Abstract
One important function of endothelial cells in glioblastoma (GBM) is to create a niche that helps promote self-renewal of cancer stem-like cells (CSLCs). However, the underlying molecular mechanism for this endothelial function is not known. Since activation of Notch signaling has been found to be required for propagation of GBM CSLCs, we hypothesized that the GBM endothelium may provide the source of Notch ligands. Here we report a corroboration of this concept with a demonstration that Notch ligands are expressed in endothelial cells adjacent to Nestin and Notch receptor-positive cancer cells in primary GBMs. Co-culturing human brain microvascular endothelial cells (hBMECs) or Notch ligand with GBM neurospheres promoted GBM cell growth and increased CSLC self-renewal. Notably, RNAi-mediated knockdown of Notch ligands in hBMECs abrogated their ability to induce CSLC self-renewal and GBM tumor growth, both in vitro and in vivo. Thus, our findings establish that Notch activation in GBM CSLCs is driven by juxtacrine signaling between tumor cells and their surrounding endothelial cells in the tumor microenvironment, suggesting that targeting both CSLCs and their niche may provide a novel strategy to deplete CSLCs and improve GBM treatment.
Keywords: Endothelial cell, niche, Notch ligand, glioblastoma, cancer stem cell
Introduction
GBM is the most lethal malignant brain tumor in adults without revolutionary improvement in treatment during the past 30 years (1,2). Any treatment that can significantly prolong patients' overall survival for more than three months, which is the best achievement so far to treat GBM when using surgery, radiation therapy and temozolomide, can be considered as a success (3). Emerging evidence shows that a small population of CSLCs within neoplasms is responsible for tumor propagation (4), including GBM (5,6). Therapies targeting CSLCs bring hope for brain tumor patients (7-10). We have demonstrated that activation of the NOTCH pathway in GBM CSLC is required for their growth in vitro and in vivo (10). However, the molecular mechanism by which NOTCH is activated in GBM CSLCs and if CSLCs, like their normal congnates, also need a niche to self-renew is unclear.
The stem cell niche is a microenvironment where stem cells reside. It is composed of stem cells, neighboring supportive cells, extracellular matrix, and other factors required for stem cell self-renewal (11). In the CNS, neural stem cells (NSCs) are located at the subventricular zone (SVZ) of the lateral ventricle and subgranular zone of the dentate gyrus in the hippocampus (12-14). Some astrocytes and neuroblasts as well as endothelial cells in the SVZ are thought to contribute to these NSC niches by providing growth factors or membrane-bound ligands to NSCs for self-renewal (15). Although normal stem cells need to reside within a niche to self-renew (11), what a CSLC niche is composed of is largely unknown. As CSLCs share many properties with normal stem cells, CSLCs may also need a CSLC niche to self-renew. A recent report showed that endothelial cells function as a CSLC niche to promote CD133+ CSLC self-renewal in medulloblastoma and GBM (16). However, signaling pathways regulating the CSLC niche are still unclear.
The notch locus was first described by Morgan in a strain of Drosophila with notched wing blades (17). Seventy years later, the gene was cloned as a cell surface receptor (18) playing a key role in the development of many different cell types and tissues, including neuron and glia (19-22). Notch signaling is initiated when transmembrane ligands on one cell bind Notch receptors on an adjacent cell and cause a gamma secretase-mediated proteolytic release of the Notch intracellular domain (NICD) (23). NICD then translocates into the nucleus where it interacts with the transcriptional co-factor CBF1 and activates targets such as the Hes and Hey genes to modulate cell fate (20, 21). In vertebrates, four NOTCH receptors (NOTCH 1-4), five ligands (Jagged1,2, Delta1,3,4), and multiple effector molecules (Hes1-6, Hey1,2,L) have been identified (24). During normal development, ligand expressing cells (signal-sending cell) generally have reduced NOTCH activity, whereas NOTCH receptor-expressing cells (signal-receiving cell) have elevated NOTCH activity (25). In general, signal-sending cells will undergo differentiation, whereas signal-receiving cells remain in an undifferentiated state (stem cell state). This phenomenon is called “lateral specification” (Figure S1) (25). We and others have recently demonstrated that GBM CSLCs have elevated NOTCH activity (10, 26, 27), and that NOTCH pathway blockade with a gamma-secretase inhibitor (GSI) depletes GBM CSLCs, inhibits tumor growth, and prolongs survival of mice bearing intracranial xenografts (10). In the current study, we investigated if acquired NOTCH activity in GBM CSLCs comes from endothelial cells, which function as niche cells to promote GBM CSLC self-renewal by providing NOTCH ligands to NOTCH receptors expressed in GBM CSLCs.
Materials and Methods
GBM samples
Fresh primary GBM samples were obtained from the University of Michigan University Hospitals with approval from the Internal Review Board.
Cell Culture
Given the evidence that only some GBM cells fall in the CSLC hypothesis (28), we only choose GBM neurospheres that we have demonstrated to fall in the CSLC hypothesis for this study (10, 29). GBM neurosphere cells (HSR-GBM1, HSR-GBM2, and HSR-GBM3) derived from three different GBM patients were cultured and maintained in NeuroCult proliferation medium (Stem Cell Technologies, Vancouver, Canada) supplemented with 10 ng/ml EGF (PeproTech, Rocky Hill, NJ), 10 ng/ml FGFb (PeproTech), and 2 μg/ml heparin (Sigma, Saint Louis, MO) (10). hBMECs and human umbilical vein endothelial cells (HUVECs) were purchased from Cell Systems Corporation (catalog #ACBRI-376, Kirkland, WA) and ATCC (catalog #CRL-1730, Manassas, VA), respectively. hBMECs were maintained in CSC complete medium (Cell Systems) and HUVECs were cultured in F12K with 10% FBS, 0.1 mg/ml heparin, and 0.03 mg/ml endothelial cell growth supplement (ECGS). For co-culture system, neurospheres were seeded on top of the attached endothelial cells and maintained in the serum-free endothelial cells medium (Invitrogen, Carlsbad, CA) supplemented with 10 ng/ml EGF, 20 ng/ml FGFb, and 10 μg/ml heparin. For GBM cell differentiation, plates were coated with 15 ug/ml polyornithine for a minimum of 3h at 37 °C. GBM cells were then cultured on the coated plates at the density of 1×105 cells/cm2 in NeuroCult differentiation medium (Stem Cells Technology). The differentiated GBM cells were utilized 7-14 days later.
Lentivirus Production and shRNA Transduction
Lentiviruses were produced by transfecting BOSC 23 cells (ATCC, catalog #CRL-11270) with 16 μg total of pSicoR (Addgene, catalog #11579, Cambridge, MA), psPAX2 (Addgene, catalog #12260), and PMD.2G (Addgene, catalog #12259) plasmids with a ratio of 5: 3.5: 1.75 in a 10-cm dish, using lipofectamine 2000 (Invitrogen) (30). Tranfected cell media containing viruses were collected 48h later and filtered through a 0.45-μm PVDF membrane. Virus-containing media were used immediately or stored at -80°C. Cells were transduced with lentiviruses by the centrifugation method (1000g, 1.5h) with addition of 4-8 μg/ml polybrene. The medium was changed the next day, and gene expression was analyzed one week later(31). Transduction efficiency was monitored by GFP intensity with greater than 90% of cells being infected (30). shRNA sequences were designed by pSicoOligomaker 1.5 (courtesy of Dr. T. Jacks, MIT). The siRNA sequences used to knock down NOTCH ligands were 5′-GTGAGTGGTTGAATATGAT-3′ for JAG1 and 5′-GGAGAGAGGGGGCCAATGA-3′ for DLL4.
Orthotopic Xenograft Implantation
Mice were obtained and experiments were performed in accordance with guidelines from the University Animal Care and Use Committee (UCUCA) at the University of Michigan. 4-5 week-old athymic nude-Foxn1nu mice were purchased from Harland Laboratories (Indianapolis, IN) for intracranial xenograft implantation (8, 10). 50,000 single cells from GBM neurospheres with or without 50,000 endothelial cells were stereotactically injected into the brain (2 mm right, 1 mm back, 2 mm deep from the bregma) (8, 10). Tumor growth was monitored by MRI (supplementary methods).
Statistical analysis
Statistical analyses were done using GraphPad Prism 4 (GraphPad Software, San Diego, CA). Data graphed with error bars represent mean and SE from experiments done in triplicate unless otherwise noted. A two-sided Student's t test was used to determine the significance of any differences.
Results
NOTCH receptor-expressing cells are co-localized with NESTIN-positive tumor cells in primary GBM and have elevated level of NOTCH activity
Recently, we have demonstrated that Notch pathway blockade depletes GBM CSLCs in vitro and prevents their propagation in vivo (10). To investigate if NOTCH receptor-expressing cells in primary GBMs are CSLCs, we first examined NOTCH1 and NESTIN expression by immunofluorescence staining in primary GBM frozen sections. We found that NOTCH1-expressing cells co-localized with NESTIN-expressing cells in three different primary GBM samples (Figure 1A). NOTCH2-expressing cells also co-localized with NESTIN-expressing cells in primary GBMs (Figure S2). Furthermore, NOTCH ligand JAG1- or DLL1-expressing cells are adjacent to the cells with NOTCH pathway activation as indicated by Hes5 expression (Figure 1B). In addition, Hes5-expressing cells are co-localized with NOTCH1- and NOTCH2-expressing cells in primary GBMs (Figure 1C). These results suggest that the NESTIN-positive GBM CSLCs expresses NOTCH receptors and shows elevated level of NOTCH activity. These data indicate that NOTCH ligand-expressing cells within the tumor may function as a CSLC niche by providing ligands to NOTCH receptors expressed in GBM CSLCs to activate the NOTCH pathway for their self-renewal, replicating the lateral specification phenomenon seen in normal tissue development (Figure S1).
Figure 1. NOTCH receptor-expressing cells are co-localized with NESTIN-positive tumor cells in primary GBM and have elevated level of NOTCH activity adjacent to NOTCH-ligand expressing cells.
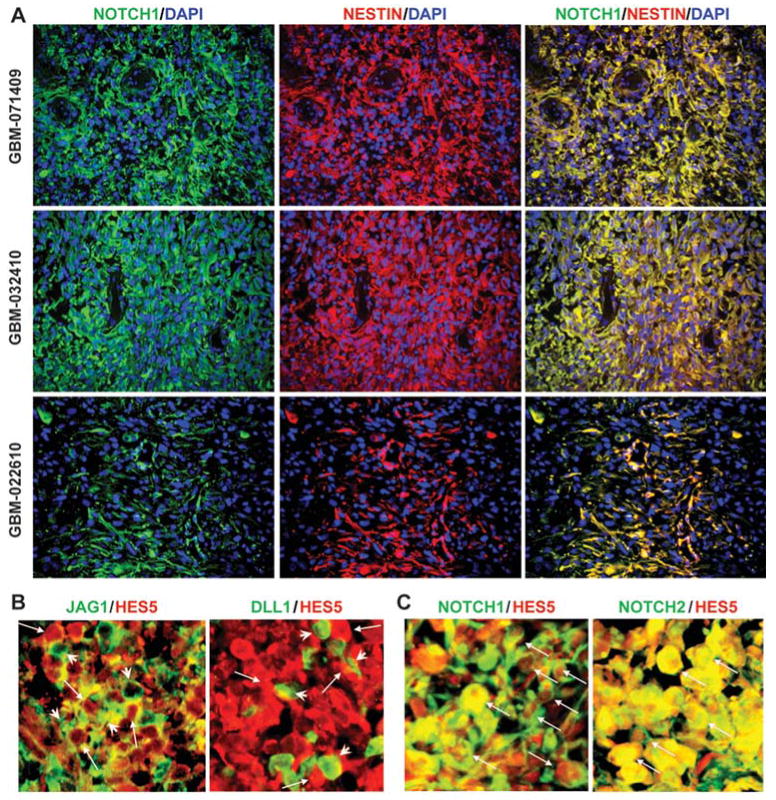
(A) Expression of NOTCH1 and NESTIN was co-localized in the same cells in the primary GBM tumors (GBM-071409, GBM-032410, and GBM-022610). (B) JAG1-expressing cells or DLL1-expressing cells (green, arrowhead) were adjacent to HES5-expressing cells (red, arrow) in primary GBM samples. (C) NOTCH1- or NOTCH2-expressing cells were co-localized with HES5-expressing cells (arrow) in primary GBM samples.
NOTCH ligands are expressed in the endothelial cells surrounded by tumor cells in primary GBMs
Next, we sought to identify the nature of NOTCH ligand-expressing cells within GBM. First, we examined NOTCH ligands DLL1, DLL4, JAG1 and JAG2 expression in frozen sections of primary GBM samples using immunofluorescence. We found that DLL1 is expressed in most tumor cells, whereas DLL4 is exclusively expressed in endothelial cells (CD31+) within GBM (Figure 2A). JAG1 or JAG2 is expressed in both endothelial cells and some tumor cells (Figure 2A and data not shown). Interestingly, using Western blot we found that cells from GBM neurosphere line HSR-GBM1 also expressed DLL1 and JAG1, but not DLL4 (Figure 2C). hBMECs expressed DLL4 and JAG1, but no detectable DLL1 (Figure 2C). A second type of endothelial cell, HUVEC, also expressed JAG1 and DLL4, but not DLL1 (data not shown). The expression pattern of these NOTCH ligands detected by Western blot is consistent with our findings in primary GBMs by immunofluorescence (Figure 2A). Furthermore, the NOTCH1 receptor and a CSLC marker, NESTIN, are expressed in the tumor cells adjacent to JAG1-expressing endothelial cells or tumor cells (Figure 2B), recapitulating the “lateral inhibition” pattern of cell distribution commonly seen in NOTCH-regulated normal development (Figure S1) (25). Taken together, these data suggest that endothelial cells within GBM may function as a CSLC niche by providing NOTCH ligands to NOTCH receptors expressed in CSLCs to activate the NOTCH pathway in CSLCs.
Figure 2. Expression pattern of NOTCH ligands in primary GBMs.
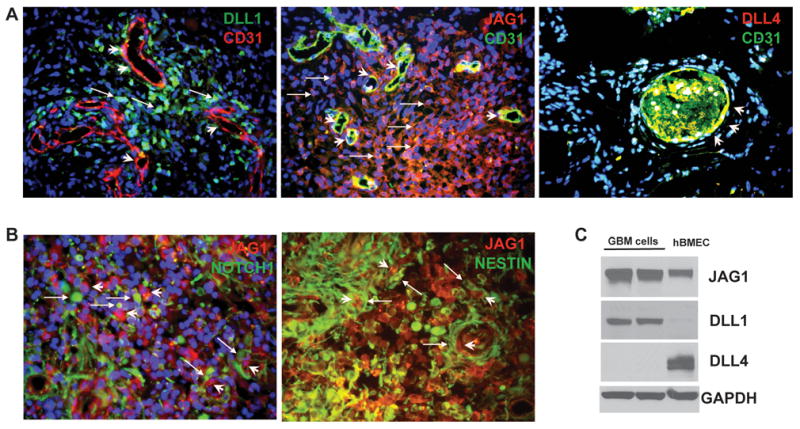
(A) NOTCH ligand DLL1 (green) was expressed in some GBM cells (arrow) and in some endothelial cells (CD31+) around the blood vessels (arrow head). JAG1 was expressed in both tumor cells (arrow) and blood vessels (arrow head). DLL4 was expressed in the blood vessels and co-localized with CD31 staining in endothelial cells (arrow head). (B) NOTCH1 receptor and CSLC marker NESTIN were expressed in tumor cells (arrow) adjacent to JAG1-expressing endothelial cells or tumor cells (arrowhead). (C) Western blots showed expression of JAG1, DLL1 and DLL4 in GBM neurosphere cells (Lane 1 and 2: same GBM cells) and hBMECs (Lane 3), consistent with immunohistochemistry results. GAPDH was used as a loading control.
Better differentiated GBM cells express higher level of NOTCH ligands and reduced level of CD133, and are less tumorigenic in mouse
The fact that NOTCH ligand JAG1 is expressed in tumor cells adjacent to the NOTCH receptor- and NESTIN-expressing cells in primary GBMs (Figure 2B) indicates some tumor cells may also function as a stem cell niche to provide NOTCH ligands to NOTCH receptors expressed in adjacent CSLCs. To test this, we first forced GBM neurosphere to differentiate, grow as mono-layer, and attach to the culture plate by switching to the differentiation medium. We found that the proliferation rate detected by Ki-67 staining was reduced and three lineage differentiation markers GFAP (glia), Tuj1 (Neuron), and Gal-C (oligodendrocyte) were induced in the differentiated GBM cells (Figure 3A). GFAP-positive cells were increased from 84.7±4.8% to 95.9±1.3%, Tuj-1-positive cells from 20.5±6.1% to 88.6±1.8%, and Gal-C-positive cells from 0 to 1.5±0.9% (p<0.01, t-test). Expression of DLL1 was also induced in the differentiated cells (Figure 3B). Furthermore, DLL1 is up-regulated in all neurosphere lines HSR-GBM1-3 (Figure 3B, S3). Interestingly, HSR-GBM2 showed up-regulation of all the ligands (Figure S3). In addition, expression of CSLC marker CD133 and NOTCH target HES1 were reduced in differentiated GBM cells (Figure 3B), suggesting that GBM CSLCs lose NOTCH activity and “stemness” when they are differentiated. Consistent with these data, CD133-positive population was reduced in differentiated GBM cells (Figure 3C). The CD133-negative GBM population also had elevated levels of NOTCH ligand DLL1 expression compared with the CD133-positive population (Figure 3C). Finally, when we injected 50,000 neurosphere cells or 50,000 differentiated GBM cells into the forebrain of immunodeficient mice, 100% (5 out of 5) mice receiving GBM neurosphere cells formed large intracranial xenografts as detected by MRI (Figure 3D), whereas only 40% (2 out of 5) mice injected with differentiated GBM cells formed tumors. Brain pathology confirmed the existence of intracranial xenografts and formation of larger tumor in the neurosphere-derived xenografts (Figure 3D). GBM intracranial xenograft also shows a similar feature as human primary tumor (Figure S4). These data demonstrate that GBM neurospheres enrich tumor-initiating CSLCs and that they can be used as a CSLC-enrichment model whereas forced differentiated cells can be used as a CSLC-depletion model, consistent with previous reports (6, 8, 29). Taken together, these data suggest that GBM neurospheres enrich CSLC population and that differentiated GBM cells may also provide NOTCH ligands to NOTCH receptors expressed in adjacent GBM CSLCs.
Figure 3. Differentiated GBM cells express NOTCH ligands and have reduced ability to form intracranial xenograft in mice.
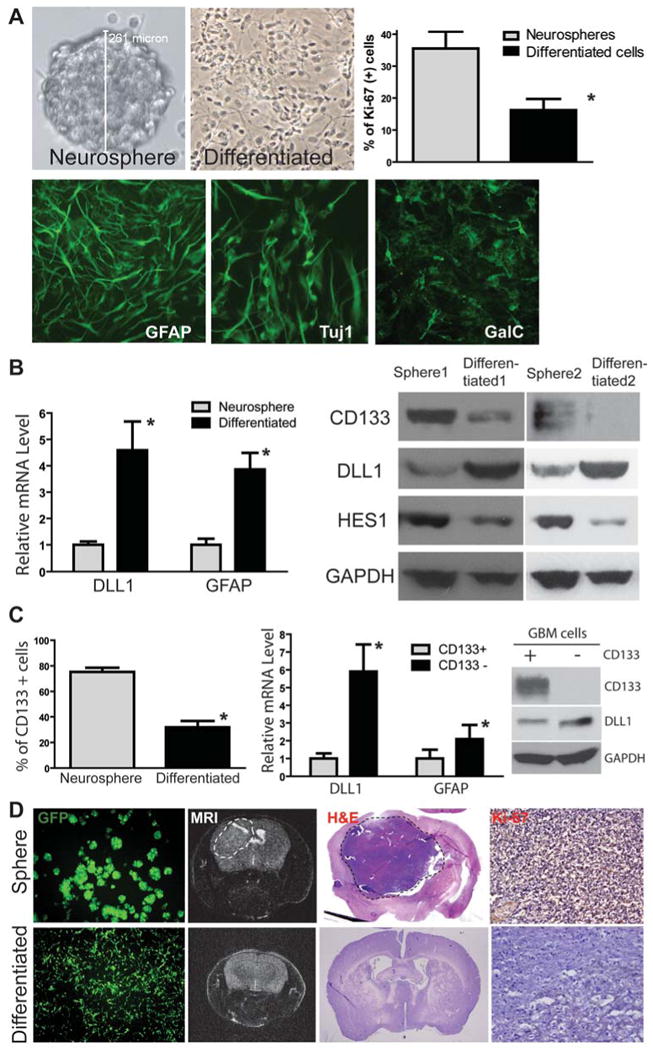
(A) HSR-GBM1 neurosphere line was forced to differentiate and grow as a mono-layer, which had reduced proliferation (*: p<0.05, t-test) and induced expression of GFAP (glial), Tuj1 (neuronal) and GalC (oligodendrocyte) markers. (B) Differentiated HSR-GBM1 cells expressed higher levels of DLL1 and GFAP at the mRNA level detected by quantitative RT-PCR (left panel). In addition, differentiated cells expressed a higher level of DLL1 and lower levels of NOTCH target HES1 and stem cell marker CD133 at the protein level as detected by Western blot in two different GBM neurosphere line HSR-GBM1 and HSR-GBM2. (C) CD133-positive population was significantly reduced in differentiated tumor cells compared with GBM neurospheres (HSR-GBM1) detected by flow cytometry (left panel). In addition, CD133-negative population in HSR-GBM1 neurospheres expressed higher levels of DLL1 and GFAP at mRNA level compared with CD133-positive population (middle panel). Up-regulation of DLL1 in CD133-negative population was also confirmed at the protein level by Western blot (right panel). (D) When GBM neurospheres or differentiated cells from HSR-GBM1 labeled with GFP using lentivirus were injected into the mouse brain, neurosphere cells formed large xenografts as detected by MRI (dash circle), whereas differentiated cells had reduced ability to form xenografts. Pathology was confirmed by H&E staining and immunostaining of the proliferation marker Ki-67.
NOTCH ligands promote GBM neurosphere growth in vitro
To examine if NOTCH ligands expressed in endothelial cells and differentiated tumor cells may have functional effects on GBM cells, we treated GBM neurosphere with soluble JAG1 or DLL1 peptide and examined GBM neurosphere growth. We found that JAG1 peptide treatment for 5 days increased HSR-GBM1 and HRS-GBM2 neurosphere growth in a dose-dependent fashion (Figure 4A). Furthermore, both JAG1 and DLL1 peptide induced HSR-GBM3 neurosphere growth in vitro (Figure 4A), suggesting that NOTCH ligand may induce NOTCH signaling in CLSCs through being immobilized by attachment to the extracellular matrix or the adjacent cells (32, 33). A weak growth response in HSR-GBM3 maybe due to the different genetic/epigenetic background among different tumors.
Figure 4. NOTCH ligand JAG1 or DLL1 promotes GBM neurosphere growth in vitro.
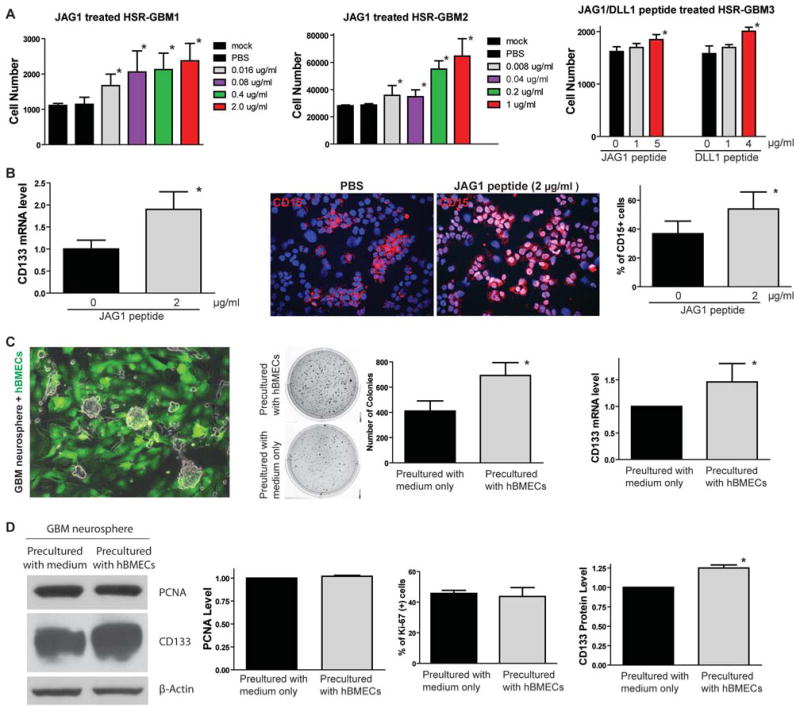
(A) Soluble ligand JAG1 peptide treatment for 5 days induced growth of HSR-GBM1 and HSR-GBM2 in a dose-dependent fashion (left and middle panels). JAG1 or DLL1 peptide also induced growth of HSR-GBM3 (right panel). (B) 2 μg/ml JAG1 peptide treatment increased CD133 mRNA expression in HSR-GBM1 (left panel). 2 μg/ml JAG1 peptide treatment also increased the CD15-positive CSLC population in GBM neurospheres detected by immunofluorescence (right panel, n=6 random fields, *: p<0.05, t-test). (C) GBM neurospheres co-cultured with GFP-labeled hBMECs were sorted by flow cytometry and examined clonogenesis by soft agar formation assay. GBM cells formed more colonies when precultured with hBMECs (n=6 wells were counted, *: p<0.01, t-test). In addition, expression of CSLC marker CD133 was also induced at mRNA level in GBM neurospheres pre-cultured with hBMECs compared to those pre-cultured with medium only (right panel, n=6 repeats of this experiment, *: p<0.01, t-test). (D) There was no proliferation change in GBM neurospheres pre-cultured with or without hBMECs, identified by PCNA protein expression using Western blot and percentage of Ki-67 positive population by immunofluorescence. However, CD133 expression was significantly induced in GBM neurospheres pre-cultured with hBMECs (right panel). p-Actin was used as a loading control.
In addition, expression of CSLC marker CD133 and CD15 were also induced in JAG1 or DLL1 peptide-treated GBM neurospheres (Figure 4B), suggesting that activation of NOTCH signaling by JAG1 or DLL1 increases self-renewal of GBM CSLCs in vitro. Finally, we use clonogenesis as a functional CSLC marker to examine if hBMECs can induce GBM CSLC self-renewal. First we labeled hBMECs with GFP by lentiviral vector and co-cultured hBMECs with unlabeled GBM neurospheres (Figure 4C) for three days. Then, we sorted GFP-negative GBM neurosphere cells and put them into soft agar to examine the changes of tumorigenicity. Medium alone cultured GBM neurospheres were used as a control. We found that hBMECs promoted clonogenesis of GBM neurospheres (Figure 4C). CD133 expression was also induced in GBM neurospheres precultured with hBMECs, indicating CSLC population is increased (Figure 4C). Interestingly, the proliferation rate of the entire GBM neurosphere culture (detected by proliferation marker PCNA expression using Western blot and percentage of Ki-67 positive population in the whole culture) did not change when precultured with hBMECs compared to precultured with medium only (Figure 4D), whereas expression of CSLC marker CD133 was significantly increased (Figure 4D). These data suggest that hBMECs only induce GBM CSLC self-renewal instead of proliferation of the whole culture (Figure S5). Taken together, these data demonstrate that NOTCH ligand JAG1 and DLL1 are sufficient to promote GBM CSLC self-renewal in vitro, suggesting that the endothelial or tumor cells that express higher levels of NOTCH ligands in GBM may provide these ligands to GBM CSLCs to activate the NOTCH pathway and promote self-renewal of GBM CSLCs.
Knockdown of NOTCH ligand expression in hBMECs by shRNA decreases CSLC population in co-cultured GBM neurospheres in vitro
To further investigate if NOTCH ligands expressed in endothelial cells are required for GBM CSLC growth, we also conducted loss-of-ligand function studies in endothelial cells co-cultured with GBM neurospheres. As JAG1 and DLL4 are the predominantly expressed NOTCH ligands in hBMECs (Figure 2), we first knocked down JAG1 expression in hBMECs using shRNA lentiviral vector pSicoR-shJAG1-GFP (Figure 5A, B). Then, shJAG1 or control lentivirus infected hBMECs were co-cultured with GBM neurospheres using serum-free endothelial cell culture medium for three days. GBM neuros-phere cells (GFP-negative) were subsequently sorted by flow cytometry, and tumor growth and the CD133-positive population were assessed by MTS assay and flow cytometry, respectively (Figure 5A). We found that GBM neurosphere growth was increased when co-cultured with hBMECs compared to neurospheres cultured by medium only, whereas knockdown JAG1 expression by shRNA in endothelial cells abrogated hBMEC-induced GBM growth (Figure 5B). CD15-positive population was also reduced in GBM neurospheres co-cultured with shJAG1-infected hBMECs (Figure 5C). Furthermore, knockdown of JAG1 expression in hBMECs decreased CD133-positive population and CD133 mRNA expression in GBM neurospheres co-cultured with hBMECs (Figure 5D). However, GBM neurosphere proliferation was not changed when co-cultured with shJAG1-infected hBMECs (Figure 5D). Taken together, these data demonstrated that reduced CSLCs in GBM neurospheres when co-cultured with shJAG1-infected hBMECs was due to reducing CSLC self-renewal, but not due to reduced proliferation of the whole culture (Figure S5), suggesting that NOTCH ligands expressed in hBMECs are essential for the maintenance of GBM CSLCs in vitro.
Figure 5. Knocking down JAG1 expression in hBMECs by shRNA decreases CSLC population in co-cultured GBM neurospheres in vitro.
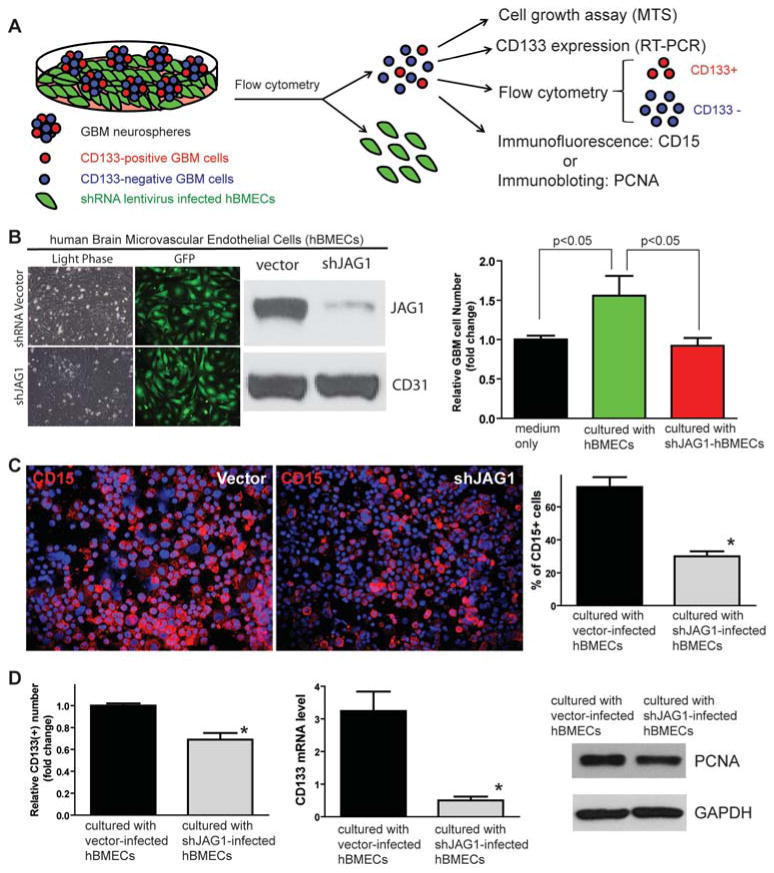
(A) Schematic showing the experimental approach used to examine if NOTCH ligands expressed in endothelial cells are essential for co-cultured GBM neurosphere growth. (B) Western blot showed that JAG1 expression was significantly knocked down by shJAG1 lentiviruses in hBMECs to be co-cultured with GBM neurospheres. CD31 was served as an internal control. HSR-GBM1 growth was increased when co-cultured with hBMECs compared to neurospheres cultured by medium only, whereas knockdown of JAG1 expression in hBMECs abrogated hBMEC-induced HSR-GBM1 growth. (C) CD15-positive CSLC population detected by immunofluorescence was also reduced in GBM neurospheres co-cultured with shJAG1-infected hBMECs compared to GBM neurospheres co-cultured with empty vector-infected hBMECs. (D) Knockdown of JAG1 expression in hBMECs decreased CD133-positive population in HSR-GBM1 neurospheres co-cultured with hBMECs compared to HSR-GBM1 neurospheres cultured with empty vector-infected hBMECs (left). mRNA expression of CD133 was also lowered in HSR-GBM1 neurosphere cells co-cultured with sh-JAG1-infected hBMECs compared to HSR-GBM1 cells co-cultured with empty vector-infected hBMECs (middle). There was no proliferation change in HSR-GBM1 neurospheres co-cultured with empty vector- or shJAG1-infected hBMECs, identified by PCNA protein expression using Western blot (right).
Knocking down JAG1 or DLL4 expression in hBMECs by shRNA inhibits growth of GBM intracranial xenografts derived from the mixture of GBM neurosphere cells and hBMECs
To examine if expression of NOTCH ligands in endothelial cells is required for GBM CSLCs growth in vivo, shJAG1- or shDLL4-infected hBMECs were mixed with GBM neurosphere cells (1:10) and injected into the brain of immunodeficient mice (Figure 6A). Tumor growth was monitored by MRI and tumor volume calculated (Figure 6A). We found that GBM neurospheres co-cultured with shJAG1- or shDLL4-infected hBMECs developed significantly smaller intracranial xenograft compared with those derived from a mixture of GBM neurospheres and empty vector-infected hBMECs (Figure 6B, C). Furthermore, we examined the cell specificity of the NOTCH ligand effect by using another type of human endothelial cells, HUVECs, and found that knockdown of JAG1 expression in HUVECs also reduced growth of intrancranial xenografts (Figure 6D). These data demonstrate that NOTCH ligands expressed in endothelial cells contribute to the growth of intracranial xenografts derived from the mixture of GBM neurosphere cells and endothelial cells, indicating that endothelial cells may function as a niche for GBM CSLCs by providing NOTCH ligands to NOTCH receptors expressed in GBM CSLCs.
Figure 6. Knocking down JAG1 or DLL4 expression by shRNA in hBMECs inhibits growth of GBM intracranial xenografts derived from the mixture of GBM neurospheres and hBMECs.
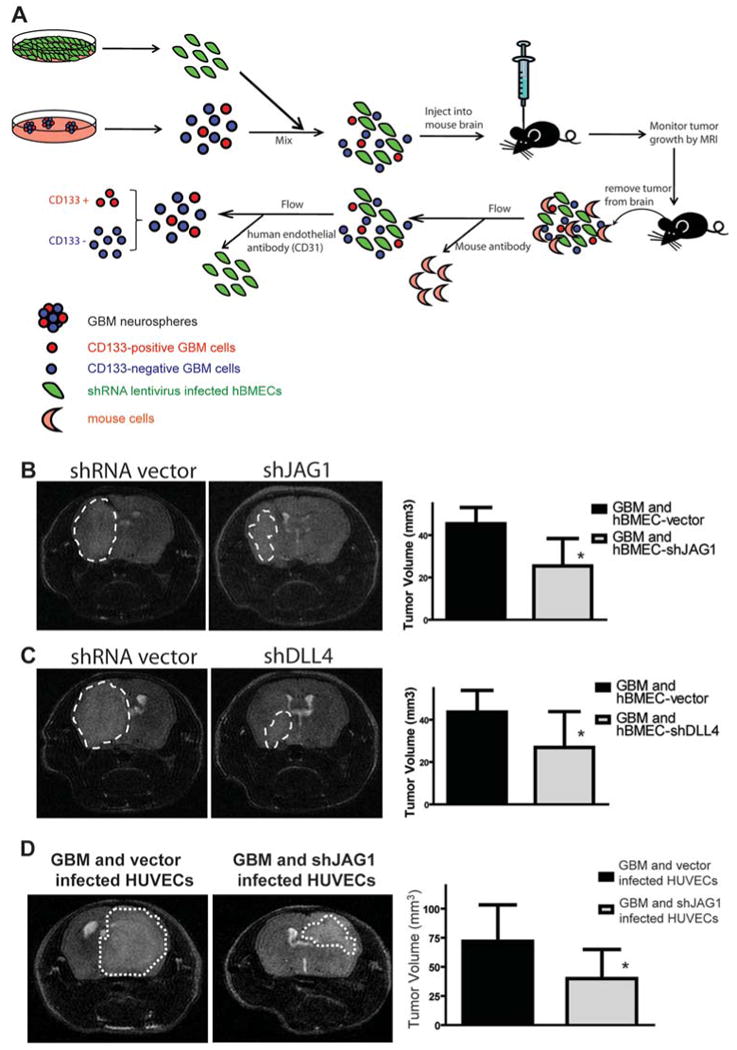
(A) Schematic showing the experimental approach used to examine if NOTCH ligand expressed in endo-thelial cells are essential for the growth of GBM intracranial xenografts derived from the mixture of GBM neurospheres and endothelial cells. (B) MRI showed that the growth of GBM intracranial xeno-grafts derived from a mixture of GBM neurospheres and shJAG1 infected hBMECs was much slower than those derived from a mixture with empty vector infected hBMECs (n=10 per cohort, *: p<0.01, t-test). (C) MRI scanning showed that the growth of GBM intracranial xenografts derived from a mixture of GBM neurospheres and shDLL4 infected hBMECs was much slower than those derived from a mixture with empty vector infected hBMECs (n=10 per cohort, *: p<0.05, t-test). (D) Growth of GBM in-tracranial xenografts derived from a mixture of GBM neurospheres and a second type of endothelial cells (HUVECs) infected with shJAG1, was also significantly slower than those derived from a mixture with empty vector infected HUVECs (n=5 per cohort, *: p<0.05, t-test).
CD133-positive population was reduced in GBM xenografts derived from mixture of GBM neurospheres and hBMECs with ligand knockdown
To examine the CSLC population changes in GBM xenografts derived from a mixture of GBM neurosphere and endothelial cells with or without ligand knockdown, first we sorted human GBM cells using mouse cell surface antigen (to remove mouse cells) and GFP (to remove human endothelial cells as they were infected with pSicoR-GFP lentiviral vector with or without shRNA against NOTCH ligand) (Figure 6A). Then we examined the CD133-positive GBM cell changes (Figure 6A). We found that CD133-positive GBM CSLCs were significantly reduced in GBM xenografts mixed with shJAG1 or shDLL4-infected hBMECs (Figure 7A). Similarly, CD133-positive population was also decreased in GBM xenografts mixed shJAG1-infected HUVECs (Figure 7A). Consistent with these data, CD15-positive GBM CSLC population was also reduced in GBM xenografts mixed with shJAG1-infected hBMECs (Figure 7A). However, there was no significant change in tumor proliferation (Figure 7B) and apoptosis (Figure 7C) in the xenografts derived from the mixture of GBM neurospheres and hBMECs with JAG1 or DLL4 knockdown, suggesting that reduced GBM intracranial xenograft growth was due to loss of CSLCs (Figure S5), which need to receive NOTCH ligands from endothelial cells to activate NOTCH for their self-renewal. These data are consistent with previous findings that the endothelial niche mainly affects CSLCs instead of the proliferation and apoptosis of the whole tumor (16). Taken together, our results demonstrate that NOTCH ligands expressed in endothelial cells are critical for the propagation of GBM CSLCs.
Figure 7. CSLC population is reduced in GBM xenografts derived from mixture of GBM neuros-pheres and hBMECs with ligand knockdown.
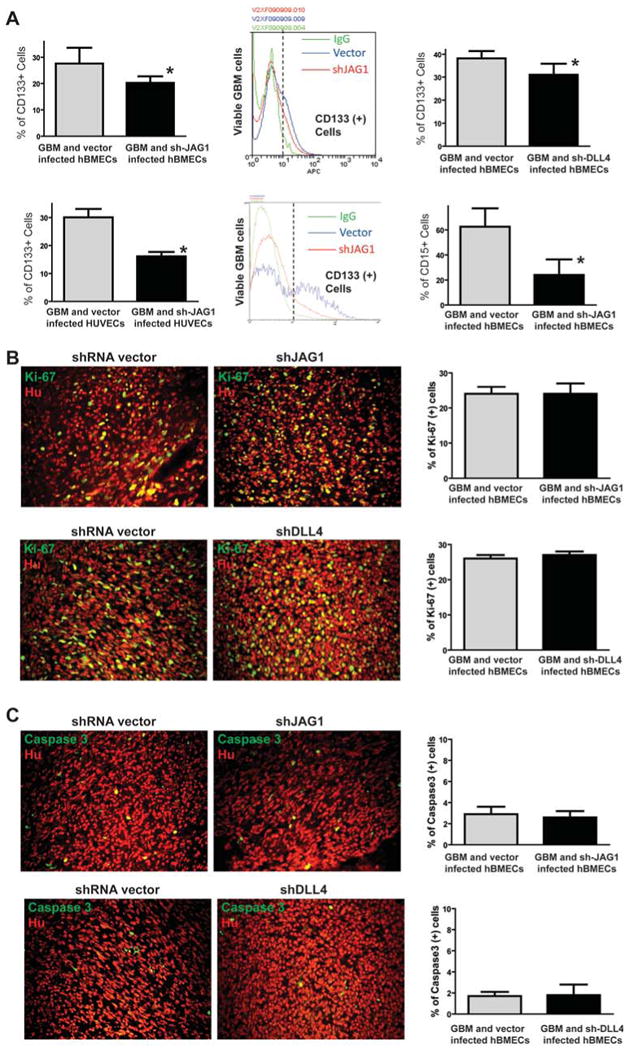
(A) Intracranial xenografts derived from the mixture of GBM neurosphere and hBMECs were dissected and dissociated into single cell suspension to examine the CD133-positive population by flow cytometry. CD133-positive population was significantly decreased in the xenograft from GBM neurospheres and sh-JAG1 or sh-DLL4 infected hBMECs compared to the xenograft from GBM neurospheres and vector infected hBMECs (upper panels). CD133-positive population was also significantly decreased in the xenograft from GBM neurospheres and sh-JAG1-infected HUVECs compared to the xenograft from GBM neurosphere and vector-infected HUVECs (lower left and middle panels). In addition, CD15-positive population was also significantly decreased in the xenograft from GBM neurospheres and sh-JAG1 infected hBMECs compared to the xenograft from GBM neurospheres and vector infected hBMECs (lower right panel). (B) Immunofluorescent staining of Ki-67 in the xenografts derived from the mixture of GBM neurospheres and hBMECs with JAG1 knockdown (upper panels) or DLL4 knockdown (lower panels) showed no proliferation changes. (C) Immunofluorescent staining of cleaved caspase 3 in the xenografts derived from the mixture of GBM neurospheres and hBMECs with JAG1 knockdown (upper panels) or DLL4 knockdown (lower panels) showed no apoptosis changes.
Disscussion
In the current study, we explored the molecular mechanism underlying NOTCH activation in GBM CSLCs. We found that endothelial cells within the tumor function as a CSLC niche to promote CSLC self-renewal by providing NOTCH ligands to the NOTCH receptors expressed in GBM CSLCs (Figure S6).
To our knowledge, there is still no published documentation addressing whether cell-contact dependent signaling pathways contribute to CSLC self-renewal within their niche in GBM. Here, we first show that NOTCH ligands are expressed by endothelial cells and some tumor cells around the NESTIN- and NOTCH receptor-positive CSLCs in primary GBM samples. Then, we demonstrate that knockdown of NOTCH ligand JAG or DLL in endothelial cells decreases CD133-positive population in GBM neurospheres in vitro and inhibits co-cultured GBM neurosphere propagation in vivo. Finally, we show that the reduced GBM xenograft growth is due to decreased GBM CSLC population. Taken together, our data provide experimental evidence that endothelial cells function as CSLC niche by providing NOTCH ligands to activate NOTCH signaling in GBM CSLCs (Figure S6). Therefore, several novel and innovative approaches can be developed based on interrupting the interaction between CSLCs and their niche(s). For example, a NOTCH ligand blocking antibody or peptide, in combination with chemo- and radiation-therapy as well as CSLC-targeting therapy, may result in improved GBM treatment.
In the CNS, that endothelial cells can function as a stem cell niche was initially found in normal NSCs, with endothelial cells promoting asymmetric self-renewal of NSCs from the SVZ (15). Later, it was confirmed that the vascular niche regulates self-renewal of NSCs in vivo (34) at both embryonic and adult stages (35). In addition, it has been shown that the NOTCH signaling pathway plays a critical role in regulating niche-dependent self-renewal of NSC in SVZ (36, 37). Furthermore, there is some experimental evidence showing that GBM may originate from SVZ NSCs (38). Consistent with the normal stem cell niche, it has also been shown that endothelial cells function as a cancer stem cell niche to promote CD133+ CSLC growth in GBM and medulloblastoma (16). Here, we demonstrate that the endothelial niche provides NOTCH ligands to GBM CSLCs to activate NOTCH signaling for their self-renewal. Together, these data indicate that GBM CSLCs share a common feature with their normal cognate in not only being endothelial niche dependent, but also NOTCH pathway dependent.
Although, providing NOTCH ligands could be a mechanism by which endothelial niche maintains GBM CSLCs, the cause of expression of NOTCH ligands in the endothelial niche in GBM is still unclear. It has been shown that CD133-positive GBM CSLCs induce VEGF expression which may contribute to angiogenesis or endothelial niche formation in GBM (39). Previous studies have also shown that VEGF induces DLL4 expression in both physiological and pathological angiogenesis (40, 41). It is, therefore, possible that expression of NOTCH ligands in endothelial cells originates from GBM CSLCs through the VEGF pathway. In addition, endothelial cells may not be the only source of NOTCH ligands to GBM CSLCs. Our data show that some primary GBM cells also express DLL1 and differentiated tumor cells have elevated DLL1 expression compared to GBM neurospheres, indicating that differentiated tumor cells may also function as niche cells for the maintenance of CSLCs. Further experimental evidence is needed to address this point.
The endothelial niche maintenance of GBM CSLC self-renewal could be through multiple ways. In this study, we demonstrate that endothelial cells function as a stem cell niche for GBM CSLCs by directly providing NOTCH ligands. However, a recent study shows that nitric oxide (NO) released from tumor endothelium diffuses to neighboring glioma stem-like cells and activates the NOTCH pathway within these stem-like cells in a PDGF-induced mouse GBM model (42). It is, therefore, possible that activation of NOTCH signaling in GBM CSLCs maybe through both NOTCH ligand dependent and independent ways. In addition, two recent studies report that activation of HIF2a and PI3K pathway are required for CSLC growth within the endothelial niche in GBM and medulloblastoma (43, 44). These data not only further support the hypothesis that CSLCs share a common feature with their normal cognates in terms of dependence on an endothelial niche, but also suggest that the interaction between CSLCs and their endothelial niche could be through multiple mechanisms.
If GBM CSLCs reside within endothelial niche to self-renew, targeting both CSLC and its niche may be necessary in order to eliminate CSLCs. Indeed, inhibition of angiogenesis by VEGF blocking antibody, Bevacizumab, has shown encouraging results in recurrent GBM patients (45). One possible reason is that Bevacizumab depletes tumor blood vessels and self-renewing CSLCs in GBM (16). However, given the evidence of heterogeneity of GBM (46, 47), VEGF resistant GBM cells may develop. Therefore, targeting endothelial stem cell niche by NOTCH ligand blocking antibodies may be another alternative approach to treat GBM patients. Indeed, recent reports show that DLL4 blocking antibody reduces growth of tumor which are resistant to VEGF blocking antibody therapy (48-50). Thus, targeting the NOTCH ligand-dependent endothelial niche may be a novel way to treat GBM patients, particularly for those patients who are resistant to VEGF inhibitor therapy.
In summary, we have demonstrated that endothelial cells function as a CSLC niche by providing NOTCH ligands to CSLCs for self-renewal. Targeting of CSLCs and stem cell niche via NOTCH inhibition and NOTCH ligand blockade may provide an innovative approach for GBM treatment, as niche cells and the NOTCH signaling pathway are essential for self-renewal of GBM CSLCs. Although the current study focuses on GBM, this approach may have relevance to multiple forms of cancer.
Supplementary Material
Acknowledgments
We would like to acknowledge grant support to Dr. Fan from Accelerate Brain Cancer Cure Project Award, American Brain Tumor Association Translational Grant, and Voices Against Brain Cancer Research Grant.
References
- 1.Louis D, Ohgaki H, Wiestler O, Cavenee W. WHO Classification of Tumours of the Central Nervous System. Lyon (France): IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006 Mar 10;24(8):1253–65. doi: 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001 Nov 1;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004 Nov 18;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006 Aug 1;66(15):7445–52. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 8.Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006 Dec 7;444(7120):761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 9.Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008 Jun 10;26(17):2821–7. doi: 10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010 Jan;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell. 2004 Mar 19;116(6):769–78. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 12.Temple S, Alvarez-Buylla A. Stem cells in the adult mammalian central nervous system. Curr Opin Neurobiol. 1999 Feb;9(1):135–41. doi: 10.1016/s0959-4388(99)80017-8. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J Neurobiol. 1998 Aug;36(2):105–10. [PubMed] [Google Scholar]
- 14.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999 Jun 11;97(6):703–16. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004 May 28;304(5675):1338–40. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 16.Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007 Jan;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Morgan T. The Theory of the Gene. The American Naturalist. 1917;51:513–44. [Google Scholar]
- 18.Artavanis-Tsakonas S, Muskavitch MA, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1977–81. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000 May 26;101(5):499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 20.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006 Feb;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 21.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005 Jun;8(6):709–15. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 22.Taylor MK, Yeager K, Morrison SJ. Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development. 2007 Jul;134(13):2435–47. doi: 10.1242/dev.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003 Sep 29;22(42):6598–608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 24.Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002 Sep-Oct;1(5):466–76. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 25.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999 Apr 30;284(5415):770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 26.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005 Mar 15;65(6):2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006 May;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008 Dec 4;456(7222):593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004 Oct 1;64(19):7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 30.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10380–5. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004 Nov 1;64(21):7787–93. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 32.Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000 Dec;113(Pt 23):4313–8. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Milner LA, Deng Y, Iwata M, Banta A, Graf L, et al. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998 Jan;8(1):43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 34.Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008 Sep 11;3(3):279–88. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, et al. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. 2008 Sep 11;3(3):289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyfeler Y, Kirch RD, Mantei N, Leone DP, Radtke F, Suter U, et al. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. Embo J. 2005 Oct 5;24(19):3504–15. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreu-Agullo C, Morante-Redolat JM, Delgado AC, Farinas I. Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat Neurosci. 2009 Dec;12(12):1514–23. doi: 10.1038/nn.2437. [DOI] [PubMed] [Google Scholar]
- 38.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005 Aug 25;353(8):811–22. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 39.Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006 Aug 15;66(16):7843–8. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 40.Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, et al. Deltalike ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3219–24. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003 Jan;23(1):14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charles N, Ozawa T, Squatrito M, Bleau AM, Brennan CW, Hambardzumyan D, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010 Feb 5;6(2):141–52. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009 Jun 2;15(6):501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008 Feb 15;22(4):436–48. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Marcello J, Reardon DA, Quinn JA, et al. Bevacizumab plus irinotecan in recurrent glioblastoma multiforme. J Clin Oncol. 2007 Oct 20;25(30):4722–9. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- 46.TCGA_Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 Sep 26;321(5897):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006 Dec 21;444(7122):1083–7. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 49.Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006 Dec 21;444(7122):1032–7. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 50.Hoey T, Yen WC, Axelrod F, Basi J, Donigian L, Dylla S, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009 Aug 7;5(2):168–77. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


