Abstract
Genome-wide association studies for breast cancer have identified over 40 single-nucleotide polymorphisms (SNPs), a subset of which remains statistically significant after genome-wide correction. Improved strategies for mining of genome-wide association data have been suggested to address heritable component of genetic risk in breast cancer. In this study, we attempted a two-stage association design using markers from a genome-wide study (stage 1, Affymetrix Human SNP 6.0 array, cases=302, controls=321). We restricted our analysis to DNA repair/modifications/metabolism pathway related gene polymorphisms for their obvious role in carcinogenesis in general and for their known protein–protein interactions vis-à-vis, potential epistatic effects. We selected 22 SNPs based on linkage disequilibrium patterns and high statistical significance. Genotyping assays in an independent replication study of 1178 cases and 1314 controls were attempted using Sequenom iPLEX Gold platform (stage 2). Six SNPs (rs8094493, rs4041245, rs7614, rs13250873, rs1556459 and rs2297381) showed consistent and statistically significant associations with breast cancer risk in both stages, with allelic odds ratios (and P-values) of 0.85 (0.0021), 0.86 (0.0026), 0.86 (0.0041), 1.17 (0.0043), 1.20 (0.0103) and 1.13 (0.0154), respectively, in combined analysis (N=3115). Of these, three polymorphisms were located in methyl-CpG-binding domain protein 2 gene regions and were in strong linkage disequilibrium. The remaining three SNPs were in proximity to RAD21 homolog (S. pombe), O-6-methylguanine-DNA methyltransferase and RNA polymerase II-associated protein 1. The identified markers may be relevant to breast cancer susceptibility in populations if these findings are confirmed in independent cohorts.
Keywords: genome-wide association study, breast cancer, epistasis, genetic risk, DNA repair
Introduction
Breast cancer is a multi-factorial, polygenic disease resulting from the interplay of genetic, environmental and lifestyle risk factors. Linkage studies have revealed that breast cancer tends to cluster in families and disease prevalence is two-fold higher among the first-degree relatives of affected individuals.1 Familial clustering is characterized by early onset of disease often mediated by high-to-moderate penetrance mutations in genes, such as those encoding breast cancer (BRCA1 and BRCA2),2, 3 ataxia telangiectasia mutated (ATM),4 cell cycle checkpoint kinase 2 (CHEK2),5 tumor protein 53 (TP53),6 partner and localizer of BRCA2,7 BRCA1-interacting protein C-terminal helicase 1 (BRIP1)8 and phosphatase and tensin homolog (PTEN).9 Nonetheless, these genes in aggregate account for <25% of the observed familial genetic risk.10 A polygenic model has been proposed to explain the remaining genetic risk in non-BRCA familial and sporadic breast cancer cases.11 Single-nucleotide polymorphisms (SNPs)-based genome-wide association studies (GWAS) have identified low-risk conferring common variants in several complex diseases. For European, Ashkenazi Jewish and Asian population-based GWAS, more than 40 breast cancer susceptibility loci in several genes and intergenic regions have already been reported and a subset of these associations have reached genome-wide significance level.12, 13, 14 These variants account for a small proportion of overall genetic risk of breast cancer, leaving open the question of hidden or missing heritability. Current debates suggest that this may be further explained by rare variants, epistasis, epigenetics, gene–environment interactions and copy number variations.15, 16
In a typical GWAS, the frequencies for each SNP (single-locus tests for association)17 are compared between cases and controls to catalogue polymorphisms potentially associated with the phenotype of interest. The most promising SNPs, sorted based on P-value ranking (highest significance) and/or showing significance in haplotype association analysis,18 are selected and replicated in a larger but independent set of cases and controls. In this process, SNPs that are not top ranked because of their modest P-values are ignored, and as a result potentially informative markers may have been missed. It has been proposed by others19, 20 that even modest associations (P-value based), if highly reproducible in independent cohorts, may still be pertinent to the phenotypes under investigation presumably through epistatic interactions (interactions of alleles or genes), a phenomenon strongly implicated in the etiology of breast cancer and the heritable component of genetic risk. Because the majority of the published GWAS concentrate on single-locus strategies to identify novel breast cancer susceptibility loci, a candidate gene approach restricted to specific pathway related gene polymorphisms to more effectively mine GWAS data is presented considering moderately associated SNPs. If reproduced in further independent studies, these may serve as putative candidates for epistatic effects.
Previously reported studies focused on common variants in the genes involved in DNA repair/metabolism pathways and cell cycle regulation, and the markers were selected based on candidate gene approaches.21, 22 In this study, we extend this premise using SNPs in or flanking the DNA repair, modifications and metabolism pathway-related genes from the Affymetrix 6.0 array (Santa Clara, CA, USA) (stage 1 of GWAS23) for independent replication, stage 2 of the association study design) to identify additional breast cancer susceptibility loci not previously reported.
Materials and methods
Study population and DNA isolation
We used stage 1 results of our published breast cancer GWAS, described elsewhere.23 Briefly, sporadic breast cancer cases (n=348), characterized by late onset of disease and controls (n=348) who had no documented history of breast cancer in the first- and second-degree relatives were selected for stage 1 of the GWAS.23 All subjects were predominantly of Caucasian origin. Breast cancer cases (median age=51 years; age range=26–90 years, with number of cases <40 years=35; 40–60 years=241; >60 years=72) were from Alberta, Canada, recruited by the PolyomX Program24 and the Canadian Breast Cancer Foundation-Tumor Bank, (CBCF-TB)24 during the years 2001–2005 and since 2005–2008, respectively. The two projects PolyomX Program and CBCF-TB are funded by different granting agencies, and nomenclature adopted merely indicates this and in no way reflects bias in sampling of population. All cases had a histologically confirmed diagnosis of invasive ductal breast carcinoma at the time of enrolment in the study. Gender-matched apparently healthy controls (median age=50 years; age range=36–70 years, with number of controls <40 years=50; 40–60 years=226; >60 years=72), also from Alberta, Canada (accessed from the Tomorrow Project25), were frequency matched to cases based on age. The proportions of cases and controls for three different age groups (<40, 40–60 and >60 years) were not statistically significant (two tailed z-test; data not shown). All control subjects' enrolled here were free from cancer at the time of recruitment in the study. Potential population confounders were removed, leaving cases (n=302) and controls (n=321) for association analysis.23 Informed consents were obtained from all study participants, and the study was approved by Research Ethics Board of Alberta Health Services. Genomic DNA was extracted from the peripheral blood samples of both cases and controls using commercially available Qiagen (Mississauga, ON, Canada) DNA isolation kits.
SNP selection, genotyping and platform-specific genotype concordance
Data filtering and call rate clean up (Hardy–Weinberg equilibrium (HWE) P>0.001 and SNPs call rate >99%) were carried out as described earlier.23 Of the 906 600 SNPs genotyped using Affymetrix SNP 6.0, a total of 782 838 SNPs qualified for the downstream analysis. The associations of SNPs with breast cancer were evaluated using correlation/trend tests with one degree of freedom (df). Correlation/trend test is similar to χ2-test of independence, except that it is also believed to be a trend test that evaluates correlation of a minor allele with the case status using Pearson's correlation coefficient. The allelic tests with 782 838 SNPs (stage 1) showed that a total of 35 519 SNPs statistically significantly associated with breast cancer at P<0.05. Of the 35 519 SNPs, we identified 215 polymorphisms (minor allele frequency (MAF)>10%) within or in close proximity to 49 gene regions implicated in pathways or of relevance to DNA repair, modifications and metabolism based on National Center for Biotechnology Information human genome build 37. In all, six of 215 SNPs were statistically significantly associated with breast cancer at P<0.001 (correlation/trend tests with one df) and were included for stage 2 replication study. To reduce the redundancy among the remaining 209 SNPs, we then calculated the pairwise LD (r2) among the markers and found that 73 SNPs were strongly correlated (r2≥0.8). Of these 73 short-listed SNPs, 16 were in strong LD (r2≥0.8), with at least one SNP contained within the identified 3903 haplotype blocks (P<0.05) in haplotype association analysis. All haplotypes at a frequency threshold of 1% or more were tested together against the reference haplotype for their associations with breast cancer. The haplotype association analysis per se was carried out as described elsewhere.23 As our primary objective in this study was to evaluate the moderately associated SNPs from stage 1 GWAS results, we relaxed the significance threshold in haplotype association analysis to P<0.05 as compared with our previous study (P<0.001).23 Overall, we used allelic tests and haplotype association tests to select SNPs for replication study in an independent set of 1178 invasive breast cancer cases and 1314 apparently healthy individuals serving as controls (stage 2).
Genotyping assays were performed on Sequenom iPLEX Gold platform (San Diego, CA, USA) (services from the McGill University, Genome Quebec Innovation Center, Montreal, Canada). Within- (Sequenom only) and cross-platform (Affymetrix vs Sequenom) SNP concordances for 22 SNPs were assessed using 205 and 551 duplicate samples, respectively.
Statistical considerations
Allelic associations were evaluated using correlation/trend tests with one df, and their corresponding odds ratios (ORs) and 95% confidence intervals (CIs) were estimated using unconditional logistic regression implemented in the SNP & Variation Suite v7.3.1 (Helix Tree Software).26 Genotypic associations were also considered for gaining insights in to relative contributions from individual genotypes to breast cancer risk using unconditional logistic regression with two df using the freeware, SNPstats,27 and the results from codominant models were summarized in the study. A combined analysis with all samples from stages 1 and 2 (a total of 1480 cases and 1635 controls) was performed to increase the statistical power. The associations for the allelic tests in combined analysis were further examined with 1000-times permutation tests and false discovery rates (FDRs) to identify observations by chance alone (type 1 error) using Helix Tree software. Helix Tree calculates FDR using the original P-value times the number of tests divided by the number of tests minus the rank order of the original P-value in the descending order.
Subgroup analyses were attempted (correlation/trend tests with 1 df) to identify associations with subphenotypes within the combined breast cancer cases using a common reference (combined controls) as described previously.28 The subphenotypes examined were family history of breast cancer, menopausal status and luminal A status. Subgroup analyses help interrogate potential confounding influence of disease heterogeneity on the observed associations. Tumors were classified as luminal A based on estrogen and progesterone receptor status (ER+/PR+, ER−/PR+ and ER+/PR−) and human epidermal growth factor receptor-2 status (HER2−).29 All the remaining cases were classified as non-luminal A tumors.
Our sample size conferred more than 80% power to detect associations using a codominant model for a SNP with 10% MAF, disease prevalence at 1/10 in population for breast cancer, a relative risk of 1.3, type I error of 0.05 and with the LD between markers at r2 of 0.8.30
The LD patterns for regions showing the strongest and consistent associations across stages 1 and 2 and combined analyses were examined using Haploview v4.2.31 For the three methyl-CpG-binding domain protein 2 (MBD2) SNPs, haplotype frequencies were estimated using SNPstats.27 The software implements the expectation-maximization algorithm coded into haplo.stats package to calculate the estimated relative frequencies for each haplotype.32 Haplotype association analyses for MBD2 SNPs were performed with unconditional logistic regression using the default setting of a log-additive model and expressed in terms of ORs and 95% CIs (feature available in SNPstats).
Results
Initial assessment of the data quality
Of the 22 SNPs selected for replication in stage 2, genotyping for one SNP (rs17519016) was not successful. The cross-platform (Affymetrix vs Sequenom) SNP call concordance for the remaining 21 SNPs using 551 duplicate samples from stage 1 was more than 98%. Within-platform (Sequenom) SNP call concordance among the 205 duplicates used in stage 2 was more than 99.4%. Per sample and per SNP call rates for stage 2 were >98.3 and >98.4%, respectively, and all 21 SNPs were in HWE proportion at P>0.001 in controls (Table 1). Cross-platform and within-platform discordances were very low (<2%) and are in agreement with previously reported GWAS studies.12, 23 Further, the MAFs were consistent among the two stages and also comparable to HapMap Central Europeans (CEU) population (data not shown), indicating that the scope of false-positive associations due to genotyping errors (systematic or random) was effectively minimized.
Table 1. Characteristics of the SNPs used in the study.
| Stage 1b | Stage 2c | Stages 1+2d | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs (Chr.) | Positiona | Associated gene | Gene relationship | Gene distance (bp) | Minor allele | MAF | HWE Pcontrols | Call ratee | MAF | HWE Pcontrols | Call ratee | MAF | HWE Pcontrols | Call ratee |
| rs17622933 (4p15.2) | 24 790 680 | DHX15 | Upstream | 204 496 | T | 0.30 | 0.06 | 1.00 | 0.31 | 0.52 | 1.00 | 0.31 | 0.89 | 1.00 |
| rs7200108 (16p13.12) | 13 438 884 | ERCC4 | Upstream | 575 129 | G | 0.12 | 0.61 | 1.00 | 0.12 | 0.94 | 1.00 | 0.12 | 0.66 | 1.00 |
| rs7317643 (13q33.3) | 108 536 028 | LIG4 | Downstream | 323 765 | A | 0.12 | 0.95 | 1.00 | 0.14 | 0.53 | 1.00 | 0.13 | 0.52 | 1.00 |
| rs1646807 (18q21.2) | 51 388 197 | MBD2 | Downstream | 289 773 | T | 0.17 | 0.80 | 0.99 | 0.19 | 1.00 | 1.00 | 0.18 | 0.91 | 1.00 |
| rs4041245 (18q21.2) | 51 685 525 | MBD2 | Intron | 0 | G | 0.42 | 0.68 | 1.00 | 0.41 | 0.01 | 1.00 | 0.41 | 0.04 | 1.00 |
| rs656923 (18q21.2) | 51 701 796 | MBD2 | Intron | 0 | G | 0.19 | 0.71 | 1.00 | 0.20 | 0.74 | 1.00 | 0.20 | 0.60 | 1.00 |
| rs7239408 (18q21.2) | 51 432 800 | MBD2 | Downstream | 245 170 | A | 0.23 | 0.61 | 1.00 | 0.24 | 0.51 | 1.00 | 0.24 | 0.43 | 1.00 |
| rs7614 (18q21.2) | 51 681 244 | MBD2 | 3′ UTR | 0 | C | 0.42 | 0.27 | 1.00 | 0.41 | 0.03 | 1.00 | 0.41 | 0.17 | 1.00 |
| rs8094493 (18q21.2) | 51 700 391 | MBD2 | Intron | 0 | G | 0.42 | 0.68 | 1.00 | 0.41 | 0.02 | 1.00 | 0.41 | 0.05 | 1.00 |
| rs904276 (18q21.2) | 51 434 340 | MBD2 | Downstream | 243 630 | C | 0.22 | 0.47 | 1.00 | 0.23 | 0.39 | 1.00 | 0.23 | 0.29 | 1.00 |
| rs2044760 (2q23.1) | 148 989 731 | MBD5 | Intron | 0 | T | 0.39 | 0.23 | 1.00 | 0.35 | 0.21 | 1.00 | 0.36 | 0.51 | 1.00 |
| rs1556459 (10q26.3) | 130 810 711 | MGMT | Upstream | 454 766 | C | 0.16 | 0.80 | 1.00 | 0.15 | 0.07 | 1.00 | 0.15 | 0.12 | 1.00 |
| rs3996018 (3q13.13) | 108 163 202 | MYH15 | Intron | 0 | G | 0.24 | 0.43 | 1.00 | 0.23 | 0.28 | 1.00 | 0.23 | 0.18 | 1.00 |
| rs13250873 (8q24.11) | 117 806 169 | RAD21 | Downstream | 52 004 | G | 0.31 | 0.52 | 1.00 | 0.32 | 0.27 | 1.00 | 0.32 | 0.20 | 1.00 |
| rs2297381 (15q15.1) | 41 827 655 | RPAP1 | Intron | 0 | G | 0.50 | 0.84 | 1.00 | 0.48 | 0.67 | 1.00 | 0.48 | 0.63 | 1.00 |
| rs6893184 (5q23.1) | 118 730 867 | TNFAIP8 | Downstream | 574 | G | 0.34 | 0.63 | 1.00 | 0.33 | 0.0017 | 1.00 | 0.33 | 0.00240 | 1.00 |
| rs7721752 (5q23.1) | 118 746 110 | TNFAIP8 | Downstream | 15 817 | G | 0.33 | 0.77 | 1.00 | 0.30 | 0.01 | 1.00 | 0.31 | 0.04 | 1.00 |
| rs6795465 (3q28) | 189 537 521 | TP63 | Intron | 0 | C | 0.12 | 0.42 | 1.00 | 0.13 | 0.27 | 0.99 | 0.13 | 0.17 | 0.99 |
| rs7636114 (3q28) | 189 088 705 | TPRG1 | Downstream | 45 612 | C | 0.11 | 0.80 | 1.00 | 0.11 | 0.14 | 1.00 | 0.11 | 0.21 | 1.00 |
| rs7700025 (4q34.3) | 177 814 863 | VEGFC | Upstream | 100 968 | G | 0.28 | 0.50 | 1.00 | 0.30 | 0.79 | 1.00 | 0.30 | 0.91 | 1.00 |
| rs9992272 (4q34.3) | 177 737 136 | VEGFC | Upstream | 23 241 | C | 0.16 | 0.71 | 1.00 | 0.17 | 0.99 | 0.98 | 0.16 | 0.93 | 0.99 |
Abbreviation: Chr., chromosome.
From NCBI human genome build 37; DHX15, DEAH (Asp–Glu–Ala–His) box polypeptide 15; ERCC4, excision repair cross-complementing rodent repair deficiency, complementation group 4; LIG4, ligase IV, DNA, ATP-dependent; MBD2, methyl-CpG-binding domain protein 2; MBD5, methyl-CpG-binding domain protein 5; MGMT, O-6-methylguanine-DNA methyltransferase; MYH15, myosin, heavy chain 15; RAD21, RAD21 homolog (S. pombe); RPAP1, RNA polymerase II-associated protein 1; TNFAIP8, tumor necrosis factor, alpha-induced protein 8; TP63, tumor protein 63; TPRG1, tumor protein p63 regulated 1; VEGFC, vascular endothelial growth factor C.
302 cases and 321 controls.
1178 cases and 1314 controls.
1480 cases and 1635 controls; MAF, combined minor allele frequency in both cases and controls.
Combined SNP call rate in both cases and controls.
Stage 2 analysis
In stage 2, six SNPs showed suggestive associations with breast cancer (Table 2). Three SNPs (rs8094493, rs4041245 and rs7614) were from MBD2 gene regions and were marginally associated with reduced risk for breast cancer (ORs: 0.90, 0.91 and 0.92, respectively; Table 2). The other three SNPs rs13250873, rs1556459 and rs2297381 were located in or close proximity of RAD21 homolog (S. pombe; RAD21), O-6-methylguanine-DNA methyltransferase (MGMT) and RNA polymerase II-associated protein 1 (RPAP1) gene regions, respectively, and showed suggestive associations with increased risk for breast cancer.
Table 2. Six SNPs with the strongest and consistent associations with breast cancer susceptibility across stages 1, 2 and in combined analysis.
| Stage 1a | Stage 2b | Stages 1+2 (combined analysis)c | |||||||
|---|---|---|---|---|---|---|---|---|---|
| SNPs | Allele or genotype | OR, 95% CI | P-valued | OR, 95% CI | P-valued | OR, 95% CI | P-valued | FDR | Permutation P-valuee |
| rs8094493 | G (minor allele) | 0.68 (0.54, 0.85) | 0.0009 | 0.90 (0.81, 1.01) | 0.0773 | 0.85 (0.77, 0.94) | 0.0021 | 0.045 | 0.038 |
| GT | 0.66 (0.46, 0.94) | 0.0044 | 0.80 (0.67, 0.95) | 0.0410 | 0.77 (0.66, 0.90) | 0.0019 | ND | ND | |
| GG | 0.48 (0.31, 0.77) | 0.86 (0.68, 1.09) | 0.76 (0.61, 0.94) | ND | ND | ||||
| rs4041245 | G (minor allele) | 0.68 (0.54, 0.85) | 0.0009 | 0.91 (0.81, 1.02) | 0.0893 | 0.86 (0.77, 0.95) | 0.0026 | 0.027 | 0.048 |
| GA | 0.66 (0.46, 0.94) | 0.0044 | 0.79 (0.67, 0.94) | 0.0340 | 0.76 (0.65, 0.89) | 0.0018 | ND | ND | |
| GG | 0.48 (0.31, 0.77) | 0.87 (0.69, 1.10) | 0.77 (0.62, 0.95) | ND | ND | ||||
| rs7614 | C (minor allele) | 0.67 (0.54, 0.84) | 0.0006 | 0.92 (0.82, 1.03) | 0.1356 | 0.86 (0.78, 0.95) | 0.0041 | 0.029 | 0.069 |
| CT | 0.70 (0.49, 0.99) | 0.0038 | 0.81 (0.68, 0.97) | 0.0690 | 0.79 (0.67, 0.92) | 0.0053 | ND | ND | |
| CC | 0.47 (0.30, 0.74) | 0.89 (0.70, 1.12) | 0.77 (0.63, 0.95) | ND | ND | ||||
| rs13250873 | G (minor allele) | 1.29 (1.01, 1.64) | 0.0383 | 1.14 (1.01, 1.28) | 0.0306 | 1.17 (1.05, 1.30) | 0.0043 | 0.023 | 0.07 |
| GA | 1.34 (0.96, 1.87) | 0.1100 | 1.10 (0.93, 1.30) | 0.0910 | 1.14 (0.98, 1.33) | 0.0190 | ND | ND | |
| GG | 1.56 (0.91, 2.68) | 1.33 (1.02, 1.73) | 1.37 (1.08, 1.74) | ND | ND | ||||
| rs1556459 | C (minor allele) | 1.50 (1.10, 2.04) | 0.0102 | 1.13 (0.97, 1.32) | 0.1151 | 1.20 (1.04, 1.37) | 0.0103 | 0.043 | 0.161 |
| CT | 1.49 (1.03, 2.14) | 0.0390 | 1.05 (0.88, 1.26) | 0.0740 | 1.13 (0.96, 1.32) | 0.0120 | ND | ND | |
| CC | 2.21 (0.80, 6.07) | 1.89 (1.07, 3.32) | 1.96 (1.20, 3.20) | ND | ND | ||||
| rs2297381 | G (minor allele) | 1.27 (1.01, 1.58) | 0.0368 | 1.10 (0.98, 1.23) | 0.0986 | 1.13 (1.02, 1.25) | 0.0154 | 0.054 | 0.234 |
| GA | 1.28 (0.87, 1.89) | 0.1100 | 1.02 (0.85, 1.24) | 0.1800 | 1.07 (0.90, 1.27) | 0.0430 | ND | ND | |
| GG | 1.60 (1.03, 2.50) | 1.21 (0.97, 1.50) | 1.28 (1.05, 1.55) | ND | ND | ||||
Abbreviations: CI, confidence interval; FDR, false discovery rate; ND, not determined; OR, odds ratio.
302 cases and 321 controls.
1178 cases and 1314 controls.
1480 cases and 1635 controls.
Individual P-values across stages and combined analysis are indicated. P-values for minor allele were calculated using the correlation/trend test with one df, whereas the P-values for the heterozygote and variant genotypes (codominant genotypic model) were calculated using unconditional logistic regression with two df; FDR for observed associations in combined analysis using allelic association tests.
1000-times permutation P-value for observed associations in combined analysis using allelic association tests; P-values shown in bold indicate the combined analysis from allelic association tests.
The association test results for the remaining 15 SNPs are summarized in Supplementary Table 1. Fourteen of these showed no statistical significance and one SNP (rs7636114) showed suggestive association trend in stage 2 (but in opposite direction to the stage 1 results) and is therefore not considered for further analysis.
Combined analysis (stages 1 and 2)
We combined the results for six SNPs from stages 1 and 2, and conducted a combined analysis and found not only similar direction of risk but also stronger association signals for all six variants (Table 2). The MBD2 SNPs rs8094493 (OR: 0.85, P<0.0021), rs4041245 (OR: 0.86, P<0.0026) and rs7614 (OR: 0.86, P<0.0041) were significantly associated with reduced risk of breast cancer. The observed FDR of 0.045, 0.027 and 0.029, respectively, for the allelic associations in combined analysis provided confidence in the study findings. We also subjected the data to permutation testing (1000 times) and observed permutation P-values of 0.038, 0.048 and 0.069, respectively, an indication that the reported findings may not be attributed to associations by chance alone. The heterozygote and variant homozygote genotypes of MBD2 SNPs from codominant models also conferred similar trends of reduced risks of breast cancer (ORs: 0.76–0.79).
The remaining polymorphisms analyzed (rs13250873, rs1556459 and rs2297381, Table 2) also showed significant associations, except the direction of risk for breast cancer (allelic ORs: 1.13–1.20) was in opposite direction to the ones observed for MBD2 SNPs. The association signals for all three SNPs were characterized by low FDR values (0.023–0.054); the 1000-times permutation tests also showed marginal significance for rs13250873. In the codominant genotypic models, variant homozygotes (OR≥1.28) showed stronger associations than heterozygotes (OR: 1.07–1.14) in the combined analysis for rs13250873, rs1556459 and rs2297381.
Subgroup analyses
Owing to potential for genetic risk determinants to be associated with specific clinical and molecular subtypes of breast cancer, we reviewed clinicopathological characteristics of the cases in both stages 1 and 2, and conducted stratified analyses (Table 3). We evaluated allelic associations for six SNPs with the following subgroups: without and with family history of breast cancer, pre- and postmenopausal status and luminal A and non-luminal A (ie, good and poor prognostic groups, respectively) breast cancer status of the tumors, using correlation/trend tests with one df. We found associations between clinicopathological characteristics and the polymorphisms considered, and the observed ORs were consistent across subgroups (Table 3). None of the observed associations were stronger than the single-locus effects, and hence it is less likely that these clinicopathological characteristics (potential confounders) have significant effects on initial observed associations with unstratified cases (Table 2).
Table 3. Subgroup analyses based on family history of breast cancer, menopausal status and luminal A tumors.
| Premenopausal womena | Postmenopausal womenb | Cases with family historyc | Cases without family historyd | Luminal A tumorse | Non-luminal A tumorsf | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | OR, 95% CI | P-value | OR, 95% CI | P-value | OR, 95% CI | P-value | OR, 95% CI | P-value | OR, 95% CI | P-value | OR, 95% CI | P-value |
| rs8094493 | 0.84 (0.74, 0.96) | 0.011 | 0.86 (0.76, 0.97) | 0.016 | 0.86 (0.75, 0.99) | 0.031 | 0.85 (0.75, 0.96) | 0.009 | 0.88 (0.78, 0.99) | 0.039 | 0.80 (0.68, 0.94) | 0.006 |
| rs4041245 | 0.85 (0.74, 0.97) | 0.015 | 0.86 (0.76, 0.97) | 0.016 | 0.87 (0.76, 0.99) | 0.041 | 0.85 (0.75, 0.96) | 0.009 | 0.88 (0.78, 1.00) | 0.043 | 0.80 (0.68, 0.94) | 0.006 |
| rs7614 | 0.85 (0.75, 0.97) | 0.018 | 0.87 (0.77, 0.98) | 0.027 | 0.88 (0.77, 1.01) | 0.079 | 0.85 (0.75, 0.96) | 0.010 | 0.89 (0.79, 1.01) | 0.064 | 0.80 (0.68, 0.94) | 0.007 |
| rs13250873 | 1.22 (1.06, 1.40) | 0.005 | 1.12 (0.98, 1.27) | 0.085 | 1.18 (1.02, 1.36) | 0.024 | 1.15 (1.01, 1.31) | 0.029 | 1.11 (0.97, 1.26) | 0.117 | 1.20 (1.02, 1.41) | 0.030 |
| rs1556459 | 1.27 (1.07, 1.51) | 0.008 | 1.16 (0.99, 1.37) | 0.074 | 1.20 (1.00, 1.45) | 0.047 | 1.19 (1.01, 1.40) | 0.037 | 1.17 (0.99, 1.39) | 0.062 | 1.20 (0.97, 1.48) | 0.092 |
| rs2297381 | 1.22 (1.07, 1.39) | 0.003 | 1.09 (0.97, 1.22) | 0.165 | 1.19 (1.04, 1.36) | 0.012 | 1.13 (1.00, 1.27) | 0.044 | 1.15 (1.02, 1.30) | 0.024 | 1.17 (1.00, 1.36) | 0.050 |
Abbreviations: CI, confidence interval; OR, odds ratio.
623 premenopausal women.
829 postmenopausal women.
575 cases with family history of breast cancer in their first- or second-degree relatives.
808 cases without family history of breast cancer.
761 cases with luminal A tumors.
397 cases with non-luminal A tumors; associations of SNPs with each of these subgroups were assessed using common 1635 controls and expressed in terms of OR and 95% CI; the P-values were obtained from a correlation/trend test with one df
Pairwise LD profiling between markers
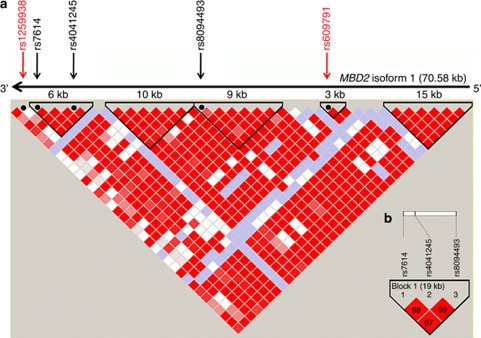
We examined LD profiles for the six identified variants (Table 2) using HapMap CEU genotype data (available from http://www.hapmap.org). We found that three MBD2 SNPs (rs8094493, rs4041245 and rs7614) in intron 3, intron 6 and the 3′-untranslated region, respectively, were in strong LD with D′=1 (Figure 1a), and these profiles were also observed in our study population (Figure 1b). rs7614 and rs4041245 were located in a LD block spanning ∼6 kb region, and rs8094493 was located in a LD block of ∼9 kb region.
Figure 1.
Pairwise LD profiles between SNPs from MBD2 gene region. (a) LD profile of whole MBD2 isoform 1 spanning ∼70.58 kbp. The gene is in reverse orientation (3′–5′) on chromosome 18q arm. Five SNPs (three from our study that shown in black and two from Zhu et al35 that are shown in red) in MBD2 gene regions are shown based on their relative position on HapMap CEU data set (phase 1 and 2-full data set). LD blocks were defined using ‘CI' method as explained by Gabriel et al.42 D′ values are given for LD between the markers. The darker the cell, the greater the D′ value between the SNPs. (b) LD profile for three MBD2 SNPs from our study based on our study population.
We also analyzed the remaining three SNPs (rs13250873, rs2297381 and rs1556459) that were associated with breast cancer in our study population (Table 2) and found that these SNPs belong to different blocks/regions and were not correlated with each other (data not shown). The LD blocks containing rs13250873 and rs1556459 did not show annotated genes. However, we observed UTP23 (∼19 kb downstream) and RAD21 (∼52 kbp downstream) as the nearest genes flanking rs13250873 and for rs1556459, the closet gene was MGMT at ∼450 kb upstream. On the other hand, the polymorphism rs2297381 was located in intron 5 of RPAP1 gene.
Haplotype analysis for MBD2 gene polymorphisms
We reasoned that the highly correlated SNPs from the MBD2 gene region may form distinct haplotypes that could potentially explain the population diversity. Polymorphisms rs8094493, rs4041245 and rs7614 formed two major haplotypes, one with common alleles (major allele) and other with variant alleles (minor allele). The common haplotype had a population frequency of 0.58 (0.60 for cases and 0.56 for controls), and the variant haplotype had a population frequency of 0.40 (0.38 for cases and 0.42 for controls). The variant form was significantly associated with the reduced risk of breast cancer (OR: 0.86, P<0.0029; Table 4). The population diversity that could be explained by the two major haplotypes identified in this analysis was 98%.
Table 4. Haplotypes for three MBD2 SNPs and their associations with breast cancer risk.
| Haplotype | Frequency | |||||||
|---|---|---|---|---|---|---|---|---|
| rs4041245 | rs8094493 | rs7614 | Cases | Controls | Total | OR | 95% CI | P-value |
| T | A | T | 0.601 | 0.5624 | 0.5823 | 1 | — | — |
| C | G | G | 0.385 | 0.4232 | 0.4053 | 0.86 | 0.77–0.95 | 0.0029 |
| * | * | * | Rare (<1%) | Rare (<1%) | 0.0124 | 1.11 | 0.70–1.78 | 0.66 |
Denotes alleles forming the rare haplotypes (<1% frequency).
P-value shown in bold indicates statistically significant association at alpha, 0.05.
Discussion
In this study, we identified SNPs associated with breast cancer among genes related to DNA repair, modifications and metabolism. A total of six loci were identified using a two-stage association study design, and these were not previously reported in published GWAS for breast cancer12, 13, 14, 23 as putative markers for breast cancer susceptibility. The identified loci were highly reproducible in an independent study (stage 2), and the statistical significance of the findings was consistent across study stages, in the combined analysis and across clinicopathological subtypes of breast cancer. These loci are promising markers and warrant independent validation in Caucasian population or in diverse ethnic cohorts to evaluate the generalizability of our findings.
The six loci identified were from four chromosomes 18, 15, 10 and 8. Both single-locus and haplotype association analyses indicated that MBD2 gene loci (rs8094493, rs4041245 and rs7614) conferred protection against breast cancer. The magnitude and the direction of the association signals in both stages were consistent between allelic and genotypic models (Table 2). The allelic risk effects were enriched in combined analysis with stronger association of P-values of <10−3. Low FDR values and permutation testing provided further confidence in our findings by ruling out the observations as false positives. Mechanistic relationships to breast carcinogenesis are suggested because MBD2 is a well-characterized gene and the encoded protein binds to methylated promoter regions and mediates transcriptional repression of tumor suppressor genes.33 DNA (cytosine-5)-methyltransferase 1 (DNMT1) is reported to interact with the methyl-CpG-binding protein complex, MBD2 and MBD3 at late S-phase replication foci, and as such these interactions may direct DNMT1 to hemimethylated sequences following DNA replication and silencing of genes in the S phase.34
Earlier, Zhu et al35 reported the associations of two SNPs (rs1259938 and rs609791) in MBD2 gene regions with the reduced risk of breast cancer in premenopausal Caucasian women.35 We evaluated for possible LD between the distinct MBD2 SNPs reported here and those reported by Zhu et al.35 The polymorphisms reported by earlier investigators were not in LD with the SNPs reported here (Figure 1a). The notable differences between our study and those by Zhu et al35 are (i) the SNPs rs1259938 and rs609791 in the previous study did not show association with the breast cancer phenotype in unstratified cases, although they showed statistical significance when cases were stratified by pre- and postmenopausal status; (ii) we identified distinct MBD2 gene SNPs and these were all statistically significantly associated with breast cancer as a phenotype even in both unstratified (Table 2) and stratified cases (Table 3); and (iii) sample sizes were substantially larger in our study (total sample size of 1480 cases and 1615 controls) as opposed to 393 cases and 436 controls from the nested case–control study with a Caucasian population reported by Zhu et al.35 In summary, observations with a larger sample size (this study) showed association with breast cancer even without stratification of cases, and the haplotypes associated were also distinct. However, it is important to note that the magnitude and direction of risk and the gene identified are similar in both studies. We did not genotype the polymorphisms reported by Zhu et al35 at this time, and may therefore require independent validation. The SNPs analyzed by Zhu et al35 were not present in the Affymetrix SNP 6.0 array.
Other genes/loci were identified for breast cancer risk in this study. rs2297381 was located in intron 5 of RPAP1 and was associated with the risk of breast cancer. RPAP1 is a poorly understood gene possibly involved in the interaction of RNA polymerase II and its regulators of protein complex formation.36 To our knowledge, this is the first report on RPAP1 gene SNP associated with breast cancer risk. rs13250873 and rs1556459, located ∼52 kbp downstream of RAD21 and ∼454 kbp upstream of MGMT, respectively, were significantly associated with the risk of breast cancer across both stages and in combined analysis. Both RAD21 and MGMT are well-studied genes with significant roles in carcinogenesis. The RAD21 protein is involved in double-strand breaks repair as well as chromatid cohesion during mitosis.37, 38 Intronic polymorphisms in RAD21 gene have been associated with breast cancer in high-risk population.39 Similarly, MGMT repairs the alkylated guanine due to carcinogenic effects induced by alkylating agents.40 Coding SNPs of MGMT gene are reported to be associated with breast cancer risk.41 MGMT SNP reported in this study is ∼454 kb upstream of the MGMT gene. Although rs13250873 and rs1556459 were not located in the gene regions, further replication of these findings and fine mapping of these loci are required to determine whether the identified polymorphisms exert their action through regulation of the nearby RAD21 and MGMT genes.
None of the associations reached genome-wide significance level in this two-stage association study with the combined sample size of 1480 cases and 1635 controls. However, confidence in the reported associations stems from the stringent quality control parameters employed (>98% SNP and sample call rates, HWE P>0.001 in controls and >98% SNP concordance in replicates and good call rate concordance across platforms). Furthermore, the low FDR values and results from permutation testing should favor considering the reported polymorphisms for replication in independent studies. In summary, we identified additional breast cancer susceptibility loci in Caucasian women by focusing on genes related to DNA repair, modifications and metabolism. Our study supports the concept of investigating moderate association signals from stage 1 GWAS using a candidate gene approach restricted to specific pathway-related gene polymorphisms. In this study, we did not consider all related DNA repair/modifications/metabolism pathway gene polymorphisms or their potential associations with other subtypes of breast cancer (basal, HER2+ and luminal B) due to limitations in sample size. Other reported DNA repair/modifications/metabolism gene polymorphisms (which did not reach genome-wide significance) in previously published studies, if replicated in independent cohorts, should also be considered along with the six reported variants here as putative candidates for epistatic models to gain insights to the missing heritability of sporadic breast cancer.
Acknowledgments
We thank Kathryn Calder, Adrian Driga, Jennifer Dufour, Diana Carandang, and Lillian Cook for assistance and technical help. We acknowledge Dr Yutaka Yasui for critical reading of the manuscript. The PolyomX Program and the CBCF-Tumor Bank were funded by the Alberta Cancer Foundation and Alberta Cancer Prevention and Legacy Fund managed by Alberta Innovates-Health Solutions; and the Canadian Breast Cancer Foundation – Prairies/NWT Region, respectively. Funding support for this project was provided by Alberta Cancer Research Institute (ACRI), Alberta Cancer Board (ACB) operating grants to SD and an operating grant from the Canadian Breast Cancer Foundation – Prairies/NWT Region to SD and JRM. PR is supported by the Canadian Partnership against Cancer and the Alberta Cancer Foundation for the Tomorrow Project. We thank the anonymous reviewers for their suggestions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Byrne C, Brinton LA, Haile RW, Schairer C. Heterogeneity of the effect of family history on breast cancer risk. Epidemiology. 1991;2:276–284. doi: 10.1097/00001648-199107000-00007. [DOI] [PubMed] [Google Scholar]
- Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–875. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- CHEK2 Breast Cancer Case-Control Consortium CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10 860 breast cancer cases and 9065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Easton DF. How many more breast cancer predisposition genes are there. Breast Cancer Res. 1999;1:14–17. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Thomas G, Ghoussaini M, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. Common disease, multiple rare (and distant) variants. PLoS Biol. 2010;8:e1000293. doi: 10.1371/journal.pbio.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu SA, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28:207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. doi: 10.1086/344780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SH, Chernoff H, Cong L, Ding Y, Zheng T. Discovering interactions among BRCA1 and other candidate genes associated with sporadic breast cancer. Proc Natl Acad Sci USA. 2008;105:12387–12392. doi: 10.1073/pnas.0805242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musani SK, Shriner D, Liu N, et al. Detection of gene x gene interactions in genome-wide association studies of human population data. Hum Hered. 2007;63:67–84. doi: 10.1159/000099179. [DOI] [PubMed] [Google Scholar]
- Smith TR, Levine EA, Perrier ND, et al. DNA-repair genetic polymorphisms and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:1200–1204. [PubMed] [Google Scholar]
- Cunningham JM, Vierkant RA, Sellers TA, et al. Cell cycle genes and ovarian cancer susceptibility: a tagSNP analysis. Br J Cancer. 2009;101:1461–1468. doi: 10.1038/sj.bjc.6605284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrawat B, Sridharan M, Ghosh S, et al. Potential novel candidate polymorphisms identified in genome-wide association study for breast cancer susceptibility. Hum Genet. 2011;130:529–537. doi: 10.1007/s00439-011-0973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PolyomX 2001 and CBCF-TB 2005 , http://www.abtumorbank.com/?about .
- Tomorrow project 2001 , http://www.albertahealthservices.ca/ tomorrowproject.asp .
- Golden helix, inc.bozeman, MT, USA HelixTree® software. http://www.goldenhelix.com .
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Mavaddat N, Dunning AM, Ponder BA, Easton DF, Pharoah PD. Common genetic variation in candidate genes and susceptibility to subtypes of breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:255–259. doi: 10.1158/1055-9965.EPI-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L, Lacey JV., Jr Receptors, associations, and risk factor differences by breast cancer subtypes: positive or negative. J Natl Cancer Inst. 2011;103:451–453. doi: 10.1093/jnci/djr046. [DOI] [PubMed] [Google Scholar]
- Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. BMC Genet. 2008;9:36. doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem Soc Trans. 2005;33:1537–1540. doi: 10.1042/BST0331537. [DOI] [PubMed] [Google Scholar]
- Tatematsu KI, Yamazaki T, Ishikawa F. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells. 2000;5:677–688. doi: 10.1046/j.1365-2443.2000.00359.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Brown HN, Zhang Y, Holford TR, Zheng T. Genotypes and haplotypes of the methyl-CpG-binding domain 2 modify breast cancer risk dependent upon menopausal status. Breast Cancer Res. 2005;7:R745–R752. doi: 10.1186/bcr1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Zeghouf M, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay MJ, Troelstra C, van der Spek P, et al. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsusaka T, Morrison C, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell. 2001;1:759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- Sehl ME, Langer LR, Papp JC, et al. Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clin Cancer Res. 2009;15:2192–2203. doi: 10.1158/1078-0432.CCR-08-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Han J, Tranah GJ, Hankinson SE, Samson LD, Hunter DJ. Polymorphisms in O6-methylguanine DNA methyltransferase and breast cancer risk. Pharmacogenet Genomics. 2006;16:469–474. doi: 10.1097/01.fpc.0000215065.21718.4c. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.