.Qin J, Chen HG, Yan Q, Deng M, Liu J, Doerge S, et al. Protein phosphatase-2A is a target of epigallocatechin-3-gallate and modulates p53-Bak apoptotic pathway. Cancer Res. 2008;68:4150–62. doi: 10.1158/0008-5472.CAN-08-0839. [DOI] [PubMed] [Google Scholar]
- 39.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–9. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 40.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–30. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]