Abstract
Plants constantly survey the surrounding environment using several sets of photoreceptors. They can sense changes in the quantity (=intensity) and quality (=wavelength) of light and use this information to adjust their physiological responses, growth, and developmental patterns. In addition to the classical photoreceptors, such as phytochromes, cryptochromes, and phototropins, ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), and LOV KELCH PROTEIN 2 (LKP2) proteins have been recently identified as blue-light photoreceptors that are important for regulation of the circadian clock and photoperiodic flowering. The ZTL/FKF1/LKP2 protein family possesses a unique combination of domains: a blue-light-absorbing LOV (Light, Oxygen, or Voltage) domain along with domains involved in protein degradation. Here, we summarize recent advances in our understanding of the function of the Arabidopsis ZTL/FKF1/LKP2 proteins. We summarize the distinct photochemical properties of their LOV domains and discuss the molecular mechanisms by which the ZTL/FKF1/LKP2 proteins regulate the circadian clock and photoperiodic flowering by controlling blue-light-dependent protein degradation.
Keywords: blue light, LOV domain, ZTL, FKF1, LKP2, photoperiodic flowering, circadian clock, Arabidopsis
INTRODUCTION
Plants have developed many sensor mechanisms to monitor various changes in the surrounding environment. One of the most crucial environmental factors for plants is light. Plants use light not only as a primary energy source for photosynthesis, but also as a way to judge changes in their surroundings. Most plants possess multiple sets of photoreceptors that cover a broad range of wavelengths (=colors) as well as intensities of light. These photoreceptors enable plants to accurately survey ambient light conditions (Chen et al., 2004) and adjust their development, morphology, and metabolic rates to the specific environment in which they live. Plants have acquired three major classes of photoreceptor molecules: a red-/far-red-light-reversible photoreceptor called phytochrome (Franklin and Quail, 2010), and two types of blue-light photoreceptors, cryptochrome (Chaves et al., 2011) and phototropin (Christie, 2007). Phytochrome and cryptochrome signals coordinate to regulate various developmental processes throughout the plant’s life, such as seed germination, hypocotyl elongation, greening, and flowering (Chen et al., 2004). In contrast, phototropin signals mainly control directional movement (Christie, 2007).
In Arabidopsis thaliana, there are two phototropins, phot1 and phot2, that share partially overlapping functions. Both phototropins regulate phototropism (Sakai et al., 2001), chloroplast relocation movement (Kagawa et al., 2001; Sakai et al., 2001), light-induced stomatal opening (Kinoshita et al., 2001), cotyledon and leaf expansion (Sakamoto and Briggs, 2002; Takemiya et al., 2005), and hypocotyl growth (Folta and Spalding, 2001). Phototropins have two photosensory domains called Light, Oxygen, or Voltage (LOV) domains in the N-terminal half and a Ser/Thr kinase at the C-terminal half (Christie et al., 1998). Through the LOV domains, blue light regulates the kinase activity of phototropins and blue-light-dependent phosphorylation is an important initial process of phototropin signaling (Pedmale and Liscum, 2007; Christie et al., 2011). In addition to the phototropins, Arabidopsis has two additional types of protein that possess LOV domains (Figure 1A). One such family comprises three proteins that each possess a single LOV domain: ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), and LOV KELCH PROTEIN 2 (LKP2) (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001) (Figure 1A). The ZTL/FKF1/LKP2 proteins play roles in the circadian clock and photoperiodic flowering (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). The other type of protein is referred to as PAS/LOV protein (PLP) (also called LOV/LOV protein (LLP)), which contains two LOV domains, although the physiological function of PLP is largely unknown (Ogura et al., 2008; Kasahara et al., 2010) (Figure 1A).
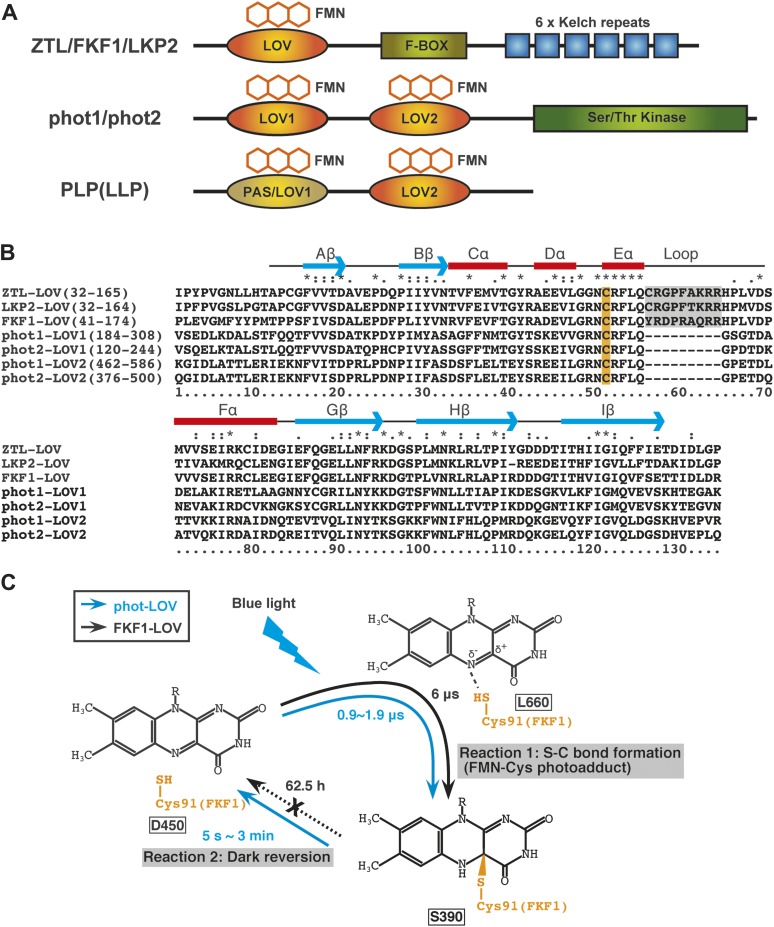
Figure 1.
The Domain Structures of ZTL/FKF1/LKP2 and Related Proteins, Amino Acid Sequence Alignments of LOV Domains, and Photochemical Properties of Phototropin and FKF1 LOV Domains.
(A) Schematic illustration of functional domains of ZTL/FKF1/LKP2 family proteins, phototropin proteins, and PAS/LOV proteins. LOV domain bound to an FMN molecule functions as a blue-light-sensing domain. The ZTL/FKF1/LKP2 family proteins possess one LOV domain at the N-terminus region followed by an F-box domain and six Kelch repeats in the C-terminal region. The phototropins contain two FMN-binding LOV domains in their N-terminal region (LOV1 and LOV2) and a serine/threonine kinase domain at the C-terminus. The PAS/LOV proteins (PLP or also called LLP) contain two LOV domains; however, the cysteine residue essential for the cysteinyl adduct formation within the first LOV domain is not always conserved in some plant species, including Arabidopsis (Kasahara et al., 2010), indicating that there is no light-induced photocycle of the LOV domain. The LOV2 domains of PLP proteins usually contain the conserved cysteine and show the blue-light-induced photocycle in vitro.
(B) Sequence alignment of several LOV domains. The alignment includes Arabidopsis thaliana ZTL, LKP2, FKF1, phot1-LOV1, phot1-LOV2, phot2-LOV1, and phot2-LOV2. Identical and similar amino acids are marked on top of the alignment by asterisks (*) and (: or .), respectively. The conserved cysteines for FMN binding are highlighted in orange. The characteristic loop regions of ZTL/FKF1/LKP2 LOV are gray-shaded. The predicted secondary structure elements are shown on top of the alignment. Arrows and boxes indicate β-strands and α-helices, respectively.
(C) Schematic representation of LOV-domain photochemistry. In darkness, the FMN chromophore is non-covalently bound in the LOV domain (L450). Light triggers the production of a reactive triplet-state flavin (L660) that leads to formation of a covalent bond between the FMN and a conserved cysteine residue in the LOV domain (S390). The photoreaction process of the phot-LOV protein (right blue arrow circuit) is fully reversible in the dark. In contrast, the FKF1–LOV protein (a black dotted arrow) shows a slow dark reversion rate. The dark reversion rate (half-lives; t1/2) of each protein is shown.
Based on amino acid sequence similarities between the ZTL/FKF1/LKP2 LOV domains and the phototropin LOV domains, it was proposed that the ZTL/FKF1/LKP2 proteins may be blue-light photoreceptors as well (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). Since then, we have acquired ample photochemical evidence of the blue-light-absorbing properties and blue-light-specific functions of the ZTL/FKF1/LKP2 LOV domains. It is now clear that the ZTL/FKF1/LKP2 proteins fulfill the following criteria for being photoreceptors: (1) they possess chromophores required for light absorption, (2) light absorption induces functional changes at the molecular level, and (3) they regulate light-dependent responses.
All ZTL, FKF1, and LKP2 proteins possess two additional functional domains: an F-box domain and a Kelch repeat domain (Figure 1A). The combination of these domains suggests that they are involved in protein stability regulation (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). Recent results indicate that these proteins function as E3 ubiquitin ligases and mediate proteasome-dependent protein degradation in a light-dependent manner (Más et al., 2003; Imaizumi et al., 2005; Kim et al., 2007; Sawa et al., 2007). They are involved in regulation of the circadian clock and day-length-dependent flowering by controlling accumulation of key regulator proteins in the clock and flowering pathway (Más et al., 2003; Kiba et al., 2007; Fornara et al., 2009). Here, we summarize recent advances in our understanding of the molecular properties of the LOV domains of ZTL, FKF1, and LKP2 proteins and discuss the mechanisms by which these three proteins control the photoperiodic flowering pathway and the circadian clock in a light-dependent manner.
STRUCTURE AND PHOTOCHEMICAL PROPERTIES OF THE LIGHT-SENSING FKF1 LOV DOMAIN
The crystal structures and photochemical properties of the LOV domains have been well characterized using phototropin LOV domains (Christie, 2007; Matsuoka et al., 2007; Tokutomi et al., 2008). Although there are fewer papers describing the ZTL/FKF1/LKP2 LOV domains, our knowledge of the photochemical reactions of the ZTL/FKF1/LKP2 LOV domains and potential structural changes has been expanding, mainly using the FKF1 LOV domain (Imaizumi et al., 2003; Nakasako et al., 2005; Zikihara et al., 2006; Kikuchi et al., 2009; Nakasone et al., 2010). The FKF1 LOV domain has features conserved in all LOV domains as well as properties unique from the phot-LOV domains. This unique property of the LOV domain may be beneficial for the light-dependent degradation of target proteins, since FKF1 functions as an E3 ubiquitin ligase.
The LOV domain is a small photo-sensing module that belongs to a subclass of the Per-ARNT-Sim (PAS) domain super-family with members that possess diverse functions as versatile sensors (Taylor and Zhulin, 1999). In addition to the recent discovery of the blue-light-activated LOV-domain-containing histidine-kinases in various prokaryotes (Swartz et al., 2007), LOV domains are found in blue-light-sensing proteins of organisms ranging from archaea to eukaryotes. In plants, the LOV domains generally non-covalently bind a flavin cofactor, flavin mononucleotide (FMN), as a chromophore (Taylor and Zhulin, 1999). The core of the LOV domain consists of four α-helixes and five β-sheets. According to recently proposed nomenclature (Harper et al., 2003), the following secondary structural elements for the LOV domain core have been assigned: Aβ-Bβ-Cα-Dα-Eα-Fα-Gβ-Hβ-Iβ (Figure 1B).
When blue light is perceived by the phototropin LOV domain, it undergoes a unique photochemical reaction cycle (Figure 1C) (Kasahara et al., 2002). The ground state of the FMN in the LOV domains, called D450, shows an absorption spectrum typical of flavin with an absorption maximum of around 450 nm. The D450 is elevated to a singlet-excited state by blue light and then inter-converted to a triplet-excited state, called L660 (Figure 1C). A stable adduct then forms between the FMN chromophore and a cysteine conserved within the LOV domains localized in the Eα helix (Figure 1B), converting the LOV domain to the S390 state that has an absorption maximum of around 390 nm (Reaction 1, Figure 1C) (Salomon et al., 2000; Swartz et al., 2001). This is the so-called FMN–Cys photoadduct, and it occurs in 0.9–1.9 μs—almost instantly—after the FMN absorbs light energy. Reversion of S390 back to D450 also occurs rapidly, with time constants from several seconds to a few minutes, depending on the LOV domain (Reaction 2, Figure 1C) (Kasahara et al., 2002). Rapid reversion to D450 is surprising, because it means that the covalent bond between cysteine and FMN is easily broken in the phototropin LOV domain, requiring relatively higher energy to do so. Thus, FMN in the phototropin LOV domain can be repeatedly activated by blue-light exposure.
Recombinant FKF1, ZTL, and LKP2 LOV domains all bind the FMN chromophore and undergo a light-induced photochemical reaction similar to that of the phototropin LOV domain in vitro (Salomon et al., 2000; Corchnoy et al., 2003; Imaizumi et al., 2003). The absorption spectrum of the FKF1 LOV domain is also similar, showing a typical flavoprotein spectrum with a peak maximum at 450 nm. After light irradiation, absorption of around 450 nm decreases, while absorption of around 390nm increases, indicating formation of the S390 state. Recently, FMN–Cys adduct formation of FKF1 LOV polypeptides was characterized using the pulsed laser-induced transient grating (TG) method and the D450 to S390 change took place with a time constant of 6 μs (Nakasone et al., 2010). This is slow relative to phot1–LOV2 and phot2–LOV2 (1.9 and 0.9 μs, respectively) (Figure 1C) (Eitoku et al., 2005; Nakasone et al., 2006). Substitution of the conserved cysteine to alanine (C91A) abolished FMN–Cys photoadduct formation, although binding of FMN to the FKF1 LOV domain was unaffected. Absorption of D450 was similar to that of the non-mutated LOV domain (Imaizumi et al., 2003).
The FKF1 and phototropin LOV domains differ most profoundly in their rates of dark reversion. In contrast to the phototropin LOV domains, the FKF1 LOV domain does not show appreciable dark recovery in short-term experimental conditions (Imaizumi et al., 2003) (Figure 1C). The FKF1–LOV polypeptides revert from S390 to the D450 ground state with a half-life of 62.5 h at room temperature (Zikihara et al., 2006). This extremely slow dark recovery may be caused by an additional 9-amino acid insertion, which forms a loop structure between the Eα helix near the conserved cysteine residue and the Fα helix (Figure 1B). This insertion is only found in the slow dark-recovery class of LOV domains (Zikihara et al., 2006). Dark-recovery rates using an FKF1–LOV domain lacking the loop region (FKF1–LOV–NL) revealed that losing the loop region accelerates dark recovery up to approximately threefold (20.9 h) (Zikihara et al., 2006). Further, conformational change between dark and light conditions was detected for FKF1–LOV but not for FKF1–LOV–NL (Nakasone et al., 2010). This observation suggests that the loop region of FKF1–LOV is important for conformational changes. However, dark reversion of FKF1–LOV–NL is still significantly slower than that of the phototropin LOV domains, indicating that conformational differences of other residues surrounding the chromophore also affect the stability of the light-adapted state. A series of site-directed mutagenesis studies in LOV domains from several organisms supports this notion (Christie et al., 2007; Yamamoto et al., 2008; Jentzsch et al., 2009; Zoltowski et al., 2009). Most FKF1 protein synthesized within a day disappears by the end of the day (Imaizumi et al., 2003). It seems then that, once a conformational change of FKF1 is triggered by blue-light absorption, FKF1 remains in its light-activated form until it is degraded. Of course, we cannot exclude the possibility that the full length of FKF1 has a different dark-recovery rate in vivo.
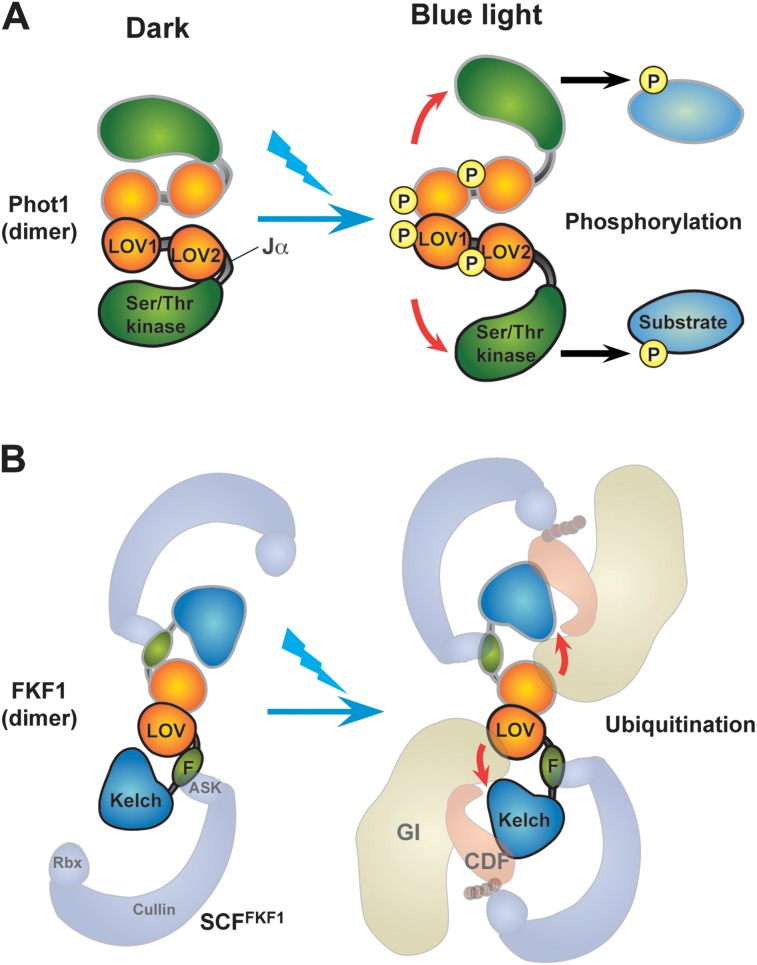
Phototropins have two tandemly aligned LOV domains. In vitro, LOV1 homodimerizes regardless of light conditions (Figure 2A) (Salomon et al., 2004; Eitoku et al., 2007), indicating that the LOV1 domain functions as a dimerization site. The LOV2 domain interacts with its neighboring α-helix linker region (Jα) in the dark but dissociates upon illumination and plays a key role in regulating C-terminal kinase activity (Figure 2A) (Eitoku et al., 2005, 2007; Nakasone et al., 2007). Functionally, LOV2 is more important for photoreceptor function than LOV1 in plants (Christie et al., 2002). The FKF1 LOV domain forms a stable dimer with an anti-parallel configuration regardless of the light conditions (Figure 2B) (Nakasako et al., 2005; Nakasone et al., 2010). Since FKF1 has only one LOV domain, the FKF1 LOV domain seems to have two functions. One role is interdomain interactions including homo- or hetero-dimerization among ZTL/FKF1/LKP2 members (Yasuhara et al., 2004), and the other is signal transduction activity caused by blue-light-induced conformational changes around the loop region (Figures 1 and 2B).
Figure 2.
Tentative Structure Changes of FKF1 Triggered by Blue-Light Absorption.
(A) Proposed structural changes of phot1. Two phot1 molecules dimerize through their LOV1 domains. The LOV2 domain is important for light-dependent function. Once phot1 absorbs blue light, the Jα-helix located adjacent to the LOV2 domain detaches from the LOV2 domain. This structural change is thought to activate the C-terminal kinase. The kinase phosphorylates N-terminal regions as well as substrate proteins to send phosphorylation signals.
(B) FKF1 also forms at least a homodimer with anti-parallel configuration in vitro. In the dark, FKF1 may be incorporated in the SCFFKF1 complex (which consists of FKF1 F-box protein, ASK, Cullin, and Rbx). After blue-light exposure, FKF1 interacts with GI through the LOV domain. Light may cause a similar structural change to phot1 to enable FKF1 to bind to GI. GI also binds to CDF1, a substrate of FKF1 for ubiquitination-dependent protein degradation. FKF1 binds to CDF1 through the Kelch repeat domain and is involved in protein degradation.
OTHER DOMAIN STRUCTURES IN ZTL/FKF1/LKP2 PROTEINS
In addition to the LOV domain, ZTL/FKF1/LKP2 proteins possess two other functional domains: F-box and Kelch repeat. An F-box protein is a component of the SKP–Cullin–Rbx–F-box (SCF) complex (Figure 2B). The F-box motif of the ZTL family of proteins interacts with Arabidopsis SKP1-like (ASK) proteins, indicating the formation of SCF E3 ubiquitin ligases in vivo (Han et al., 2004; Yasuhara et al., 2004). The Kelch repeat domain forms a β-propeller structure and functions as a protein–protein interacting domain that binds substrates for ubiquitin-mediated protein degradation (Andrade et al., 2001). These domain structures clearly indicate that the ZTL/FKF1/LKP2 proteins mediate ubiquitin-dependent protein degradation, possibly in a light-dependent manner. Since these proteins share high degrees of amino-acid sequence homologies (70–80% identities throughout the entire protein) (Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001), it was predicted that these proteins may have overlapping functions and may even degrade the same target proteins. Based on loss-of-function mutant phenotypes, at least ZTL and FKF1 play different roles in the circadian clock and photoperiodic flowering regulation (Nelson et al., 2000; Somers et al., 2000). These results indicate that ZTL and FKF1 must have different targets for degradation as well. In the following sections, we summarize our current understanding of the molecular roles of light-absorbing domains and the protein degradation function of the ZTL/FKF1/LKP2 protein family in the regulation of photoperiodic flowering and circadian clock oscillation.
FUNCTIONS OF ZTL/FKF1/LKP2 PROTEINS IN THE REGULATION OF PHOTOPERIODIC FLOWERING
Plants use photoperiod-sensing mechanisms in order to control timing of seasonal flowering to maximize their reproductive success. In Arabidopsis, expression of the FLOWERING LOCUS T (FT) gene regulated by CONSTANS (CO) protein is a crucial aspect of photoperiodic flowering (Suárez-López et al., 2001; Valverde et al., 2004; Abe et al., 2005; Wigge et al., 2005; Sawa et al., 2007). Mutations in CO and FT genes cause a strong delay in flowering under inductive long-day (LD) conditions, whereas overexpression of CO and FT strongly accelerates flowering regardless of day length (Samach et al., 2000). The CO/FT module is highly conserved not only in LD plants such as Arabidopsis, wheat, and barley, but also in short-day (SD) plants such as rice (Song et al., 2010). Therefore, studies of the regulation of the Arabidopsis CO/FT module may contribute to our understanding of the general mechanism of seasonal flowering in plants.
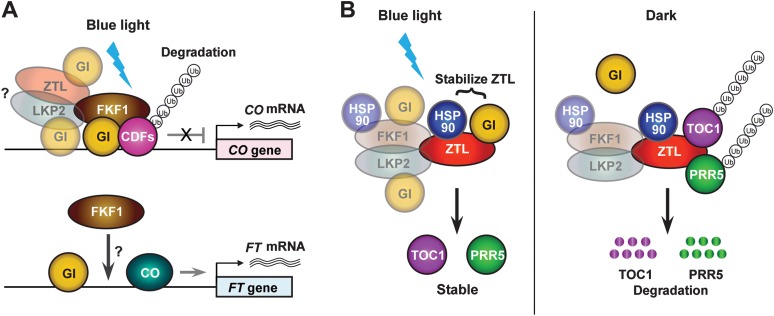
All three LOV-containing F-box proteins (ZTL, FKF1, and LKP2) are involved in the control of flowering time through the regulation of the CO/FT module (Imaizumi et al., 2003; Somers et al., 2004; Takase et al., 2011). Three functional domains in the ZTL/FKF1/LKP2 proteins are important for their roles in flowering and the circadian clock—especially light-activated LOV domains, which determine characteristic features of the proteins (Kim et al., 2007; Sawa et al., 2007). The function of FKF1 in flowering regulation is the most characterized among the three F-box members. A mutation in the FKF1 gene strongly delays flowering under LD conditions (Nelson et al., 2000; Imaizumi et al., 2003). The FKF1 Kelch repeat domain interacts with CYCLING DOF FACTOR (CDF) transcriptional repressors for poly-ubiquitination-dependent degradation (Imaizumi et al., 2005) (Figure 3A). The CDF1 protein represses transcription of the CO gene by direct binding to Dof binding sites in the CO promoter (Imaizumi et al., 2005). GIGANTEA (GI) is a large nuclear protein and a positive regulator of CO and FT gene expression (Fowler et al., 1999; Huq et al., 2000). GI also physically interacts with CDF1 on the CO promoter (Sawa et al., 2007). When the FKF1 LOV domain absorbs blue light, FKF1 interacts with GI through its LOV domain to form a protein complex in the late afternoon under LD conditions (Sawa et al., 2007). Then, FKF1 in the FKF1–GI complex degrades the CDF1 protein on the CO promoter, resulting in activation of CO transcription at the end of the day (Figure 3A). In contrast, the timing of FKF1 and GI expression is out of phase under SD conditions and FKF1 is mainly expressed in the dark. Thus, little of the FKF1–GI complex forms in light under SD conditions causing a low abundance of CO mRNA during the day (Sawa et al., 2007). CO protein is stabilized at the end of the day in LD by phytochrome A and cryptochrome photoreceptor signaling. This time- and day-length-dependent stabilization of CO is thought to be important for FT induction in LD (Valverde et al., 2004). Therefore, an alignment of CO expression with the timing of CO protein stabilization is a crucial component of this pathway. These findings establish the importance of FKF1 in a day-length measurement mechanism through regulation of the timing of daytime CO expression.
Figure 3.
Light-Mediated Proteolysis Controlled by ZTL, FKF1, and LKP2 in the Regulation of Flowering Time and the Circadian Clock.
(A) The function of FKF1 in the control of flowering time. GI forms protein complexes with CDFs on the CO promoter. In the afternoon, FKF1 is recruited to the CO promoter region through blue-light-dependent interaction with GI. FKF1 degrades CDFs, facilitating expression of CO. Since CDF2 is stabilized further in the ztl fkf1 lkp2 triple mutants than in the fkf1 mutants, the ZTL–GI and LKP2–GI complexes may also contribute to the degradation of some CDF proteins. CO and GI bind to the FT promoter and activate FT transcription, which induces flowering. FKF1 may also bind to the FT promoter; however, the function of this interaction is unknown.
(B) The function of ZTL in regulation of the circadian clock. Under blue light, ZTL interacts with GI. ZTL also forms a protein complex with HSP90. The interaction of ZTL with GI and HSP90 stabilizes ZTL protein. The ZTL–GI complex formation may sequester ZTL from interaction with TOC1 or PRR5. This enables TOC1 and PRR5 proteins to accumulate in the late afternoon. The FKF1–GI and LKP2–GI complex may also have a similar role (left side). In the dark, ZTL (FKF1 and LKP2) interact with TOC1 and PRR5 proteins and degrade them through a proteasome pathway (right side).
In addition to CO transcriptional regulation, a recent study predicts another role for FKF1 in photoperiodic flowering. Using a computational model for the photoperiodic gene circuit, Salazar et al. (2009) predicted that FKF1 may control FT expression in addition to the role of CO transcriptional activation (Salazar et al., 2009). In their simulation model, the current molecular mechanism by which FKF1 induces FT through activation of CO transcription cannot entirely explain the low levels of FT mRNA in fkf1 mutant plants; therefore, FKF1 may influence FT expression directly. This prediction is supported by the evidence that FKF1 associates with FT chromatin in vivo (Sawa and Kay, 2011). GI interacts with the FT repressors (SHORT VEGETATIVE PHASE (SVP), TEMPRANILLO (TEM) 1, and TEM2), which directly bind to the FT promoter regions where GI exists (Sawa and Kay, 2011). Although FKF1 and GI bind to similar FT promoter regions, FKF1 may not be involved in target degradation of these proteins, since no interaction between FKF1 and these FT repressors was observed (Sawa and Kay, 2011). This suggests that GI has an additional role in the activation of FT expression through complexes with the FT repressors, although the functions of the complexes remain to be revealed. The potential role of FKF1 in FT expression, if any, has yet to be addressed (Figure 3A).
The ZTL and LKP2 proteins are also involved in control of flowering time and CO expression; however, unlike FKF1, ZTL and LKP2 do not seem to be simple positive regulators of CO. A ztl mutant shows a weak early-flowering phenotype under SD conditions (Somers et al., 2004; Takase et al., 2011). An lkp2 mutation enhances the phenotype of the ztl mutant under both LD and SD conditions, even though the lkp2 mutant by itself has little effect on the regulation of flowering (Takase et al., 2011). Introducing ztl and lkp2 mutations into an fkf1 background further reduces the CO expression level (Fornara et al., 2009). One explanation for the ztl fkf1 lkp2 triple mutant phenotype is that FKF1, ZTL, and LKP2 are all involved in protein stability regulation of the CO repressor, CDF2, and possibly other CDFs (Figure 3A). Introduction of ztl and lkp2 mutations to the fkf1 mutant further increases the abundance of CDF2 protein, which may lead to lower levels of CO transcript in the mutants (Fornara et al., 2009).
Interestingly, overexpression of ZTL or LKP2 also down-regulates CO transcription, which results in a late-flowering phenotype similar to the ztl fkf1 lkp2 triple mutant under LD conditions (Schultz et al., 2001; Somers et al., 2004). The severely depressed CO expression levels in ZTL and LKP2 overexpressors also resemble that of the gi mutant. Since ZTL and LKP2 are mainly expressed in the cytosol (Kim et al., 2007; Takase et al., 2011), one possible explanation for the flowering phenotype of the ZTL/LKP2 overexpressor is that overexpression of ZTL or LKP2 may sequester GI in the cytosol by forming a ZTL(/LKP2)–GI complex. This may reduce the amount of GI–FKF1 complex in the nucleus, causing stabilization of the CDF CO repressors.
A second possibility is that overexpression of ZTL and LKP2 may enhance degradation of their substrates, TIMING OF CAB EXPRESSION 1 (TOC1) and PSEUDO RESPONSE REGULATOR 5 (PRR5). Both TOC1 and PRR5 are core clock components and the degradation of these clock proteins by the ZTL family is an important regulation in the circadian clock (Más et al., 2003; Kiba et al., 2007) (see details in the next section). Both TOC1 and PRR5 levels might be very low in the ZTL/LKP2 overexpressors. Coincidentally, the toc1 prr5 double mutant flowering phenotype resembles the late-flowering phenotypes of the ZTL and LKP2 overexpressors (Ito et al., 2008). In addition, both TOC1 and PRR5 indirectly affect expression of CO (Yanovsky and Kay, 2001; Nakamichi et al., 2007) and overexpression of PRR5 represses CDF1 transcription (Nakamichi et al., 2007), indicating that ZTL/LKP2 may regulate CO transcription through the functions of TOC1 and/or PRR5. Interestingly, each toc1 and prr5 single mutant phenotype (the early- and late-flowering phenotypes, respectively) is different from the ZTL/LKP2 overexpressor phenotypes (Nakamichi et al., 2007; Niwa et al., 2007). This suggests that both TOC1 and PRR5 levels should be low in the ZTL/LKP2 overexpressors in order to explain their flowering phenotype by TOC1 and PRR5 flowering function.
Recently, Takase et al. (2011) reported yet another possible explanation for the ZTL and LKP2 overexpression phenotype. The Kelch repeat domains in ZTL and LKP2 interact with FKF1 in yeast and in vitro (Takase et al., 2011). The authors showed that ZTL and LKP2 exclude FKF1 from the nucleus in Arabidopsis protoplast. In addition, overexpression of the LKP2 Kelch repeats is sufficient to reduce CO and FT expression under LD conditions, leading to late flowering. Since repression of CO expression in fkf1 mutants is less severe than that in ZTL and LKP2 overexpressors, it is difficult to explain the phenotype of the overexpressors by this mechanism alone. Thus, several mechanisms may regulate CO expression in ZTL and LKP2 overexpressors.
Since loss of function of the entire ZTL family and overexpression of ZTL and LKP2 both cause CO mRNA levels to be low throughout the day, the mechanisms of ZTL and LKP2-dependent CO regulation could be indirect and achieved by several different mechanisms. These results indicate that specific stoichiometries of ZTL, LKP2, FKF1, and possibly GI may be important for balancing the proper ratio for the formation of each complex, each of which has a different function, and regulating this balance may be crucial for achieving the proper expression of CO.
REGULATION OF THE CIRCADIAN CLOCK BY ZTL/FKF1/LKP2 PROTEINS
Similarly to photoperiodic flowering regulation, all ZTL/FKF1/LKP2 proteins are involved in the regulation of the circadian clock in Arabidopsis as well. However, among these, protein turnover of clock components mediated by ZTL is a principal mechanism for ZTL/FKF1/LKP2-dependent progress of the circadian clock. The ztl mutants exhibit a longer period phenotype under constant light conditions (Somers et al., 2000) and this long-period phenotype is mainly caused by the increased stability of the TOC1 core clock protein (Más et al., 2003) (Figure 3B). The ZTL protein also targets the PRR5 core clock protein for proteasome-dependent degradation through SCFZTL (Kiba et al., 2007) (Figure 3B). The ZTL LOV domain plays a crucial role in the degradation of TOC1 and PRR5 proteins (Más et al., 2003; Kiba et al., 2007). Despite amino acid sequence similarities with TOC1 and PRR5, the other PRRs (PRR3, PRR7, and PRR9), all of which are involved in circadian clock progression, are not targets of ZTL for degradation (Fujiwara et al., 2008). As with FKF1, blue light absorbed by the ZTL LOV domain enables ZTL to form a protein complex with GI during the day (Kim et al., 2007). As GI protein abundance robustly oscillates with the peaks in the afternoon, the ZTL–GI complex reaches its maximum quantity in the afternoon (Kim et al., 2007). This interaction stabilizes ZTL protein; therefore, ZTL protein is highly stable in the afternoon (Kim et al., 2007). Fujiwara et al. (2008) have proposed a possible mechanism in which the light-dependent ZTL–GI interaction separates ZTL protein from TOC1 and PRR5 and consequently protects both TOC1 and PRR5 from ZTL-dependent degradation from the active SCFZTL complex in the afternoon (Fujiwara et al., 2008).
In addition to the GI-dependent stabilization of ZTL, Kim et al. (2011) reported another mechanism by which HEAT SHOCK PROTEIN 90 (HSP90) affects ZTL protein stability. HSP90 functions as a molecular chaperone and binds to ZTL to facilitate maturation of the ZTL protein (Kim et al., 2011). This interaction affects TOC1, PRR5, and ZTL stabilities in a way that is not light-dependent. Reduction in HSP90 activity by geldanamycin (HSP90 inhibitor) treatment reduces ZTL accumulation and simultaneously increases TOC1 and PRR5 levels (Kim et al., 2011). It seems that GI and HSP90 function in the same ZTL-stabilization mechanism, because reduced HSP90 activity does not further decrease ZTL levels in the gi mutants. In addition, FKF1 protein levels are reduced when HSP90 activity is lower, indicating that the same mechanism may stabilize FKF1 and possibly LKP2 (Figure 3B).
Unlike the ztl mutants, lkp2 and fkf1 single mutants do not exhibit an obvious long-period clock phenotype (Baudry et al., 2010). Using all possible combinations of ztl, fkf1, lkp2 double and triple mutants, Baudry et al. (2010) described the overlapping roles of FKF1 and LKP2 with ZTL in the circadian clock. The fkf1 mutation enhances the longer period phenotype of the ztl mutant when these two mutations are combined. In addition, compared to the ztl single mutant, adding the lkp2 and fkf1 mutations to the ztl mutant background reduces expression of morning clock genes, such as LATE ELONGATED HYPOCOTYL (LHY) and PRR9 (Baudry et al., 2010). This is most likely due to increased levels of TOC1 and PRR5 stability. Indeed, significant stabilization of PRR5 and TOC1 proteins in the ztl lkp2, ztl fkf1 double, and ztl lkp2 fkf1 triple mutants was reported (Baudry et al., 2010; Wang et al., 2010). These data indicate that, together with ZTL, both LKP2 and FKF1 may also contribute to the ubiquitin-dependent degradation of TOC1 and PRR5 (Figure 3B). It is noteworthy that the ZTL promoter-driven ZTL and LKP2, but not FKF1, rescued the ztl circadian phenotype (Baudry et al., 2010), indicating that the molecular mechanisms by which ZTL and FKF1 regulate the stabilities of the circadian clocks might not be simply comparable.
FUTURE PERSPECTIVES
Knowledge of the molecular function of ZTL/FKF1/LKP2 proteins has accumulated in the past decade. In the area of photochemistry, only the FKF1 LOV domain has been analyzed during recent years. The ZTL LOV and LKP2 LOV domains should be analyzed as well. It will be interesting to determine whether various combinations of LOV dimers have similar photochemical properties, or whether unique pairings serve unique functions. Although the crystal structures of the LOV, F-box, and Kelch repeat domains within the ZTL/FKF1/LKP2 protein family are known, we do not understand how these domains are spatially localized within a molecule and whether there are intra-molecular interactions between domains. Analysis of the blue-light-induced photocycle using the ZTL/FKF1/LKP2 recombinant proteins containing not only the LOV domain but also other domains may facilitate further understanding of the molecular properties of the ZTL/FKF1/LKP2 protein family.
The ZTL/FKF1/LKP2 proteins seem to possess both overlapping and contrasting roles in the circadian clock and photoperiodic flowering. The contributions of these three proteins to either regulation of period length or degradation of CDF proteins may be partially explained by the absolute amount (=copy number) of protein (Fornara et al., 2009; Baudry et al., 2010). However FKF1 protein cannot rescue the ztl long-period phenotype and the flowering phenotype of ZTL overexpressors is similar to that of fkf1 mutants (Somers et al., 2004; Baudry et al., 2010). These results clearly indicate that ZTL and FKF1 possess distinct functions that are not inter-changeable. In addition, FKF1, LKP2, and ZTL proteins all interact with GI through their LOV domains in planta (Kim et al., 2007). It will be exciting to determine how affinities between each light-activated LOV domain and GI differ, and whether these affinities contribute to their unique functions in regulation of photoperiodic flowering or/and the circadian clock.
In addition, our knowledge of the light-dependent protein–protein interaction facilitated the creation of an in vivo molecular light switch. Blue-light-dependent FKF1–GI interaction was recently utilized in an optogenetic application to regulate gene expression and protein subcellular localization in a light-dependent manner in mammalian cells (Yazawa et al., 2009). This relatively long-lasting molecular switch mediated by the stable FKF1–GI interaction could be a useful tool in cell biology and other fields, in contrast to other, more transient, photoreceptor-based tools (Moglich and Moffat, 2010).
During the last decade, we have learned a lot about the molecular nature and functions of the newly identified light-regulated ZTL, FKF1, and LKP2 proteins. Since the spatial expression patterns of ZTL, FKF1, and LKP2 largely overlap (Kiyosue and Wada, 2000; Nelson et al., 2000; Yasuhara et al., 2004) and their LOV domains form homo- and heterodimers (Takase et al., 2011), the next challenge could be to figure out the potential functional differences of different dimer combinations and whether certain dimers are preferentially formed under certain conditions or in a specific pathway. In addition, it is likely that accessory proteins besides GI and components of the SCF machinery exist that affect their function in vivo. Moreover, detailed analyses of the intercellular distribution of these proteins in vivo would aide deciphering of the role of each protein. Finally, to accurately understand the function of the ZTL/FKF1/LKP2 proteins, we need to describe when and where these proteins and their complexes are formed at the whole plant level as well as specific tissues.
FUNDING
S.I. was supported by the JSPS Postdoctoral Fellowship. Y.H.S. is partly supported by a grant from the Next Generation Biogreen 21 Program (SSAC, PJ008109), Rural Development Administration, Republic of Korea. This work was supported by an NIH grant (GM079712) to T.I.
Acknowledgments
We thank Hannah Kinmonth-Shultz for critical reading of manuscript. No conflict of interest declared.
References
- Abe M, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Gonzalez-Guzman M, Serrano R, Rodriguez PL. A combination of the F-box motif and Kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol. Biol. 2001;46:603–614. doi: 10.1023/a:1010650809272. [DOI] [PubMed] [Google Scholar]
- Baudry A, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Christie JM. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- Christie JM, et al. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Christie JM, et al. Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry. 2007;46:9310–9319. doi: 10.1021/bi700852w. [DOI] [PubMed] [Google Scholar]
- Christie JM, et al. phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 2011;9:e1001076. doi: 10.1371/journal.pbio.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 2003;278:724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- Eitoku T, Nakasone Y, Matsuoka D, Tokutomi S, Terazima M. Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. J. Am. Chem. Soc. 2005;127:13238–13244. doi: 10.1021/ja052523i. [DOI] [PubMed] [Google Scholar]
- Eitoku T, Nakasone Y, Zikihara K, Matsuoka D, Tokutomi S, Terazima M. Photochemical intermediates of Arabidopsis phototropin 2 LOV domains associated with conformational changes. J. Mol. Biol. 2007;371:1290–1303. doi: 10.1016/j.jmb.2007.06.035. [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP. Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 2001;26:471–478. doi: 10.1046/j.1365-313x.2001.01038.x. [DOI] [PubMed] [Google Scholar]
- Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fowler S, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 2004;40:291–301. doi: 10.1111/j.1365-313X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ito S, Niwa Y, Nakamichi N, Kawamura H, Yamashino T, Mizuno T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:201–213. doi: 10.1093/pcp/pcm178. [DOI] [PubMed] [Google Scholar]
- Jentzsch K, et al. Mutual exchange of kinetic properties by extended mutagenesis in two short LOV domain proteins from Pseudomonas putida. Biochemistry. 2009;48:10321–10333. doi: 10.1021/bi901115z. [DOI] [PubMed] [Google Scholar]
- Kagawa T, et al. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- Kasahara M, et al. Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 2002;129:762–773. doi: 10.1104/pp.002410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Torii M, Fujita A, Tainaka K. FMN binding and photochemical properties of plant putative photoreceptors containing two LOV domains, LOV/LOV proteins. J. Biol. Chem. 2010;285:34765–34772. doi: 10.1074/jbc.M110.145367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Unno M, Zikihara K, Tokutomi S, Yamauchi S. Vibrational assignment of the flavin-cysteinyl adduct in a signaling state of the LOV domain in FKF1. J. Phys. Chem. B. 2009;113:2913–2921. doi: 10.1021/jp808399f. [DOI] [PubMed] [Google Scholar]
- Kim TS, et al. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc. Natl Acad. Sci. U S A. 2011;108:16843–16848. doi: 10.1073/pnas.1110406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Wada M. LKP1 (LOV Kelch protein 1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J. 2000;23:807–815. doi: 10.1046/j.1365-313x.2000.00850.x. [DOI] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- Matsuoka D, Iwata T, Zikihara K, Kandori H, Tokutomi S. Primary processes during the light-signal transduction of phototropin. Photochem. Photobiol. 2007;83:122–130. doi: 10.1562/2006-03-29-RA-861. [DOI] [PubMed] [Google Scholar]
- Moglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- Nakasako M, Matsuoka D, Zikihara K, Tokutomi S. Quaternary structure of LOV-domain containing polypeptide of Arabidopsis FKF1 protein. FEBS Lett. 2005;579:1067–1071. doi: 10.1016/j.febslet.2004.12.078. [DOI] [PubMed] [Google Scholar]
- Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Kinetic measurement of transient dimerization and dissociation reactions of Arabidopsis phototropin 1 LOV2 domain. Biophys. J. 2006;91:645–653. doi: 10.1529/biophysj.106.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Dynamics of conformational changes of Arabidopsis phototropin 1 LOV2 with the linker domain. J. Mol. Biol. 2007;367:432–442. doi: 10.1016/j.jmb.2006.12.074. [DOI] [PubMed] [Google Scholar]
- Nakasone Y, Zikihara K, Tokutomi S, Terazima M. Kinetics of conformational changes of the FKF1–LOV domain upon photoexcitation. Biophys. J. 2010;99:3831–3839. doi: 10.1016/j.bpj.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Niwa Y, et al. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1, and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:925–937. doi: 10.1093/pcp/pcm067. [DOI] [PubMed] [Google Scholar]
- Ogura Y, et al. Blue light diminishes interaction of PAS/LOV proteins, putative blue light receptors in Arabidopsis thaliana, with their interacting partners. J. Plant Res. 2008;121:97–105. doi: 10.1007/s10265-007-0118-8. [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- Sakai T, et al. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl Acad. Sci. U S A. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JD, et al. Prediction of photoperiodic regulators from quantitative gene circuit models. Cell. 2009;139:1170–1179. doi: 10.1016/j.cell.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Salomon M, Lempert U, Rudiger W. Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 2004;572:8–10. doi: 10.1016/j.febslet.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Samach A, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 2010;13:594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Swartz TE, et al. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- Swartz TE, et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- Takase T, et al. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011;67:608–621. doi: 10.1111/j.1365-313X.2011.04618.x. [DOI] [PubMed] [Google Scholar]
- Takemiya A, Inoue S, Doi M, Kinoshita T, Shimazaki K. Phototropins promote plant growth in response to blue light in low light environments. Plant Cell. 2005;17:1120–1127. doi: 10.1105/tpc.104.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutomi S, Matsuoka D, Zikihara K. Molecular structure and regulation of phototropin kinase by blue light. Biochim. Biophys. Acta. 2008;1784:133–142. doi: 10.1016/j.bbapap.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Iwata T, Tokutomi S, Kandori H. Role of Phe1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 2001;4:429–435. doi: 10.1016/s1369-5266(00)00196-5. [DOI] [PubMed] [Google Scholar]
- Yasuhara M, et al. Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J. Exp. Bot. 2004;55:2015–2027. doi: 10.1093/jxb/erh226. [DOI] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein–protein interactions in live cells using light. Nat. Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- Zikihara K, Iwata T, Matsuoka D, Kandori H, Todo T, Tokutomi S. Photoreaction cycle of the light, oxygen, and voltage domain in FKF1 determined by low-temperature absorption spectroscopy. Biochemistry. 2006;45:10828–10837. doi: 10.1021/bi0607857. [DOI] [PubMed] [Google Scholar]
- Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]