The identification of signal-transduction components in plant cells is proceeding on an almost daily basis. In this issue, Kudla et al. (1) continue this important process by describing the isolation and characterization of regulatory subunits of calcineurin, a Ca2+/calmodulin-dependent protein phosphatase. Since the pioneering work of Cohen (2) and collaborators, biologists have classified protein phosphatases into four types: PP1, PP2A, PP2B, and PP2C. Although this classification was sensibly designed to bring order out of chaos, it had one major drawback. Division into just four types implied a limited diversity and perhaps a lack of specificity in the mechanism of protein dephosphorylation. Consequently, investigators have tended to concentrate attention on protein kinases where, it was surmised, all the action was to be found. Indeed, several hundred protein kinases have now been cloned or purified from plant tissues (3). Estimates suggest there will be nearly a thousand identified when the Arabidopsis genome sequence is finally published. However, the observations of Kudla et al. (1) suggest that the regulation of dephosphorylation by calcineurin (classified as PP2B) might be more complex than is currently appreciated.
Calcineurin is a heterodimeric phosphatase. A conserved “A” type catalytic subunit combines with a class of variable and regulatory “B” type subunits. It is the latter subunit class that is the subject of the paper by Kudla et al. (1). Originally, calmodulin was thought to be the only B class subunit. Most early studies in plant cells suggested calmodulin to be of very limited diversity—one or two sequences at the most. It was assumed, therefore, that specificity in dephosphorylation would have to reside in some unique characteristics of individual calcium signals. Indeed, a multiplicity of hormonal, physical, chemical, and stress signals induces transient elevations in cytosolic Ca2+ in plant cells, and each signal induces unique kinetics in the generated Ca2+ transient (4). However, unlike animal cells, plants now are known to contain many isoforms of calmodulin, with the current record being 13 in wheat (5). These isoforms have cell and tissue specificity in expression, distribution, and cell location (6), and they provide the potential for at least 13 specific calcineurin activities. However, the description by Kudla et al. (1) of a distinct class of calcineurin B subunits increases the potential number of specific calcineurin activities yet again. These B subunits can be used interchangeably with calmodulin to regulate calcineurin. Some reassessment of the emphasis placed on protein-kinase control of plant-cell phosphorylation is probably long overdue.
The identification of this class of calcineurin regulatory subunits in Arabidopsis is secure. Kudla et al. (1) have used the now familiar complementation of yeast mutants (in this case lacking calcineurin) and dihybrid technologies (with rat A calcineurin subunits) to characterize subunit function and binding properties. This gene family contains six members. Sequencing identifies the presence of four EF hands or calcium-binding regions in each subunit. So these Arabidopsis calcineurins retain their dependence on signal-initiated changes in cytosolic Ca2+. However, strikingly, the B subunit sequence is most similar to frequenin, a neuronal cytosol Ca2+ sensor involved with vesicle fusion and neurotransmitter-release functions (7). Of the three B subunits investigated in detail, the expression of one subunit increased greatly when plants were signaled by cold, wounding, or drought, as determined by Northern analysis.
What function can be deduced for this class of subunits? Specifying a cellular location is one likely possibility. It has been apparent from the earliest days of phosphorylation research that the spatial location of kinases and phosphatases is important in defining substrate specificity (8). For example, in the absence of spatial localization, many proteins, not usually endogenous substrates for protein kinase, can be illegitimately phosphorylated and potentially activated; similarly for dephosphorylation. The simplest way for a signal to navigate its way through the cellular forest of kinases and phosphatases safely and induce specific responses is by a clearly defined path. This perspective sees the specificity in signal transduction as the result of a kind of hard wiring of the cell: a defined, spatially structured path involving the cytoskeleton, membranes, or wall. The function of transduction switches, like cytosolic Ca2+, is to connect and then direct information flow though one or another well structured path; much as identically constructed switches are used to connect and direct electrical current separately to the lights, the television, or the computer.
Probably one function of this class of calcineurin B subunits will be to pin phosphatase activity either to specific regions of the cytoskeleton or to membranes. Many Ca2+-binding proteins are attached to the cytoskeleton (4), and calcineurin might be among them. However, the primary difficulty with involving the cytoskeleton is its lability; the hard wiring would have to be constantly remade as the cytoskeleton changes structure.
Given our current understanding, attaching calcineurin to membranes is a much more credible function for the B subunits. Kudla et al. (1) indicate that their sequence contains a conserved myristoylation site. Transient attachment of calcineurin to membranes controlled by myristyl transferases can be anticipated. But even stronger evidence comes from one well characterized function of calcineurin: the regulation of the inward flux of K+, Na+, and Ca2+ ions through plasma membrane-located channels.
Immunophilins are receptors for immunosuppressive drugs such as cyclosporin and FK506. Binding of the drug to its immunophilin receptor generates a potent inhibitor of calcineurin that modifies the immune response by impairing T cell function. In a series of ground-breaking articles, Luan and coworkers (9–11) showed that plant cells also contained several active immunophilins. When the plant immunophilins combined with FK506, calcineurin activity was blocked. Furthermore, treatment of leaf guard cells with FK506 antagonized the inhibitory effect of Ca2+ on inward K+ flux. The implication is that inward K+ channels are active when phosphorylated; calcineurin blocks inward K+ flux by dephosphorylating channel proteins. Because plant cells contain several immunophilins, these data also might suggest that there are other, as-yet undiscovered endogenous controls of calcineurin activity. A search for a plant cyclosporin analogue might well prove productive.
Salination is a major environmental stress that is a substantial constraint on crop production (12). Salt tolerance could be improved by inhibiting the inflow of Na+ and other ions into the cytoplasm. If calcineurin blocks the inward fluxes of the major soil cations, enhancing endogenous calcineurin could help to block Na+ entry and thus engender salt tolerance. By overexpressing yeast calcineurin in tobacco, Pardo et al. (12) did indeed generate plants that were substantially more tolerant to salt application than the wild type, confirming the reality of the prediction. Salination signals also transiently elevate cytosolic Ca2+ (13), implicating Ca2+-dependent proteins, such as calcineurin, in the response.
If calcineurin regulates inward cation flux in all plant tissues, important consequences follow. Plant movements, such as heliotropism, sleep movements, rapid touch responses, and stomatal closing or opening, involve massive changes in K+ and Cl− flux into and out of motor cells, altering turgor pressure. Calcineurin becomes an essential ingredient in plant-movement regulation. Calcineurin could also control inward ion flux in the root, a process crucial for growth, development, and plant survival.
The B subunit sequence analogue, frequenin, has four Ca2+ binding regions (7). It is in these regions that most sequence similarity between the two proteins occurs. Frequenin is involved in neurotransmitter release, although its precise mode of action is not understood. However, the domain on the cytoplasmic surface of the plasma membrane of nerve cells can reach Ca2+ concentrations as high as 100 μM during an action potential (7). Frequenin may be designed to regulate within these very high Ca2+ ranges. The Ca2+ concentration in the cytoplasmic domain of the plant plasma membrane during signaling is not known. Near the mouth of any Ca2+ channel, however, transient Ca2+ concentrations as high as 100 μM can be predicted. These calcineurin B subunits might be designed to regulate at high Ca2+ concentrations and perhaps even attach calcineurin to the Ca2+ channel mouth, ensuring an instant dephosphorylation response when the cell is signaled. Regulating Ca2+ channel activity itself is a good possibility. Calmodulin would be less suitable for this purpose, because it is usually fully activated by 2–5 μM Ca2+. The Ca2+ binding constants for the B subunits need to be measured to resolve these possibilities.
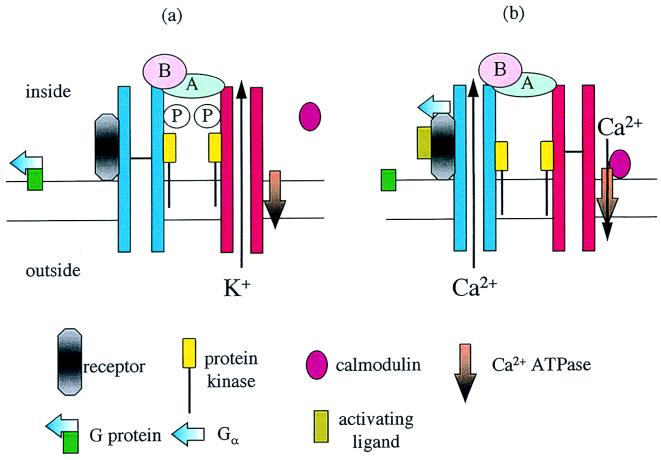
Calmodulin also is found attached to plant plasma membranes (14). Thus, there may be additional functions for calcineurin on the plasma membrane. How cells ensure that calcineurin ends up in the right place when there is a choice of different kinds of B subunit as well as calmodulin available will clearly become a primary problem. Most cellular calmodulin is soluble, highlighting the difficulty. One possible explanation is that calcineurin forms part of a much larger protein complex, a transducon (14, 15), containing receptors, K+ and Ca2+ channels, kinases, phosphatases, calmodulin or B subunit, and calcium ATPase. Selection of a particular B subunit or calmodulin would then be specified by the overall structure of the transducon and the interactions with the other main protein constituents. A suggested transducon structure is illustrated in a K+-transporting mode (Fig. 1a) and in a signaling mode (Fig. 1b).
Figure 1.
Suggested structure of a transducon on the plasma membrane. In the K+-transporting mode, (a), the major constituents remain bound together, except for calmodulin. When a ligand binds to the receptor (b), it is suggested that the receptor and Gαa subunit from a G protein remain bound together, briefly activating the Ca2+ channel. The B subunit of calcineurin situated at the Ca2+ channel mouth responds to the high Ca2+ concentrations and activates the A subunit. Dephosphorylation inhibits the K+ channel, and dephosphorylation of the Ca2+ channel prolongs opening. Calmodulin binds to the ATPase regulating Ca2+ outflow from the cell but only at a suitable distance from the channel mouth and where Ca2+ concentrations are lower.
The observation that mRNA for one B subunit accumulates substantially after cold, wounding, and drought signals (1) is of considerable importance. Each of these signals is known to elevate cytosolic calcium (4). Furthermore, it is known that changes in the expression of some other genes induced by these signals are regulated by cytosolic calcium (4, 13). However, the elevation of calcineurin-subunit expression after signaling is becoming part of a much more general pattern in plant cells.
Calmodulin, phospholipase C, Ca2+-ATPase, Ca2+-dependent protein kinase, and other protein kinases are all critical proteins concerned with the transduction of signals through a cytosolic Ca2+ pathway. The expression of each of these proteins (as well as calcineurin) is increased by orders of magnitude when unstimulated plants are exposed to cold, drought, or mechanical signals. Within several hours, the cellular concentrations of these proteins rise (16–19). Similar results have emerged for second messengers that are concerned with Ca2+ signaling. In unstimulated plant cells, the Ca2+-mobilizing signal, IP3, the IP3 precursor PIP2, and the Ca2+-mobilizing cyclic ADP-ribose are present at virtually undetectable levels. A signaling episode greatly increases the resting levels of these second messengers for many hours, but elevation itself takes hours to accomplish (20, 21). Exposing unstimulated plants to one or more signals elevates critical elements of the transduction apparatus involving Ca2+ by an order of magnitude.
The rate of information flow, the flux, through a signal-transduction pathway will be constrained by the concentration of signal-transduction constituents. In an unstimulated plant, information flow from a signal through Ca2+-dependent pathways will be slow; in a stimulated plant, information flow from the self-same signal will be enormously faster. However these data are viewed, they represent a form of cellular learning. No doubt, anticipation of further signals is the reason for cellular learning. In any case, some intriguing questions have been raised. Does enhanced expression of the Ca2+ apparatus occur in all cells? Or is the elevation limited to the recruitment of new cells into a full blown signaling mode? On current understanding, the latter possibility is more likely.
Signal-transduction networks share properties with neural networks, and the learning parallels can be drawn easily (22). Neural networks learn by increasing the numbers of connections (and the strength of the connections) between the neurons representing the chosen path to connect signal and response. The result of learning (reinforcement) is to accelerate the information flux rates between the signal and the response. Elevating Ca2+ transduction constituents is analogous to increasing the numbers of connections between neurons in a neural network. The increased information flow that results represents a kind of cellular learning. This cellular learning, coupled with the memory built into signal-transduction systems (22), suggests an unexpected form of cellular intelligence (23).
Some important questions remain to be answered. What are the identities and locations of proteins that bind B subunits? Are there other proteins that regulate calcineurin? How does calcium regulate B subunit expression? Does the elevation of B subunits involve uniform increases in all cells or are there simply more cells responding? Could tissue Ca2+ waves specify the spatial pattern of response of B subunit accumulation? How can we test the learning capacity of the plant-cell signal-transduction network? The important observations made by Kudla et al. (1) generate important questions. As always, important advances generate more questions than answers.
Footnotes
The companion to this Commentary begins on page 4718.
References
- 1.Kudla J, Xu Q, Harter K, Gruissem W, Luan S. Proc Natl Acad Sci USA. 1999;96:4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen P. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 3.Stone J M, Walker J C. Plant Physiol. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahlo R, Moutinho A, Van der Luit A, Trewavas A J. Philos Trans R Soc London B. 1998;353:1463–1473. [Google Scholar]
- 5.Yang T, Segal G, Abbo S, Feldman M, Fromm H. Mol Gen Genet. 1996;252:684–694. doi: 10.1007/BF02173974. [DOI] [PubMed] [Google Scholar]
- 6.Zielinski R E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:697–725. doi: 10.1146/annurev.arplant.49.1.697. [DOI] [PubMed] [Google Scholar]
- 7.Olaffson P, Wang T, Lu B. Proc Natl Acad Sci USA. 1995;92:8001–8005. doi: 10.1073/pnas.92.17.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faux M C, Scott J D. Trends Biochem Sci. 1996;21:312–315. [PubMed] [Google Scholar]
- 9.Luan S, Li W, Rusnak F, Assmann S M, Schreiber S L. Proc Natl Acad Sci USA. 1993;90:2202–2206. doi: 10.1073/pnas.90.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luan S, Lane W S, Schreiber S L. Plant Cell. 1994;6:885–892. doi: 10.1105/tpc.6.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan S, Kudla J, Gruissem W, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardo J M, Reddy M P, Yang S, Maggio A, Huh G H, Matsumoto T, Coca M A, Paino-Durzo M, Koiwa H, Yun D J, et al. Proc Natl Acad Sci USA. 1998;95:9681–9686. doi: 10.1073/pnas.95.16.9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight H, Trewavas A J, Knight M R. Plant J. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 14.Trewavas A J, Malho R. Plant Cell. 1997;9:1181–1195. doi: 10.1105/tpc.9.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 16.Evans D E, Williams L E. Biochim Biophys Acta. 1998;1376:1–25. doi: 10.1016/s0304-4157(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 17.Braam J, Davis R W. Cell. 1990;63:357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- 18.Hirayama T, Ohto C, Mizoguchi T, Shinozaki K. Proc Natl Acad Sci USA. 1995;92:3903–3907. doi: 10.1073/pnas.92.9.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urao T, Katagirri T, Mizoguchi T, Yamaguchi-Shinozaki K, Shinozaki K. Mol Gen Genet. 1994;244:331–340. doi: 10.1007/BF00286684. [DOI] [PubMed] [Google Scholar]
- 20.Drobak B K, Watkins P A C. Biochem Biophys Res Commun. 1994;205:739–745. doi: 10.1006/bbrc.1994.2727. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Kuzma J, Marechal E, Graeff R, Lee H C, Foster R, Chua N M. Science. 1997;278:2127–2130. doi: 10.1126/science.278.5346.2126. [DOI] [PubMed] [Google Scholar]
- 22.Bhalla U S, Iyengar R. Science. 1999;283:381–386. doi: 10.1126/science.283.5400.381. [DOI] [PubMed] [Google Scholar]
- 23.Trewavas, A. J. (1999) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]