Abstract
Antibodies are among the most powerful tools in biological and biomedical research and are presently the fastest growing category of new bio-pharmaceutics. The most common format of antibody applied for therapeutic, diagnostic and analytical purposes is the IgG format. For medical applications, recombinant IgGs are made in cultured mammalian cells in a process that is too expensive to be considered for producing antibodies for diagnostic and analytical purposes. Therefore, for such purposes, mouse monoclonal antibodies or polyclonal sera from immunized animals are used. While looking for an easier and more rapid way to prepare full-length IgGs for therapeutic purposes, we recently developed and reported an expression and purification protocol for full-length IgGs, and IgG-based fusion proteins in E. coli, called “Inclonals.” By applying the Inclonals technology, we could generate full-length IgGs that are genetically fused to toxins. The aim of the study described herein was to evaluate the possibility of applying the “Inclonals” technology for preparing IgG-fluorophore fusion proteins. We found that IgG fused to the green fluorescent proteins enhanced GFP (EGFP) while maintaining functionality in binding, lost most of its fluorescence during the refolding process. In contrast, we found that green fluorescent Superfolder GFP (SFGFP)-fused IgG and red fluorescent mCherry-fused IgG were functional in antigen binding and maintained fluorescence intensity. In addition, we found that we can link several SFGFPs in tandem to each IgG, with fluorescence intensity increasing accordingly. Fluorescent IgGs made in E. coli may become attractive alternatives to monoclonal or polyclonal fluorescent antibodies derived from animals.
Keywords: IgG, IgG-fluorophore fusion protein, inclusion bodies, Refolding, Superfolder GFP
Introduction
Fluorescently labeled antibodies are indispensable tools in almost every field of biomedical diagnosis and research, allowing the detection of antibodies, antigens, and virtually any antigenic protein in cells and tissues. The fluorescence detection can be done by staining with either direct (primary) or indirect (secondary) antibodies. Direct fluorescence staining makes use of monoclonal antibodies from hybridoma cell lines, possibly propagated inside the abdomen of a pristane-primed mouse (“the mouse ascites method”), even though this operation may cause discomfort, distress, and pain to the mice.1 Next, chemical coupling is used to attach fluorescent proteins or fluorescent chemical compounds (fluorophores) to the antibodies. Although widely used for over 60 y, chemically conjugated fluorophores and the resulting fluorescent antibody conjugates have several drawbacks. First of all, the conjugation reaction is indiscriminate with respect to the location of the target amino acid residue; if conjugation occurs within the binding site of the antibody, partial or even complete loss of antigen binding activity can occur. Moreover, the chemical conjugation reaction results in a heterogeneous mixture of antibodies having a different number of conjugated fluorescence molecules per antibody, attached at different locations. Finally, the presence of some fluorophores in close proximity can decrease fluorescence intensity via quenching mechanisms. Typically, no more than about three to five dyes can be attached to an antibody without self-quenching of fluorescence or inactivating the antibody.2,3
A fusion protein comprising an antibody and a fluorescent protein could offer several advantages over the conventional labeling method. Fluorescent proteins have been genetically fused to many proteins in various species to produce stable chimeras that retain their original biological activity as well as retaining the fluorescent properties of the fluorescent protein. However, only a limited number of studies describing the production of antibodies fused with fluorescent proteins have been published to date. This is probably due to the different folding requirements antibodies and fluorescent proteins have. GFP-related fluorescent proteins are known to fold correctly under the reducing conditions found in the cytoplasm of Aequorea and other species in which they have been recombinantly expressed.4 Because antibodies, either full-length IgGs or antibody fragments, contain disulfide bonds, they require an oxidizing environment for their correct folding. The endoplasmic reticulum (ER) lumen of eukaryotic cells favors disulfide bridge formation and so does the bacterial periplasm. scFv-GFP fusions have been purified under native conditions from the bacterial periplasm,5,6 from the bacterial cytoplasm7,8 or expressed as bacterial cytosolic inclusion bodies.9 Though successful, low yields of a bifunctional fusion protein were obtained in these studies. In another case of bacterial cytoplasmic expression, Olichon et al. used llama VHH as the antibody scaffold.10 The use of llama VHH (which has only one disulfide bond) along with co-expression of DsbC (a disulfide bond isomerase) yielded substantial amounts of fusion protein having both binding and fluorescence activities. However, VHH and scFv antibody fragments- being monovalent- usually have lower functional affinity compared with a bivalent, full length IgGs. In addition, small antibody fragments are usually less stable than full size IgG molecules and are rarely used as reagents. This is quite a drawback for a protein designated to be used for detection in a research or diagnostics setting. Haas et al. recently reported the production of full length IgG fusion to the fluorescent protein citrine in mammalian cells.11 They have managed to attach an IgG with up to two citrine molecules by adding citrine to the C-terminus of each one of the antibody light chains. E. coli based expression systems, however, are usually superior to any other expression systems in terms of costs and are therefore more likely to provide an actual cost-efficient alternative to the ascites method than cell culture production methods.
Superfolder GFP (SFGFP) is a green GFP variant that has been evolved in vitro for folding robustness.12 By using the “Inclonals” protocol recently developed by us for efficient bacterial production of monoclonal antibodies13 we were able to produce SFGFP-fused full length antibodies having both binding and fluorescence activities. In addition, by attaching two SFGFP proteins in tandem to each chain of the antibodies we were able to generate antibodies carrying up to eight fluorescent proteins. Their immunofluorescence abilities were demonstrated using both FACS and fluorescence microscopy. This is the first report describing the production of IgG fused to fluorescent proteins in E. coli. This is also the first report describing the production of any antibody format carrying fluorescent proteins in tandem.
Results
Design and production of SFGFP-fused IgGs
After successfully applying the Inclonals protocol for the production of a novel IgG-toxin fusion protein,13 we examined the possibility of applying the protocol for the production of a fusion protein comprising a full-length antibody and a fluorescent protein. The Inclonals protocol is a refolding based method for the production of full length IgGs. According to the protocol, the heavy and light chains of the desired antibody are expressed as cytoplasmic inclusion bodies in two different bacterial cultures. Following a denaturation step, the chains are mixed and refolding is performed.
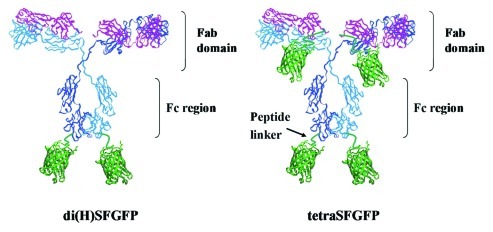
The fluorescent protein SFGFP was fused to the C-terminus of FRP5 (anti-ErbB2) antibody's heavy and light chains via a flexible linker. Initially, two SFGFP-fusion antibody formats carrying two and four SFGFP molecules were constructed (Fig. 1). Di(H)SFGFP carries two SFGFP molecules, one SFGFP fused to each of the antibody’s heavy chains while tetraSFGFP has a SFGFP molecule attached to each one of its four chains, thus carrying four SFGFP molecules. Later, to make the fluorescence of fusion-Inclonals stronger, we resolved to label each antibody with more than merely four fluorescent molecules. That was accomplished by attaching each chain at its C-terminus with two fluorescent proteins fused in tandem. Two additional IgG-fluorophore fusion proteins were then constructed, designated di(H)tanSF and tetra-tanSF, respectively carrying four and eight SFGFP molecules. Each SFGFP molecule was preceded by a short flexible linker.
Figure 1. Schematic representation of SFGFP-fusion Inclonals. The molecules are not drawn to scale.
An early attempt to construct a fluorescent antibody using EGFP as the fluorescent moiety resulted with antibodies having full binding capabilities but little fluorescence activity. Upon induction, the EGFP-fused antibody chains accumulated as green fluorescent cell pellets. However, the fluorescence was lost upon denaturation, and was not recovered during refolding (data not shown). This was not the case for SFGFP that remained fluorescent throughout the process. We were also successful in producing IgG-mCherry red-fluorescent fusion proteins that specifically stained antigen-positive cells as shown be FACS analysis (Supplementary Figure 2)
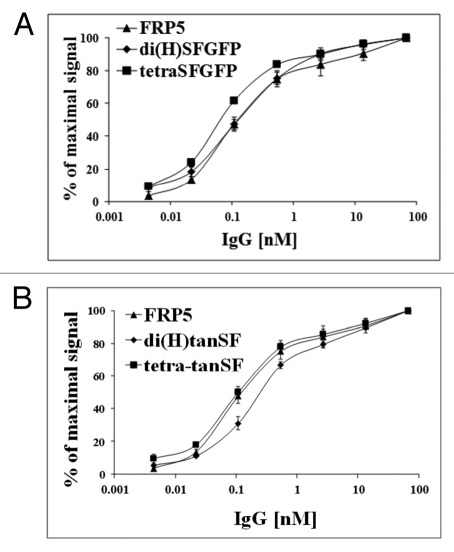
Figure 2. Binding capacity evaluation by ELISA. Binding of di(H)SFGFP and tetraSFGFP (A) and of di(H)tanSF and tetra-tanSF (B) in comparison to the parental Inclonal FRP5 IgG was evaluated on soluble ErbB2 using HRP-conjugated secondary antibodies. Error bars represent standard deviations of triplicate antibody samples that were tested in this ELISA.
All the IgG-fluorophore fusion proteins showed the expected molecular mass when analyzed by SDS-PAGE (data not shown). Expression yields were roughly similar for all the molecules, regardless of the number of fused fluorophore molecules and its identity, and were similar to the yields we previously reported for named IgGs and toxin-fused IgGs.13 In fact, it seemed that increasing the number of SFGFP molecules, although making the molecule larger and seemingly complex, improved refolding yields to some extent. Refolding was performed at 200 ml scale with an input of 20 mg solubilized and reduced inclusion bodies protein and, after protein A purification, ~3–5 mg of pure IgG-fluorophore fusion protein were obtained. As previously described,13 we typically carry out growth and induction in 500 ml of medium from which we recover about 6 g wet cell paste. We then recover ~100–200 mg of washed inclusion bodies from the cell paste.
Regarding refolding kinetics, we found that the antibody part takes at least 24 h of refolding to reach a plateau in functional antibody recovery (in our standard protocol we refold for 36 h) (unpublished observations). As for SFGFP, we found that it refolds very rapidly and regains full fluorescence within less than 20 min. We also found that SFGFP containing solubilized inclusion bodies had to be reduced prior to refolding, as un-reduced inclusion bodies had a low constant fluorescence level that did not increase upon refolding (Supplementary Figure 3). No further increase in fluorescence could be observed after longer refolding time periods (not shown).
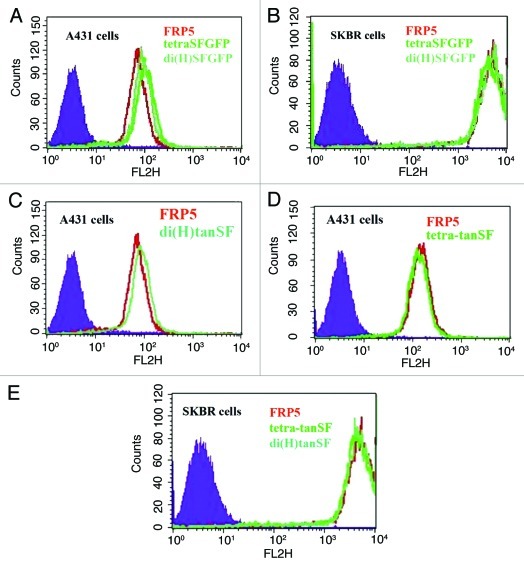
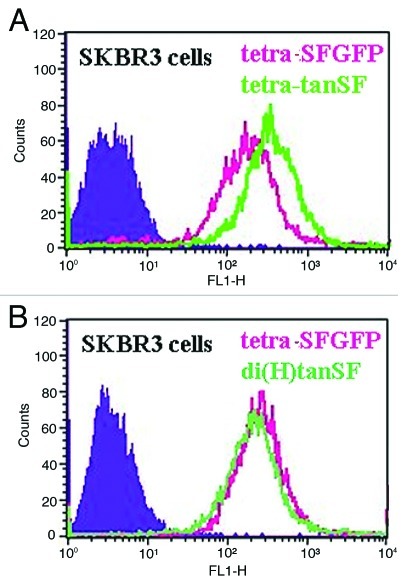
Figure 3. Binding capacity evaluation by FACS. Cellular ErbB2 binding of SFGFP-fusion antibodies in comparison to the parental Inclonal FRP5 IgG was evaluated on human epithelial carcinoma cell line A431 and human breast adenocarcinoma cell line SKBR3 using red-fluorescent phycoerythrin-labeled secondary antibodies. (A) FRP5, di(H)SFGFP and tetraSFGFP binding to A431 cells. (B) FRP5, di(H)SFGFP and tetraSFGFP binding to SKBR3 cells. (C) FRP5 and di(H)tanSF binding to A431 cells. (D) FRP5 and tetra-tanSF binding to A431 cells. (E) FRP5, di(H)tanSF and tetra-tanSF binding to SKBR3 cells. Filled areas, negative control (secondary antibody alone).
SFGFP-fused antibodies' binding capabilities
The ability of SFGFP-fusion antibodies to bind their target antigen was at first evaluated on recombinant ErbB2 by ELISA. Bound Inclonals were detected using HRP labeled anti-human secondary antibodies. The results (Fig. 2) show a dose-dependent increase in the binding of all SFGFP-fused antibodies to the recombinant antigen that is very similar to that observed for the parental Inclonal FRP5 IgG. Next, the ability of SFGFP-fusion antibodies to bind cellular antigen was evaluated by FACS. Bound Inclonals in this case were detected by means of red fluorescence obtained using phycoerythrin -labeled anti-human secondary antibodies. The analysis was performed using the two human tumor cell lines A431 and SKBR3 that express approximately 2 × 104 and 1.5 × 106 ErbB2 receptors per cell, respectively.14,15 As shown in Figure 3, the fluorescence intensity obtained with the SFGFP-fused antibodies was in each case almost identical to the fluorescence intensity obtained with the parent Inclonal FRP5 on both cell lines. Taken together, the ELISA and FACS results suggest that the fusion of SFGFP molecules to the C-terminus of FRP5′s heavy and light chains does not interfere with antigen binding capabilities of the IgG portion of the fusion protein. These results also suggest that the addition of the fluorescent molecules to the antibody’s chains does not interfere with standard secondary detection methods. FACS analysis of fluorescence intensity using SFGFP-fused antibodies.
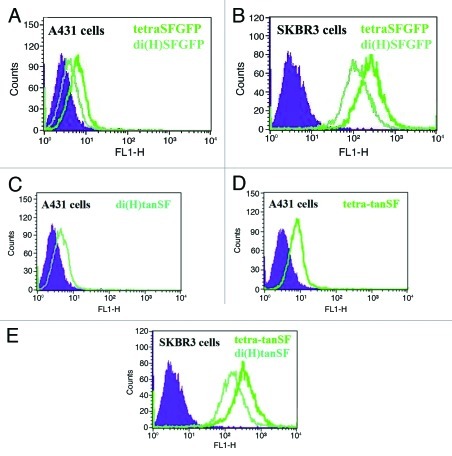
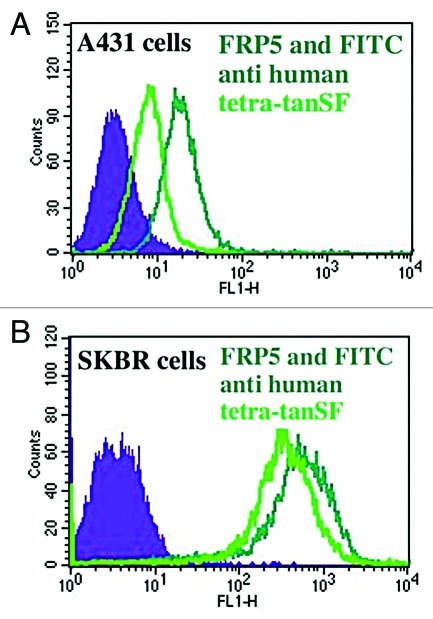
The ability of SFGFP-fused antibodies to stain antigen positive cells was evaluated on both A431 and SKBR3 cell lines by FACS without the use of labeled secondary antibodies (thus relying on SFGFP’s fluorescence alone). As shown in Figure 4, all the SFGFP-fused antibodies showed bright green fluorescent staining on both the cell lines. The ability to successfully label A431 cells demonstrates good sensitivity since A431 cells express 100 fold less ErbB2 receptors per cell than SKBR3 cells. No staining could be observed when tested on ErbB2-negative cells (FRP5 tetra-tan SF is shown as an example in Supplementary Figure 1). Each of the fusion antibodies exhibited quantitative staining by generating different signal intensities on the two cell lines according to their antigen density and number of fused SFGFP molecules per IgG. When comparing di(H)SFGFP to tetraSFGFP, tetraSFGFP’s stain is approximately twice more intense than di(H)SFGFP’s stain (344 vs. 160 fluorescence signal in the case of SKBR3 cells), suggesting that SFGFP generates a similar intensity of fluorescence whether it is fused at the C-terminus of the light chain, or at the C-terminus of the heavy chain. When comparing di(H)tanSF with tetra-tanSF, we get equivalent results regarding tanSF (a 2-fold difference that is proportional to the number of fused SFGFP molecules per IgG).
Figure 4. Evaluation of fluorescence intensity of SFGFP fused antibodies by FACS. Cellular ErbB2 binding of SFGFP-fusion antibodies was evaluated on human epithelial carcinoma cell line A431 and human breast adenocarcinoma cell line SKBR3 without a secondary antibody (measuring the green fluorescence of the antibodies). (A) di(H)SFGFP and tetraSFGFP binding to A431 cells. (B) di(H)SFGFP and tetraSFGFP binding to SKBR3 cells. (C) di(H)tanSF binding to A431 cells. (D) tetra-tanSF binding to A431 cells. (E) di(H)tanSF and tetra-tanSF binding to SKBR3 cells. Filled areas, negative control (auto fluorescence of cells).
Further evaluation of the contribution of each SFGFP molecule to the fluorescence intensity of IgG-SFGFP fusion proteins
FACS analysis was performed to further compare the signal intensity obtained in staining antigen positive cells by tetra-tanSF relatively to tetraSFGFP. As shown in Figure 5A, tetra-tanSF staining is roughly twice more intense than tetraSFGFP staining (fluorescence signals of 421 vs. 276), suggesting that the C-terminal SFGFP and the N-terminal SFGFP in the tandem pair contribute equally to the fluorescence intensity of the entire molecule. This was further supported by comparing di(H)tanSF staining with tetraSFGFP staining (Fig. 5B), where similar signal intensities were obtained using either di(H)tanSF (2 tandem pairs fused to the heavy chains) or tetraSFGFP (4 single SFGFP molecules fused, one to each chain of the IgG).
Figure 5. Evaluating the relative contribution of each SFGFP to the overall fluorescence intensity by FACS. SFGFP-Fused antibodies’ ability to stain antigen positive cells was evaluated on human breast adenocarcinoma SKBR3 cells without labeled secondary antibodies. (A) tetraSFGFP compared with tetra-tanSF (B) tetraSF compared with di(H)tanSF. Filled areas, negative control (cell auto fluorescence).
Fluorescence microscopy using SFGFP-fused antibodies
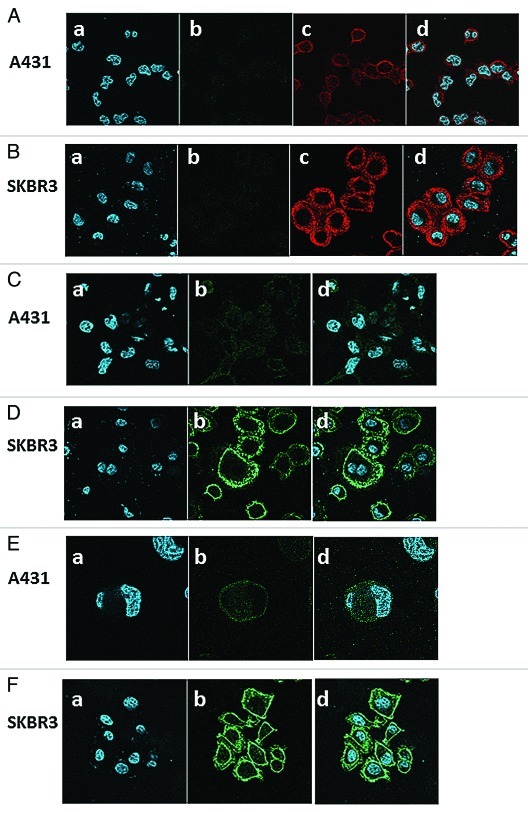
TetraSFGFP and tetra–tanSF were further tested for their ability to stain antigen positive cells for visualization by fluorescence microscopy. A431 and SKBR3 cells were probed with the fluorescent antibodies and analyzed by confocal fluorescence microscopy. For comparative purposes, both cell lines were also probed with the parent antibody FRP5. As shown in Figure 6A and 6B, when either one of the cell lines was probed with FRP5 IgG alone, no green fluorescence could be detected. Cell bound FRP5 antibodies could be visualized by Cy3-labeled anti human IgG. Red fluorescence emitted by Cy3 secondary antibodies illustrates the different levels of ErbB2 receptors on the two different cell lines, exhibiting weak signal on A431 cells and strong signal on SKBR3 cells. Images of A431 and SKBR3 cells stained with tetraSFGFP are shown in Figure 6C and 6D. Images of the cell lines stained with tetra–tanSF are shown in Figure 6E and 6F. As in the case of the Cy3 labeled polyclonal antibodies, SFGFP-fused antibodies also exhibited a much stronger fluorescent signal on SKBR3 than on A431 cells.
Figure 6. Confocal fluorescence microscopy images. Shown are images of human epithelial carcinoma cell line A431 (A, C, E) and human breast adenocarcinoma cell line SKBR3 (B, D, F) stained with nucleic acid dye Hoechst and antibodies; FRP5 IgG followed by Cy3-conjugated anti-human IgG in A and B; TetraSFGFP in C and D; Tetra-tanSF in E and F. Panels a show Hoechst staining, panels b show the green fluorescent channel, panels Ac and Bc show the red fluorescent channel and panels d show the merge of Hoechst staining with red fluorescence in A and B and with green fluorescence in C, D, E and F.
SFGFP-fused antibodies' binding and fluorescence stability analysis
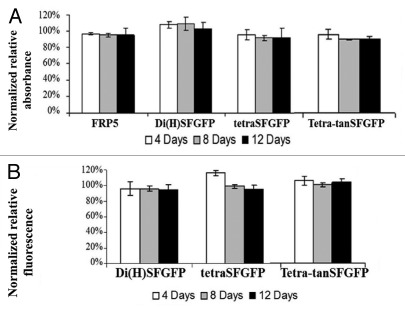
SFGFP-fused antibodies and the parental antibody FRP5 IgG were incubated for varying time periods at 37°C in PBST in order to evaluate the stability of SFGFP-fused antibodies’ binding and fluorescence activities over time. Stability was estimated by measuring residual binding by ELISA and residual fluorescence intensity by fluorescence spectrophotometer after incubation. As shown in Figure 7A, all the SFGFP-fused IgGs retained their binding potential after spending up to 12 d at 37°C, matching the stability of the parental antibody FRP5 under these conditions. Fluorescence measurements done by fluorescence spectrophotometer showed that SFGFP-fused antibodies also retained their fluorescence potential after spending up to 12 d at 37°C (Fig. 7B). These results suggest that the antibodies are stable and do not deteriorate for at least 12 d at 37°C.
Figure 7. Stability analysis of antibodies following incubation at 37°C. (A) Stability of binding capability: ELISA analysis following incubation of the antibodies at 37°C for 4, 8 or 12 d. Binding to ErbB2 of di(H)SFGFP, tetraSFGFP and tetra-tanSF in relation to parental Inclonal FRP5 IgG was evaluated using HRP-conjugated secondary antibodies. The results are presented as the percentage of the signal obtained for 0 d- incubation of each antibody, which was normalized to be 100%. (B) Stability of fluorescence intensity: Fluorimeter analysis following incubation at 37°C for 4, 8 or 12 d. Fluorescence (485-nm excitation and 528-nm emission) of di(H)SFGFP, tetraSFGFP and tetra-tanSF was measured. The results are presented as the percentage of the fluorescence obtained for 0 d- incubation of each antibody, which was normalized to be 100%. Error bars represent standard deviations of triplicates.
Evaluating how the fluorescence intensity of SFGFP is affected by the oxidative conditions of the “Inclonals” refolding protocol
GFP and derived fluorescent proteins are known to fold correctly under the reducing conditions found in the cytoplasm of E. coli.4 Therefore, when an in vivo folded SFGFP was required to be used as a reference for correctly folded SFGFP, a bacterial vector for cytoplasmic expression of SFGFP was constructed. The bacterial vector contained SFGFP fused at the C-terminus of a ZZ domain. ZZ domain is an engineered repeat of Z domain, a mutated version of Staphylococcus aureus protein A IgG binding domain.16,17 Since ZZ domain is also known for its ability to improve solubility of proteins it is fused to,18 ZZ domain was not expected to interfere with SFGFP’s folding.
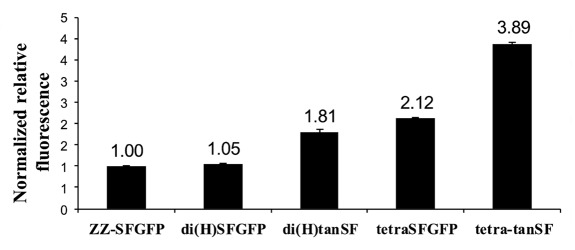
As shown in Figure 8, the intensity of the signal obtained from ZZ-SFGFP fusion protein (which carries one SFGFP molecule) was found to be similar to the intensity of the signal obtained from di(H)SFGFP (which carries two SFGFP molecule) when both proteins were applied to fluorescence spectrophotometer at equal molar concentrations. This implies that the in vivo folded SFGFP is approximately twice more fluorescent than the Inclonals technology product (refolded) SFGFP (assuming fluorescence “burden” is equally shared between the two SFGFP molecules carried by each of the heavy chains). The fluorescent signal of tetra-SFGFP was measured to be approximately twice more intense than di(H)SFGFP and ZZ-SFGFP signals. This data supports our interpretation of the FACS experiments shown in Figure 4 that the ability of the refolded SFGFP to generate fluorescence is very similar whether it is fused at the C-terminus of the light chain or at the C-terminus of the heavy chain. The fluorescent signal measured for di(H)tanSF is approximately twice more intense than the signal of di(H)SFGFP, further supporting the assumption made while discussing Figure 5A that C-terminal SFGFPs contribute to the fluorescence activity approximately the same as the N-terminal SFGFPs do (within the tandem pair). di(H)tanSF's signal is also about twice more intense than the signal of ZZ-SFGFP, suggesting the fluorescence of its four SFGFPs molecules is equivalent to the fluorescence of two natively folded ones. Taken together, these data indicate that each one of the tandemly-linked SFGFP domains of IgG-tanSF proteins is about 50% less efficient than natively folded SFGFP. This trend is maintained when tetra-tanSF is compared with tetraSFGFP (Fig. 8).
Figure 8. Fluorescence (485-nm excitation and 528-nm emission) of ZZ-SFGFP, SFGFP-fused antibodies and IgG-tanSF antibodies. The fluorescence was measured by fluorescence spectrophotometer at equivalent molar protein concentrations. Antibodies values are presented as the relative fluorescence with respect to ZZ-SFGFP signal, which was set as 1.00.
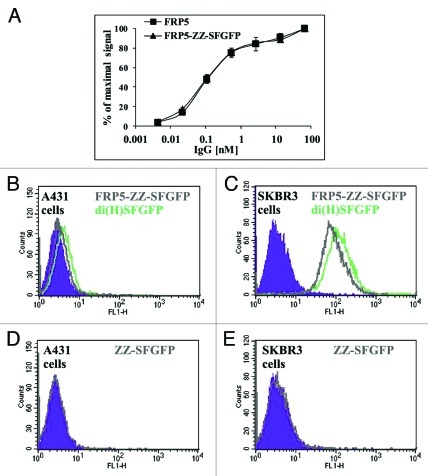
As mentioned earlier, ZZ domain is a tandem repeat of a mutated version of Staphylococcus aureus protein A IgG binding domain. Like the domain it was derived from, ZZ possesses a strong binding affinity to the Fc region of human IgG1.19 The ZZ-SFGFP fusion protein was mixed with FRP5 IgG to yield FRP5-ZZ-SFGFP immunocomplex. After the immunocomplex was shown to retain similar binding capability to that of FRP5 IgG (Fig. 9A), FACS was performed to examine its ability to stain A431 and SKBR3 cell lines in comparison to SFGFP-fused antibodies. FRP5-ZZ-SFGFP staining was found to be roughly as intense as di(H)SFGFP staining on both cell lines (see Figure 9B and 9C), confirming our interpretation of the results presented in Figure 8 that in vivo folded SFGFP is approximately twice more fluorescent than the SFGFP produced by the Inclonals technology (refolded). It is notable that, although theoretically there are two binding sites for ZZ on an IgG, we found that only a single ZZ-based fusion protein binds to each IgG molecule.19
Figure 9. Comparing cytoplasmically folded SFGFP to refolded SFGFP. (A) Binding capacity evaluation by ELISA. Binding of FRP5-ZZ-SFGFP in comparison to the parental Inclonal FRP5 IgG was evaluated on soluble ErbB2 using HRP-conjugated secondary antibodies. Error bars represent standard deviations of triplicates. (B-E) FACS analysis: di(H)SFGFP and FRP5-ZZ-SFGFP ability to stain antigen positive cells was evaluated on human epithelial carcinoma cell line A431 (B) and human breast adenocarcinoma cell line SKBR3 (C). Cells that were incubated with free ZZ-SFGFP alone are shown in D and E as a negative control. Filled areas in all panes; negative control (cells auto fluorescence).
Comparison of tetra-tanSF and FITC-labeled secondary antibodies performance
We conducted a FACS experiment to examine the ability of the most intense antibody we prepared in the described study to stain cells in comparison to conventional green staining obtained by staining with a commercial secondary green fluorescent antibody, We compared the fluorescent signal obtained by probing cells with the anti ErbB2 antibody FRP5-tetra-tanSF, with the fluorescent signal obtained by probing cells with FRP5 IgG followed by FITC labeled secondary antibodies. Fluorescein was chosen for this experiment because it is the most commonly used green fluorescent dye in FACS analysis. As shown in Figure 10A and 10B, staining with FITC-conjugated polyclonal antibodies was found to be 1.5 fold more intense than tetra-tanSF direct staining as demonstrated by FACS examinations using both A431 and SKBR3 cells.
Figure 10. FACS comparison of tetra-tanSF with FITC polyclonal antibodies. (A) A431 cells stained with FRP5 and FITC-labeled secondary antibodies (mean fluorescence intensity 26) or with FRP5-tetra-tanSF fusion protein (mean fluorescence intensity 17). (B) SKBR3 cells stained with FRP5 and FITC-labeled secondary antibodies (mean fluorescence intensity 626) or with FRP5-tetra-tanSF (mean fluorescence intensity 422). Filled areas, negative control (Cell auto fluorescence).
Discussion
We have shown the successful design, production and analysis of a chimeric antibody fused with SFGFP, a derivative of GFP. The different design variants we made enabled us to prepare IgG formats with either two, four or eight SFGFP molecules per IgG. Their utility as staining reagents was demonstrated by both FACS and fluorescence microscopy. Their binding capacity was shown to be virtually identical to that of their parental antibody in both ELISA and FACS examinations. Both their binding and fluorescence activities were found to be stable. Furthermore, the antibody carrying eight fluorescent proteins, although monoclonal, was found to yield a fluorescent signal in cell staining comparable to the signal obtained by using conventional green-fluorescence (FITC labeled) secondary polyclonal antibodies staining in FACS.
These kind of fluorophore-fused antibodies can potentially be used for research and diagnosis applications where monoclonal antibodies are desired (that is, for direct immunofluorescence staining), possibly giving cost-efficient alternative to the ascites method, as well as overcoming some of the disadvantages associated with the conventional labeling method. Conjugation of fluorophores to antibodies is typically performed by attaching the desired fluorophore with isothiocyanate, sulfonyl chloride, succinimidyl ester, or other reactive group that readily react with primary amines (that is, lysine residues) on proteins.20 The indiscriminate nature of such conjugation reactions with respect to the lysine residues that ultimately become tagged leads to a heterogeneous population of labeled antibodies, with some having diminished antigen-binding capabilities. Moreover, interactions between closely positioned fluorophores may reduce fluorescence intensity through the phenomenon of self-quenching.21 No more than about three to five organic dyes can be attached to an antibody. In the case of fluorescein, for example, maximum brightness is attained with two to four fluorophores per antibody. Higher number of fluorophores will result in a dimmer reagent.2,3
A recombinant fusion protein comprised of an antibody and a fluorescent moiety has the advantages of defined number and location of fluorophores. In the context of a fusion protein, the stoichiometry and location of fluorophores attached to the antibody are predetermined by the engineered DNA sequence, and as a result, are invariant. On top of that, fluorescent proteins have much less intrinsic environmental sensitivity than organic dyes because their fluorophore is encapsulated within an insulating layer; as a result, fluorescent proteins are much less susceptible to self-quenching.2 This is well demonstrated in our work as all the data we collected from FACS and fluorescence spectrophotometery measurements indicates a linear increase in fluorescence (rather than a decrease) with increasing number of SFGFP molecules per IgG up to eight SFGFP molecules attached to a single antibody. The ability to attach eight fluorophores to a single IgG molecule without self-quenching of fluorescence or inactivating the antibody enables the formation of a robust staining reagent which, although monoclonal, gives a fluorescence signal comparable to the signal obtained using polyclonal secondary antibodies labeled with the most commonly used green dye, FITC. GFP proteins have the added advantage of being more resistant to photobleaching than fluorescein.22
The construction of an antibody-fluorophore fusion protein having both binding and fluorescence activities posed an interesting challenge as antibodies and fluorescent proteins have different folding requirements. Completely assembled and correctly folded immunoglobulin molecules are naturally produced in B cells or plasma cells, where their correct folding, assembly and formation of disulfide bonds takes place in the oxidizing environment of the ER. Similarly, IgGs that were made in E. coli as soluble secreted proteins were expressed and exported to the bacterial periplasm, where oxidation of disulfide bonds can take place.23,24 GFP on the other hand is a cytoplasmic protein that is known to fold correctly under the reducing conditions found in the cytoplasm of the jellyfish Aequorea victoria and other species in which it has been expressed as a recombinant protein.4 An early attempt we made to construct a fluorescent antibody using EGFP as the fluorescent moiety resulted in antibodies with fine binding capabilities, but negligible fluorescence activity. Upon induction, the EGFP-fused antibody chains accumulated as green fluorescent inclusion bodies. However, the green fluorescence was lost during the denaturation step, and was apparently not recovered during the refolding process. EGFP’s inability to fold correctly under the conditions of a refolding protocol that has been optimized for IgG production is in agreement with the impaired folding behavior GFP-like proteins have previously exhibited under equivalent oxidizing environments such as the eukaryotic ER or the bacterial periplasm.4,25 To overcome this obstacle, we took advantage of the highly efficient folding capabilities of SFGFP, a GFP variant that has been evolved in vitro for folding robustness.12 As opposed to EGFP, SFGFP-fused antibody chains exhibited fluorescence under denaturing conditions as solubilized inclusion bodies in 6M guanidine hydrochloride-based buffer for a substantial amount of time. Upon completion of refolding and protein-A purification of the IgG-SFGFP fusion proteins, the products were brightly fluorescent with the refolded SFGFP having approximately half the fluorescence activity exhibited by natively folded SFGFP. Interestingly, the refolding kinetics of SFGFP fused to an antibody chain are quite rapid (Supplementary Figure 3), and efficient refolding of SFGFP required reduction prior to refolding. These observations and the fact that solubilized and reduced SFGFP-containing inclusion bodies are fluorescent (although significantly less than refolded protein, Supplementary Figure 3) may suggest the presence of an intermediate state having less fluorescence intensity. This is not characteristic of SFGFP, which demonstrates 100% fluorescence recovery upon dilution from concentrated urea and GndHCl solutions,12,26 but is conceivable considering this time the fluorescent protein folding was attempted while it was fused to another protein, as well as under the action of a refolding protocol that has been optimized for IgG production, i.e., under oxidizing conditions. It is conceivable that the “Inclonals” protocol that was optimized for antibodies and antibody-based fusion proteins that require oxidizing condition for efficient refolding can be further optimized to provide a solution to cases where the fusion partner prefers reducing conditions in the refolding step. A case in point is our experience with red fluorescent mCherry fused IgGs that initially yielded poorly fluorescent antibodies on completion of the production protocol. We found that it was possible to modify the protocol in a way that made it possible to purify brightly fluorescent IgG-mCherry fusion proteins that could stain cells in FACS (Supplementary Figure 2). It seems that the robustness of the fluorescent protein is predictive of the possibility to produce it as an IgG-fluorophore fusion protein in our system.27,28 Hence it is conceivable that we can expand our collection of “fluorescent Inclonals” to a wide range of fluorescent colors.
Most reports dealing with fluorescent antibodies describe the use of antibody fragments such as scFv or llama VHH to which fluorescent proteins were fused.5-10 Unfortunately, monovalent antibody fragments usually have lower functional affinity compared with full length IgGs. This should not rule out the use of exceptionally robust fragments such as VHH from being applied in parallel to IgGs for such applications. When comparing IgGs with fragments, one should carefully weigh the advantages of IgG-based molecules, mostly robustness, with the possibility to produce fragments by secretion, which is less costly and time consuming than refolding. Very recently, Haas et al. reported the production of full length IgG fused to the yellow fluorescent protein citrine in mammalian cells.11 The expression system they used was a CMV promoter-based plasmids introduced by transient transfection into HEK293-F cells, where the IgG-citrine fusion protein was secreted and purified from the conditioned medium. These authors managed to attach their IgG with up to two citrine molecules by adding a citrine to the C-terminus of each one of its light chains. Before successfully attaching their IgG with citrine, Haas et al. tried to fuse the IgG with the fluorescent protein EGFP. Their attempt resulted, as in our case, in a substantial amount of improperly folded protein with poor fluorescence.
When using fluorescence-based devices such as confocal microscopy or flow cytometry, one should bear in mind that the excitation these devices provide is restricted to a few wavelengths produced by their lasers. A green variant is usually superior to a yellow variant in being much more efficiently excited by the most common laser line (488 nm argon ion line). Moreover, E. coli based expression systems are usually superior to other expression systems in terms of production cost and are therefore more likely to provide an actual cost-effective alternative to the polyclonal antibodies from immunized animals or to monoclonal antibodies produced by the ascites method than mammalian cells culture production methods.
Antibody-based fluorescence detection can be done using either direct (primary) or indirect (secondary) antibody staining. Each of these procedures has its strengths and weaknesses. Direct staining using fluorescent primary antibodies is rapid, easily performed and permits parallel detection of different targets (multiparametric analysis). The main disadvantages of the direct approach is the higher cost of individually prepared fluorescent antibodies required for each target and lack of signal amplification that is offered by using secondary fluorescent antibodies. Indirect staining is technically more complex and time consuming, but offers the advantage of signal amplification using an assortment of antibodies (in the case of polyclonal secondary antibodies) labeling one primary antibody. Multiparametric analysis using indirect staining is less practical as it requires the use of antibodies obtained from different animal species.29
As mentioned earlier, the antibody carrying eight fluorescent proteins (tetra-tanSF), although monoclonal, was found to yield a fluorescent signal in cell staining comparable to the signal obtained by using conventional green-fluorescence (FITC labeled) secondary polyclonal antibodies staining in FACS. When taking into consideration that five or six secondary antibodies usually bind to a single primary antibody,30 this observation implies tetra-tanSF signal to be a robust one that would probably not fall short of FITC monoclonal staining. Moreover, by attaching to each chain of the antibody more than two SFGFP molecules, greater signal can possibly be obtained. Pack et al. for instance constructed a protein comprised of five tandemly linked EGFP molecules, which showed increasing fluorescence with increasing number of linked EGFP molecules.31 Needless to say, it is possible to prepare recombinant secondary antibodies using our approach, by attaching SFGFP to a species-specific monoclonal antibody. We are now preparing such molecules.
Materials and Methods
Construction of FRP5 IgG expression vectors
All the PCR primers that were used in this study are listed in supplementary Table 1. The VH gene of murine anti-ErbB2 antibody FRP5 was recovered from plasmid pMAZ-IgH-FRP532 by PCR using primers FRP5VH-NdeI-FOR and FRP5VH-NheI-REV. The PCR product (375 bp) was digested with NdeI and NheI restriction enzymes and introduced into T7-based, IPTG-inducible bacterial expression vector pHAK-IgH-T42713 that was cut with the same enzymes (4064 bp vector fragment), resulting with the bacterial plasmid pHAK-IgH-FRP5 for the cytoplasmic expression of anti-ErbB2 FRP5 IgG1 chimeric antibody heavy chain. The VL gene of murine anti-ErbB2 antibody FRP5 was recovered from plasmid pMAZ-IgL-FRP532 by PCR using primers FRP5VL-NdeI-FOR and FRP5VL-BsiWI-REV. The PCR product (339 bp) was digested with NdeI and BsiWI restriction enzymes and introduced into T7-based, IPTG-inducible bacterial expression vector pHAK-IgL-T42713 that was cut with the same enzymes (3411 bp vector fragment), resulting with the bacterial plasmid pHAK-IgL-FRP5 for the cytoplasmic expression of anti-ErbB2 FRP5 IgG1 chimeric antibody light chain.
Table 1. PCR primers (restriction sites are underlined).
| Primer name |
Sequences |
| FRP5VH-NdeI-FOR |
5'ATATATCATATGCAGGTACAACTGCAGCAGTCT |
| FRP5VH-NheI-REV |
5'ATATATGCTAGCAGAGGAAACGGTGACCGTGGTCC |
| FRP5VL-NdeI-FOR |
5'ATATATCATATGGACATCCAGCTGACCCAGTCTCAC |
| FRP5VL-BsiWI-REV |
5′AGCCACCGTACGTTTGATCTCCAATTTTGTCCCCGAGC |
| RGD/TAT-BsrGI-FOR |
5’GACGTGAGCCACGAAGACCCTGAGGTC |
| CH3-HindIII-EcoRI-REV |
5’AAATTTGAATTCACCTCCGGAAGCTTTACCCGGGGACAGGGAG |
| BsiWI-Back-IgL |
5’AAACGGCGTACGGTGGCTGCACCATCTGTCTTC |
| Cκ -HindIII-EcoRI-REV |
5’AAATTTGAATTCACCTTCGGAAGCTTTTCCACCGCCACACTCTCCCCTGTTGAAG |
| tandem-HindIII-FOR |
5'ATTACGAGTAAAGCTTCCGCTGGCTCCGCTGCTGGTTCTGGCGCAGCGGCAGTGAGCAAGGGCGAGGAGCTG |
| tandem-NotI-REV | 5'AATTCTCACCGCGGCCGCGCTGCCGCCGCCGCCAGAACCACCACCACCCTTGTACAGCTCGTCCATGCC |
Construction of fluorescent IgG fusion proteins expression vectors
A DNA fragment encoding the amino acid linker SAGSAAGSGAAA followed by a SFGFP gene (motif order: HindIII site – linker – NotI site – SFGFP ORF – Stop codon – EcoRI site, 747 bp) was prepared by total gene synthesis (GeneArt®, Germany).
The pHAK-IgH-FRP5 expression vector was modified by insertion of HindIII/EcoRI cloning site at the C-terminus of the antibody by PCR using the vector itself as a template and primers RGD/TAT-BsrGI-FOR and CH3-HindIII-EcoRI-REV. The PCR product was digested with BsrGI and EcoRI and cloned into pHAK-IgH-FRP5 that was linearized using the same enzymes. The resulting vector was digested with HindIII and EcoRI and ligated with SFGFP DNA fragment that was cut with the same enzymes, resulting with pHAK-IgH-FRP5-SFGFP expression vector for heavy chain fused via linker to SFGFP. The pHAK-IgL-FRP5 expression vector was modified by insertion of HindIII/EcoRI cloning site at the C-terminus of the antibody by PCR using the vector itself as a template and primers BsiWI-Back-IgL and Cκ-HindIII-EcoRI-REV. The PCR product was digested with SacI and EcoRI and cloned into pHAK-IgL-FRP5 that was linearized using the same enzymes. The resulting vector was digested with HindIII and EcoRI and ligated with SFGFP DNA fragment that was cut with the same enzymes, resulting with pHAK-IgL-FRP5-SFGFP expression vector for light chain fused via linker to SFGFP.
A DNA fragment encoding the amino acid linker SAGSAAGSGAAA followed by a SFGFP gene and then another amino acid linker GGGGSGGGGSAAA was obtained by PCR amplification using the plasmid pHAK-IgH-FRP5-SFGFP as template with primers tandem-HindIII-FOR and tandem-NotI-REV. The resulting PCR product (795 bp) was digested with HindIII and NotI and was ligated with vector fragments that were recovered after digesting plasmids pHAK-IgH-FRP5-SFGFP and pHAK-IgL-FRP5- SFGFP with the same enzymes. The resulting plasmids were named pHAK-IgH-FRP5-tanSF and pHAK-IgL-FRP5-tanSF, respectively, encoding heavy and light chains fused to two tandem copies of SFGFP.
Construction of ZZ-SFGFP expression vector
The ZZ domain encoding sequence (motif order: NdeI site – 6 histidine codons – NcoI site – ZZ ORF – NotI site, 365 bp) was obtained from the plasmid pET22b-H6T-ZZ-EGFP19 by digestion with NdeI and NotI. The sequence was cloned between the corresponding sites in pHAK-IgH-FRP5-SFGFP, generating the plasmid pH6T-ZZ-SFGFP for the cytoplasmic expression of ZZ-SFGFP fusion protein.
Production and purification of IgGs and IgG fusion proteins (Inclonals)
All the antibodies were made according to the Inclonals protocol as previously described. 33 Briefly, the heavy and light chains of the desired antibody or antibody-fluorophore fusion protein were expressed as cytoplasmic inclusion bodies in two different bacterial cultures. The purified chains were denatured in 6 M Guanidine hydrochloride buffer, mixed together and reduced. Refolding was next initiated by dilution into a refolding buffer containing arginine as an aggregation suppressor and oxidized glutathione. The refolded antibodies were finally purified using protein A chromatography (GE healthcare, USA).
Cytoplasmic expression of soluble ZZ-SFGFP fusion protein
ZZ-SFGFP was purified from E. coli BL21 (DE3) Rosetta cells transformed with pH6T-ZZ-SFGFP plasmid essentially as described for ZZ-PE38.19 Briefly, the protein was recovered from cytoplasmic extracts of induced cells by Talon (cobalt) chromatography.
FRP5-ZZ-SFGFP immunocomplex preparation
The FRP5-ZZ-SFGFP immunocomplex was made by mixing FRP5 IgG with ZZ-SFGFP fusion protein in a 1:3 molar ratio at 4°C for 16 h. Excess ZZ-SFGFP was not removed from the final product; however had no effect on the examinations made, as shown in the results section.
ELISA. ELISA plates were coated overnight at 4°C with a recombinant fusion protein comprised of the E. coli maltose binding protein (MBP) fused to the extracellular domain of ErbB2 receptor (5 μg/ml in PBS), washed once with PBST, blocked with 3% milk powder in PBS for 2 h at 37°C and washed once more with PBST. Protein A purified IgG were added in triplicate to the wells at concentrations ranging from 4.35 × 10−3 to 68.5 nM and the plates were incubated for 1 h at room temperature. Unbound IgG was removed, the wells were washed three times with PBST and HRP-conjugated anti-human antibodies (Jackson Immunoresearch Laboratories, West Grove, PA, USA) (diluted 1:5000 in PBST) were added to the wells for 1 h at room temperature. Following the removal of unbound secondary antibodies, the wells were washed three times with PBST. ELISA was developed with TMB substrate for 10–30 min at RT. Color development was stopped with 1M H2SO4. Absorbance was then read at 450 nm (Spectramax 190, Molecular Devices). Each ELISA was performed at least 5 independent times and representative ones are shown in the figures. Since there is a difference between the binding of the secondary antibody to the paternal to the SFGFP fusion proteins (probably due to reduced accessibility by adding the fluorescent protein), results are shown as % maximal binding.
FACS
Approximately 1.0 × 106 cells were taken for each FACS experiment. The cells were trypsin-detached, washed with FACS buffer (2% FCS in PBS) and incubated with Protein A-purified IgGs (68.5 nM/3% BSA in PBS) at 4°C for 1 h. The cells were then washed twice with FACS buffer to remove unbound antibodies. For binding capacity evaluation, the cells were next incubated with R-phycoerythrin-conjugated F(ab')2 Fragment-specific goat anti-human IgG (H+L) (Jackson ImmunoResearch Laboratories West Grove, PA, USA) (diluted 1:200/3% BSA in PBS) at 4°C for 30 min. After another washing step, 10,000 cells were analyzed by flow cytometer (FACSort, Becton Dickinson) using 488 nm argon ion laser (EGFP, mCherry and SFGFP excitation) and 635 nm red diode laser (R-phycoerythrin excitation).
Fluorescence microscopy
For microscopy experiments, the cells were grown to 50–70% confluence on poly-l-lysine coated coverslips. The cells were gently washed twice (2% BSA in PBS), fixed with 4% paraformaldehyde in PBS at RT for 20 min, washed twice more and blocked with 90% fetal calf serum/2% BSA in PBS at RT for 30–60 min. Following the removal of the blocker, Protein A purified IgGs were added (68.5 nM/2% BSA in PBS) for 1 h incubation at RT. Following the removal of unbound antibodies, cells were washed two times and incubated with Hoechst 33258 (5μg/ml) at RT for 30 min. The cells were then washed two times and incubated with Cy3-conjugated F(ab')2 Fragment-specific goat anti-human IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) (1:100/ 2% BSA in PBS) at RT for 30 min. Following the removal of unbound secondary antibodies, the slides were washed twice with PBS, mounted with Fluorescent Mounting Medium, and examined using laser scanning confocal microscope (LSM META, Zeiss).
Comparative fluorescence analysis
SFGFP-fusion antibodies and ZZ-SFGFP fusion protein were diluted with PBS to a concentration of 137 nm and added to the wells of a black 96 well plate. SFGFP fluorescence was measured by fluorescence spectrophotometer (Synergy, Biotek) at 528 nm following 485 nm excitation.
Functional stability
SFGFP-fused IgGs were diluted to a concentration of 137 nm in PBST and incubated at 37°C up to 12 d. Residual binding and fluorescence activities were evaluated by ELISA and fluorescence spectrophotometer, respectively, as described above.
Supplementary Material
Acknowledgments
We thank Prof. Winfried Wels (Georg Speyer Haus, Frankfurt, Germany) for the FRP5 clone. This study was supported in part by research grant No. 08–3013-PG from the Israel Cancer Research fund (ICRF) and by grant No. 3–00000–6067 from the Israel Ministry of Health.
Glossary
Abbreviations
- GFP
green fluorescent protein (originating from the jellyfish Aequorea victoria)
- mCherry
monomeric cherry (red fluorescent protein)
- SFGFP
superfolder GFP
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/19581
References
- 1.Institute of Laboratory Animal Resources (U.S.). Committee on Methods of Producing Monoclonal Antibodies. Ebrary Inc. Monoclonal antibody production. Washington, D.C.: National Academy Press, 1999. [Google Scholar]
- 2.Pawley JB. Handbook of biological confocal microscopy. New York: Springer, 2006. [Google Scholar]
- 3.Vira S, Mekhedov E, Humphrey G, Blank PS. Fluorescent-labeled antibodies: Balancing functionality and degree of labeling. Anal Biochem. 2010;402:146–50. doi: 10.1016/j.ab.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J Bacteriol. 2000;182:4068–76. doi: 10.1128/JB.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey JL, Coley AM, Tilley LM, Foley M. Green fluorescent antibodies: novel in vitro tools. Protein Eng. 2000;13:445–52. doi: 10.1093/protein/13.6.445. [DOI] [PubMed] [Google Scholar]
- 6.Griep RA, van Twisk C, van der Wolf JM, Schots A. Fluobodies: green fluorescent single-chain Fv fusion proteins. J Immunol Methods. 1999;230:121–30. doi: 10.1016/S0022-1759(99)00131-3. [DOI] [PubMed] [Google Scholar]
- 7.Schwalbach G, Sibler AP, Choulier L, Deryckère F, Weiss E. Production of fluorescent single-chain antibody fragments in Escherichia coli. Protein Expr Purif. 2000;18:121–32. doi: 10.1006/prep.1999.1185. [DOI] [PubMed] [Google Scholar]
- 8.Morino K, Katsumi H, Akahori Y, Iba Y, Shinohara M, Ukai Y, et al. Antibody fusions with fluorescent proteins: a versatile reagent for profiling protein expression. J Immunol Methods. 2001;257:175–84. doi: 10.1016/S0022-1759(01)00462-8. [DOI] [PubMed] [Google Scholar]
- 9.Oelschlaeger P, Srikant-Iyer S, Lange S, Schmitt J, Schmid RD. Fluorophor-linked immunosorbent assay: a time- and cost-saving method for the characterization of antibody fragments using a fusion protein of a single-chain antibody fragment and enhanced green fluorescent protein. Anal Biochem. 2002;309:27–34. doi: 10.1016/S0003-2697(02)00290-7. [DOI] [PubMed] [Google Scholar]
- 10.Olichon A, Surrey T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J Biol Chem. 2007;282:36314–20. doi: 10.1074/jbc.M704908200. [DOI] [PubMed] [Google Scholar]
- 11.Haas AK, von Schwerin C, Matscheko D, Brinkmann U. Fluorescent Citrine-IgG fusion proteins produced in mammalian cells. MAbs. 2010;2:648–61. doi: 10.4161/mabs.2.6.13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 13.Hakim R, Benhar I. “Inclonals”: IgGs and IgG-enzyme fusion proteins produced in an E. coli expression-refolding system. MAbs. 2009;1:281–7. doi: 10.4161/mabs.1.3.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes NE, Gerber HA, Saurer S, Groner B. Overexpression of the c-erbB-2 protein in human breast tumor cell lines. J Cell Biochem. 1989;39:167–73. doi: 10.1002/jcb.240390208. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M, Hynes NE, Groner B, Wels W. A bivalent single-chain antibody-toxin specific for ErbB-2 and the EGF receptor. Int J Cancer. 1996;65:538–46. doi: 10.1002/(SICI)1097-0215(19960208)65:4<538::AID-IJC24>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, et al. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–13. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 17.Jendeberg L, Tashiro M, Tejero R, Lyons BA, Uhlén M, Montelione GT, et al. The mechanism of binding staphylococcal protein A to immunoglobin G does not involve helix unwinding. Biochemistry. 1996;35:22–31. doi: 10.1021/bi9512814. [DOI] [PubMed] [Google Scholar]
- 18.Inouye S, Sahara Y. Soluble protein expression in E. coli cells using IgG-binding domain of protein A as a solubilizing partner in the cold induced system. Biochem Biophys Res Commun. 2008;376:448–53. doi: 10.1016/j.bbrc.2008.08.149. [DOI] [PubMed] [Google Scholar]
- 19.Mazor Y, Noy R, Wels WS, Benhar I. chFRP5-ZZ-PE38, a large IgG-toxin immunoconjugate outperforms the corresponding smaller FRP5(Fv)-ETA immunotoxin in eradicating ErbB2-expressing tumor xenografts. Cancer Lett. 2007;257:124–35. doi: 10.1016/j.canlet.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Mullins JM. Fluorochromes: properties and characteristics. Methods Mol Biol. 2010;588:123–34. doi: 10.1007/978-1-59745-324-0_15. [DOI] [PubMed] [Google Scholar]
- 21.Chen RF, Knutson JR. Mechanism of fluorescence concentration quenching of carboxyfluorescein in liposomes: energy transfer to nonfluorescent dimers. Anal Biochem. 1988;172:61–77. doi: 10.1016/0003-2697(88)90412-5. [DOI] [PubMed] [Google Scholar]
- 22.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–90. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, et al. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263:133–47. doi: 10.1016/S0022-1759(02)00036-4. [DOI] [PubMed] [Google Scholar]
- 24.Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol. 2007;25:563–5. doi: 10.1038/nbt1296. [DOI] [PubMed] [Google Scholar]
- 25.Jain RK, Joyce PB, Molinete M, Halban PA, Gorr SU. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem J. 2001;360:645–9. doi: 10.1042/0264-6021:3600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews BT, Schoenfish AR, Roy M, Waldo G, Jennings PA. The rough energy landscape of superfolder GFP is linked to the chromophore. J Mol Biol. 2007;373:476–90. doi: 10.1016/j.jmb.2007.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyawaki A, Nagai T, Mizuno H. Mechanisms of protein fluorophore formation and engineering. Curr Opin Chem Biol. 2003;7:557–62. doi: 10.1016/S1367-5931(03)00097-8. [DOI] [PubMed] [Google Scholar]
- 28.Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nat Cell Biol. 2003;(Suppl):S1–7. [PubMed] [Google Scholar]
- 29.McClatchey KD. Clinical laboratory medicine. Philadelphia: Lippincott Wiliams & Wilkins, 2002. [Google Scholar]
- 30.Shapiro HM. John Wiley & Sons. Practical flow cytometry. New York: Wiley-Liss, 2003. [Google Scholar]
- 31.Pack C, Saito K, Tamura M, Kinjo M. Microenvironment and effect of energy depletion in the nucleus analyzed by mobility of multiple oligomeric EGFPs. Biophys J. 2006;91:3921–36. doi: 10.1529/biophysj.105.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazor Y, Barnea I, Keydar I, Benhar I. Antibody internalization studied using a novel IgG binding toxin fusion. J Immunol Methods. 2007;321:41–59. doi: 10.1016/j.jim.2007.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.