Abstract
Rapidly expanding aging populations and a concomitant increase in the prevalence of age-related diseases are global health problems today. Over the past three decades, a large body of work has led to the identification of genes and regulatory networks that affect longevity and health span, often benefiting from the tremendous power of genetics in vertebrate and invertebrate model organisms. Interestingly, many of these factors appear linked to lipids, important molecules that participate in cellular signaling, energy metabolism, and structural compartmentalization. Despite the putative link between lipids and longevity, the role of lipids in aging remains poorly understood. Emerging data from the model organism Caenorhabditis elegans suggest that lipid composition may change during aging, as several pathways that influence aging also regulate lipid metabolism enzymes; moreover, some of these enzymes apparently play key roles in the pathways that affect the rate of aging. By understanding how lipid biology is regulated during C. elegans aging, and how it impacts molecular, cellular, and organismal function, we may gain insight into novel ways to delay aging using genetic or pharmacological interventions. In the present review we discuss recent insights into the roles of lipids in C. elegans aging, including regulatory roles played by lipids themselves, the regulation of lipid metabolic enzymes, and the roles of lipid metabolism genes in the pathways that affect aging.
Keywords: C. elegans, lipids, lipase, ascarosides, fatty acids, nuclear hormone receptors, mitochondria, N-acylethanolamine
Introduction
One of the most desirable goals in biomedical research is to understand the molecular mechanisms that promote healthy aging. Identifying the processes that lead to extended longevity in laboratory settings may allow the exploitation of that knowledge to generate pharmacological treatment regimens that delay the onset and/or reduce the severity of age-associated diseases, or even extend life span per se. In model organisms, several genetic and nutritional conditions have been identified that not only extend mean and/or maximal life span but also postpone the onset of phenotypes associated with aging, such as a loss of mobility, a decline in cognitive ability, and others.
It has been almost 30 years since single gene mutations were first found to affect aging in the nematode Caenorhabditis elegans (reviewed in Kenyon, 2010a). Since then, C. elegans has become a powerhouse for studies on the mechanisms that affect longevity. Numerous labs are identifying an ever-increasing number of genes, molecules, and regulatory networks that influence aging. Several excellent reviews summarize recent progress in this field and the reader is referred to these (Fontana et al., 2010; Gallo and Riddle, 2010; Kenyon, 2010b; Zhou et al., 2011).
Despite the identification of several discrete pathways that affect longevity, the molecular mechanisms that actually result in life span extension remain obscure. Of note, many pro-longevity signaling pathways affect lipid biology and/or nutrient sensing/signaling. For example, insulin/insulin-like growth factor (IGF) signaling (IIS), which affects aging in many model organisms, integrates nutritional cues, and regulates fat storage in many animals (Fontana et al., 2010; Kenyon, 2010b). Other pathways known to influence aging and lipid biology and/or nutrient sensing include dietary restriction mechanisms including intermittent fasting paradigms; the electron transport chain (ETC) of the mitochondria; signaling following genetic or physical germline removal; target of rapamycin (TOR) signaling, which governs nutrient sensing and is linked to dietary restriction; and translation inhibition, which interacts with TOR and dietary restriction in yet poorly understood ways.
One simple explanation would be that all longevity-controlling pathways equivalently affect storage lipid levels, thus evoking similar overall changes in energy balance to extend life span. This is clearly not the case, as mutants in various aging pathways differentially alter overall fat storage. Thus, some C. elegans mutants are long-lived and exhibit increased levels of triacylglycerides (TAGs, i.e., stored fat); these include worms carrying a mutation in the daf-2 gene, which encodes the C. elegans insulin/IGF-1 receptor (Kenyon et al., 1993; Ashrafi et al., 2003), germline-less glp-1 mutants (Arantes-Oliveira, 2002; O’Rourke et al., 2009), and tub-1 mutants (Mukhopadhyay et al., 2005). In contrast, eat-2 mutants, which experience dietary restriction due to mechanically reduced pharyngeal pumping, are long-lived but have reduced fat storage (Lakowski and Hekimi, 1998; Srinivasan et al., 2008; Brooks et al., 2009). Thus, long-lived mutants can be high or low in fat. Moreover, in the tub-1 mutants, two genetically distinct pathways influence fat storage and aging (Mukhopadhyay et al., 2005). Therefore, there is no strict link between aging and global fat storage, at least when assessing fat deposition by techniques that monitor global lipid stores in whole animals or whole animal extracts. Clearly, if lipids indeed modulated longevity, their role must be more diverse and complex than initially anticipated.
The Central Roles of Lipids in Cellular and Molecular Biology
Lipids are broadly defined as hydrophobic or amphiphilic molecules formed in whole or in part by ketoacyl and/or isoprene groups (Fahy et al., 2009; Subramaniam et al., 2011). Lipids play a central role in metazoan physiology, as they affect cellular and organismal functions in three principal, non-exclusive ways: (i) in signaling cascades, both directly as signaling molecules (e.g., diacylglycerol, fatty acids, phosphatidylinositols, sterols, ceramides, and sphingolipids), and indirectly by reversibly and irreversibly tethering signaling proteins to cellular membranes (e.g., via prenylation or palmitoylation); (ii) as structural elements in cellular membranes, providing cellular, and subcellular compartmentalization; and (iii) as key molecules in energy metabolism. Lipids are also integral components in lipophilic vitamins, which influence a large number of cellular processes, and it is conceivable that vitamin bioavailability may influence longevity. In line with their central role in physiology, lipids, or at least genes directly involved in lipid metabolism, have recently been mapped into several signaling pathways that affect longevity; yet, the role of lipids in the modulation of aging remains enigmatic (Ackerman and Gems, 2012). Here, we review recent reports that suggest important roles for lipids and lipid metabolizing enzymes in aging.
Lipid Signaling in Aging: Nuclear Hormone Receptor Action
Lipids and their derivatives can act in various signaling pathways to affect aging, including cell membrane receptor signaling or Nuclear Hormone Receptor (NHR) signaling (Germain et al., 2006). NHR signaling is intriguing, as lipids do not only modulate NHRs but some NHRs in turn affect lipid biology through their downstream regulatory actions. NHRs are ligand-gated transcription factors that are conserved in metazoans, and they play important roles in the regulation of physiology and development (Germain et al., 2006).
The C. elegans genome encodes a large family of 284 NHRs, far exceeding the number of NHRs in the human, mouse, or fly genomes (Taubert et al., 2011). The vast majority of these NHRs appear to be derived from an ancestor related to Hepatocyte Nuclear Receptor 4 (HNF4; Robinson-Rechavi et al., 2005), which plays important functions in the development and function of liver and pancreas in mammals (Maestro et al., 2007; Gonzalez, 2008), whereas fly HNF4 regulates lipid mobilization and fatty acid β-oxidation (Palanker et al., 2009). Multiple studies suggest that mammalian HNF4 can be modulated by interactions with fatty acids and/or fatty acid derivates, suggesting that lipids act as ligands for HNF4 (Hertz et al., 2003; Yuan et al., 2009). Although the in vivo consequences of lipid-HNF4 interactions remain controversial, it is possible that lipids may similarly bind and/or regulate some C. elegans NHRs. However, no ligands have been identified for any C. elegans NHRs save for DAF-12, an NHR unrelated to HNF4. DAF-12 binds a steroid-like ligand with nanomolar affinity (Motola et al., 2006), and regulates development and aging in C. elegans (Antebi et al., 1998, 2000). Individual daf-12 alleles affect aging in distinct ways, depending on whether they cause a complete loss of daf-12 activity or abrogate only specific molecular functions, e.g., the capacity to bind ligands (reviewed in Gáliková et al., 2011). Notably, DAF-12 is required for the life span extension in worms that lack the germline (see below); these worms are thought to be long-lived due to a concomitant absence of germline-derived signals that restrict life span (Arantes-Oliveira, 2002). Thus, lipid-derived hormones play a key role in this pathway, albeit how DAF-12 acts in this context is still poorly understood.
A Role for NHR Signaling and Fatty Acid Desaturases in Aging
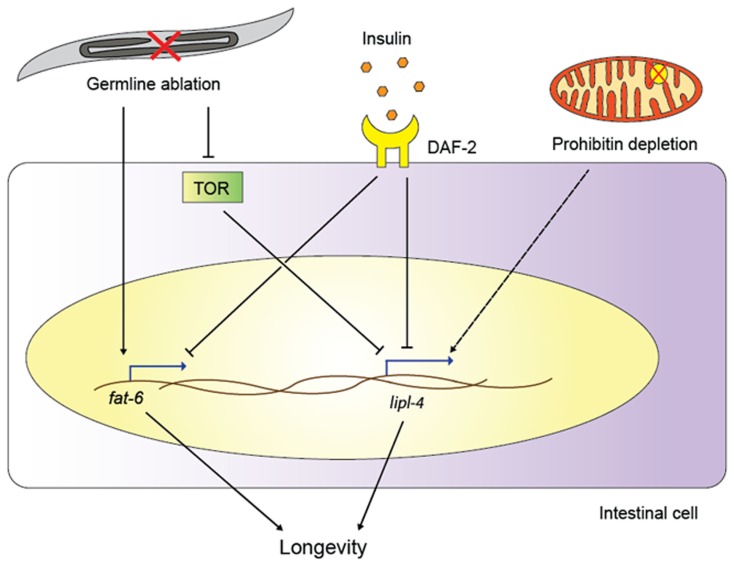
Adding to the role for DAF-12 in longevity, a new study reports a key role for another NHR and for one downstream target, a fatty acid desaturase (Goudeau et al., 2011); this study thus directly links NHRs, lipid metabolism, and long life span. Specifically, in germline-less glp-1 mutants, NHR-80 is upregulated, and transgenic NHR-80 overexpression further extends animal life span. One key regulatory target of NHR-80 turns out to be fat-6, a stearoyl-CoA desaturase that converts stearic acid into oleic acid; like nhr-80, fat-6 is required for the pro-longevity effect of genetic germline ablation (Figure 1). In contrast to germline-less mutants, nhr-80 is dispensable for the longevity of other long-lived mutants, including IIS pathway mutants, mitochondrial mutants, and worms grown under dietary restriction (Goudeau et al., 2011). Thus, NHR-80, DAF-12, and FAT-6 constitute a lipid signaling pathway that is essential for the long lifespan specifically in germline-less animals. However, the pathway is obviously more complex, as oleic acid supplementation rescues the loss of fat-6, but cannot complement for the loss of nhr-80. This suggests that NHR-80 may regulate other (lipid biology) genes in this context. Similarly, the specific role of oleic acid remains to be determined; Brock et al. (2006) previously showed that nhr-80 mutants exhibit several changes in their fatty acid profile, likely as a consequence of reduced oleic acid levels. Any of these fatty acid species could in principle be relevant for lifespan extension in germline-less animals.
Figure 1.
Functions of lipid remodeling enzymes in various aging pathways. Germline ablation activates the transcription of the fatty acid desaturase fat-6 and, through TOR signaling, of the lipase gene lipl-4. Insulin signaling also upregulates lipl-4 and fat-6, and lipl-4 is required for longevity of both germline-less worms and daf-2/insulin receptor mutants. Prohibitin depletion extends the life spans of various C. elegans longevity models, possibly also relying on lipase action (as indicated by the dashed line; for details, see main text).
The specificity of NHR-80 and FAT-6 in this particular pathway is noteworthy. The ability of NHR-80 to drive fat-6 expression is apparently restricted to germline-less animals, as nhr-80 single mutants exhibit near wild-type levels of fat-6 (Brock et al., 2006). In contrast, nhr-80 mutants show a significant decreased fat-5 and fat-7 expression (Brock et al., 2006), whereas in germline-less worms, nhr-80 mutation does not affect fat-7 and only mildly affects fat-5 expression (Goudeau et al., 2011). In line with a specific regulatory effect of NHR-80 on fat-6, only fat-6, but not fat-5 or fat-7 are required for life span extension in germline-less animals. Mechanistically, NHR-80 must assemble distinct regulatory complexes at the fat-5, -6, and -7 promoters in the different genetic contexts to achieve appropriate gene expression. Moreover, NHR-80 and the closely related NHR-49 both regulate fatty acid desaturase gene expression (Van Gilst et al., 2005b; Brock et al., 2006), but only NHR-80 targets fat-6 (Goudeau et al., 2011); due to this difference, NHR-49 is probably not required for the life span extension in germline-less animals, although this has not yet been directly tested using germline-less nhr-49 mutants.
Goudeau et al. (2011) did not find a requirement for nhr-80 in other longevity pathways such as IIS or dietary restriction, but fatty acid desaturases have been implicated in several such circuits (Figure 1). For example, fat-1, -2, -3, -6, and -7 are all induced in daf-2 mutants, and this regulation is at least partially dependent on the downstream transcription factor DAF-16, a forkhead box O transcription factor that plays key roles in longevity assurance within multiple pathways (Murphy et al., 2003; Halaschek-Wiener et al., 2005; Budovskaya et al., 2008). These data suggest that increasing the synthesis of mono- and/or poly-unsaturated fatty acids (MUFAs and PUFAs, respectively) may be important for long-lived mutants; indeed, fat-6 or fat-7 depletion shortens the life span of long-lived daf-2 mutants, albeit only mildly (Murphy et al., 2003). Taken together, the above data indicate that at least some fatty acid desaturases contribute to the life span extension downstream of reduced insulin signaling or germline removal.
The roles of fatty acid desaturases and their regulators have also been studied in wild-type worms, and although the roles of these genes in wild-type worms are likely distinct from the lifespan modulation in long-lived strains, the data are nevertheless informative. For example, depletion of fat-7 by RNA interference (RNAi; Fire et al., 1998) shortens the life span of wild-type worms (Van Gilst et al., 2005b). Similarly, depletion of the transcription factors NHR-49 or SBP-1 (the ortholog of mammalian sterol response element binding protein, a master regulator of lipogenesis and adipogenesis) or their coregulator MDT-15 reduces fatty acid desaturase expression, increases the ratio of stearic acid to oleic acid, and shortens lifespan (Van Gilst et al., 2005b; Taubert et al., 2006, 2008; Yang et al., 2006). In the case of mdt-15 depletion, the short lifespan can be partially rescued by exogenous PUFAs, suggesting a requirement for PUFAs to maintain normal life span (Taubert et al., 2006). mdt-15 is also required for the longevity of other long-lived mutants, including IIS pathway mutants, mitochondrial mutants, and the eat-2 mutants that mimic dietary restriction (Rogers et al., 2011), although it is not clear whether these effects relate to MDT-15’s impact on fatty acid metabolism (Taubert et al., 2006; Yang et al., 2006). fat-3 mutants also live shorter than wild-type worms, further supporting the notion that reduced levels of PUFAs may shorten life span (Hillyard and German, 2009). Contrasting these studies, nhr-80 mutants exhibit an increased stearic acid to oleic acid ratio, yet a normal lifespan (Brock et al., 2006; Goudeau et al., 2011). Similarly, fat-5, -6, or -7 single mutants do not show a shortened life span, albeit these mutants upregulate other desaturases, perhaps in a compensatory fashion (Brock et al., 2006). Lastly, fat-4 mutants exhibit an increased lifespan (Lucanic et al., 2011), possibly through their effects on N-acylethanolamine (NAE) levels and endocannabinnoid signaling (see below). Clearly, fatty acid desaturases affect the life span of wild-type worms and long-lived mutants alike; yet, the requirements for individual enzymes and their upstream regulators are not the same in all genetic (and perhaps environmental) conditions.
A recent study suggests that fatty acid desaturation per se may not be the only determinant of how these molecules affect life span, but that parameters such as chain-length matter as well (Shmookler Reis et al., 2011). The authors used lipid profiling in various long-lived mutants to generate correlations between longevity and fatty acid composition. Several parameters correlate well with longevity; perhaps most strikingly, overall fatty acid oxidation susceptibility (as expressed by the peroxidation index) was strongly anti-correlated with longevity. Although the study focused on a set of IIS pathway mutants with extreme longevity phenotypes (all mutants were either close to wild-type life span, or very long-lived, as opposed to representing a more continuous spectrum), these data suggest that high PUFA levels may be disadvantageous for a long life. This somewhat contradicts the findings that, e.g., nhr-49 mutants, nhr-49 (RNAi), or fat-7 (RNAi) worms are short-lived, as these worms show changes in stearic to oleic acid ratio, but only relatively minor changes in the distribution of other fatty acid species, including most PUFAs (Van Gilst et al., 2005b). However, nhr-49 mutants, nhr-49 (RNAi), or fat-7 (RNAi) worms are severely short-lived, and perhaps the same fatty acid properties do not apply equally in long-lived worms and in severely short-lived worms. In any case, the findings by Shmookler Reis et al. add to an existing body of data suggesting that fatty acid composition is regulated in aging worms and that it may influence life span.
These above studies suggest that fatty acid desaturation is likely important for normal and for extended lifespan, with context-specific requirements for individual regulators and metabolic enzymes. However, the above studies analyzed total fatty acids obtained from whole worm extracts. Thus, the question remains as to whether changes in fatty acid profiles may affect longevity through a role in metabolism, signaling, or perhaps membrane structure (Hulbert, 2011). Given that the observed changes in fatty acid abundance are rather large in some instances, it may seem likely that certain types of unsaturated fatty acids are structurally relevant, e.g., in the membranes of specific organelles, which in turn may affect nematode physiology. Affected organelles could be the mitochondria or the endoplasmic reticulum (ER), both of which can influence lifespan (Wong et al., 1995; Lakowski and Hekimi, 1996; Feng et al., 2001; Henis-Korenblit et al., 2010). Perhaps, membrane lipids may also influence aging by altering the properties of the nuclear envelope, which in turn may influence gene transcription or other nuclear processes. Notably, nuclear structure is linked to aging, as mutations of lamin A cause a progeria syndrome in humans (De Sandre-Giovannoli, 2003), a phenotype that is reflected in C. elegans lmn-1 mutants (lmn-1 is the only C. elegans lamin a gene), which also exhibit a short life span and altered nuclear architecture (Bank and Gruenbaum, 2011; Bank et al., 2011). Without doubt the elucidation of the mechanisms by which unsaturated fatty acids affect life span will remain a challenging and fascinating topic for future research.
Lipase Action and a Link to Autophagy
Two recent studies suggest that lipid-remodeling enzymes other than the desaturases can also influence longevity, namely the lipases. Lipases are esterases that catalyze the hydrolysis of fat, thus producing mono- or diglycerides, glycerol, and free fatty acids (Branicky et al., 2010). Wang et al. (2008) found that the lipase LIPL-4 is required for germline-removal induced longevity, suggesting that lipid hydrolysis plays a critical role in life span extension (Figure 1). Moreover, LIPL-4 expression is increased in animals without a germline (Wang et al., 2008), and intestinal overexpression of LIPL-4 alone not only reduces fat storage (Wang et al., 2011) but is also sufficient to extend the life span in wild-type worms (Wang et al., 2008).
How does LIPL-4 action contribute to increased life span? Several mechanisms appear possible. For one, lipase action can change overall fat levels, and such changes could alter animal lifespan. However, as pointed out above, fat levels and longevity do not correlate in C. elegans, and thus simple reduction of overall storage lipids is unlikely to cause lifespan extension. Instead, given that intestine-specific LIPL-4 is sufficient to extend the lifespan of germline-less animals, it is tempting to speculate that LIPL-4 action could generate lipids that act in endocrine fashion to modulate downstream pathways. In this context we reiterate the role of DAF-12 in the longevity of germline-less animals (Hsin and Kenyon, 1999). Perhaps, LIPL-4 products directly or indirectly contribute to altered abundance of DAF-12 ligands.
Another way for LIPL-4 to modulate lifespan could involve autophagy (Lapierre et al., 2011), a catabolic process that degrades cellular organelles and macromolecular complexes, thus recycling their molecular building blocks (Kundu and Thompson, 2008). Autophagy is a highly regulated process that participates in normal growth, development, and homeostasis, and it is known to modulate aging in C. elegans (Meléndez et al., 2003; Hansen et al., 2008). Lapierre et al. found that reduced TOR activity induces autophagy and LIPL-4 lipase activity in long-lived, germline-less animals. Autophagy and lipase action are co-dependent, and hence the authors suggested that LIPL-4 activity promotes autophagy. It will be interesting to determine whether it indeed does so, and if yes, by what mechanism, e.g., by providing a specific type of lipid required for autophagosome formation, or possibly by generating a regulatory ligand for a (membrane or nuclear) receptor that promotes autophagy. As described above, LIPL-4 is required for lifespan extension in IIS signaling mutants (Wang et al., 2008), and autophagy is required for lifespan extension due to impaired TOR signaling, IIS signaling, or mitochondrial function (Hansen et al., 2008; Tóth et al., 2008). Thus, autophagy is emerging as an important player in several longevity pathways, and may perhaps broadly contribute to lifespan extension in C. elegans. This could involve LIPL-4 or possibly other lipases.
Lipases other than LIPL-4 may also contribute to life span modulation and/or are age-regulated in C. elegans. Most strikingly, lips-7 is induced in long-lived ctbp-1 mutants, and lips-7 depletion prevents this longevity, whereas another lipase is dispensable; lips-7 induction also correlates with lower fat levels in ctbp-1 mutants (Chen et al., 2009). Again, the functional consequence of lips-7 induction is not clear, but it is possible that lips-7 induced autophagy contributes to the longevity of ctbp-1 mutants. Vice versa, it would be useful to test whether lips-7 plays a role in the long life span of germline-less animals. Although both LIPL-4 and LIPS-7 affect overall fat storage, it would be intriguing if they were differentially required for life span extension in different genetic backgrounds.
LIPS-7 and LIPL-4 are members of protein families, and their homologs may also affect aging. In line with this notion, LIPL-1, -2, -5, and -7, and LIPS-4, -14, and -17 expression is altered in aging worms and/or in long-lived mutants (Budovskaya et al., 2008; Golden et al., 2008; McCormick et al., 2012; Youngman et al., 2011), suggesting that they may contribute to lipid remodeling in these contexts. However, whether these regulations are cause or consequence of aging remains to be determined, and none of these lipases have yet been studied genetically to determine their roles in longevity pathways. Nevertheless, taken together with the fact that intestinal LIPL-4 expression is sufficient to extend worm life span, these data suggest that temporal and spatial changes in lipase expression may influence aging. In summary, lipases are likely to act selectively and in a tightly controlled fashion within certain signaling pathways to affect longevity.
Signaling by Lipid-Derived Small Molecules
An exciting recent study has revealed that signaling by lipid-derived small molecules can directly modulate life span in C. elegans. Lucanic et al. (2011) identified N-acylethanolamines (NAEs) as signaling molecules that link dietary restriction to longevity. NAEs are lipid derivatives that participate in endocannabinoid signaling (Matias and Di Marzo, 2007). In mammals, endocannabinoids have profound effects on energy homeostasis by influencing food intake and by affecting energy metabolism in adipose, liver, pancreas, and skeletal muscle (Banni and Di Marzo, 2010). Lucanic et al. show that NAEs function in the C. elegans pharynx to signal nutrient availability; thus, compared to ad libitum fed worms, wild-type worms under dietary restriction show reduced NAE levels. Artificial reduction of NAEs under ad libitum conditions extends life span, and this effect is refractory to dietary restriction; conversely, exogenous NAEs suppress dietary restriction-induced lifespan extension. Together, these data demonstrate that NAEs and dietary restriction act via overlapping pathways (Figure 2). Further demonstrating a direct link between lifespan and lipid biology, Lucanic et al. find that fat-4 mutants show reduced NAE levels and a concomitant lifespan extension, although molecules other than NAEs may also contribute to this effect. Lastly, the synthesis of one particular NAE species is reduced in long-lived worms carrying a mutation in the ribosomal protein S6 kinase gene rsks-1, and pharmacological supplementation with this NAE reverts the mutant’s longevity. RSKS-1 is known to influence life span, and it is a critical downstream target of TOR signaling (Hansen et al., 2007; Pan et al., 2007). Thus, the data described in Lucanic et al. also suggest a link between the nutrient sensor TOR and NAE signaling.
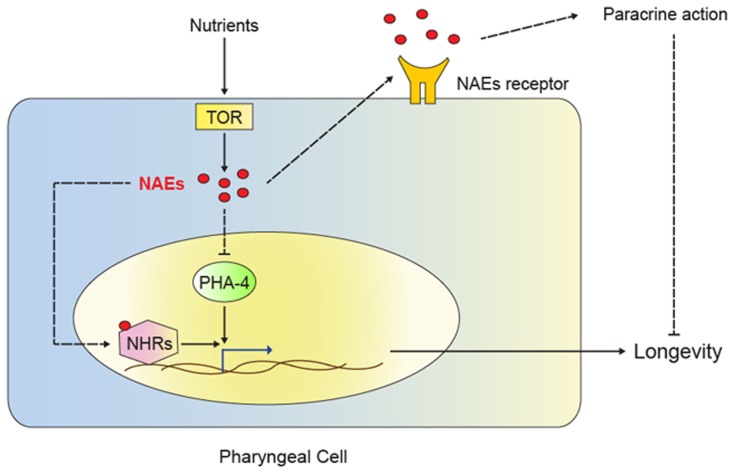
Figure 2.
Model for NAE action in dietary restriction-mediated longevity. The TOR signaling pathway senses nutrient availability. Activation of TOR triggers NAE synthesis in the pharyngeal cell, leading to several possible mechanisms of action. NAEs could act in paracrine fashion to stimulate signaling through unidentified receptors, thus promoting growth, reproduction and inhibiting longevity. NAEs could also inhibit the transcription factor PHA-4 and therefore repress genes required for dietary restriction-induced longevity. Lastly, NAEs could act as ligands for nuclear hormone receptors (NHRs) and result in the regulation of genes required for dietary restriction-mediated longevity. Low NAE levels signal dietary restriction and result in activation of the transcription of genes that contribute to extension of lifespan.
The novel role of NAEs in longevity is exciting. Yet, as noted by Lucanic et al. (2011), the C. elegans genome lacks obvious orthologs to the endocannabinoid receptors that bind NAEs in mammals (McPartland and Glass, 2003); thus, the molecular mode of NAE action in worms remains unclear. The C. elegans genome encodes a large number of G-protein coupled receptors, and it is possible that some of these (or some other membrane receptors) bind NAEs and mediate their effects in paracrine fashion (Figure 2). Alternatively, NAEs may act as NHR ligands, akin to the role of two endogenous NAEs as ligands for the mouse NHR peroxisome proliferator activated receptor alpha (PPARα; Fu et al., 2003; Verme, 2005). By activating PPARα, these NAEs modulate feeding behavior. Perhaps, NAEs similarly modulate NHR activity in C. elegans; although PPARα orthologs are unrecognizable in the C. elegans genome, NHR-49 acts in a PPARα-like fashion to regulate fatty acid β-oxidation and the fasting response(Van Gilst et al., 2005a,b), and it would be interesting to determine whether it participates in NAE signaling and/or dietary restriction (Figure 2). Other candidates include the aforementioned NHR-80 (Goudeau et al., 2011), NHR-69, which was recently shown to modulate longevity (Park et al., 2012), and other NHRs that regulate lipid metabolism in C. elegans (Ashrafi et al., 2003; Arda et al., 2010; Wang et al., 2011). Clearly, much remains to be discovered about the NAEs and their role in the regulation of metabolism, behavior, and aging.
In addition to the NAEs, another class of lipid-derived small molecules is emerging as candidate life span modulators: the ascarosides, a family of small molecules whose founding members were identified based on their capacity to induce the formation of dauer larvae (also collectively referred to as dauer pheromones). The dauer is a specialized C. elegans larva that is long-lived and stress resistant, and its formation is induced by overcrowding, starvation, or high temperatures (Hu, 2007). Parallels between dauer larvae and long-lived mutants had been noted a long time ago: not only are dauer larvae long-lived, but many genetic pathways (e.g., IIS) and dietary stimuli (e.g., starvation) that control dauer formation also affect life span. Recent work has identified the molecular identity of the ascarosides (Jeong et al., 2005; Butcher et al., 2007). These molecules are linked to lipid metabolism in two ways. Firstly, the ascaroside core structure is composed of a sugar (ascarylose) and a lipid moiety. Thus, the abundance of individual ascarosides may directly reflect the availability of certain lipids. Secondly, ascarosides are intricately linked to lipid metabolism through their biosynthetic pathway, as several ascaroside biosynthesis enzymes are involved in fatty acid β-oxidation; these include the acyl-CoA oxidase ACOX-1, the enoyl CoA-hydratase MAOC-1, the β-hydroxyacyl-CoA dehydrogenase DHS-28, and DAF-22, a homolog of sterol carrier protein SCPx (Butcher et al., 2009; Pungaliya et al., 2009; Joo et al., 2010; von Reuss et al., 2012). In line with an important role for these enzymes in fatty acid catabolism, daf-22, dhs-28, and maoc-1 mutants all accumulate excess fat; yet, daf-22 and dhs-28 mutants are short-lived, whereas worms with depleted maoc-1 are long-lived (Hansen et al., 2005; Joo et al., 2009; Zhang et al., 2010). As maoc-1, daf-22, dhs-28, and acox-1 are expected to act in a linear biosynthetic pathway (von Reuss et al., 2012), the differential effects of individual gene deletions on life span is somewhat unexpected. Perhaps, distinct ascarosides differentially affect lifespan, and the enzymes outlined above are not equivalently required for the synthesis of each ascaroside subspecies. Structurally diverse members of the ascaroside family continue to be identified (von Reuss et al., 2012), and thus such functional specialization may not be too surprising. Lastly, it is also conceivable that the enzymes outlined above influence lifespan through a mechanisms that does not involve ascarosides.
In addition to the identification of the ascarosides and relevant biosynthetic enzymes, two recent studies describe the identification of dauer pheromone receptors (Kim et al., 2009; McGrath et al., 2011). Worms carrying mutations in the pheromone receptor genes srg-36, srg-37, srbc-64, and srbc-66, or in gpa-3, a key downstream effector, are compromised for dauer formation upon ascaroside treatment; alas, none of the studies report life span phenotypes. It will be interesting to test whether these mutants, or other mutants defective for dauer pheromone sensing and/or signaling exhibit an increase or decrease in longevity; perhaps, different pheromone receptor combinations will emerge that specifically affect life span but not other phenotypes, akin to the combinatorial and differential roles of ascarosides in development and behavior (Srinivasan et al., 2012).
Mitochondrial Pathways to Longevity and Lipid Metabolism
Mutations in mitochondrial ETC genes extend lifespan in C. elegans and in other organisms, and lipids play critical structural and energetic roles in mitochondria (Nicholls, 2002; Marchi et al., 2012). Mitochondria produce energy by means of oxidative phosphorylation, which generates a proton gradient across the inner mitochondrial membrane that is used to generate ATP. Oxidative phosphorylation also produces reactive oxidative species (ROS) that react with and can damage macromolecules such as DNA, proteins, and lipids. The mitochondrial free radical theory of aging first proposed by Harman (1956) suggested that ROS-induced damage accumulation would gradually lead to a decrease in many cellular functions and eventually result in aging. However, several studies have cast doubt on this model (reviewed in Hekimi et al., 2011), and a recent study found that mitochondrially produced superoxide signals may in fact contribute to life span extension (Yang and Hekimi, 2010), possibly by signaling the occurrence of damage to relevant repair pathways. In line with this notion, moderate and transient ROS levels trigger autophagy to remove damaged organelles, sustaining cell life, and therefore promoting longevity (Marchi et al., 2012). Alternative explanations for the increased longevity of ETC mutants include the notion that reduced mitochondria function generally slows down metabolism (the rate-of-living theory) or the possibility that specific metabolic changes cause delayed aging (Cristina et al., 2009). Below, we discuss several recent studies that provide new insights into how mitochondria may affect life span, hinting at roles for energy metabolism, homeostasis, and signaling.
Mitochondria are intricately linked to lipid biology because triglycerides and fatty acids provide acetyl-CoA, the substrate for the citric acid cycle that is directly coupled to mitochondrial oxidative phosphorylation. A recent study elegantly linked fat metabolism and mitochondrial energetics to longevity in C. elegans (Artal-Sanz and Tavernarakis, 2009). The authors investigated the prohibitin complex, which is formed at the inner mitochondrial membrane. Depletion of the prohibitins extends life span in several genetic backgrounds, including IIS pathway mutants, mitochondrial mutants, and the eat-2 mutants mimicking dietary restriction; in contrast, reduced prohibitin levels shorten the lifespan in wild-type animals, suggesting that prohibitins act in a context-specific fashion. Because of the direct link between mitochondria and fat catabolism, the authors assessed fat storage in worms with depleted prohibitins. Strikingly, they found that in the long-lived mutants, prohibitin depletion correlates with lower fat levels, reduced mitochondria content, and increased ATP production, whereas in wild-type worms, prohibitin depletion results in only a slight decrease in fat content, an increase in mitochondrial level, and no change in ATP synthesis. Prohibitin depletion also decreases fat levels and increases life span in fat-7 and nhr-49 mutants, which store excess fat. These data suggest that fat-7 and nhr-49 are not required for the life span extension in this context, and that prohibitin depletion reduces fat levels and achieves life span extension even in high-fat backgrounds. Although it is not certain that reduced fat storage is causally linked to lifespan extension, these data are especially intriguing in view of the roles for lipases in the long life span of germline-less and IIS pathway mutants (see above). Indeed, it is tempting to speculate that lipases may be required for the increase in life span following prohibitin depletion (Figure 1), which would be supported by the concomitant decrease in fat levels.
Prohibitins localize to the inner membrane of the mitochondria, which is the only eukaryotic membrane to contain the bacterial lipid cardiolipin (Marchi et al., 2012). Given the shared localization of prohibitins and cardiolipin it is tempting to speculate that cardiolipin abundance could alter lifespan by affecting the properties of the inner mitochondrial membrane and its associated proteins. A recent study on wild-type worms found that cardiolipin abundance decreases with worm age, as does mitochondrial number (Gruber et al., 2011). Another study found that cardiolipin synthase mutant worms exhibit a decreased mitochondrial membrane potential and developmental phenotypes, but no lifespan phenotype was reported (Sakamoto et al., 2012). It would be informative to find out whether cardiolipin synthase mutation, or perhaps cardiolipin depletion in adult worms, affects aging in wild-type worms, or in long-lived mutants.
In addition to the requirement for prohibitins in several longevity pathways, mitochondrial membrane potential – the driving force for ATP generation – may broadly impact aging. Lemire et al. (2009) found that reduced mitochondrial membrane potential correlates with long life span in several mutants and RNAi treated worms. Moreover, the artificial reduction of membrane potential with a chemical uncoupler – which dissipates the proton gradient across the inner mitochondrial membrane and thus prevents ATP synthesis – directly increases lifespan. Given the effect of chemical uncoupling, one might expect that channels alleviating the protein gradient, such as the uncoupling proton UCP-4, might also positively influence aging. However, ucp-4 deficiency does not alter the lifespan of wild-type worms (Iser et al., 2005), albeit it is possible that ucp-4 is required for the long lifespan of certain mutants. Together, the above studies suggest that mitochondria, and perhaps especially membrane potential and the function of the inner mitochondrial membrane, may play important roles in multiple pathways that extend lifespan.
Recent discoveries have also shed new light on the mechanisms by which mitochondrial ETC mutants may achieve an extended life span. An exciting study by Durieux et al. (2011) found that ETC loss is required specifically in the intestine to delay aging. A signal emanating from the intestine – termed a mitokine – is proposed to set the rate of aging throughout the body. The molecular nature of the proposed mitokine is unclear, but ROS come immediately to mind, given the recent identification of superoxide as a signaling molecule in long-lived mitochondrial mutants (Yang and Hekimi, 2010). Lipid-derived molecules could also play such a role, especially given that the C. elegans intestine is the major organ involved in lipid metabolism. NAEs, or perhaps ascarosides, could in principle perform such functions. Whatever the molecule, these studies all reinforce the notion that mitochondria play a key role in influencing life span, and provide new evidence that lipids are paramount for the longevity-affecting role of mitochondria.
Sphingolipid and Ceramide Signaling
Sphingolipids and ceramides constitute a diverse class of lipids that play important roles in many processes, including cellular proliferation, differentiation, and apoptosis (Kolesnick, 2002). Ceramides are composed of a sphingosine group and a fatty acid, and they represent important components of cellular structures, especially membranes, while also participating in cellular signaling pathways. Notably, one of the first genes found to affect yeast longevity, Longevity Assurance Gene 1 (LAG1), encodes a ceramide synthase (D’Mello et al., 1994; Guillas et al., 2001). LAG1 has four homologs in C. elegans, and a recent paper investigated the potential of these genes to influence life span in C. elegans (Tedesco et al., 2008). However, neither mutation nor overexpression of hyl-1, the closest LAG1 ortholog, results in life span extension, and hyl-1 depletion only causes a mild lifespan extension. These conflicting results may relate to the fact that different E. coli strains were used as food sources in mutant and RNAi studies; alternatively, the RNAi clone may have off target effects. Depletion of two hyl-1 homologs, hyl-2 and lagr-1, results in a shortened lifespan. In summary, whether and how ceramides and/or sphingolipids affect longevity in C. elegans remains obscure and needs to be studied further.
Conclusion
Lipids are key for many biological processes, acting in structural, metabolic, and/or signaling capacities. It is thus of little surprise that lipids, and the enzymes involved in lipid synthesis and remodeling, are materializing as key players in aging. We have highlighted recent studies in C. elegans that depict exciting new connections between aging and lipid biology, including emerging roles for lipases and fatty acid desaturases, and the identification of lipid-derived signaling molecules that influence – or may influence – aging. Some of the challenges that lie ahead include defining in which tissues and by what molecular mechanisms these newly discovered molecules act (including the identification of their receptors). It will also be important to determine whether these molecules and enzymes such as lipases act more broadly or in restricted fashion.
The recent identification of novel lipid-derived signaling molecules that affect aging suggests that our insight into lipid signaling is likely incomplete. Similar limitations may apply to our views of structural and metabolic contributions of lipids to aging. To gain better insight into lipid action, and to identify the key molecules/metabolites linked to aging we may benefit from new technologies such as (lipid) metabolomics. Recent publications suggest that metabolomics is a powerful approach to identify novel regulatory relationships (Walker et al., 2011), and to reveal metabolite changes in aging worms (Fuchs et al., 2010). However, metabolomic approaches have to date primarily been used to analyze whole-worm extracts, which provides little insight into tissue-restricted or subcellular roles of individual lipids. Perhaps, organelle purification may be required to yield information about spatially restricted roles for certain lipids. Similarly, sophisticated analysis of individual lipid synthesis pathways as performed by Perez and Van Gilst (2008) can be used to delineate activity changes of certain pathways over time, e.g., in aging worms.
In summary, the studies reviewed here provide exciting new directions for future research on the roles of lipids in aging. Given that many genes and molecules are conserved in mammals, there is a great potential that such pathways may be developed as targets for drugs to ameliorate age-related diseases and/or slow aging.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. M. Hansen and D. Park for critical comments on the manuscript, and Jennifer M. Grants for help with illustrations. Stefan Taubert holds the Canada Research Chair in Transcriptional Regulatory Networks, and obtains research support from the Canadian Institute of Health Research (MOP-93713 and IAB-112231), the Canada Foundation of Innovation, UBC, CMMT, and CFRI.
References
- Ackerman D., Gems D. (2012). The mystery of C. elegans aging: an emerging role for fat: distant parallels between C. elegans aging and metabolic syndrome? BioEssays. 10.1002/bies.201100189 [DOI] [PubMed] [Google Scholar]
- Antebi A., Culotti J. G., Hedgecock E. M. (1998). daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125, 1191–1205 [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh W. H., Tait D., Hedgecock E. M., Riddle D. L. (2000). daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14, 1512–1527 [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N. (2002). Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505 10.1126/science.1065768 [DOI] [PubMed] [Google Scholar]
- Arda H. E., Taubert S., Macneil L. T., Conine C. C., Tsuda B., Van Gilst M., Sequerra R., Doucette-Stamm L., Yamamoto K. R., Walhout A. J. M. (2010). Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol. Syst. Biol. 6, 367. 10.1038/msb.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M., Tavernarakis N. (2009). Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature 461, 793–797 [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. (2003). Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 10.1038/nature01279 [DOI] [PubMed] [Google Scholar]
- Bank E. M., Ben-Harush K., Wiesel-Motiuk N., Barkan R., Feinstein N., Lotan O., Medalia O., Gruenbaum Y. (2011). A laminopathic mutation disrupting lamin filament assembly causes disease-like phenotypes in Caenorhabditis elegans. Mol. Biol. Cell 22, 2716–2728 10.1091/mbc.E11-01-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank E. M., Gruenbaum Y. (2011). Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2, 350–357 10.4161/nucl.2.5.17838 [DOI] [PubMed] [Google Scholar]
- Banni S., Di Marzo V. (2010). Effect of dietary fat on endocannabinoids and related mediators: consequences on energy homeostasis, inflammation and mood. Mol. Nutr. Food Res. 54, 82–92 10.1002/mnfr.200900516 [DOI] [PubMed] [Google Scholar]
- Branicky R., Desjardins D., Liu J.-L., Hekimi S. (2010). Lipid transport and signaling in Caenorhabditis elegans. Dev. Dyn. 239, 1365–1377 [DOI] [PubMed] [Google Scholar]
- Brock T. J., Browse J., Watts J. L. (2006). Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2 e108. 10.1371/journal.pgen.0020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks K. K., Liang B., Watts J. L. (2009). The influence of bacterial diet on fat storage in C. elegans. PLoS ONE 4, e7545. 10.1371/journal.pone.0007545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya Y. V., Wu K., Southworth L. K., Jiang M., Tedesco P., Johnson T. E., Kim S. K. (2008). An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134, 291–303 10.1016/j.cell.2008.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R. A., Fujita M., Schroeder F. C., Clardy J. (2007). Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3, 420–422 10.1038/nphys583 [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Li W., Ruvkun G., Clardy J., Mak H. Y. (2009). Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc. Natl. Acad. Sci. U.S.A. 106, 1875. 10.1073/pnas.0906762106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Whetstine J. R., Ghosh S., Hanover J. A., Gali R. R., Grosu P., Shi Y. (2009). The conserved NAD (H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc. Natl. Acad. Sci. U.S.A. 106, 1496. 10.1073/pnas.0903497106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina D., Cary M., Lunceford A., Clarke C., Kenyon C. (2009). A Regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 5, e1000450 10.1371/journal.pgen.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A. (2003). Lamin a truncation in Hutchinson-Gilford progeria. Science 300, 2055–2055 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- D’Mello N. P., Childress A. M., Franklin D. S., Kale S. P., Pinswasdi C., Jazwinski S. M. (1994). Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem. 269, 15451–15459 [PubMed] [Google Scholar]
- Durieux J., Wolff S., Dillin A. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R. H., Shimizu T., Spener F., van Meer G., Wakelam M. J. O., Dennis E. A. (2009). Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50(Suppl.), S9–S14 10.1194/jlr.R800095-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bussière F., Hekimi S. (2001). Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644 10.1016/S1534-5807(01)00071-5 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S. Q., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fontana L., Partridge L., Longo V. D. (2010). Extending healthy life span – from yeast to humans. Science 328, 321–326 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodríguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., Piomelli D. (2003). Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature 425, 90–93 10.1038/nature01921 [DOI] [PubMed] [Google Scholar]
- Fuchs S., Bundy J. G., Davies S. K., Viney J. M., Swire J. S., Leroi A. M. (2010). A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 8, 14. 10.1186/1741-7007-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gáliková M., Klepsatel P., Senti G., Flatt T. (2011). Steroid hormone regulation of C. elegans and Drosophila aging and life history. Exp. Gerontol. 46, 141–147 10.1016/j.exger.2010.08.021 [DOI] [PubMed] [Google Scholar]
- Gallo M., Riddle D. L. (2010). Regulation of metabolism in Caenorhabditis elegans longevity. J. Biol. 9, 7. 10.1186/jbiol215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P., Staels B., Dacquet C., Spedding M., Laudet V. (2006). Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 58, 685–704 10.1124/pr.58.4.4 [DOI] [PubMed] [Google Scholar]
- Golden T. R., Hubbard A., Dando C., Herren M. A., Melov S. (2008). Age-related behaviors have distinct transcriptional profiles in Caenorhabditis elegans. Aging Cell 7, 850–865 10.1111/j.1474-9726.2008.00433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. J. (2008). Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab. Pharmacokinet. 23, 2–7 10.2133/dmpk.23.2 [DOI] [PubMed] [Google Scholar]
- Goudeau J., Bellemin S., Toselli-Mollereau E., Shamalnasab M., Chen Y., Aguilaniu H. (2011). Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 9, e1000599. 10.1371/journal.pbio.1000599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J., Ng L. F., Fong S., Wong Y. T., Koh S. A., Chen C.-B., Shui G., Cheong W. F., Schaffer S., Wenk M. R., Halliwell B. (2011). Mitochondrial changes in ageing Caenorhabditis elegans – what do we learn from superoxide dismutase knockouts? PLoS ONE 6, e19444. 10.1371/journal.pone.0019444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. (2001). C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 20, 2655–2665 10.1093/emboj/20.11.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaschek-Wiener J., Khattra J. S., McKay S., Pouzyrev A., Stott J. M., Yang G. S., Holt R. A., Jones S. J. M., Marra M. A., Brooks-Wilson A. R., Riddle D. L. (2005). Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 15, 603–615 10.1101/gr.3274805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Chandra A., Mitic L. L., Onken B., Driscoll M., Kenyon C. (2008). A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24. 10.1371/journal.pgen.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A.-L., Dillin A., Kenyon C. (2005). New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, e17. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J., Kenyon C. (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Harman D. (1956). Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11, 298–300 [DOI] [PubMed] [Google Scholar]
- Hekimi S., Lapointe J., Wen Y. (2011). Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 21, 569–576 10.1016/j.tcb.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S., Zhang P., Hansen M., McCormick M., Lee S.-J., Cary M., Kenyon C. (2010). Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. U.S.A. 107, 9730–9735 10.1073/pnas.1002575107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz R., Ben-Haim N., Petrescu A. D., Kalderon B., Berman I., Eldad N., Schroeder F., Bar-Tana J. (2003). Rescue of MODY-1 by agonist ligands of hepatocyte nuclear factor-4alpha. J. Biol. Chem. 278, 22578–22585 10.1074/jbc.M212138200 [DOI] [PubMed] [Google Scholar]
- Hillyard S. L., German J. B. (2009). Quantitative lipid analysis and life span of the fat-3 mutant of Caenorhabditis elegans. J. Agric. Food Chem. 57, 3389–3396 10.1021/jf8031414 [DOI] [PubMed] [Google Scholar]
- Hsin H., Kenyon C. (1999). Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 10.1038/20694 [DOI] [PubMed] [Google Scholar]
- Hu P. J. (2007). Dauer. WormBook 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert A. (2011). Longevity, lipids and C. elegans. Aging (Albany N. Y.) 3, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iser W. B., Kim D., Bachman E., Wolkow C. (2005). Examination of the requirement for ucp-4, a putative homolog of mammalian uncoupling proteins, for stress tolerance and longevity in C. elegans. Mech. Ageing Dev. 126, 1090–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong P.-Y., Jung M., Yim Y.-H., Kim H., Park M., Hong E., Lee W., Kim Y. H., Kim K., Paik Y.-K. (2005). Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433, 541–545 10.1038/nature03201 [DOI] [PubMed] [Google Scholar]
- Joo H. J., Kim K. Y., Yim Y. H., Jin Y. X., Kim H., Kim M. Y., Paik Y. K. (2010). Contribution of the peroxisomal acox gene to the dynamic balance of daumone production in Caenorhabditis elegans. J. Biol. Chem. 285, 29319–29325 10.1074/jbc.M110.122663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H. J., Yim Y.-H., Jeong P.-Y., Jin Y. X., Lee J. E., Kim H., Jeong S. K., Chitwood D. J., Paik Y.-K. (2009). Caenorhabditis elegansutilizes dauer pheromone biosynthesis to dispose of toxic peroxisomal fatty acids for cellular homoeostasis. Biochem. J. 422, 61–71 10.1042/BJ20090513 [DOI] [PubMed] [Google Scholar]
- Kenyon C. (2010a). The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Proc. R. Soc. Lond. B Biol. 366, 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Kenyon C. J. (2010b). The genetics of ageing. Nature 464, 504–512 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Kim K., Sato K., Shibuya M., Zeiger D. M., Butcher R. A., Ragains J. R., Clardy J., Touhara K., Sengupta P. (2009). Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326, 994–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnick R. (2002). The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J. Clin. Invest. 110, 3–8 10.1172/JCI200216127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M., Thompson C. B. (2008). Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 3, 427–455 10.1146/annurev.pathmechdis.2.010506.091842 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S. (1996). Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272, 1010–1013 10.1126/science.272.5264.1010 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S. (1998). The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 95, 13091–13096 10.1073/pnas.95.22.13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L. R., Gelino S., Meléndez A., Hansen M. (2011). Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 21, 1507–1514 10.1016/j.cub.2011.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire B. D., Behrendt M., DeCorby A., Gášková D. (2009). C. elegans longevity pathways converge to decrease mitochondrial membrane potential. Mech. Ageing Dev. 130, 461–465 10.1016/j.mad.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Lucanic M., Held J. M., Vantipalli M. C., Klang I. M., Graham J. B., Gibson B. W., Lithgow G. J., Gill M. S. (2011). N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans. Nature 473, 226–229 10.1038/nature10007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestro M. A., Cardalda C., Boj S. F., Luco R. F., Servitja J. M., Ferrer J. (2007). Distinct roles of HNF1beta, HNF1alpha, and HNF4alpha in regulating pancreas development, beta-cell function and growth. Endocr. Dev. 12, 33–45 10.1159/000109603 [DOI] [PubMed] [Google Scholar]
- Marchi S., Giorgi C., Suski J. M., Agnoletto C., Bononi A., Bonora M., De Marchi E., Missiroli S., Patergnani S., Poletti F., Rimessi A., Duszynski J., Wieckowski M. R., Pinton P. (2012). Mitochondria-ros crosstalk in the control of cell death and aging. J. Signal Transduct. 2012, 1–17 10.1155/2012/807682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias I., Di Marzo V. (2007). Endocannabinoids and the control of energy balance. Trends Endocrinol. Metab. 18, 27–37 10.1016/j.tem.2006.11.006 [DOI] [PubMed] [Google Scholar]
- McCormick M., Chen K., Ramaswamy P., Kenyon C. (2012). New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell 11, 192–202 10.1111/j.1474-9726.2011.00768.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. T., Xu Y., Ailion M., Garrison J. L., Butcher R. A., Bargmann C. I. (2011). Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477, 321–325 10.1038/nature10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland J. M., Glass M. (2003). Functional mapping of cannabinoid receptor homologs in mammals, other vertebrates, and invertebrates. Gene 312, 297–303 10.1016/S0378-1119(03)00638-3 [DOI] [PubMed] [Google Scholar]
- Meléndez A., Tallóczy Z., Seaman M., Eskelinen E.-L., Hall D. H., Levine B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 [DOI] [PubMed] [Google Scholar]
- Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T., Li Y., Suino-Powell K., Xu H. E., Auchus R. J., Antebi A., Mangelsdorf D. J. (2006). Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124, 1209–1223 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A. J. M., Tissenbaum H. A. (2005). C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab. 2, 35–42 10.1016/j.cmet.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C. (2003). Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Nicholls D. G. (2002). Mitochondrial function and dysfunction in the cell: its relevance to aging and aging-related disease. Int. J. Biochem. Cell Biol. 34, 1372–1381 10.1016/S1357-2725(02)00077-8 [DOI] [PubMed] [Google Scholar]
- O’Rourke E. J., Soukas A. A., Carr C. E., Ruvkun G. (2009). C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10, 430–435 10.1016/j.cmet.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L., Tennessen J. M., Lam G., Thummel C. S. (2009). Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 9, 228–239 10.1016/j.cmet.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., Lithgow G. J., Kapahi P. (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 10.1111/j.1474-9726.2006.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Jones K. L., Lee H., Snutch T. P., Taubert S., Riddle D. L. (2012). Repression of a potassium channel by nuclear hormone receptor and TGF-β signaling modulates insulin signaling in Caenorhabditis elegans. PLoS Genet. 8, e1002519. 10.1371/journal.pgen.1002519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C. L., Van Gilst M. R. (2008). A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 8, 266–274 [DOI] [PubMed] [Google Scholar]
- Pungaliya C., Srinivasan J., Fox B. W., Malik R. U., Ludewig A. H., Sternberg P. W., Schroeder F. C. (2009). A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 106, 7708. 10.1073/pnas.0811918106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Maina C. V., Gissendanner C. R., Laudet V., Sluder A. (2005). Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J. Mol. Evol. 60, 577–586 10.1007/s00239-004-0175-8 [DOI] [PubMed] [Google Scholar]
- Rogers A. N., Chen D., Mccoll G., Czerwieniec G., Felkey K., Gibson B. W., Hubbard A., Melov S., Lithgow G. J., Kapahi P. (2011). Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 14, 55–66 10.1016/j.cmet.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Inoue T., Otomo Y., Yokomori N., Ohno M., Arai H., Nakagawa Y. (2012). Deficiency of cardiolipin synthase causes abnormal mitochondrial function and morphology in germ cells of Caenorhabditis elegans. J. Biol. Chem. 287, 4590–4601 10.1074/jbc.M111.314823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Xu L., Lee H., Chae M., Thaden J. J., Bharill P., Tazearslan C., Siegel E., Alla R., Zimniak P., Ayyadevara S. (2011). Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany N. Y.) 3, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., von Reuss S. H., Bose N., Zaslaver A., Mahanti P., Ho M. C., O’Doherty O. G., Edison A. S., Sternberg P. W., Schroeder F. C. (2012). A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 10, e1001237. 10.1371/journal.pbio.1001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Sadegh L., Elle I. C., Christensen A. G. L., Faergeman N. J., Ashrafi K. (2008). Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 7, 533–544 10.1016/j.cmet.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Fahy E., Gupta S., Sud M., Byrnes R. W., Cotter D., Dinasarapu A. R., Maurya M. R. (2011). Bioinformatics and systems biology of the lipidome. Chem. Rev. 111, 6452–6490 10.1021/cr200295k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Hansen M., Van Gilst M. R., Cooper S. B., Yamamoto K. R. (2008). The mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 4, e1000021. 10.1371/journal.pgen.1000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Van Gilst M. R., Hansen M., Yamamoto K. R. (2006). A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20, 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Ward J. D., Yamamoto K. R. (2011). Nuclear hormone receptors in nematodes: evolution and function. Mol. Cell. Endocrinol. 334, 49–55 10.1016/j.mce.2010.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco P., Jiang J., Wang J., Jazwinski S. M., Johnson T. E. (2008). Genetic analysis of hyl-1, the C. elegans homolog of LAG1/LASS1. Age (Dordr.) 30, 43–52 10.1007/s11357-008-9046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth M. L., Sigmond T., Borsos E., Barna J., Erdélyi P., Takács-Vellai K., Orosz L., Kovács A. L., Csikós G., Sass M., Vellai T. (2008). Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338 [DOI] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R. (2005a). A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 102, 13496–13501 10.1073/pnas.0506234102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Jolly A., Yamamoto K. R. (2005b). Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3, e53. 10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verme J. L. (2005). The nuclear receptor peroxisome proliferator-activated receptor- mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 67, 15–19 10.1124/mol.104.006353 [DOI] [PubMed] [Google Scholar]
- von Reuss S. H., Bose N., Srinivasan J., Yim J. J., Judkins J. C., Sternberg P. W., Schroeder F. C. (2012). Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J. Am. Chem. Soc. 134, 1817–1824 10.1021/ja210202y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K., Finnegan D. M., Shioda T., Hansen M., Yang F., Niebergall L. J., Vance D. E., Tzoneva M., Hart A. C., Näär A. M. (2011). A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147, 840–852 10.1016/j.cell.2011.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. C., Min W., Freudiger C. W., Ruvkun G., Xie X. S. (2011). RNAi screening for fat regulatory genes with SRS microscopy. Nat. Methods 8, 135–138 10.1038/nmeth.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. C., O’Rourke E. J., Ruvkun G. (2008). Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960 10.1126/science.1162011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A., Boutis P., Hekimi S. (1995). Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Vought B. W., Satterlee J. S., Walker A. K., Jim Sun Z.-Y., Watts J. L., DeBeaumont R., Saito R. M., Hyberts S. G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A. C., Wagner G., Naeaer A. M. (2006). An ARC/mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 10.1038/nature04942 [DOI] [PubMed] [Google Scholar]
- Yang W., Hekimi S. (2010). A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 8, e1000556. 10.1371/journal.pbio.1000556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman M. J., Rogers Z. N., Kim D. H. (2011). A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 7, e1002082. 10.1371/journal.pgen.1002082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Ta T. C., Lin M., Evans J. R., Dong Y., Bolotin E., Sherman M. A., Forman B. M., Sladek F. M. (2009). Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS ONE 4, e5609. 10.1371/journal.pone.0005609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. O., Box A. C., Xu N., Le Men J., Yu J., Guo F., Trimble R., Mak H. Y. (2010). Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 4640–4645 10.1073/pnas.0910179107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. I., Pincus Z., Slack F. J. (2011). Longevity and stress in Caenorhabditis elegans. Aging (Albany N. Y.) 3, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]