Abstract
A set of Centre d'Étude du Polymorphisme Humain (CEPH) cell lines serves as a large reference collection that has been widely used as a benchmark for allele frequencies in the analysis of genetic variants, to create linkage maps of the human genome, to study the genetics of gene expression, to provide samples to the HapMap and 1000 Genomes projects, and for a variety of other applications. An explicit feature of the CEPH collection is that these multigenerational families represent reference panels of known relatedness, consisting mostly of three-generation pedigrees with large sibships, two parents, and grandparents. We applied identity-by-state (IBS) and identity-by-descent (IBD) methods to high-density genotype data from 186 CEPH individuals in 13 families. We identified unexpected relatedness between nominally unrelated grandparents both within and between pedigrees. For one pair, the estimated Cotterman coefficient of relatedness k1 exceeded 0.2, consistent with one-eighth sharing (eg, first-cousins). Unexpectedly, significant IBD2 values were discovered in both second-degree and parent–child relationships. These were accompanied by regions of homozygosity in the offspring, which corresponded to blocks lacking IBS0 in purportedly unrelated parents, consistent with inbreeding. Our findings support and extend a 1999 report, based on the use of short tandem-repeat polymorphisms, that several CEPH families had regions of homozygosity consistent with autozygosity. We benchmarked our IBD approach (called kcoeff) against both RELPAIR and PREST software packages. Our findings may affect the interpretation of previous studies and the design of future studies that rely on the CEPH resource.
Keywords: pedigrees, inbreeding, single-nucleotide polymorphisms, identity-by-descent
Introduction
A set of cell lines developed by the Centre d'Étude du Polymorphisme Humain (CEPH) serves as one of the most widely used resources in cell biology. These lymphoblastoid cell lines were derived from 809 individuals in 62 three-generation pedigrees. In the 1990s, these lines were used extensively for human genome mapping studies1, 2, 3 (reviewed in Prescott et al4). These maps were created at relatively low resolution, with polymorphic markers identified at ∼5 to 15 centimorgan (cM) distances (∼5 to 15 Mb). Subsequently, 180 CEPH family samples were used as part of the International HapMap project, consisting of 60 trios that comprise grandfather/grandmother/parent members of three-generation pedigrees.5, 6 The CEPH collection has been utilized for a broad range of other applications such as assessing genetic variation underlying gene expression, studying allelic variation,7 and identifying cis or trans expression quantitative trait loci (eg Morley et al8 and Monks et al9).
Consanguinity is known to occur with varying frequencies among populations due to geographical and cultural factors.10 The offspring of consanguineous parents may have regions of homozygosity due to autozygosity. An individual is autozygous at a chromosomal locus if he or she inherits two copies of a single ancestral allele from consanguineous parents. The risk of recessive disorders is higher within inbred families because of the increased probability for two deleterious alleles that are identical-by-descent (IBD). At the population level, the consequences of consanguinity include changes in allele frequencies and the appearance of regions of homozygosity, with the potential to impact measurements of genetic variation in population data. One fundamental assumption of the CEPH project has been that pedigree structures are correct as annotated. Recently, evidence for inbreeding was established within a subset of the HGDP-CEPH panel.11
A variety of methods are available to determine relatedness between individuals based on genotype data. We adopted three complementary approaches. First, we used identity-by-state (IBS) methods in which SNP genotypes are compared between a pair of individuals at each chromosomal position, typically involving ∼900 000 comparisons per pair. We plotted IBS sharing and generated characteristic profiles for a variety of relationship types.12, 13 Furthermore, pairwise relationships between all members of a study population can be plotted to distinguish relatedness according to the autosome-wide amount of IBS2* (defined as AB/AB sharing) divided by the sum of IBS2* and IBS0. Such a ratio (IBS2*_ratio), suggested by Lee14 and related to an approach by Rosenberg,15 reduces to a value of 1 for parent–child and identical samples (for whom there are essentially no IBS0 calls), and to a value of 2/3 for unrelated individuals. We previously validated Lee's approach, showing that IBS2*_ratio values of 2/3 corresponded to related individuals, as expected.16 We note that other factors such as heterozygosity rates can impact these measurements, with some pairs of individuals having IBS2*_ratios of 2/3 being related.
A second approach for determining relatedness involves IBD estimation. IBD0, IBD1, and IBD2 states can be inferred from a subset of their corresponding IBS states. There has been a large amount of work in relationship estimation using IBD. Recent algorithms include FastIBD,17 a revised maximum likelihood estimator that includes Fst as a parameter,18 and ERSA.19 We define K0, K1 and K2 as estimates of Cotterman coefficients of relatedness (k0, k1, k2), which allow the inference of the degree of relatedness between individuals.20, 21 We recently validated our IBD approach (kcoeff)16 against the IBD estimates given by PLINK.22 In this study, we compare kcoeff's IBD estimates to those provided by PREST23 and annotations of relationships given by the likelihood-based methods of RELPAIR.24
A third approach involves determining regions of homozygosity based on the absence of AB genotype calls in contiguous stretches. Our study extends work by Broman and Weber25 who measured short tandem-repeat polymorphisms in 134 individuals from eight CEPH families. They identified long stretches of homozygous markers, particularly in CEPH/Venezuelan pedigree 102 and CEPH/Amish pedigree 884. They interpreted these as due to the mating of closely related individuals (autozygosity) rather than linkage disequilibrium in the population. The present study includes analyses of three of the same extended families. Broman and Weber25 propose that long regions of homozygosity due to autozygosity are common in human genomes. Our combined analyses indicate the presence of relatedness both within and between pedigrees and shows that the majority of individuals with homozygosity are from inbred populations. This suggests that there are relatively few protracted regions of homozygosity in outbred populations.
Materials and methods
CEPH genotype data
Genomic DNA from 186 individuals in CEPH pedigrees (families 35, 66, 102, 104, 884, 1331, 1356, 1400, 1416, 1424, 1427, 1477, 1582) was previously obtained by the Coriell Institute for Medical Research (CIMR) and used to generate data on the Affymetrix 6.0 genotyping platform (n=934 940 SNPs). We obtained these data and filtered them to include only autosomal SNPs (n=871 166). SNP data for 181 samples were deposited by CIMR in the NIH Database of Genotypes and Phenotypes under study accession phs000268.v1.p1. Nine of the samples (NA07340, NA12248, NA12249, NA10835, NA10845, NA11930, NA11931, NA11932, NA11933) overlap with HapMap III.26
IBS and IBD analyses
We analyzed IBS with SNPduo (a web-based program that generates plots and tables of IBS sharing across chromosomes12), SNPduo++ (a command-line program used to analyze all 17 205 pairwise comparisons between the 186 samples12, 14), and Partek Genomics Suite (version 6.5; St Louis, MO, USA) to obtain IBS2*_ratio values.14, 16
We analyzed IBD with kcoeff software to estimate Cotterman coefficients of relatedness metrics K0, K1, and K2.16 The algorithm, kcoeff, uses an IBS0_ratio (IBS0/(IBS0+IBS2*)), which is related to the IBS2*_ratio, when calculating K0, K1, and K2. Concordant homozygous SNPs were removed for each pairwise comparison resulting in an average of 419 297 informative SNPs.
Homozygosity analyses
We developed an algorithm (called hetSNP) in Perl that employed an SNP by SNP sliding window approach to identify regions of homozygosity for every individual in a population. For each window, the percentages of homozygous, heterozygous, and No Call (NC) alleles were calculated. Minimal homozygous regions were defined as windows of 200 SNPs containing ≤1% heterozygous alleles and ≤5% NCs. Overlapping homozygous regions were combined into a single region. Homozygous regions ≥3 Mb and ≥800 SNPs were reported. This region size was selected to define informative regions, facilitating SNPduo analysis.
Homozygosity and distant IBD
Our IBD method is robust for inferring relationships with an estimated K1≥0.03. Pairwise comparisons below this K1 estimate may correspond to true distantly related individuals or truly unrelated individuals. To support potential relatedness we used SNPduo to identify chromosomal regions (blocks) lacking IBS0 (implying IBD1 sharing). Truly unrelated individuals are expected to have K1 estimates of zero that may not always be exactly zero due to a window approach in which some regions of little variability have fewer IBS0 calls. To determine whether relationships were present that involved stretches of homozygosity, we applied the following criteria, of which the first four were necessary:
A region lacking heterozygous (AB) calls in a child across a segment ≥3 Mb and ≥800 SNPs.
A corresponding parental region lacking IBS0 calls, likely representing relatedness between the parents. This region must be equal to or larger than the segment lacking heterozygous calls in the child.
IBD2 sharing between a parent and a child supporting abnormal relatedness between the parents.
SNP intensity data indicating a euploid copy number.
For large sibships, such as those in CEPH families, multiple siblings (on average one quarter) are expected to have a lack of AB calls in the regions of inbreeding.
For individuals who are candidates for inbreeding, the occurrence of autozygosity on multiple chromosomal loci provides additional support.
RELPAIR and PREST analyses
We analyzed relatedness using RELPAIR as described.24 We excluded chromosomes X, Y and M (mitochondrial SNPs) before implementing the PLINK ‘-thin' command (n=25 times) to randomly select SNPs. The final output consisted of 1412 (out of 17 205) comparisons that were assigned at least one of the following relationships in 1 out of 25 runs: monozygotic twins, parent–offspring, full-siblings (FS), avuncular (AV), grandparent–grandchild (GG), half-siblings, or cousins (CO). Relationships not specified as described above were assigned unrelated (UN) status.
We also analyzed relatedness with PREST using the ‘--aped' and ‘--wped' options.23 The following quality control measures were employed using PLINK:22 (1) individuals with ≤98% genotype call rate were removed; (2) SNPs with ≤90% genotype call rate were removed; (3) SNPs failing Hardy–Weinberg equilibrium (HWE) with a P≤0.0001 were removed; (4) SNPs with a minor-allele frequency ≤0.01 were removed. Zero individuals were removed, whereas 3130 SNPs with low call rate, 114 850 SNPs with low minor-allele frequency , and 820 SNPs that failed HWE were removed. A total of 753 418 remaining SNPs were pruned within PLINK using the ‘—thin' command, providing 45 451 SNPs for the PREST analysis. We note that as some samples were duplicated, it was impossible to accurately specify annotated relationships for PREST input files.
Results
Unexpected sharing in CEPH pedigrees
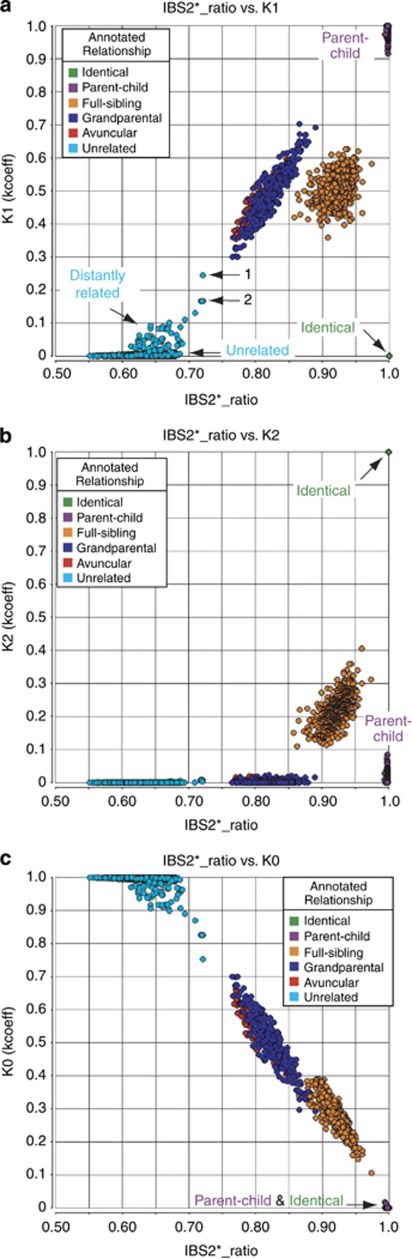
We obtained high-density SNP genotype data from a set of 186 CEPH individuals comprising 13 separate families. To determine the genetic relatedness of these individuals, we measured both IBS and IBD for every pairwise comparison (n=17 205 pairs) using autosomal data (see Materials and Methods). Each data point of an IBS2*_ratio plot consisted of a single pair of individuals (Figure 1a). The x-axis (IBS2*_ratio) included values of (IBS2*/(IBS0 +IBS2*)) where IBS2* denotes AB/AB genotypes. The y-axis included measurements of K1 (Figure 1a), K2 (Figure 1b), and K0 (Figure 1c) using the kcoeff method16 that estimates Cotterman coefficients of relatedness.
Figure 1.
Relationships among CEPH three-generation pedigree members based on IBS and IBD measurements. (a) IBS2*_ratio plot annotated by relationships. Each data point corresponds to a comparison of two individuals based on genotype data. The IBS2*_ratio consisted of autosome-wide (IBS2*/(IBS0+IBS2*)) on the x-axis measured against kcoeff's K1 (level of genome shared IBD1) on the y-axis. Clusters were expected (based on prior sample annotation) of identical, parent–child, full-siblings, 1/4 sharing (ie, AV and GG), and unrelated individuals. We also observed pairs of samples having x-axis values consistent with distant relatedness (eg, arrows 1 and 2). (b) IBS2*_ratio (x-axis) versus kcoeff's K2 (level of genome-shared IBD2; y-axis), annotated by relationships. (c) IBS2*_ratio (x-axis) versus kcoeff's K0 (level of genome shared IBD0; y-axis).
On the basis of available pedigree information, we expected to observe three identical sample pairs, 317 parent–child, 522 full-sibling, 506 one-quarter sharing divided into 386 grandparent–grandchild and 120 AV pairs (includes all AV and materternal relationships; inferred by placement of known identical samples within their respective pedigree), and unrelated individuals. Identical and parent–child relationships had expected IBS2*_ratio values of 1.0 owing to few IBS0 calls (Figures 1a–c), but were separated along the y-axis because identical samples are solely IBD2 (Figure 1b; see arrow). Unexpectedly, we observed a fourth pairwise comparison that segregated to a position indicating identical samples (Figure 1a and b; note identical samples overlap). Three pairs had been previously annotated as part of CEPH/Venezuelan pedigree 102 and 104, whereas the fourth pair was annotated as a grandmother–granddaughter relationship (NA12863 and NA12859 in CEPH/Utah pedigree 1400). We concluded that both of these CEPH/Utah samples were derived from the granddaughter (based on sibling relatedness on the IBS2* plot and K1 and K2 estimates; see below). It was subsequently confirmed that the granddaughter's DNA sample had been genotyped twice (Dr Norman Gerry, Coriell Cell Repositories, personal communication).
We confirmed 317 parent–child relationships based on IBS and IBD estimates. As expected for relationships with no IBS0, IBS2*_ratio values were near 1.0 (Figure 1a). Notably, some parent–child relationships also had appreciable levels of IBD2 (K2, Figure 1b) and IBD0 (K0, Figure 1c) that will be discussed in detail below.
Siblings who have theoretical Cotterman coefficients of 1/4 IBD0, 1/2 IBD1, and 1/4 IBD2, had IBS2*_ratio values near 0.90 and were distinct from other relationships (Figures 1a and b) because of the presence of IBD2 sharing. A total of 522 full-sibling pairs were confirmed based on IBS estimates as well as IBD1 and IBD2 estimates.
Pairwise relationships involving one-quarter sharing included AV and GG comparisons based on pedigree annotations. These pairs had IBS2*_ratio values that ranged from 0.77 to 0.86 (Figure 1a). Furthermore, IBD analysis for these 506 pairs matched expected Cotterman coefficient values with estimates centered on 1/2 IBD0 and 1/2 IBD1 (K1, Figure 1a; K0, Figure 1c). In addition to the unexpected IBD2 sharing estimated in parent–child relationships, some GG and AV relationships were also inferred to have IBD2 sharing (Figure 1b). These will be explained alongside parent–child relationships with IBD2 in detail below. We generated a complete list of IBD estimates for annotated pairwise comparisons (Supplementary Table 1).
Pairs of individuals who were annotated as unrelated are expected to have IBS2*_ratio values of 2/3.14, 16 Values >2/3 can be attributed to genetic relatedness or to elevated heterozygosity in one (or both) individuals.16 We therefore rely on the kcoeff method to identify distantly related individuals. Unexpectedly, we observed a cluster of pairwise comparisons with K1 values ≥0.03 indicating distant relatedness (Figure 1a; see arrow). In some instances K1 values ranged from 0.166 to 0.244, consistent with theoretical Cotterman coefficients of 0.25 (eg, first-cousins) or 0.125 (eg, first-cousin once removed). These included grandfather/grandmother couples (NA12977 and NA12978) from pedigree 1427 (Figure 1a, arrow 1; K1 value of 0.244) and (NA13180 (duplicate sample NA13055) and NA13181 (duplicate sample NA13057)) from pedigree 102/104 (arrow 2; four comparisons; K1 value of 0.166). Another notable pair, paternal grandmother NA11931 and maternal grandmother NA11933, from pedigree 1424 had a K1 of 0.13. It is important to note that this pair is present in HapMap 3 and represents an unannotated related pair. Each pair had regions that lacked IBS0 based on SNPduo analyses, supporting the finding of distant genetic relationships (data not shown). A complete list of individuals inferred to be related is presented in Supplementary Table 2.
Increasing relatedness between pairs of individuals was associated with decreasing K0 estimates (Figure 1c). Given that the IBS2*_ratio includes IBS0 information in the denominator, it is expected that a decrease in IBS0 results in a higher IBS2*_ratio. Furthermore, the estimated level of K0 should also decrease as the level of IBS0 is reduced. Note that some parent–child relationships have estimated IBD0 values that will be discussed below.
IBS confirmation of IBD relatedness findings
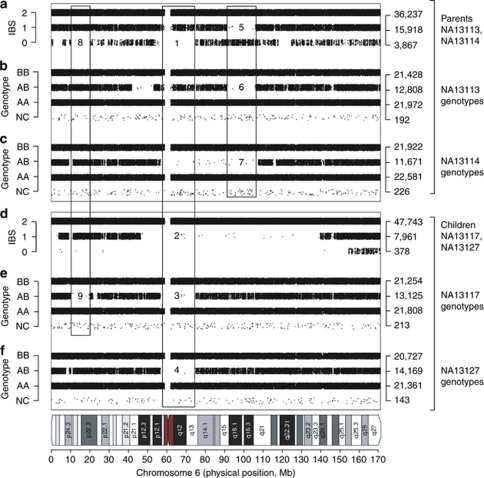
To confirm IBD1 (displayed in Figure 1a) or IBD2 sharing (Figure 1b) based on the genotype data, we analyzed relationships on a chromosome-by-chromosome basis in SNPduo to determine and visualize the extent of IBS sharing. We applied this to Amish individuals NA13113 and NA13114 from pedigree 884, in which unexpected sharing was detected (K1=0.092; Figure 2a). The IBS sharing between these two parents included many extended regions with a lack of IBS0 calls (eg, regions 1 and 8). Furthermore, each of the four grandparents (ie, the parents of NA13113/NA13114) in the pedigree shared K1 values ranging from 0.051 to 0.092 with respect to the other three (NA13111, NA13112, NA13115, and NA13116). As a consequence, the genomes of NA13113 and NA13114 had extensive tracts of homozygosity (Figures 2b and c, regions 6 and 7) as previously reported.25 These regions of homozygosity (due to autozygosity) were not shared, thus resulting in observed states of either IBS0 or IBS2 (Figure 2a, region 5).
Figure 2.
Inbreeding in a CEPH/Amish pedigree. For all panels, data for chromosome 6 are shown based on SNPduo analyses.12 Upper three panels: (a) pairwise IBS patterns are presented for parents NA13113 and NA13114. Note regions 1 and 8 in which an absence of IBS0 is shown. Region 5 corresponds to regions 6 and 7 (panels b and c) and represents two individuals with different stretches of homozygosity (lack of AB calls) compared with each other; whereas both were homozygous, the differences were evident from the occurrence of IBS0 in the region 5. (b) Genotypes of NA13113. Note homozygosity in region 6. (c) Genotypes of NA13114. Note homozygosity in region 7. (d) Pairwise IBS patterns for NA13117 and NA13127 (children of NA13113 and NA13114). Note that region 2 indicates IBD2 between the siblings NA13117 and NA13127 and overlaps a region of inferred IBD1 between the parents (region 1). (e) Genotypes for NA13117. Note that regions 3 and 9 are homozygous segments that correspond to a lack of IBS0 in regions 1 and 8 from panel A; this suggests consanguinity. (f) Genotypes of NA13127. Region 4 is a homozygous segment that corresponds to region 1 and is identical to region 3. This supports consanguinity due to lack of IBS0 between the parents (a).
Given the relatedness between the parents, we expected to observe homozygous segments in the offspring in regions where the parents were related. We plotted the results of SNPduo analysis for a representative pair of full-siblings (NA13117 and NA13127) for chromosome 6 (Figure 2d) in which there was IBD2 sharing (absence of IBS0 and IBS1, region 2) in a region corresponding to relatedness between the parents (region 1). We observed homozygosity in this same region in the children (Figures 2e and f, regions 3 and 4). We highlight a second example of homozygosity in child NA13117 (Figure 2e, region 9) caused by relatedness between the parents (Figure 1a, region 8).
Identification of distantly related individuals based on homozygosity in pedigrees
In addition to using IBS and IBD to define distantly related individuals, we further identified individuals who were inbred using analyses of homozygosity in the context of pedigrees. We applied our analysis to individuals for whom parental genotype data were available and also applied other criteria (see Materials and Methods). A notable discovery involved pedigree 1582. A region lacking IBS0 between the paternal grandfather (NA12921) and maternal grandmother (NA12924) was observed that overlaid a region of homozygosity in the grandchildren (NA12915, NA12917, NA12918, and NA12919). This occurred on chromosome 1 and spanned 5.26 Mb and involved 1600 SNPs. We summarize our homozygosity findings for all pedigrees in Figure 3 and our IBS/IBD estimates for related founders in Table 1 as well as a complete list of the 86 individuals inferred to be related (Supplementary Table 2). We confirmed a copy number state of 2 in the regions of homozygosity (data not shown).
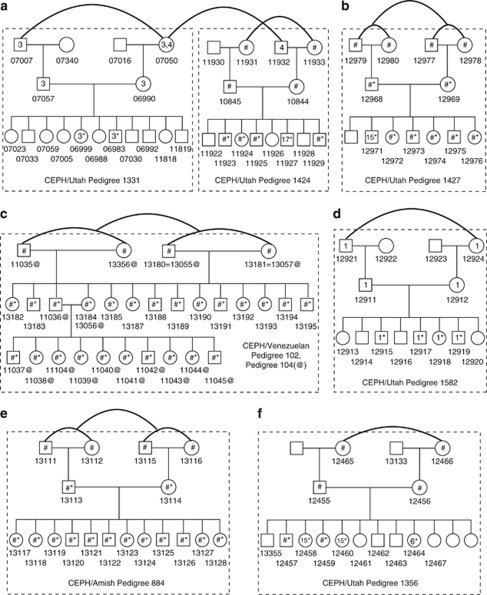
Figure 3.
Revised CEPH pedigrees. Curved lines denote significant relatedness between pairs of individuals with estimated autosome-wide K1 and pIBD1 values indicated in Table 2. Dashed rectangles correspond to pedigrees as given on the Coriell Cell Repositories website,34 except that CEPH/Venezuelan pedigrees 102 and 104 had not previously been explicitly presented as a single pedigree visually. Numbers given on the pedigrees for individuals correspond to standard Coriell designations (eg, 11930 corresponds to cell line GM11930 or DNA sample NA11930). (a) CEPH/Utah pedigrees 1331 and 1424 were interrelated based on K1 sharing between grandmother NA07050 and grandfather NA11932 on chromosome 4. Additionally, a significant K1 value was estimated between paternal grandmother NA11931 and maternal grandmother NA11933 in CEPH/Utah pedigree 1424. (b) CEPH/Utah pedigree 1427 had significantly elevated K1 between NA12977 and NA12978 and marginal K1 between all four grandparents supported by homozygosity and SNPduo analysis (see Materials and Methods). (c) CEPH/Venezuelan families 102 and 104 include numerous AV relationships. All four grandparents displayed elevated K1 levels. (d) CEPH/Utah pedigree 1582 had minimal K1 levels associated with homozygosity between NA12921 and NA12924 (supported by SNPduo analysis). (e) CEPH/Amish pedigree 884 was characterized by four grandparents with elevated K1 levels. (f) CEPH/Utah pedigree 1356 had minimal K1 levels also associated with homozygosity and supported by SNPduo analysis. *Indicates the presence of homozygosity due to autozygosity; #indicates the presence of multiple regions; any number within a square or circle indicates a unique chromosome in which relatedness or homozygosity was present; @indicates the members of CEPH/Venezuelan pedigree 104 within the combined CEPH/Venezuelan pedigrees 102 and 104.
Table 1. k1 estimates for relationships within and between families reported in Figure 3.
| Sample 1 | Sample 2 | IBS2*_ratio | K0 | K1 | K2 | CO | pIBD0 | pIBD1 | pIBD2 |
|---|---|---|---|---|---|---|---|---|---|
| NA12977 | NA12978 | 0.721 | 0.753 | 0.244 | 0.003 | 25 | 0.716 | 0.280 | 0.005 |
| NA13055 | NA13181 | 0.721 | 0.826 | 0.167 | 0.008 | 25 | 0.888 | 0.112 | 0 |
| NA13055 | NA13057 | 0.719 | 0.826 | 0.167 | 0.008 | 25 | 0.887 | 0.113 | 0 |
| NA13057 | NA13180 | 0.718 | 0.826 | 0.166 | 0.008 | 25 | 0.887 | 0.113 | 0 |
| NA13180 | NA13181 | 0.719 | 0.826 | 0.166 | 0.008 | 25 | 0.888 | 0.112 | 0 |
| NA11931 | NA11933 | 0.709 | 0.870 | 0.130 | 0 | 25 | 0.884 | 0.116 | 0 |
| NA11035 | NA13356 | 0.694 | 0.890 | 0.110 | 0 | 25 | 0.885 | 0.114 | 0.002 |
| NA13115 | NA13116 | 0.669 | 0.906 | 0.092 | 0.002 | 25 | 0.931 | 0.069 | 0 |
| NA13111 | NA13115 | 0.687 | 0.907 | 0.087 | 0.006 | 25 | 0.897 | 0.100 | 0.003 |
| NA13112 | NA13114 | 0.659 | 0.918 | 0.083 | 0 | 25 | 0.952 | 0.037 | 0.011 |
| NA13111 | NA13114 | 0.654 | 0.917 | 0.082 | 0 | 25 | 0.910 | 0.090 | 0 |
| NA11035 | NA13180 | 0.665 | 0.918 | 0.082 | 0 | 21 | 0.992 | 0.004 | 0.004 |
| NA11035 | NA13055 | 0.666 | 0.918 | 0.082 | 0 | 20 | 0.993 | 0.003 | 0.004 |
| NA13111 | NA13116 | 0.668 | 0.919 | 0.082 | 0 | 25 | 0.926 | 0.068 | 0.007 |
| NA13112 | NA13116 | 0.673 | 0.926 | 0.070 | 0.004 | 25 | 0.947 | 0.050 | 0.003 |
| NA13112 | NA13115 | 0.681 | 0.936 | 0.064 | 0 | 20 | 0.943 | 0.057 | 0 |
| NA11035 | NA13181 | 0.660 | 0.947 | 0.053 | 0 | 1 | 0.994 | 0.007 | 0 |
| NA11035 | NA13057 | 0.659 | 0.947 | 0.053 | 0 | 1 | 0.993 | 0.007 | 0 |
| NA13111 | NA13112 | 0.684 | 0.949 | 0.051 | 0 | 22 | 0.944 | 0.056 | 0 |
| NA13180 | NA13356 | 0.688 | 0.963 | 0.037 | 0 | 11 | 1 | 0 | 0 |
| NA13055 | NA13356 | 0.690 | 0.963 | 0.037 | 0 | 11 | 1 | 0 | 0 |
| NA13181 | NA13356 | 0.683 | 0.969 | 0.031 | 0 | 0 | 1 | 0 | 0 |
| NA13057 | NA13356 | 0.682 | 0.969 | 0.031 | 0 | 0 | 1 | 0 | 0 |
| NA12978 | NA12980 | 0.672 | 0.985 | 0.015 | 0 | 1 | 0.963 | 0.037 | 0 |
| NA07050 | NA11932 | 0.674 | 0.989 | 0.011 | 0 | 0 | 1 | 0 | 0 |
| NA12979 | NA12980 | 0.674 | 0.990 | 0.010 | 0 | 1 | 0.981 | 0.012 | 0.007 |
| NA12977 | NA12979 | 0.632 | 0.991 | 0.009 | 0 | 1 | 0.965 | 0.035 | 0 |
| NA12978 | NA12979 | 0.669 | 0.992 | 0.008 | 0 | 0 | 0.985 | 0.015 | 0 |
| NA12465 | NA12466 | 0.661 | 0.996 | 0.004 | 0 | 0 | 1 | 0 | 0 |
| NA12921 | NA12924 | 0.664 | 0.997 | 0.003 | 0 | 0 | 1 | 0 | 0 |
| NA12977 | NA12980 | 0.628 | 0.997 | 0.003 | 0 | 1 | 0.992 | 0.003 | 0.004 |
Abbreviations: CO, assignment of cousin status by RELPAIR per 25 trials; K0, K1, K2, estimates of k0, k1, k2, respectively; pIBD0, pIBD1, pIBD2, estimates of k0, k1, k2, respectively.
Each pairwise comparison had estimates of Cotterman coefficients given by kcoeff (K0, K1, K2), RELPAIR (CO) and PREST (pIBD0, pIBD1, pIBD2).
We compared the amounts of homozygosity we identified in all individuals to those reported by Broman and Weber25 (Table 2). We also report amounts of homozygosity in individuals not studied by Broman (listed in Table 3). Notable individuals included NA11035 from pedigree 104 who had 86 Mb of homozygous regions and NA12969 (daughter of NA12977/NA12978; Figure 1c, arrow 1) from pedigree 1427 who had 268 Mb total from 16 homozygous regions. A brief comparison of regions inferred to be homozygous by the kcoeff method and from Broman and Weber is presented in Supplementary Figure 1 in which we report comparable results, but better define the boundaries due to a greater number of markers. A complete list of individuals with the chromosome and position of each region is presented in Supplementary Table 3.
Table 2. Comparison of estimates of homozygosity in this study to Broman and Weber25.
| Our analysis | Broman and Weber (1999) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample ID | # | SNPs | Avg SNPs | Length (Mb) | Avg length (Mb) | # | Markers | Length (cM) | Avg length (cM) |
| NA06988 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 24 | 3.3 | 3.3 |
| NA07007 | 1 | 3200 | 3200 | 9.9 | 9.9 | 1 | 28 | 5.8 | 5.8 |
| NA07016 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 9 | 0.0 | 0.0 |
| NA07057 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 10 | 0.0 | 0.0 |
| NA10834 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 10 | 3.2 | 3.2 |
| NA10835 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 15 | 6.9 | 6.9 |
| NA12240 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 16 | 2.6 | 2.6 |
| NA12251 | 5 | 22769 | 4554 | 70.9 | 14.2 | 4 | 8–75 | 75.5 | 18.9 |
| NA13180 | 0 | 0 | 0 | 0.0 | 0.0 | 0 | 0 | 0.0 | 0.0 |
| NA13181 | 0 | 0 | 0 | 0.0 | 0.0 | 0 | 0 | 0.0 | 0.0 |
| NA13111 | 6 | 13507 | 2251 | 47.6 | 7.9 | 4 | 6–45 | 35.5 | 8.9 |
| NA13112 | 0 | 0 | 0 | 0.0 | 0.0 | 1 | 11 | 6.5 | 6.5 |
| NA13113 | 9 | 26832 | 2981 | 85.5 | 9.5 | 8 | 11–50 | 105.1 | 13.1 |
| NA13114 | 13 | 51291 | 3945 | 165.1 | 12.7 | 10 | 9–113 | 159.9 | 16.0 |
| NA13115 | 7 | 19537 | 2791 | 59.6 | 8.5 | 7 | 8–45 | 57.4 | 8.2 |
| NA13116 | 9 | 34527 | 3836 | 86.9 | 9.7 | 9 | 10–51 | 116.2 | 12.9 |
| NA13117 | 10 | 35841 | 3584 | 119.0 | 11.9 | 9 | 14–93 | 111.5 | 12.4 |
| NA13118 | 9 | 25224 | 2803 | 84.2 | 9.4 | 5 | 15–77 | 61.7 | 12.3 |
| NA13119 | 5 | 15700 | 3140 | 54.9 | 11.0 | 5 | 14–58 | 65.7 | 13.1 |
| NA13120 | 10 | 24824 | 2482 | 75.8 | 7.6 | 9 | 9–64 | 93.1 | 10.3 |
| NA13121 | 12 | 25879 | 2157 | 77.4 | 6.5 | 9 | 13–39 | 84.0 | 9.3 |
| NA13122 | 17 | 60736 | 3573 | 198.4 | 11.7 | 16 | 9–87 | 195.8 | 12.2 |
| NA13123 | 11 | 28567 | 2597 | 99.4 | 9.0 | 8 | 9–71 | 77.6 | 9.7 |
| NA13124 | 7 | 24387 | 3484 | 74.5 | 10.7 | 6 | 13–35 | 57.0 | 9.5 |
| NA13125 | 12 | 42154 | 3513 | 132.7 | 11.1 | 10 | 15–90 | 124.8 | 12.5 |
| NA13126 | 10 | 23843 | 2384 | 78.0 | 7.8 | 7 | 7–32 | 66.4 | 9.5 |
| NA13127 | 8 | 29861 | 3733 | 96.8 | 12.1 | 9 | 9–64 | 118.0 | 13.1 |
| NA13128 | 14 | 43883 | 3135 | 142.8 | 10.2 | 13 | 14–45 | 109.7 | 8.4 |
| NA13182 | 14 | 53545 | 3825 | 164.8 | 11.8 | 10 | 11–69 | 157.9 | 15.8 |
| NA13183 | 7 | 27214 | 3888 | 83.8 | 12.0 | 6 | 9–70 | 105.9 | 17.7 |
| NA13184 | 11 | 51703 | 4700 | 158.6 | 14.4 | 10 | 9–61 | 198.4 | 19.8 |
| NA13185 | 8 | 33980 | 4248 | 110.8 | 13.9 | 6 | 9–55 | 98.6 | 16.4 |
| NA13187 | 16 | 66355 | 4147 | 204.1 | 12.8 | 12 | 8–68 | 200.7 | 16.7 |
| NA13188 | 17 | 75916 | 4466 | 243.9 | 14.4 | 12 | 16–95 | 253.1 | 21.0 |
| NA13189 | 13 | 66482 | 5114 | 218.1 | 16.8 | 9 | 10–87 | 200.7 | 22.3 |
| NA13190 | 12 | 42965 | 3580 | 129.1 | 10.8 | 9 | 7–60 | 158.3 | 17.6 |
| NA13191 | 10 | 38273 | 3827 | 125.8 | 12.6 | 5 | 19–76 | 106.9 | 21.4 |
| NA13192 | 13 | 49941 | 3842 | 157.5 | 12.1 | 11 | 7–77 | 175.1 | 15.9 |
| NA13193 | 10 | 36398 | 3640 | 120.0 | 12.0 | 7 | 10–71 | 128.7 | 18.4 |
| NA13194 | 12 | 55427 | 4619 | 185.6 | 15.5 | 12 | 8–69 | 183.5 | 15.3 |
| NA13195 | 7 | 26082 | 3726 | 76.1 | 10.9 | 4 | 17–69 | 77.7 | 19.4 |
Abbreviations: Avg Length (cM), average length of the regions in centimorgans by Broman and Weber; Avg length (Mb), average length of the regions in megabases; Avg SNPs, average number of SNPs per region; #, number of regions reported; Length (cM), total length of all regions in centimorgans by Broman and Weber; Length (Mb), total length of all regions in megabases; Markers, range of markers reported per segment by Broman and Weber; SNPs, total number of SNPs within all regions.
We report regions of homozygosity (see Materials and Methods) that were ≥3.00 Mb and ≥800 SNPs, whereas Broman reported regions that were based on LOD scores and could be as small as 0.0 cM with only nine markers.25
Table 3. Individuals with new estimates of homozygosity.
| Sample ID | # | SNPs | Avg SNPs | Length (Mb) | Avg length (Mb) |
|---|---|---|---|---|---|
| NA12969 | 16 | 80350 | 5022 | 268.4 | 16.8 |
| NA12977 | 12 | 59460 | 4955 | 200.7 | 16.7 |
| NA13056 | 12 | 4318 | 51821 | 158.6 | 13.2 |
| NA11036 | 11 | 3462 | 38078 | 121.5 | 11.0 |
| NA11035 | 10 | 3191 | 31906 | 86.1 | 8.6 |
| NA11104 | 6 | 3773 | 22637 | 60.4 | 10.1 |
| NA11038 | 5 | 3946 | 19730 | 48.7 | 9.7 |
| NA11045 | 4 | 4415 | 17661 | 44.0 | 11.0 |
| NA11039 | 4 | 3469 | 13875 | 42.9 | 10.7 |
| NA11037 | 4 | 3006 | 12024 | 38.8 | 9.7 |
| NA11043 | 5 | 3107 | 15536 | 37.6 | 7.5 |
| NA11040 | 3 | 3023 | 9069 | 31.4 | 10.5 |
| NA11929 | 3 | 8920 | 2973 | 27.6 | 9.2 |
| NA11041 | 4 | 2130 | 8521 | 27.0 | 6.7 |
| NA11044 | 3 | 3378 | 10133 | 25.9 | 8.6 |
| NA12978 | 2 | 5836 | 2918 | 21.9 | 11.0 |
| NA11924 | 4 | 7234 | 1809 | 21.5 | 5.4 |
| NA12973 | 5 | 5884 | 1177 | 19.7 | 4.0 |
| NA11925 | 2 | 4509 | 2255 | 19.4 | 9.7 |
| NA12974 | 4 | 5461 | 1365 | 18.9 | 4.7 |
| NA11923 | 2 | 4484 | 2242 | 18.8 | 9.4 |
| NA12980 | 3 | 3650 | 1217 | 12.6 | 4.2 |
| NA11927 | 1 | 2723 | 2723 | 11.8 | 11.8 |
| NA12459 | 2 | 2678 | 1339 | 8.5 | 4.3 |
| NA12972 | 2 | 2845 | 1423 | 8.0 | 4.0 |
| NA12968 | 2 | 2183 | 1092 | 7.2 | 3.6 |
| NA12976 | 2 | 1886 | 943 | 7.0 | 3.5 |
| NA12975 | 2 | 1849 | 925 | 6.9 | 3.5 |
| NA11042 | 1 | 2914 | 2914 | 6.5 | 6.5 |
| NA12915 | 1 | 1600 | 1600 | 5.3 | 5.3 |
| NA12917 | 1 | 1600 | 1600 | 5.3 | 5.3 |
| NA12918 | 1 | 1600 | 1600 | 5.3 | 5.3 |
| NA12919 | 1 | 1600 | 1600 | 5.3 | 5.3 |
| NA12909 | 1 | 793 | 793 | 4.6 | 4.6 |
| NA12458 | 1 | 962 | 962 | 4.4 | 4.4 |
| NA12460 | 1 | 962 | 962 | 4.4 | 4.4 |
| NA13355 | 1 | 962 | 962 | 4.4 | 4.4 |
| NA12862 | 1 | 860 | 860 | 4.2 | 4.2 |
| NA12457 | 1 | 1715 | 1715 | 4.1 | 4.1 |
| NA12464 | 1 | 1716 | 1716 | 4.1 | 4.1 |
| NA06999 | 1 | 1146 | 1146 | 3.7 | 3.7 |
| NA06983 | 1 | 1141 | 1141 | 3.6 | 3.6 |
| NA12971 | 1 | 1436 | 1436 | 3.2 | 3.2 |
| NA12900 | 1 | 884 | 884 | 3.1 | 3.1 |
| NA13133 | 1 | 761 | 761 | 3.0 | 3.0 |
Abbreviations: Avg length (Mb), average length of the regions in megabases; Avg SNPs, average number of SNPs per region; #, number of regions reported; Length (Mb), total length of all regions in megabases; SNPs, total number of SNPs within all regions.
We report regions of homozygosity (see Materials and Methods) that were ≥3 Mb and ≥800 SNPs in individuals that were not studied in Broman and Weber's report25 and sorted by the total length of homozygosity.
Comparisons to RELPAIR and PREST
We compared our analysis method to that of RELPAIR,24 a leading software package that has recently been used to annotate relationships in HapMap Phase III.27 We used RELPAIR to analyze all pairwise relationships using 25 independent runs for each comparison (see Materials and Methods). We note that although RELPAIR identified all identical and full-sibling relationships, it also called several annotated parent–child and second-degree relationships as full-siblings (in particular, those that had unexpected IBD2 estimates). These apparently misclassified individuals were within those pedigrees in which inbreeding has already been shown (see above). In addition, some second-degree relationships (eg, AV) were miscalled as being a different second-degree relationship (eg, half-sibling). We summarized the relationships as annotated by RELPAIR and based on prior annotation in a confusion matrix (Table 4), and we listed the RELPAIR annotation for each pairwise comparison in Supplementary Table 1.
Table 4. Confusion matrix for relationships inferred by RELPAIR compared with authentic relationships based on annotated CEPH three-generation pedigrees.
| Relationship inferred by RELPAIR | ||||||||
|---|---|---|---|---|---|---|---|---|
| Annotated relationship | MZ | PO | FS | HS | AV | GG | CO | UN |
| Identical/MZ | 75 | NC | NC | NC | NC | NC | NC | NC |
| Parent–offspring | NC | 6697 | 1228 | NC | NC | NC | NC | NC |
| Full-sibling | NC | NC | 13050 | NC | NC | NC | NC | NC |
| Half-sibling | NC | NC | NC | NC | NC | NC | NC | NC |
| Avuncular | NC | NC | 346 | 47 | 2599 | NC | NC | NC |
| Grandparent–grandchild | 25 | NC | 603 | 3196 | 1799 | 4039 | 13 | NC |
| Cousin | NC | NC | NC | NC | NC | NC | NC | NC |
| Unrelated* | NC | NC | NC | NC | NC | NC | 808 | 395600 |
Abbreviations: AV, avuncular or materternal; CO, cousin; FS, full-sibling; GG, grandparent–grandchild; HS, half-sibling; MZ, monozygotic (ie, identical samples); NC, not called; PO, parent–offspring; UN, inferred to be unrelated based on no familial assignment from RELPAIR.
For each pairwise comparison RELPAIR generated 25 inferred relationships.*Annotated as unrelated. Samples NA12859/NA12863 (previously annotated as granddaughter/grandmother but identified as replicate samples in this study based on IBS/IBD and RELPAIR analyses) are noted in the confusion matrix analysis.
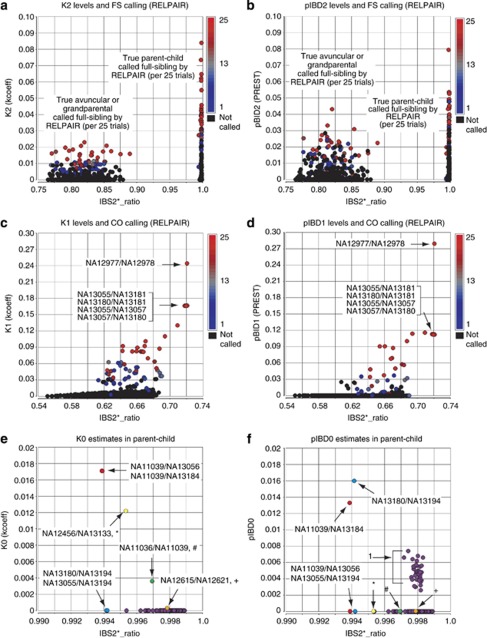
To determine whether the amount of IBD2 estimated in these relationships impacted RELPAIR's ability to differentiate full-sibling from parent–child and second-degree relationships, we plotted IBS2*_ratio values versus K2 (Figure 4a) and the corresponding IBD2 estimate of PREST, pIBD2 (Figure 4b). We annotated these plots by the number of times RELPAIR called full-sibling per 25 trials. RELPAIR called 50 (out 317 parent–child relationships) of these pairwise comparisons a full-sibling relationship in ≥13 out of 25 instances with an average K2 of 0.043 ± 0.014 and a pIBD2 of 0.027 ± 0.017. Overall, 18 parent–child comparisons were called full-sibling less than half of the time with average values 0.021±0.006 for K2 and 0.013±0.015 for pIBD2. The increase in estimated IBD2 was correlated with the increase in RELPAIR assigning full-sibling status to annotated parent–child relationships with a linear Pearson correlation of r=0.892 for K2 and r=0.719 for pIBD2. Similar results were observed for elevated IBD2 in AV or grandparental relationships that were incorrectly assigned full-sibling status by RELPAIR. A higher correlation of r=0.839 was associated with full-sibling designation and K2 as opposed to r=0.632 for pIBD2. As IBD2 estimates increased, RELPAIR had a higher likelihood of misclassifying relationships as full-sibling. Furthermore, estimates for level of IBD2 using the kcoeff method were more consistent than those of PREST.
Figure 4.
Comparison of kcoeff to PREST and RELPAIR for k1 and k2, and comparison of kcoeff and PREST for k0. An IBS2*_ratio is plotted as a function of varying IBD estimates annotated by RELPAIR for k1 and k2 estimates with a comparison of k0 between kcoeff and PREST in parent–child relationships. (a) K2 (kcoeff) estimates of true AV, GG, and parent–child relationships incorrectly assigned a FS annotation from RELPAIR. Note the positive relationship between level of K2 and number per 25 trials of RELPAIR FS annotation. The color scale for panels a and b includes black (no RELPAIR FS calls) and ranges from blue (1 FS call per 25 trials) to red (25/25 FS calls). (b) pIBD2 (PREST) estimates of relationships incorrectly assigned a FS annotation from RELPAIR. (c) K1 estimates of unrelated and distantly related individuals assigned a CO annotation from RELPAIR. Note the positive relationship between level of K1 and number per 25 trials of CO annotation. (d) pIBD1 estimates of unrelated and distantly related individuals assigned a CO annotation from RELPAIR. (e) K0 estimates in parent–child relationships with selected pairs being identified. (f) pIBD0 estimates in parent–child relationships with selected pairs highlighted. Note the discrepancies between panels e and f. Also note that as there were duplicate samples present, it was not possible to fully annotate all relationships (see arrow 1 in panel f). The x and y-axis scales are the same for panels a/b, c/d, and e/f. Arrows indicate specific pairwise comparisons (see text for details).
RELPAIR also annotates first-cousin relationships.24 We analyzed all pairwise relationships that were annotated as unrelated, plotted IBS2*_ratio values versus estimates of K1 and pIBD1, and annotated by assignment of first cousins by RELPAIR (Figures 4c and d) per 25 trials. Note than some 1/4th sharing relationships with the lowest K1 and pIBD1 estimates were occasionally called first cousins and comprised a small minority of relationships designated as first cousins (data not shown). A total of 29 comparisons were designated as first cousins in a majority of RELPAIR runs (≥13/25) with an average K1 of 0.092±0.004 (Figure 4c) and pIBD1 of 0.063±0.060 (Figure 4d). Additionally, 34 comparisons were called first cousins at least once for values of 0.034±0.016 for K1 and 0.007±0.011 for pIBD1. The level of K1 in individuals annotated as unrelated had a correlation value of r=0.853, whereas pIBD1 estimates were r=0.761. This further supports the kcoeff estimation of IBD level as being more accurate than the maximum likelihood approach of PREST.
We also compared our IBD0 estimates in parent–child relationships to those of PREST. Using the kcoeff method, we detected IBD0 in four parent–child relationships (Figure 4e; NA11039/NA13056 (duplicated 13184), see red circle; NA12456/NA13133, see yellow circle; NA11036/NA11039, see green circle; and NA12615/NA12621, see orange circle). NA11039/NA13056 (duplicated 13184) and NA11036/NA11039 represent a trio, and we analyzed them using SNPduo to visualize the IBS0 (Supplementary Figures 2a and b). We observed extensive IBS0 in two regions between mother/daughter NA13056/NA11039 (Supplementary Figure 2a, regions 1 and 2 spanning ∼48 Mb on chromosome 14) and in one region between father/daughter pair NA11036/NA11039 (Supplementary Figure 2b, region 3 spanning ∼3 Mb on chromosome 14). Consistent with the K0 estimates, PREST's pIBD0 values indicated IBD0 in mother/daughter NA13184/NA11039. Surprisingly, pIBD0 was not detected in mother/daughter pair NA13056/NA11039 involving an identical comparison (Figure 4f; see arrows). A similar splitting of identical comparisons was found between father/son NA13180/NA13194 and father/son NA13055/NA13194 (Figure 4f) for whom we estimated similar, elevated K0 values for both comparisons. pIBD0 estimates by PREST were elevated for a group of pairwise comparisons (Figure 4f; arrow 1), which could not be fully annotated because some of the parental samples were duplicated. When these samples were manually annotated as parent–child, the pIBD0 estimates reverted to zero (data not shown) suggesting they were false positive results.
Discussion
The main finding of the present study is that CEPH pedigrees include previously unreported relationships both within and between pedigrees. As expected, we uncovered many regions of homozygosity due to autozygosity in the offspring of related individuals across a subset of 13 pedigrees that included 186 individuals. A subset of our findings was broadly consistent with a 1999 report by Broman and Weber25 that many CEPH individuals have extended regions of homozygosity. We describe homozygosity due to autozygosity in almost two dozen additional individuals, based on IBD (kcoeff), IBS (SNPduo) and homozygosity analyses. A recent report suggested that 10.4% out of the 6.7 billion people in the world have an inbreeding coefficient greater or equal to second cousins (F ≥0.0156).10 An estimation of inbreeding coefficients in our samples revealed that 21% had an F ≥0.0156. An additional 9% had a coefficient of inbreeding greater than third cousins (F ≥0.0039).
Our analysis of the amount of homozygosity observed in both inbred and outbred pedigrees suggests that very few individuals outside of inbred pedigrees have long homozygous regions. In fact, 111 out of 186 individuals had no regions of homozygosity in segments >3 Mb and having more than 800 SNPs. Many of the ones in which we report segments of homozygosity were present in pedigrees that had inbreeding and support the finding from Broman and Weber.25 This assessment is somewhat contrasted by other studies that suggest that long homozygous segments are common in the human genome.28, 29, 30 However, our results indicate that small homozygous segments around 1 Mb in length are quite common in both inbred and outbred individuals with genome-wide totals in excess of 40Mb per average individual (data not shown). This result agrees with previous estimates of total homozygosity based on the length of each segment.31, 32, 33 Many of these and smaller regions may be present due to sharing of common haplotypes, which is not accounted for by the kcoeff method and is below the resolution that we report. This could result in higher total homozygosity levels that match findings from previous studies.
We benchmarked our results against two leading software programs. We reported many first and second degree relationships incorrectly called as full-siblings by RELPAIR and discovered a correlation between miscalls and unexpected IBD2 sharing. Full-sibling relationships can either be inferred from the proportion of IBD2 sharing alone or by the expected Cotterman coefficients of relatedness (ie, K0 (1/4) K1 (1/2), and K2 (1/4)). For parent–child pairs with a small amount of IBD2 that were called full-sibling by RELPAIR, we did not observe a relative increase in IBS0. We thus conclude that RELPAIR misclassified relationships that were atypical (ie, parent–child with IBD2 or second-degree with IBD2). PREST generated IBD estimates that were generally comparable to those of kcoeff for these relationships.
RELPAIR, PREST and kcoeff were comparable in detecting distant relatedness between individuals annotated as unrelated. RELPAIR was more consistent than PREST at detecting distantly related individuals. There was a higher correlation between number per 25 trials of RELPAIR cousin calls and the kcoeff K1 estimate of IBD1 relative to PREST's pIBD1, suggesting similar abilities. Although RELPAIR was not explicitly designed to be run 25 times in order to derive a consensus output,24 we did 25 runs in accordance with others' usage of the program.27
As previously noted by the developers of RELPAIR, distinguishing between the three forms of second-degree relationships is difficult.24 In the present study, many false positive half-sibling relationships were called by RELPAIR, as well as false positive GG and AV amongst the second-degree relationships. Neither our method (kcoeff) nor PREST explicitly classifies second-degree relationships (although PREST can do so, given a genetic map specified by cM distance in the map file). Instead, they provide IBD estimates that can be used to infer relationships.
It is important to note a potential limitation of the kcoeff program that produced decreased K1 and K2 estimates for individuals with homozygosity due to inbreeding. Since the kcoeff method relies on the presence of IBS2* calls (AB/AB) based on the assumption of no inbreeding, homozygosity affects how the IBD states are assigned. This applies only to pairs of individuals in whom IBD1 or IBD2 sharing is present. For example, homozygosity present in one individual that is compared with a second individual with normal heterozygosity (eg, AA/AB calls) would result in zero IBS0 and zero IBS2* calls, leaving only IBS1 and concordant homozygous IBS2 calls (eg, AA/AA, which are not informative for kcoeff analyses). The IBD state of this region would be dependent on the flanking regions. This will have a minimal effect on kcoeff IBD estimates for relationships that have significant amounts of homozygosity, as indicated by our results.
Additionally, it is worth noting a unique ability of the kcoeff software. Estimating relatedness has typically been restricted to within-group pairwise comparisons because of the impact that different allele frequencies can have on IBD estimation. The software, kcoeff, allows for a comparison of between-group individuals because of the underlying ratio it uses to infer IBD. When two people share IBD1 or IBD2 within a given segment, their IBS0_ratio (IBS0/(IBS0+IBS2*)) for that window will be zero as IBS0 does not exist, but for individuals who are unrelated (and within the same mating population) their IBS0_ratio is centered on 1/3. However, for individuals who are from different groups, unrelated segments will have IBS0_ratios >1/3 because of the increase in IBS0 (ie, there are more differences between two members of different groups).16 IBD estimates from individuals belonging to two different groups will have reduced noise because there will be fewer regions of little variability between them to confound K1 estimates, as occurs when K1 estimates are slightly higher than zero.
Acknowledgments
We thank Drs Dorit Berlin and Norman Gerry of the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research for kindly providing SNP data. TJD and JP were supported by NIH grant R43 MH86192, and JP was supported by grant HD24061.
TJD is an employee and stockholder of Partek, Inc. GH is an employee of Partek.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- NIH/CEPH Collaborative Mapping Group. A comprehensive genetic linkage map of the human genome. Science. 1992;258:67–86. [PubMed] [Google Scholar]
- NIH/CEPH Collaborative Mapping Group. A comprehensive genetic linkage map of the human genome. Science. 1992;258:148–162. [PubMed] [Google Scholar]
- Weissenbach J, Gyapay G, Dib C, et al. A second-generation linkage map of the human genome. Nature. 1992;359:794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Prescott SM, Lalouel JM, Leppert M. From linkage maps to quantitative trait loci: the history and science of the Utah genetic reference project. Annu Rev Genomics Hum Genet. 2008;9:347–358. doi: 10.1146/annurev.genom.9.081307.164441. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium: The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium: A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks SA, Leonardson A, Zhu H, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittles AH, Black ML. Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci USA. 2010;107 (Suppl 1:1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger AL, Sahbatou M, Gazal S, Cann H, Genin E. Consanguinity around the world: what do the genomic data of the HGDP-CEPH diversity panel tell us. Eur J Hum Genet. 2011;19:583–587. doi: 10.1038/ejhg.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Pevsner J. Visualization of shared genomic regions and meiotic recombination in high-density SNP data. PLoS One. 2009;4:e6711. doi: 10.1371/journal.pone.0006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JC, Roberson ED, Currier DG, Pevsner J. Locations and patterns of meiotic recombination in two-generation pedigrees. BMC Med Genet. 2009;10:93. doi: 10.1186/1471-2350-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. Testing the genetic relation between two individuals using a panel of frequency-unknown single nucleotide polymorphisms. Ann Hum Genet. 2003;67 (Pt 6:618–619. doi: 10.1046/j.1529-8817.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Stevens EL, Heckenberg G, Roberson ED, Baugher JD, Downey TJ, Pevsner J. Inference of relationships in population data using identity-by-descent and identity-by-state. Plos Genet. 2011;7:e1002287. doi: 10.1371/journal.pgen.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet. 2011;88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AD, Weir BS. A maximum-likelihood method for the estimation of pairwise relatedness in structured populations. Genetics. 2007;176:421–440. doi: 10.1534/genetics.106.063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff CD, Witherspoon DJ, Simonson TS, et al. Maximum-likelihood estimation of recent shared ancestry (ERSA) Genome Res. 2011;21:768–774. doi: 10.1101/gr.115972.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterman W.A calculus for statistico-genetics. Dissertation, Ohio State University, Columbus, OH (reprinted in Ballonoff P (ed.): Genetics and Social Structure Stroudsburg, PA: Dowden, Hutchinson & Ross; 1974157–272. [Google Scholar]
- Cotterman CW. Relationship and probability in Mendelian populations. Am J Med Genet. 1983;16:393–440. doi: 10.1002/ajmg.1320160311. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain. Am J Hum Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Wang C, Li JZ, Rosenberg NA. Inference of unexpected genetic relatedness among individuals in HapMap Phase III. Am J Hum Genet. 2010;87:457–464. doi: 10.1016/j.ajhg.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Bryc K, Boyko AR, et al. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, et al. Long contiguous stretches of homozygosity in the human genome. Hum Mutat. 2006;27:1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan R, Leutenegger AL, Abdel-Rahman R, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel M, Lu TT, Kayser M, Krawczak M. Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans. Hum Mol Genet. 2010;19:2927–2935. doi: 10.1093/hmg/ddq198. [DOI] [PubMed] [Google Scholar]
- Coriell Cell Repositories , http://ccr.coriell.org/ .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.