Abstract
The β-haemoglobinopathies are the most prevalent inherited disorders worldwide. Gene therapy of β-thalassaemia is particularly challenging given the requirement for massive haemoglobin production in a lineage-specific manner and the lack of selective advantage for corrected haematopoietic stem cells. Compound βE/β0-thalassaemia is the most common form of severe thalassaemia in southeast Asian countries and their diasporas1,2. The βE-globin allele bears a point mutation that causes alternative splicing. The abnormally spliced form is non-coding, whereas the correctly spliced messenger RNA expresses a mutated βE-globin with partial instability1,2. When this is compounded with a non-functional β0 allele, a profound decrease in β-globin synthesis results, and approximately half of βE/β0-thalassaemia patients are transfusion-dependent1,2. The only available curative therapy is allogeneic haematopoietic stem cell transplantation, although most patients do not have a human-leukocyte-antigen-matched, geno-identical donor, and those who do still risk rejection or graft-versus-host disease. Here we show that, 33 months after lentiviral β-globin gene transfer, an adult patient with severe βE/β0-thalassaemia dependent on monthly transfusions since early childhood has become trans-fusion independent for the past 21 months. Blood haemoglobin is maintained between 9 and 10 g dl–1, of which one-third contains vector-encoded β-globin. Most of the therapeutic benefit results from a dominant, myeloid-biased cell clone, in which the integrated vector causes transcriptional activation of HMGA2 in erythroid cells with further increased expression of a truncated HMGA2 mRNA insensitive to degradation by let-7 microRNAs. The clonal dominance that accompanies therapeutic efficacy may be coincidental and stochasticor resultfrom a hithertobenign cellexpansion caused by dysregulation of the HMGA2 gene in stem/progenitor cells.
The design of integrative vectors for human β-globin gene transfer has been difficult. Thegenetic elements required for high and erythroid-specific expression are complex: the β-globin gene with its introns, promoter and β-locus control region (β-LCR)3,4. Lentiviral vectors have proven capable of transferring these elaborate structures with fidelity and high titres5,6. Hence, several mouse models of the β-haemoglobinopathies have been corrected, long-term, by ex vivo transduction of haematopoietic stem cells (HSCs) with β-globin lentiviral vectors5–10. These advances have prompted the prudent initiation of a human clinical trial (Supplementary Note 1).
The general structure of the β-globin-expressing lentiviral vector has been previously described6,8 (Supplementary Fig. 1). It is a self-inactivating vector with two copies of the 250-base-pair (bp) core of the cHS4 chromatin insulator11 implanted in the U3 region. It encodes a mutated adult β-globin (βA(T87Q)) with anti-sickling properties6 that can be distinguished from normal adult β-globin (βA) by high-performance liquid chromatography (HPLC) analysis in individuals receiving red blood cell transfusions and/or β1-thalassaemia patients.
This report focuses on the first treated patient (P2) who did not receive back-up cells: a male, aged 18 years at the time of treatment, with severe βE/β0-thalassaemia. A previous patient (P1) failed to engraft because the HSCs had been compromised by the technical handling of the cells without relation to the gene therapy vector. P1 failed to engraft after 5 weeks and was thus given back-up cells (Supplementary Note 2). P2 was first transfused at age three because of poorly tolerated anaemia (6.7 g dl–1 despite residual fetal haemoglobin (HbF)) and major hepatosplenomegaly. Transfusion requirements rapidly increased to once a month (2–3 red blood cell packs each time; 157 ml of red blood cells per kg the year before transplant). He was splenectomized at age 6. In spite of this, Hb levels decreased several times to as low as 4 g dl–1, and hydroxurea therapy was ineffective. Iron chelation was initiated at age 8 by parenteral deferoxamine overnight, 5 times a week. The patient did not have a related human-leukocyte-antigen-matched donor and was thus enrolled in this trial after informed consent.
The ex vivo transduction efficiency of bulk bonemarrow CD341 cells was 0.6 vector per cell after 1 week in culture after gene transfer. The patient was conditioned by intravenous Busulfex (3.2 mg kg–1 day–1 for 4 days) without the addition of cyclophosphamide, before transplantation with autologous gene-modified and cryopreserved cells (3.9 × 106 CD34+ cells per kg). Haematopoietic reconstitution was uneventful (Supplementary Fig. 2b).
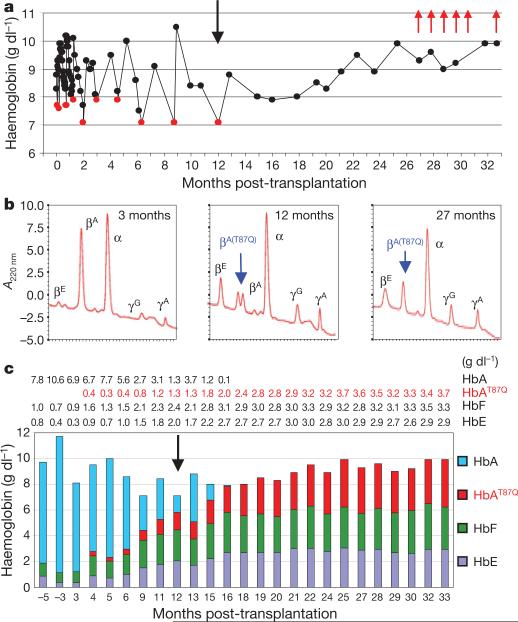
Patient P2 was transplanted on 7 June 2007. At present, approximately 3 years post-transplantation, the biological and clinical evolution is remarkable, and the patient's quality of life is good. The patient was last transfused on 6 June 2008 (Fig. 1a, c) and has stable Hb levels between 9 and 10 g dl–1 (Fig. 1a), of which approximately one-third is comprised of each of the therapeutic Hb-βA(T87Q) (αβ(T87Q) Hb dimer), HbE (αβE Hb dimer) and HbF (αγ Hb dimer) (Fig. 1b, c and Supplementary Fig. 3a), with HbA2 (αδ Hb dimer) being negligible. No trace of transfused HbA (αβA Hb dimer) has been detected in the blood for the past 17 months (Fig. 1c). The ratios of γ/βE-globin chains remain similar, 33 months post-transplantation, when compared to levels at the time of transplantation, following a transient supplemental rise in γ-globin expression shortly after cell engraftment (Supplementary Fig. 3b). These observations indicate that the concurrent increase in γ- and βE-globin blood concentration is the consequence of the discontinuation of blood transfusions, which exerted a global suppressive effect on endogenous globin gene expression, together with the downstream selection for red blood cells with the highest Hb-βA(T87Q), HbE and HbF combined content. Although there is partial correction of the anaemia with concurrent decrease in blood reticulocyte and erythroblast counts (Supplementary Fig. 2c, d), the hypererythroid state remains (Supplementary Note 3). Consistent with previous studies in mice12, it is likely that sufficiently corrected β-thalassaemic red blood cells are selected in the blood due to their prolonged lifespan. As expected from the vector design that confers erythroid-specific expression, βA(T87Q)-globin was undetectable in purified blood CD15+ cells (granulocytes-monocytes) (data not shown). Because the patient had poor tolerance to oral iron chelation (deferasirox), deferiprone was substituted and phlebotomies were performed during the past 6 months to withdraw 200 ml of blood each month (Fig.1a). Notably, blood Hb levels havebeen maintained in spite of a cumulated blood withdrawal of 1.2 l.
Figure 1. Conversion to transfusion independence.
a, Total Hb concentrations in whole blood. Red dots, transfusion time points; black vertical arrow, the last time the patient was transfused; red arrows, phlebotomies (200 ml each) to remove excess iron. b, HPLC blood globin chain profiles. Note that βA only derives from blood transfusions. c, Contribution of each Hb species, quantified by HPLC, to total blood Hb concentrations (in g dl–1). Actual numbers for each Hb species are indicated above the chart.
Twenty-one months after the last transfusion, the mean corpuscular haemoglobin (MCH) of the last 6 months averaged 28.7 pg, which is within the normal range (27–32 pg; mean 29.5 pg) and contrasts with the low values averaging at 19 pg found in non-transfused βE/β0-thalassaemia patients1. Because 36.2% of all the β-like globin chains produced during the last 6 months is βA(T87Q)-globin, the contribution of βA(T87Q)-globin to the measured MCH was 28.7 × 0.362 5 10.4 pg. The ratio of βA(T87Q)-globin output to the mean normal endogenous βA-globin output on a per-gene basis is thus 10.4/(29.5/2) 5 70.5%. Because it is unlikely that all of the circulating red blood cells derive from vector-bearing progenitors despite peripheral selection for corrected red blood cells, one can surmise that βA(T87Q)-globin expression assessed from red blood cells is anywhere between 70.5% and 100% of normal value on a per-gene basis.
The percentage of vector-bearing nucleated blood cells (NBCs) increased progressively after transplant and stabilized at ~11% (Table 1 and Supplementary Fig. 2a). Among blood subsets, 18.7% and 9.3% of granulocytes-monocytes (CD151) and B lymphocytes (CD191), respectively, contain an integrated vector at 33 months post-transplantation (Table 1). The lower level of gene marking for erythroblasts (CD45–/CD71+) of blood (~2.9%) versus bone marrow origin (9.8% at 24 months post-transplantation) (Table 1) is in agreement with the contention that genetically corrected erythroblasts appear less frequently in the blood than uncorrected erythroblasts, as the former are capable of differentiating further within the bone marrow compartment into reticulocytes and red blood cells. The low level of T-cell (CD3+) gene marking (~1.7%) (Table 1) is probably due to the omission of cyclophosphamide from the conditioning regimen. Ten to twenty per cent of bone marrow burst-forming units-erythroid (BFU-Es) and colony-forming units-granulocyte/ monocyte (CFU-GMs) were vector positive by polymerase chain reaction (PCR) (Table 1). At 24 months post-transplantation, the proportion of long-term culture-initiating cells (LTC-ICs) (~1/1,000 CD34+ cells) bearing an integrated vector was 11.1% (Table 1).
Table 1.
Percentages of vector-bearing cells in blood and bone marrow cells
| Cell fraction | Months |
||||||
|---|---|---|---|---|---|---|---|
| 21 | 24 | 25 | 27 | 30.5 | 32 | 33 | |
| Blood | |||||||
| Whole blood (%) | 6.3 | 7.0 | 7.4 | 8.5 | 10.7 | 10.9 | 10.8 |
| T lymphocytes (%) | 0.9 | 0.7 | 1.1 | 1.3 | 1.7 | 1.7 | 1.7 |
| B lymphocytes (%) | 5.3 | 5.7 | 6.6 | 9.3 | 13.5 | 9.5 | 9.3 |
| Granulocytes-monocytes (%) | 12.3 | 13.1 | 14.9 | 16.2 | 15.6 | 19.3 | 18.7 |
| Erythroblasts (%) | 1.9 | 2.1 | 2.2 | 1.8 | 2.8 | 3.3 | 2.9 |
| Bone marrow | |||||||
| Whole bone marrow (%) | 11.5 | 13.5 | – | – | – | – | – |
| BFU-E (%) | 20.0 | 10.6 | – | – | – | – | – |
| CFU-GM (%) | 18.2 | 13.0 | – | – | – | – | – |
| Erythroblasts (%) | – | 9.8 | – | – | – | – | – |
| LTC-ICs (%) | – | 11.1 | – | – | – | – | – |
Chromosomal integration sites (IS) of the βA(T87Q)-globin vector in NBCs were analysed by DNA pyrosequencing of ligation-mediated (LM)-PCR products (Fig. 2a) and subsets thereof. Twenty-four of them were found in both lymphoid and myeloid lineages. Cross-contamination was excluded by real-time quantitative (q)PCR to detect specific provirus/flanking DNA junctions in highly purified NBC subsets. Two of the most abundant and stable multilineage IS (lymphoid and myeloid) lay within the RFX3 and ZZEF1 genes. Their contribution, as monitored by qPCR, remained below 0.5% and 1% for RFX3 and ZZEF1, respectively (not shown).
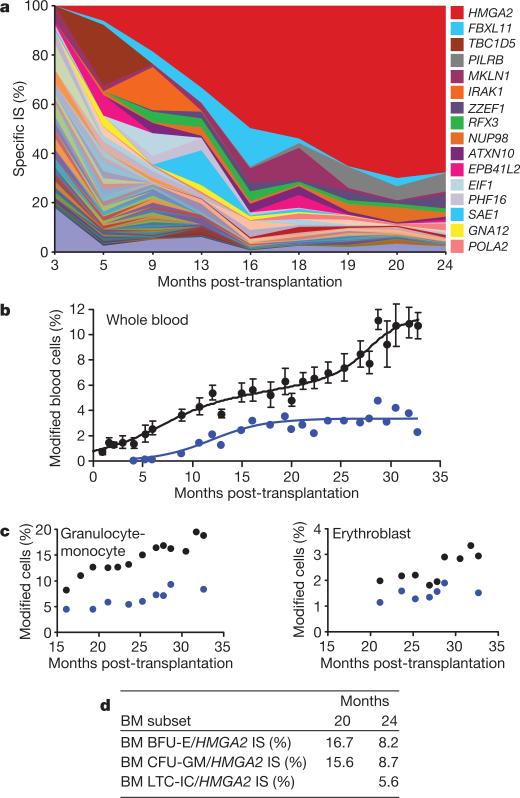
Figure 2. Genome-wide integration site (IS) distribution and HMGA2 IS clonal dominance.
a, Relative abundance of vector-bearing cell clones in NBCs, as analysed by DNA pyrosequencing of LM-PCR products (one colour per IS, with the predominant HMGA2 IS in red). b, c, Mean percentages of vector-bearing NBCs at any IS (black dots, ±s.d.) versus the specific HMGA2 IS (blue dots), as quantified by qPCR in NBCs (b) and in specific cell fractions (c). d, Percentages of vector-bearing BFU-Es, CFU-GMs and LTC-ICs at the HMGA2 IS in bone marrow (BM) harvests.
Importantly, there was dominance over time of an IS at the high mobility group AT-hook 2 (HMGA2)13 locus in both granulocytes-monocytes and erythroblasts (Fig. 2). However, the HMGA2 IS was undetectable by qPCR in fresh or expanded B or T lymphocytes. In myeloid cells, DNA sequence analysis showed that a single vector had integrated in the forward orientation within the ~113-kb third intron of the HMGA2 gene (Fig. 3d). The integrated vector was intact, except for the presence of only one of the two 250-bp cHS4 insulator cores at each end (Supplementary Fig. 1). The qPCR-mediated analysis of the HMGA2/provirus junction showed that ~45% (plateau) of vector-bearing NBCs were positive for the HMGA2 IS (Fig. 2b). However, untransduced cells continued to predominate so that only approximately 3%, 8% and 2% of circulating NBCs, granulocytes-monocytes and erythroblasts, respectively, were HMGA2 IS positive 28 months post-transplantation (Fig. 2b, c). For BFU-Es and CFU-GMs, 8.2% and 8.7%, respectively, are HMGA2 IS positive 24 months post-transplantation (Fig. 2d); this accounts for 67% and 78% of vector-bearing CFU-GMs and BFU-Es, respectively. Notably, the frequency of LTC-ICs positive for HMGA2 IS was 5.6% (Fig. 2d), thus accounting for ~50% of vector-bearing LTC-ICs (Table 1). The mean number of colonies produced by LTC-ICs, or proliferative potential, was not increased when the HMGA2 IS was present (2.6 before transplantation versus 1.5 at 24 months post-transplantation); neither was the mean size of BFU-E colonies (not shown).
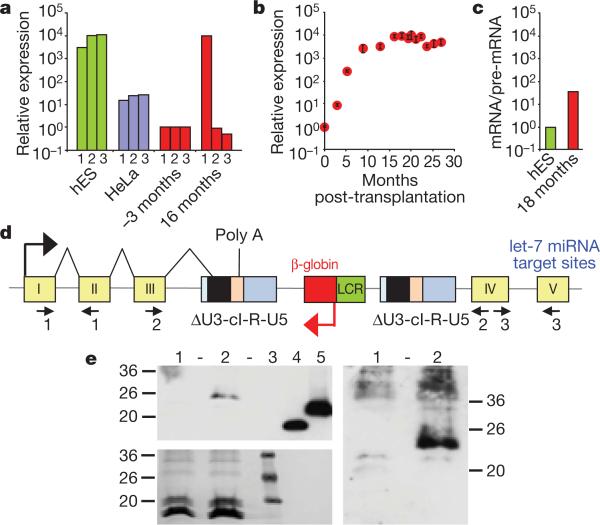
Figure 3. Elevated, erythroid-specific expression of truncated HMGA2 transcripts.
a, Staggered RT-qPCR of HMGA2 transcripts to detect the junctions between exons I–II (1), III–IV (2) and IV–V (3) before (–3 months) and after (16 months) transplantation. HeLa and human embryonic stem (hES) cells express full-length HMGA2 mRNA. b, Kinetics of HMGA2 transcripts (mean ± s.d.). c, Ratios of spliced versus unspliced HMGA2 RNA by RT-qPCR specific for either exon I–intron I or exon I–exon II junctions. d, Diagram of the vector at the HMGA2 IS, showing abnormal splicing and polyadenylation/cleavage of the truncated transcript within the vector provirus. ΔU3, self-inactivating U3; cI, cHS4. e, Western blot analysis with a polyclonal antibody against human HMGA2 and with the enhanced chemiluminescence (ECL, top left) or ECL advance (right) western blot detection systems. Numbers along the left and right edges are the weights (in kilodaltons) of the molecular markers. Bottom left shows Ponceau staining. Numbers along the top indicate blood progenitors from a β-thalassaemic control individual (1), patient P2's blood progenitors 27 months post-transplantation showing a band consistent with truncated fusion protein of 135 amino acids (2), recombinant truncated (73 amino acids) (4) and full-length (109 amino acids) (5) human HMGA2. Molecular mass markers are shown in (3). HMGA2 protein was not detected in CD15+ cells.
Expression of HMGA2 in NBCs was increased by ~10,000-fold, 16 months post-transplantation, as compared to pre-transplant levels (Fig. 3a). However, HMGA2 was only expressed in erythroblasts, consistent with β-LCR erythroid specificity. HMGA2 mRNA was undetectable in granulocytes-monocytes (not shown) despite a greater frequency of HMGA2 IS in granulocytes-monocytes versus erythro-blasts (Fig. 2c). Consistent with the asymptotic saturation of HMGA2 IS representation in NBCs at the DNA level, RT-qPCR analysis revealed that HMGA2 mRNA expression has also stabilized (Fig. 3b).
In certain benign tumours, HMGA2 is activated by chromosomal rearrangement within the long third intron, thereby deleting the two distal exons (IV and V)13. The missing distal portion of the mRNA contains multiple binding sites for let-7 microRNAs, which trigger physiological HMGA2 RNA degradation13. Staggered RT-qPCR reactions specific for each exon junction showed that only exons I to III were present in most of the HMGA2 transcripts in patient P2's NBCs (Fig. 3a). The predominant HMGA2 mRNA species expressed in the patient's cells was sequenced. It is truncated by alternative splicing of the third intron with a cryptic 3' splice signal (GTAT(C)6AG) located within the cHS4 insulator core and cleavage/polyadenylation within the adjacent R region of the left long-terminal-repeat (Fig. 3d). Western blot analysis with a polyclonal antibody against human HMGA2 revealed a reactive protein in the vicinity of the expected size of 135 amino acids, which comprises the 83 amino acids encoded by the first three HMGA2 exons fused, in-frame, to 52 amino acids corresponding to the residual downstream open reading frame of the abnormally spliced transcript (Fig. 3e). In an effort to estimate the respective contributions of β-LCR-mediated enhancer effect versus post-transcriptional mRNA stabilization by loss of let-7 microRNA target sites, we quantified the ratio of spliced mRNA versus unspliced pre-mRNA HMGA2 by RT-qPCR with differential primers and probes, on the basis of the fact that let-7 miRNAs are only functional in the cytoplasm and are thus only capable of degrading cytoplasmic HMGA2 mRNAs and not the nuclear pre-mRNA14. This assay indicated that there was a 36-fold excess of patient P2's truncated HMGA2 mRNA over pre-mRNA compared to the ratio observed in human embryonic stem cells, which express large amounts of full-length HMGA2 mRNA (Fig. 3c). With ~10,000-fold excess of HMGA2 RNA, the β-LCR thus accounted for a calculated 278-fold enhancer effect in erythroid cells.
We then asked whether dominance of the HMGA2 IS is associated with a breach of haematopoietic homeostasis. As seen above, HMGA2 mRNA expression has asymptotically stabilized for the past 15 months (Fig. 3b) in parallel to the observed stabilization of the presence of the clone (Fig. 2b, c). Bone marrow cytology and immunotyping were normal, except for bone marrow erythroid hyperplasia as seen in β-thalassaemia (Supplementary Note 3). Karyotype analysis of bone marrow cells was normal as was high-resolution array-comparative-genomic-hybridization (CGH) analysis of chromosomes. LTC-IC frequency was within the normal range (1 per 1,000 CD34+ cells), and no decrease in cytokine requirement was observed in clonogenic assays.
We also investigated whether vector integration at the HMGA2 IS was associated with altered globin gene expression with respect to cand βA(T87Q)-globin levels. As seen above, there was no increase in γ/βE-globin ratios in the blood after restoration of steady-state haematopoiesis (Supplementary Fig. 3b). In addition, we compared HbF/HbE ratios in single blood-derived BFU-E colonies during the first 6 months post-transplantation—at a time when HMGA2 IS was not yet detectable in erythroid progenitors (n = 8)—to BFU-Es harvested during a recent 12–28 months post-transplantation period when 50–80% of BFU-Es were modified at the HMGA2 IS (n = 64). HbF/HbE ratios remained constant between untransduced (n = 16) and transduced BFU-E colonies, whether the HMGA2 IS was absent or vastly predominant (Supplementary Fig. 3c).
We finally asked whether βA(T87Q)-globin expression was especially high at the HMGA2 IS compared to other IS. At 4 months post-transplantation, at a time when the HMGA2 IS copy number was too low to be quantified by qPCR (<0.06%), βA(T87Q)-globin expression in circulating red blood cells was at least 13% of the sum of all the endogenous β-like-globins (Fig. 1c), although patient P2 was still undergoing blood transfusions and accumulation of corrected red blood cells was far from complete. Furthermore, HPLC analysis performed on individual BFU-E colonies showed that the median Hb βA(T87Q)-globin concentration per colony was significantly greater when HMGA2 IS was undetectable (n = 8) than during the period of HMGA2 IS dominance (n = 64) (Supplementary Fig. 3c), thus indicating that the HMGA2 IS was not especially favourable for high expression of the transferred βA(T87Q)-globin gene.
Since the discovery of the β-LCR over 20 years ago3,4, gene therapy for the β-haemoglobinopathies in humans has been long awaited. Here we have shown that a patient with severe βE/β0-thalassaemia and lifelong transfusion requirement has converted to sustained transfusion independence for the past 21 months. The increase in steady-state Hb provided by βA(T87Q)-globin alone was up to 3.7 g dl–1, which compares favourably to the highest levels reported in mouse models with any β-globin lentiviral vector even at high copy number15. This amount is expected to provide therapeutic benefit in patients with βE/β0-thalassaemia2, where the mean spontaneous blood Hb levels between severe, transfusion-dependent, βE/β0-thalassaemia and better tolerated cases are separated by less than 2 g dl–1 (ref. 2), although the long-term prognosis remains unknown.
The emergence of a partially dominant cell clone requires careful consideration. High, erythroid-specific expression of truncated fusion HMGA2 mRNA and corresponding protein was found to be confined to erythroid cells by a β-LCR-mediated effect—predicted by certain genotoxicity studies16 but not others17—and upon increased stability of the truncated mRNA. Although a single cHS4 insulator 250-bp core has been found to be effective in lentiviral vectors18, the loss of one element in the intended cHS4 doublet may have contributed to insulator failure. The presence of the HMGA2 IS in similar proportion among erythroblasts, granulocytes-monocytes and LTC-IC subsets, but not in lymphocytes, leads us to propose that haematopoiesis deriving from the transduced clone initiating cell is myeloid-biased19,20 in this patient. The considerable departure from polyclonal distribution exhibited by the HMGA2 IS clone (~50% of all IS in myeloid cells, including LTC-ICs) can either be the consequence of a stochastic event in the context of a low initial number of transduced HSCs–1, with strictly erythroid-specific HMGA2 expression, or explained by differential expansion/production of myeloid-biased haematopoiesis triggered by dysregulated expression of HMGA2 in upstream progenitors. The truncated HMGA2 mRNA may thus be transiently expressed at abnormally high levels upon β-LCR lineage priming22, before the β-LCR enhancing capacity becomes subsequently restricted to the erythroid lineage. Although the lack of sufficient numbers of patient P2's bone marrow cells available for analysis precluded the investigation of vector-derived HMGA2 in the rare common myeloid progenitor, the latter hypothesis may have resulted in myeloid-biased cell expansion (Fig. 4). Similarly, the respective roles in HMGA2 activation of the β-LCR versus the loss of let-7 miRNA targets are not directly quantifiable in the rare upstream progenitors and stem cells that cannot be isolated to sufficient numbers and degree of purity. Notably, very high compared with undetectable levels of HMGA2 mRNA in erythroblasts versus granulocyte-monocyte cells, respectively, did not result in a substantial further expansion of erythroid cells.
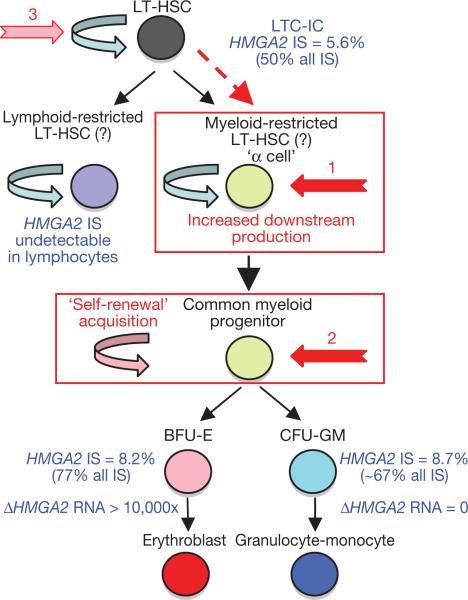
Figure 4. Homeostatic myeloid-biased cell expansion.
Assuming that HMGA2 dysregulation was causative in the onset of clonal dominance, cell expansion is likely to occur by transient HMGA2 expression upstream of the erythroid/granulocyte-monocyte bifurcation (by lineage priming22 and/or loss of let-7 microRNA control13). This is because the HMGA2 IS is represented in erythroblasts, granulocyte-monocyte and LTC-IC cells in similar proportions, whereas HMGA2 expression is only detected in erythroid cells. Because the HMGA2 IS is undetectable in lymphocytes, long-term (LT) homeostatic cell expansion is myeloid-biased. The HMGA2 IS initiating cell is likely to be a myeloid-biased LT-HSC19/α cell20 with increased downstream cell production (1, in red) or a common myeloid progenitor with acquired self-renewal capability (2, in red).
The clone may remain homeostatic or be a prelude to multistep leukaemogenesis23,24; clone exhaustion or substitution may also occur. A reassuring fact is that overexpression of a truncated HMGA2 mRNA is correlated with benign tumour phenotypes (for example, lipomas)13. In addition, the findings in this gene therapy patient bear certain similarities with paroxysmal nocturnal haemoglobinuria (PNH), a clonal yet benign haematopoietic expansion without increased risk of secondary leukaemia25. Paroxysmal nocturnal haemoglobinuria is occasionally associated with multilineage overexpression of a truncated HMGA2 mRNA together with an inactivated PIGA gene25. A paroxysmal nocturnal haemoglobinuria patient with almost complete HMGA2 clonal dominance has been observed for 17 years without leukaemic progression (T. Kinoshita, personal communication). This contrasts with overexpression of the full-length HMGA2 mRNA found in several malignancies, presumably because the primary event is the loss of let-7 microRNAs, which control the degradation of multiple oncogeneic mRNAs that include MYC and RAS26.
The frequent occurrence of homeostatic, clonal haematopoiesis initiated by retroviral activation of proto-oncogenes has been documented in mice27, whereas polyclonal haematopoietic reconstitution with clinical efficacy after lentiviral transfer has been recently reported in a human clinical trial28. Because perturbations of gene expression triggered by integrated vectors have been detected in human gene therapy trials without apparent ill effects29, our understanding of observed cases of polyclonality after integrative gene transfer would benefit from in-depth genome-wide expression studies. Interestingly, we have now identified occurrences of vector integration within the HMGA2 gene, in the absence of leukaemic progression, in several patients of the retroviral gene therapy trial for X-SCID30.
As with any other therapeutic approach, patient treatment decisions should ultimately rest on the ethical evaluation of risk/benefit ratios that would be provided by a larger series of patients with severe β-thalassaemia.
METHODS SUMMARY
Blood and marrow cells of patient P2 were purified by magnetic cell sorting with specific antibodies. Vector-positive progenitors were scored by PCR. Vector copy numbers and specific insertion sites were quantified by qPCR, RNA expression levels by RT-qPCR, and globin chains and Hb species by HPLC. Integration site analysis was performed by LM-PCR and DNA pyrosequencing. Bone marrow analysis included cytology, immunophenotyping, clonogenic assays, karyotype and high-resolution array-CGH.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Post-transplant cell purification
Cell fractions were purified by several rounds of enrichment or depletion using specific antibodies against CD15, CD19, CD3, CD71 or CD45 included in whole-blood purification microbeads and column kits (Miltenyi Biotec). High purity of resulting cell fractions was verified by FACS analysis (>98%). To achieve maximum purity for lymphocytes, purified lymphocytes were grown for 1 week in RPMI medium supplemented with 10% human serum (Institut national de transfusion sanguine) containing phytohaemagglutinin (0.5 μg ml–1; Sigma) and IL-2 (40 U ml–1; Chiron Proleukin) for CD3+ cells and CD40L (500 ng ml–1; Alexis Biochemicals) and IL-4 (100 U ml–1; Peprotech) for CD19+ cells followed by cell sorting with a Becton Dickinson FACS Aria cell sorter (99.9% purity).
qPCR analyses
Genomic DNA was prepared using NucleoSpin blood kits (Macherey Nagel). Average vector copy numbers were determined by qPCR with primers amplifying the packaging signal Ω1 gag after normalization for endogenous β-actin genes (Supplementary Table 1). Results were compared with those obtained after serial dilutions of genomic DNA from a cell line containing one copy of the integrated globin lentiviral (LentiGlobin, LG) vector per haploid genome. Estimation of the mean vector copy number per transduced cell was obtained by qPCR performed on pooled in vitro colony forming cell (CFC) obtained from the blood. The number of individual CFC colonies (used to generate the qPCR-tested pool) positive for the presence of a vector provirus was scored (Supplementary Table 2). The mean vector copy number per transduced cell was thus obtained by dividing the average vector copy number by this value. The per cent of HMGA2-specific provirus relative to total provirus was determined by comparative CT method (ΔΔCT) using the delta value derived from a LG/HMGA2 junction plasmid (containing an equimolar amount of the gag and vector/HMGA2 junction targets) as the standard. The primers for the HMGA2 TaqMan assay are listed (Supplementary Table 1). For qPCR of the LG–RFX3 and LG–ZZEF1 junctions, the same principle was applied with specific primers and probe (Supplementary Table 1).
RT-qPCR analysis
After total RNA extraction with the Purelink micro to midi total RNA purification system (Invitrogen) and cDNA synthesis using the Superscript III first strand synthesis super mix (Invitrogen), the HMGA2 and GAPDH cDNAs were quantified using TaqMan gene expression assays (Supplementary Table 1). The comparative CT method (ΔΔCT) was used to compare HMGA2 production levels between cell types. qPCR analysis of spliced versus unspliced HMGA2 transcripts were performed with specific primers (Supplementary Table 1) as described in Fig. 3. Control samples from which the reverse transcriptase or the sample had been omitted were included. All qPCR assays were performed with the 7300 ABI Prism Detection system and the TaqMan gene expression master mix containing ROX from Applied Biosystems. To demonstrate that only one integrated vector provirus was present in cells with the HMGA2 IS, three qPCR experiments were simultaneously performed on the same genomic DNA preparation with primers specific for either (1) a ‘HIV gag packaging signal’ DNA sequence (common to all vector provirus regardless of IS); (2) the HS3–HS4 junction of the β-LCR cassette included in the vector (also common to all vector provirus regardless of IS); and (3) the HMGA2 IS–vector provirus junction (with the left LTR). Quantitative controls included plasmid DNA constructs that reconstituted a full-length vector provirus either with or without the flanking HMGA2 IS junction, mixed in different proportions. Results in triplicates unambiguously indicate that only one integrant at the HMGA2 IS is present in these cells. Furthermore, a Mu transposon-based method developed by the F.B. laboratory (paper submitted) to cleave DNA with minimal bias showed that no other co-dominant IS was present in HMGA2 IS positive cells.
Western blot analysis
Five million myeloid cells were extracted from methylcellulose-containing erythroid colonies generated from patient P2's blood sample 27 months post-transplantation. Myeloid colonies obtained from the blood of a non-treated β-thalassaemic patient were used as a negative control. One-fifth of the protein volumes was separated on SDS–PAGE, transferred onto nitrocellulose and visualized by Ponceau staining for loading comparison. The chimaeric HMGA2 protein was detected by means of a rabbit polyclonal anti-human HMGA2 antibody31 revealed by the enhanced chemiluminescence (ECL) or ECL advance western blot detection systems (Amersham).
Haematopoietic colonies and PCR amplification
One million peripheral blood mononuclear cells or 7,500 CD34+ bone marrow cells were cultured over 14 days in α-MEM (PAA) based methylcellulose medium (Methocult H4230, StemCell Technologies) supplemented with 2 mM l-glutamine, antibiotics, hEpo (3 U ml–1; Assay design), hIL3 (10 ng ml–1; Peprotech) and hSCF (50 ng ml–1; Peprotech) at 3 × 105 and 2,500 per ml, respectively. The colonies were scored and collected after 14 days incubation at 37 °C and 5% CO2. Colonies were washed with PBS and kept frozen for subsequent analysis. DNA wasobtained upon proteinase K lysis and purified using the Qiaex II gel extraction beads (Qiagen). Detection of the LG vector was carried out by PCR with primers amplifying the vector (Supplementary Table 2). The presence of DNA was monitored using primers amplifying the human erythropoietin gene (Supplementary Table 2). Progenitors carrying a vector at the HMGA2 IS were identified with primers amplifying both the LG vector and the LG–HMGA2 junction (Supplementary Table 2). The PCR reactions were carried out at 94 °C, 60 °C and 72 °C for 30, 30 and 50 s, respectively, and proceeded in the Mastercycler gradient system (Eppendorf) for 35 cycles. Amplified DNA was analysed by gel electrophoresis. For evaluation of vector copy number per HMGA2 modified progenitor, genomic DNA of colonies in which the vector is inserted at the HMGA2 IS was submitted to qPCR analyses as described in the previous section.
LTC-IC assays
LTC-IC assays were performed in StemSpan SFEM medium (Stemcell technologies) on irradiated MS5 monolayers at several dilutions of CD34+ cells (2,000 to 16 per well) in 96-well plates with 12–24 replicate wells per concentration. After 5 weeks with weekly change of one half medium volume, all cells were transferred in α-MEM based methylcellulose medium (GF H4434, Stemcell technologies) to determine the total clonogenic cell content of each LTC. The frequency of LTC-IC was determined using L-Calc software (StemCell Technologies).The meannumber of colonies produced by LTC-IC, or proliferative potential, was calculated by dividing the total number of LTC-ICs by the total number of CFCs. To assess the percentage of vector-modified LTC-ICs during the readout phase of the assay, a maximum of one clonogenic colony per well was submitted to PCR-based scoring to ensure that only independent LTC-ICs were analysed.
HPLC analysis
HPLC analyses were performed with a Prominence chromatograph (Shimadzu) and its LC Solution software. Globin chains from whole blood, pooled erythroid colonies and reticulocytes were separated by reverse-phase HPLC using a 4 × 250 mm Nucleosil 300-5 C4 column (Macherey-Nagel). Samples were eluted with a gradient mixture of solution A (water/acetonitrile/trifluoroacetic acid/heptafluorobutyric acid, 700:300:0.7:0.1) and solution B (water/acetonitrile/trifluoroacetic acid/heptafluorobutyric acid, 450:550:0.5:0.1). The absorbance was measured at 220 nm. Haemoglobins from individual erythroid colonies were separated by ion-exchange HPLC on a PolyCAT A column (PolyLC Inc.). Elution was achieved with a linear gradient mixture of buffer C (Tris 40 mM, KCN 3 mM; pHadjusted at6.5 withacetic acid)and bufferD (Tris 40 mM, KCN 3 mM, NaCl 200 mM; pH adjusted at 6.5 with acetic acid) of different ionic strength. The detection wavelength was 418 nm.
Bone marrow karyotype and high-resolution array-CGH analysis
Total bone marrow cells were cultured for 17, 24 and 48 h. Metaphases were treated for Reverse Heat and Giemsa (RHG) banding, and 30 mitoses were fully analysed following the recommendations of the International System for Human Cytogenetic Nomenclature. Genomic copy number analysis was performed using high-density CGH array technology as previously described. Five-hundred nanograms of genomic DNA was labelled and co-hybridized with control DNA on the 244K Human Genome CGH Microarray (Agilent Technologies). Scanned data were processed using Feature Extraction and DNA Analytics software (Agilent Technologies). The analysis tools ADM1, ADM2 and visual inspection were used to search for copy number abnormalities.
Integration site analysis (LM-PCR and DNA pyrosequencing)
Aliquots of genomic DNA extracted from patient samples were digested using two different restriction enzymes (MseI or NlaIII). The digested samples were ligated to DNA linkers, then digested using ApoI to cleave the internal fragments derived from the vector, and amplified by nested PCR as previously described32. Each second-round LTR specific primer contains a unique 8-nucleotide barcode which indexes the amplification products32. The PCR products were gel purified, pooled and sequenced using the 454/Roche GS FLX platform. Pyrosequencing reads were decoded, trimmed to remove LTR and linker sequences, then mapped to the human genome (hg18) to yield integration sites using criteria as previously described32.
Supplementary Material
Acknowledgements
We thank S. Cross, C. Ballas and L. Duffy for cGMP vector manufacturing and QC testing; T. Andrieux, D. Bachir, C. Courne, A. Henri, A. Janin, A. Moindrot, M.-E. Noguera and F. Pinto for their experimental or medical contributions; F. Calvo, C. Eaves, K. Humphries, G. Manfioletti, R. Nagel, K. Sii Felice and A. Slanetz for discussions; and C. Berry for statistical analysis. This work was supported by NIH grants HL090921 to P.L. and AI52845 and AI082020 to F.B., and l'Association française contre les myopathies.
Footnotes
Author Contributions P.L. is the scientific director of the overall project, conceived the strategy and supervised the studies. M.C.-C. and E.G. are the principal clinical investigators. F.Be., R.G., G.S. and E.G. conducted clinical work. E.P., K.W., R.C., Y.B. and P.L. initiated the studies. M.C.-C., E.P., O.N.,G.W.,K.H., F.F.,J.D., M.D., T.B., B.G.-L., L.C., R.S., L.M.-C., J.S., J.L., N.K., B.G., K.C., Y.B., F.Bu., S.H.-B.-A. and P.L. designed or performed experiments. E.P., O.N., G.W., K.H., J.D., M.D., B.D., F.Bu. and P.L. analysed the data. All authors discussed results and conclusions. P.L. wrote the paper.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Fucharoen S, Winichagoon P. Clinical and hematologic aspects of hemoglobin E β-thalassemia. Curr. Opin. Hematol. 2000;7:106–112. doi: 10.1097/00062752-200003000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Olivieri NF, et al. Studies in haemoglobin E β-thalassaemia. Br. J. Haematol. 2008;141:388–397. doi: 10.1111/j.1365-2141.2008.07126.x. [DOI] [PubMed] [Google Scholar]
- 3.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 4.Tuan D, London IM. Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic ε-globin gene in K562 leukemia cells. Proc. Natl Acad. Sci. USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.May C, et al. Therapeutic haemoglobin synthesis in β-thalassaemic mice expressing lentivirus-encoded human β-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 6.Pawliuk R, et al. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 7.Hanawa H, et al. Extended β-globin locus control region elements promote consistent therapeutic expression of a γ-globin lentiviral vector in murine β-thalassemia. Blood. 2004;104:2281–2290. doi: 10.1182/blood-2004-03-0863. [DOI] [PubMed] [Google Scholar]
- 8.Imren S, et al. Permanent and panerythroid correction of murine β thalassemia by multiple lentiviral integration in hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2002;99:14380–14385. doi: 10.1073/pnas.212507099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levasseur DN, Ryan TM, Pawlik KM, Townes TM. Correction of a mouse model of sickle cell disease: lentiviral/antisickling β-globin gene transduction of unmobilized, purified hematopoietic stem cells. Blood. 2003;102:4312–4319. doi: 10.1182/blood-2003-04-1251. [DOI] [PubMed] [Google Scholar]
- 10.Malik P, Arumugam PI, Yee JK, Puthenveetil G. Successful correction of the human Cooley's anemia β-thalassemia major phenotype using a lentiviral vector flanked by the chicken hypersensitive site 4 chromatin insulator. Ann. NY Acad. Sci. 2005;1054:238–249. doi: 10.1196/annals.1345.030. [DOI] [PubMed] [Google Scholar]
- 11.Chung JH, Whiteley M, Felsenfeld G. A 5’ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 12.Miccio A, et al. In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of β-thalassemia. Proc. Natl Acad. Sci. USA. 2008;105:10547–10552. doi: 10.1073/pnas.0711666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions. Int. J. Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 14.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadelain M. Recent advances in globin gene transfer for the treatment of β-thalassemia and sickle cell anemia. Curr. Opin. Hematol. 2006;13:142–148. doi: 10.1097/01.moh.0000219658.57915.d4. [DOI] [PubMed] [Google Scholar]
- 16.Hargrove PW, et al. Globin lentiviral vector insertions can perturb the expression of endogenous genes in β-thalassemic hematopoietic cells. Mol. Ther. 2008;16:525–533. doi: 10.1038/sj.mt.6300394. [DOI] [PubMed] [Google Scholar]
- 17.Arumugam PI, et al. Genotoxic potential of lineage-specific lentivirus vectors carrying the β-globin locus control region. Mol. Ther. 2009;17:1929–1937. doi: 10.1038/mt.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanawa H, Yamamoto M, Zhao H, Shimada T, Persons DA. Optimized lentiviral vector design improves titer and transgene expression of vectors containing the chicken β-globin locus HS4 insulator element. Mol. Ther. 2009;17:667–674. doi: 10.1038/mt.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieburg HB, et al. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107:2311–2316. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nature Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- 22.Bottardi S, et al. Lineage-specific transcription factors in multipotent hematopoietic progenitors: a little bit goes a long way. Cell Cycle. 2007;6:1035–1039. doi: 10.4161/cc.6.9.4208. [DOI] [PubMed] [Google Scholar]
- 23.Ott MG, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1–EVI1, PRDM16 or SETBP1. Nature Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 24.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue N, et al. Molecular basis of clonal expansion of hematopoiesis in 2 patients with paroxysmal nocturnal hemoglobinuria (PNH). Blood. 2006;108:4232–4236. doi: 10.1182/blood-2006-05-025148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kustikova OS, et al. Retroviral vector insertion sites associated with dominant hematopoietic clones mark “stemness” pathways. Blood. 2007;109:1897–1907. doi: 10.1182/blood-2006-08-044156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 29.Cassani B, et al. Integration of retroviral vectors induces minor changes in the transcriptional activity of T cells from ADA-SCID patients treated with gene therapy. Blood. 2009;114:3546–3556. doi: 10.1182/blood-2009-02-202085. [DOI] [PubMed] [Google Scholar]
- 30.Wang GP, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cattaruzzi G, et al. The second AT-hook of the architectural transcription factor HMGA2 is determinant for nuclear localization and function. Nucleic Acids Res. 2007;35:1751–1760. doi: 10.1093/nar/gkl1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang GP, et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.