Abstract
The development of cellulose as an organizing structure in the plant cell wall was a key event in both the initial colonization and the subsequent domination of the terrestrial ecosystem by vascular plants. A wealth of experimental data has demonstrated the complicated genetic interactions required to form the large synthetic complex that synthesizes cellulose. However, these results are lacking an extensive analysis of the evolution, specialization, and regulation of the proteins that compose this complex. Here we perform an in-depth analysis of the sequences in the cellulose synthase (CesA) family. We investigate the phylogeny of the CesA family, with emphasis on evolutionary specialization. We define specialized clades and identify the class-specific regions within the CesA sequence that may explain this specialization. We investigate changes in regulation of CesAs by looking at the conservation of proposed phosphorylation sites. We investigate the conservation of sites where mutations have been documented that impair CesA function, and compare these sites to those observed in the closest cellulose synthase-like (Csl) families to better understand what regions may separate the CesAs from other Csls. Finally we identify two positions with strong conservation of the aromatic trait, but lacking conservation of amino acid identity, which may represent residues important for positioning the sugar substrate for catalysis. These analyses provide useful tools for understanding characterized mutations and post-translational modifications, and for informing further experiments to probe CesA assembly, regulation, and function through site-directed mutagenesis or domain swapping experiments.
Keywords: cellulose, cellulose synthase, CesA, CslD, CslF, comparative phylogenetics, cellulose synthase superfamily, sequence-based analysis
Introduction
A defining characteristic of the plant kingdom is the presence of a rigid cell wall. All plant cells are surrounded by a primary cell wall that determines cell shape by resisting internal turgor pressure, protects the cell from pathogens, and helps retain water essential for cellular function. This wall is composed of a diverse array of long polysaccharides with the main load-bearing and organizing structure provided by cellulose. The subsequent development of the first specialized conducting cells, the tracheary elements, allowed plants to grow in arid environments and increase size and tissue complexity. Tracheary elements involve a specialized secondary cell wall, enriched with cellulose and reinforced with lignin (Popper, 2008). Thus the inclusion of cellulose in cell walls enabled plants to colonize land and ultimately dominate terrestrial environments.
Cellulose consists of chains of β1 → 4 linked glucan residues between 500 and 14,000 monomers long. Individual chains hydrogen bond to neighboring chains, which creates a regular crystalline structure and densely packs the chains into a stabilized, inert core. Cellulose is an emergent compound with properties determined by the coordinated synthesis of several glucan chains whose combined properties bestow greater rigidity, tensile strength, and recalcitrance to chemical attack than isolated β1 → 4 glucan chains (Somerville, 2006).
Cellulose is produced by the cellulose synthase (CesA) proteins. These proteins act in a large synthetic complex observed in the plasma membrane (Brown and Montezinos, 1976) and cytosol (Bowling and Brown, 2008) of algae and plants. Although the complex is hypothesized to be composed of a hexamer of hexamers, with 36 independent subunits, the number of CesAs in a complex, as well as their stoichiometry, remain unknown (Doblin et al., 2002). Arabidopsis contains 10 CesA isoforms or classes. Of these, CesAs 4, 7, and 8 are implicated in synthesizing the secondary cell wall, while the other CesAs are implicated in primary cell wall synthesis (Somerville, 2006). Because mutations to any of CesAs 4, 7, or 8 cause identical phenotypes and double mutants do not have additive phenotypes, it is believed that CesAs 4, 7, and 8 act in the same complex and each make some essential contribution to the function of that complex (Taylor et al., 2003). Similar requirements are observed in rice (Tanaka et al., 2003). Null mutations to the primary CesAs 1 or 3 are lethal at the stage of pollen development, while null mutants to CesAs 2, 5, 6, or 9 suffer less severe effects on root and hypocotyl elongation or seed coat development (Beeckman et al., 2002; Desprez et al., 2002; Ellis et al., 2002; Stork et al., 2010). However, the quadruple knockout to CesAs 2, 5, 6, and 9 is lethal at the pollen stage, indicating that these gene products are partially redundant with each other and that the presence of at least one is essential (Desprez et al., 2007; Persson et al., 2007). This demonstrates that, like the secondary CesA complex, the primary complex is composed of three essential classes of CesAs.
The genes of the CesA family lie within a larger gene superfamily called the cellulose synthase-like (Csl) superfamily which are found throughout the plant kingdom and synthesize many of the non-cellulosic polysaccharides of the plant cell wall. Genes in this superfamily share a common origin and show distant similarity to CesAs identified in bacteria, which form their own clade sister to the plant Csl superfamily. All genes in the plant Csl superfamily fall into the category of type 2 processive glycosyltransferases, and each contains the active residues D, D, D, QxxRW. The amino acid sequences around these residues differ between families. Although all proteins in this superfamily have several transmembrane domains, the length and topology of the proteins also differs between families. Most Csl proteins localize to the Golgi and are presumably active there, while the CesA proteins localize to both the Golgi and plasma membrane and are presumably active only at the membrane.
In addition to the CesA family, the Csl superfamily has been divided into families CslA/B/C/D/E/F/G/H/, and J (Yin et al., 2009). Of these, CslD and CslF are most similar in sequence to the CesA genes and share the same membrane topology as the CesAs (Yin et al., 2009). CslFs are responsible for the production of β1 → 3, β1 → 4 mixed glucan linkages found in grasses (Burton et al., 2006). The product of the CslDs remains unknown.
Despite the importance of cellulose synthesis to the cell wall, a number of key questions about the CesAs remain unanswered. While it is clear from genetic evidence that there is a requirement for three distinct genetic CesAs positions in Arabidopsis primary and secondary complexes, it is not clear how broadly this division is shared across land plants. Since members of one CesA class cannot rescue mutants to a separate class, it would be expected that there are class-specific contributions that each CesA makes either to the architecture of the complex, its catalytic activity, or through the recruitment of interacting accessory and regulatory partners. However, at this time it is not understood what the class-specific contributions of each genetic position might be.
At the same time, it is not clear how the activity or assembly of the complex is regulated. Since mechanical or osmotic stress causes internalization of CesA proteins within minutes (Crowell et al., 2009; Gutierrez et al., 2009), there must be very rapid and coordinated changes to CesAs in the complex and recruitment of endocytosis factors in the presence of certain stimuli. It is also likely that some mechanism prevents cellulose synthesis from occurring in the Golgi, where the presence of crystalline cellulose could be disruptive to normal Golgi architecture and trafficking. This aspect sets CesAs apart from several other Csl family proteins, including CslFs, which are active in the Golgi (Carpita and McCann, 2010). Since CslFs are closely related to CesAs, there should be some region or modification that explains how CslFs can be active in the Golgi, while CesA activity is restricted there.
Here we use an in-depth analysis of the sequences of the CesA family to identify regions, amino acids, and modifications of potential importance to the assembly, function, and regulation of CesA proteins. Using CesA sequences from eight complete and three incomplete plant genomes representing the evolutionary distance from moss to eudicots (Table 1), we determine that CesA sequences diverged into six clades representing each essential component of the primary or secondary complex sometime after the divergence of lycophytes but prior to evolution of seed plants.
Table 1.
Genome information used.
| Name | Abbreviation | Classification | Genome completed | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | AT | Eudicot (rosid) | Yes | Arabidopsis Genome Institute (2000) |
| Arabidopsis lyrata | AL | Eudicot (rosid) | Yes | |
| Populus trichocarpa | Po | Eudicot (rosid) | Yes | Tuskan et al. (2006) |
| Vitis vinifera | VV | Eudicot (rosid) | Yes | Jaillon et al. (2007) |
| Oryza sativa | OS | Monocot | Yes | Goff et al. (2002) |
| Zea mays | ZM | Monocot | Yes | Schnable et al. (2009) |
| Sorghum bicolor | SB | Monocot | Yes | Paterson et al. (2009) |
| Pinus radiata | PR | Gymnosperm | No | |
| Pinus taeda | PT | Gymnosperm | No | |
| Physcomitrella patens | PP | Moss | Yes | Rensing et al. (2008) |
| Selaginella moellendorffii | SM | Lycophyte | Yes |
Cellulose synthase genes were identified from the following genomes listed here with the abbreviation used in this study and the classification of the organism.
Our results identify a number of regions and residues with the potential to explain differences in activity, interaction, and regulation between the CesAs. They will prove useful for the interpretation of future mutants and experimental results. They can also be used to inform further investigation of the CesA family through site-directed mutagenesis or domain swapping.
Materials and Methods
Acquiring and aligning sequences
Sequences were obtained by using the BLASTP resource from NCBI (Altschul et al., 1997) using each of the 10 Arabidopsis CesAs (Table 2) and comparing against the available plant genomes (Table 1) with the genome resources available from JGI (http://www.phytozome.net). All hits were recorded, totaling 88 CesA sequences. The same was done with 62 CslD sequences from Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, Oryza sativa, Selaginella moellendorffii, and Physcomitrella patens and 8 CslF sequences from Oryza sativa. A general BLAST using each Arabidopsis CesA sequence against bacterial genomes was performed to acquire 12 bacterial CesA sequences that could be used as an outgroup to root the phylogenetic analysis for all included plant Csl sequences. Specifically, the bacterial sequences were used to test whether the aligned plant sequences grouped appropriately into their corresponding CesA, CslD, and CslF families, and to ensure that no sequences falling outside these three families had been included in the analyses.
Table 2.
Arabidopsis genes used as base of BLAST search.
| AT identifier | Arabidopsis gene name |
|---|---|
| AT4G32410.1 | CesA1 |
| AT4G39350.1 | CesA2 |
| AT5G05170.1 | CesA3 |
| AT5G44030.1 | CesA4 |
| AT5G09870.1 | CesA5 |
| AT5G65740.1 | CesA6 |
| AT5G17420.1 | CesA7 |
| AT4G18780.1 | CesA8 |
| AT2G21770.1 | CesA9 |
| AT2G25540.1 | CesA10 |
| AT2G33100.1 | CslD1 |
| AT5G16910.1 | CslD2 |
| AT3G03050.1 | CslD3 |
| AT4G38190.1 | CslD4 |
| AT1G02730.1 | CslD5 |
| AT1G32180.1 | CslD6 |
The following CesA and CslD sequences of Arabidopsis were as the starting point of a BLAST search to identify homologs in other organisms.
An alignment was generated of the plant CesAs using the alignment program MUSCLE (Edgar, 2004). In addition, an alignment of the CesA, CslD, and CslF sequences was generated by MUSCLE for comparisons among the clades. These alignments are available in Supplementary Material.
The sequence of each alignment was parsed by individual residue into a MySQL database relating the amino acid in each protein to its position in the protein and in relation to the clade and global alignments. This database allowed for rapid, automated comparisons between CesA sequences and served as the basis for subsequent analysis.
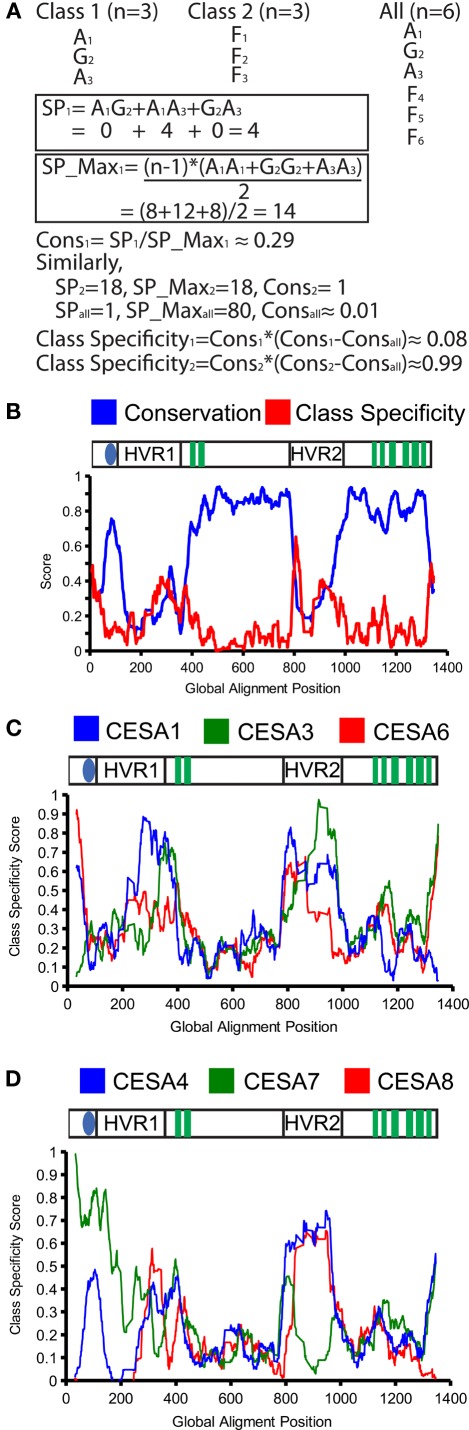
Further annotations were added based on transmembrane prediction, phosphorylation, and mutation data as well as all subsequent calculations. This database was queried against for all subsequent analysis comparing and calculating conservation at residues. It was also used in the calculation of class-specificity as shown in Figure 2. The annotations were also used to determine the position of all features used in schematics of the CesA protein shown.
Figure 2.
Conservation and class-specificity in the cellulose synthase family. An example calculation of conservation (Cons) and class specificity using the sum-of-pairs (SP) and max-sum-of-pairs (SP_max). SP and SP_Max are determined from the BLOSUM62 matrix (A). The conservation and class specificity averaged for all CesA genes with a sliding window of 10 amino acids is plotted. The x-axis represents the position in the global alignment. The cartoon above the graph plots the major CesA features against alignment positions. Transmembrane domains are represented as green boxes, the zinc finger as the blue oval, and both hypervariable regions are listed (HVR) (B). The class specificity for each CesA family is plotted for the primary CesAs (C) and secondary CesAs (D).
Construction of phylogeny
A phylogeny of the CesA sequences was constructed using maximum likelihood performed with the program PhyML (Guindon and Gascuel, 2003). The final phylogenetic tree was generated using PhyML from Phylogeny.fr with 100 bootstraps on the alignments of the CesA family.
Identifying class-specific regions
To develop a measure for class-specificity, we used the global alignment of all plant CesAs and the BLOSUM62 substitution matrix (Henikoff and Henikoff, 1992). First we developed a metric to score conservation at an alignment position ranging from 0 to 1. We calculated conservation by comparing the sum-of-pairs (SP) score from the BLOSUM62 matrix, a measure of amino acid similarity commonly used to score multiple alignments (Gupta et al., 1995), against the “theoretical maximum” score (SP_Max) that could be observed at that position. SP_Max is determined by calculating SP as though each amino acid shared perfect conservation with all the others in the column. We calculated conservation at each position by dividing SP by SP_Max. Effectively, this is a normalized value of conservation at each position, with perfect conservation at an aligned position giving a value of 1 (Figure 2A).
We calculated the conservation score for each position in the alignment for each CesA clade, as well as at the conservation at each alignment position across the entire CesA family.
To calculate a score for class-specificity, we compare the conservation within a clade to the conservation in the overall family. The conservation of the clade is subtracted by the conservation across all clades and multiplied by the conservation across all clades (negative values are fixed at 0). When there is strong conservation within a clade at an alignment position, but not strong conservation across all clades the class specificity score will be close to 1. When there is perfect conservation across all clades or no conservation within a clade, the score will be 0. Figure 2A provides an example of these calculations for a position in a set of two clades with six total sequences.
Representing sequence conservation in a region
To represent the conservation of sequences in each family, we created frequency plots of the amino acids found at each position in the global alignment. The height of an amino acid character at a given position is proportional to the frequency at which that amino acid occurs in the alignment. We used WebLogo to generate these plots (Crooks et al., 2004).
Determining phosphorylation sites
The Arabidopsis Protein Phosphorylation Site Database (PhosPhAt) compiles phosphorylation sites in Arabidopsis that have been specifically observed through mass spectrometry (Heazlewood et al., 2008). Each of the positions observed to be phosphorylated in any of the Arabidopsis proteins was manually annotated in the database as an observed phosphorylation site. These annotations also indicated whether the phosphorylation had been specifically observed at a site, or whether it was an ambiguous event due to the presence of a phosphate in a peptide with multiple serines or threonines. Phosphorylation sites observed specifically in CesA7, but not present in the PhosPhAt database were also included (Taylor, 2007).
Assigning transmembrane regions
To predict the location of the transmembrane domains of the protein, we used the module TMAP available from the European Molecular Biology Open Software Suite (Persson and Argos, 1994; Rice et al., 2000). TMAP uses an alignment of the protein family as the input for its predictions. The alignment of the CesAs was used. The resulting prediction contained eight transmembrane domains at positions agreeing with the established literature (Somerville, 2006).
Identifying known mutations
To map the known mutations, the literature of published mutations was screened. The position of each mutation was annotated in the database and the conservation of that position within CesA clades, within the full CesA family, and across the CesA, CslD, and CslF clades was manually investigated.
Identifying unique residue requirements
To identify positions with strong conservation of amino acid property but lacking conservation of specific amino acid identity, we divided the amino acids into categories based on the properties in Table 8. We screened for all sites with greater than 80% of the amino acids in the alignment column of the CesA family having the same property, but with no dominant amino acid in the alignment column (i.e., the most frequently observed amino acid in the column occurred less than 50% of the time). Each of the candidate columns was manually evaluated to judge whether any potentially interesting information could be determined.
Table 8.
Amino acid properties probed for conservation of property without identity.
| Property | Amino Acids |
|---|---|
| Charged | D, E, H, K, R |
| Positive | H, K, R |
| Negative | D, E |
| Small | A, G |
| Containing Aromatic | F,H,W,Y |
Amino acids are denoted by the standard single letter amino acid abbreviations. This division of amino acids was used to characterize the properties found in each alignment position in the CesAs.
Results
Phylogeny of the CesA family
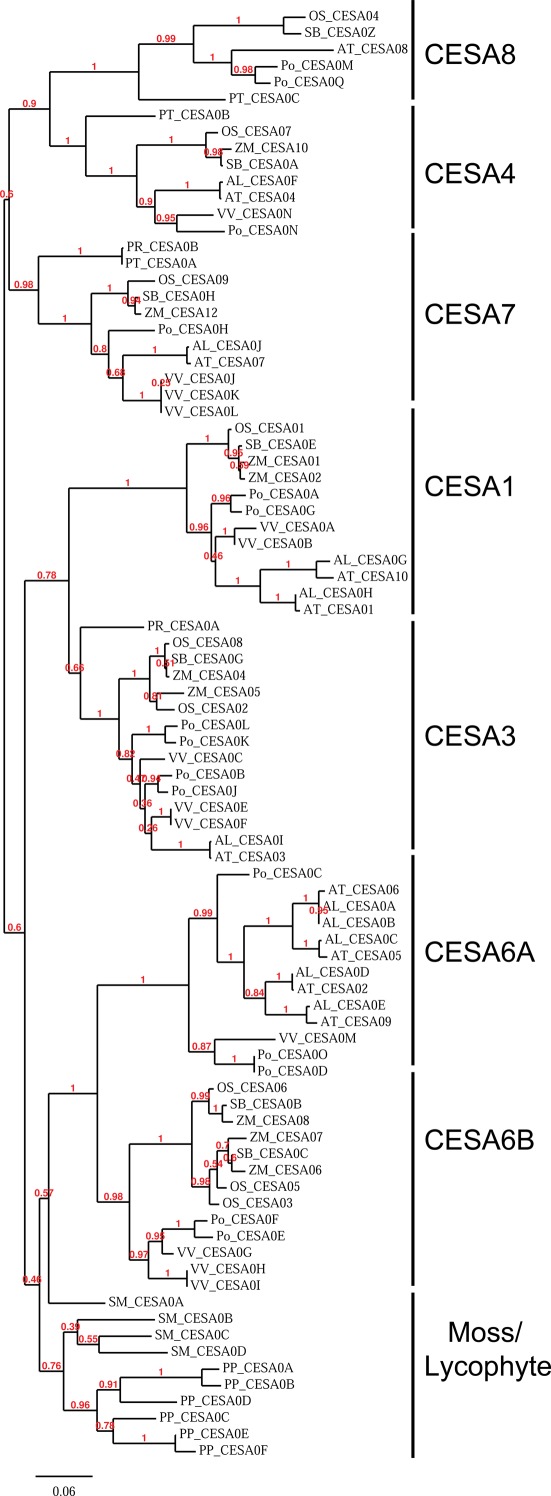
To better understand the evolution of the CesA family, we aligned the CesA sequences of the 11 genomes used for the study with MUSCLE and built a maximum likelihood phylogeny using PhyML (Guindon and Gascuel, 2003; Edgar, 2004). This phylogeny demonstrates that the CesA sequences of P. patens and S. moellendorffii form their own clades, while in the seed plants the phylogeny is separated into six major clades, reflecting ancient gene duplication events (Figure 1). Each of the clades represents one of the six required CesA classes identified in A. thaliana and O. sativa mutant studies. Each completed genome of the seed plants contains at least one CesA in each of the six major clades, suggesting that the requirement of a representative at each of the six genetic positions is shared across the seed plants. In addition, the CesA6 clade is divided into two subclades: one subclade (6A) is found only in the sampled eudicot species, while the other (6B) is found in both monocots and eudicots. Arabidopsis appears to have lost its CesA6B sequence.
Figure 1.
Phylogeny of the cellulose synthase family in land plants. A phylogenetic tree built using maximum likelihood by the program PhyML on the alignment of the CesA family. The phylogeny shows that the CesA family is divided into clades which correspond to the required CesAs in Arabidopsis. The clades are labeled based on their similarity to the identified Arabidopsis CesA. Numbers reported represent bootstrap values with 100 total bootstraps run. Organism abbreviations are listed in Table 1.
For convenience, we will refer to the CesA clades here based on their similarities to the A. thaliana (At) CesAs, e.g., “CesA1 clade” is the set of CesA genes most closely related to At CesA1.
The phylogenetic subdivision of the family is also reinforced by similarities in the expression patterns of CesA genes between Arabidopsis and rice. The secondary CesA genes of Arabidopsis (AtCesA4, AtCesA7, and AtCesA8) and rice (OsCesA4, OsCesA7, and OsCesA9) are expressed highly and specifically in vasculature. There is a class of highly and broadly expressed primary CesAs (AtCesAs 1, 3, and 6 and OsCesAs 1, 3, and 8). In both rice and Arabidopsis, the members of the CesA6 clade(AtCesAs 2, 5, and 9 and OsCesAs 5 and 6) are expressed differentially and in specialized tissues (Wang et al., 2010).
Analysis of sequence conservation identifies five class-specific regions
Because each clade reflects its own required and non-redundant isoform in the CesA complex, we decided to identify class-specific regions within the CesA sequence that could explain this specialization. We developed an algorithm to score the degree of class-specificity in a clade or across the CesA family at a given alignment position. This algorithm gives a score of 0 when there is either perfect conservation across all CesAs conservation at all at an alignment position. When a position shows strong conservation within a clade and this differs from the residues found across all CesAs, the class specificity score will be close to 1. For an example calculation see Figure 2A.
To present these data, we graphed the overall conservation and class-specificity scores as an average value of a sliding window over the sequence (Figure 2B). The overall conservation across all CesAs is similar to those published from the initially identified sequences (Pear et al., 1996).
Considering class-specificity across all CesA clades, five particular regions stand out: the far N-terminus and C-terminus are highly class-specific, there are two peaks of class-specificity within the second cytoplasmic loop, and the region at the end of the first cytoplasmic loop has a local peak in both conservation and class-specificity (Figure 2B). Plotting the class-specificity score for each family individually revealed interesting differences between CesAs. For the far N-terminus, only the motifs for CesA clades 1, 6, and 7 were significantly class-specific. For the far C-terminus, only the motifs for CesA clades 1, 3, 4, and 7 were class-specific (Figures 2C,D).
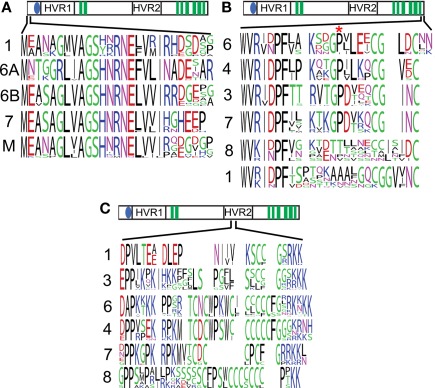
To investigate these class-specific regions more closely, we generated frequency plots of the conserved sequences found in each CesA clade. The far N-terminus of the CesAs shows a strongly conserved pattern L(V/I)AGSHNRNE(F/L)V in the clades containing CesA1, 6, and 7. Interestingly, moss and lycophyte CesA sequences have strong similarity to the sequences seen in CesA clades 1, 6, and 7, while CesA clades 3, 4, and 8 appear to have lost this region (Figure 3A).
Figure 3.
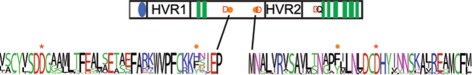
Sequences found in selected class-specific regions. Frequency plots of the sequences found in CesA clades represent the diversity of amino acids found at an alignment position, with more frequent amino acids drawn larger. The cartoon above each plot represents the location of that sequence in the global alignment. The numbers to the left of each sequence indicate which CesA clade the sequence was derived from, M represents moss plus lycophyte. A region at the far N-terminus is conserved in CesAs 1, 6, 7, and mosses and lycophytes, but lost from CesAs 3, 4, and 8 (A). The C-terminus shows strong conservation within CesAs 1, 3, 4, and 6, but not CesAs 1 and 8. The site of the rsw5 (P → S) is marked by an asterisk and is also not conserved in CesAs 1 and 8 (B). A region with conserved cysteines is flanked by charges in the second hypervariable region of the CesAs (C).
The far C-terminal end of the CesA proteins is a ∼23 amino acid long putatively cytoplasmic region following the eighth transmembrane domain. CesA clades 1, 3, 4, and 7 show strong conservation, while the sequences in CesA clades 1 and 8 are more divergent (Figure 3B). The second strongly conserved proline in this region is particularly noteworthy, because it is mutated to serine in the Arabidopsis CesA3 mutant allele rsw5. This mutant suffers radial swelling and reduced elongation when grown at the restrictive temperature of 31°C (Baskin et al., 1992) and the rsw5 mutation was shown to impair the ability of CesA3 to competitively incorporate into the CesA complex (Wang et al., 2006).
The first class-specific region in the second cytoplasmic loop occurs at the beginning of the hypervariable region. Though there are strongly class-specific residues, there is a similar overall pattern. In each case a cysteine-rich region is flanked on either side by a highly charged (usually positive) region (Figure 3C). We will describe and discuss the results from the remaining class-specific regions in the section on CesA phosphorylation.
Sequence conservation and putative phosphorylation sites
The PhosPhAt compiles phosphorylation sites in Arabidopsis that have been specifically observed through mass spectrometry (Heazlewood et al., 2008). A number of phosphorylation events have been observed in several Arabidopsis CesAs. One challenge in interpreting these phosphorylation data is that it is often difficult to determine whether an observed phosphorylation event is physiologically relevant. By mapping reported phosphorylation events onto our alignment, we show that 14 of the 23 sites show strong conservation (>70%) of serines and threonines (Table 3).
Table 3.
Conserved phosphorylation sites.
| Observed Arabidopsis residue | Conserved in |
|---|---|
| CesA3 S3 | CesA6A (100%) |
| CesA3 S148 | CesA3 eudicots (100%) |
| CesA3 S151 | CesA1 (78%), CesA3 (100%) |
| CesA1 S162 | CesA1 (100%), CesA3 eudicots (100%) |
| CesA1 T165 | CesA1 (100%), CesA3 angiosperm (100%) |
| CesA1 S167 | CesA1 (78%) |
| CesA7 S181 | CesA7 (100%) |
| CesA3 S211 | CesA1 (89%), CesA3 (93%), Moss (80%) |
| CesA3 T227 | CesA3 angiosperm (100%) |
| CesA1 S686 | CesA1 (100%) |
| CesA1 S688 | CesA1 (100%) Moss (100%) |
| CesA3 S671 | |
| CesA3 T672 | CesA1 (76%), CesA3 (86%), CesA6A (82%) |
| CesA2 T715 | CesA4 (88%), Moss (80%) |
A list of observed phosphorylation sites in Arabidopsis from PhosPhAt which occur at positions that are strongly conserved (>70% S/T) in at least one CesA family. The percentage of serines/threonines in each family are listed under conservation. When serines/threonines are highly conserved in one clade, but absent from others, the diresidues are universal among one clade.
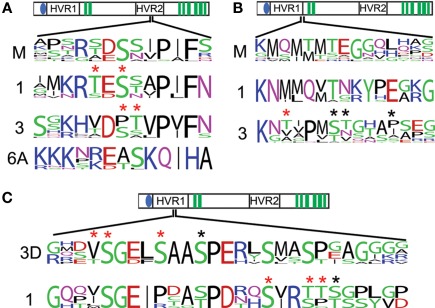
The phosphorylation sites occur in two distinct regions. There is a broad region in the hypervariable region in the first cytoplasmic loop. This region contains several sites with strong conservation in CesA1 and CesA3 clades. There is also a single site in this region corresponding to the Arabidopsis CesA3 S211 site that is generally a serine/threonine in CesA1, CesA3, and the moss/lycophyte clade and may represent an important ancestral site of phosphorylation (Figure 4B). This region corresponds to the second peak in class-specificity found in the CesAs (Figure 2B). With the exception of the site corresponding to CesA3 S211, the gymnosperm sequences are quite different from the angiosperm sequences in this region, suggesting that the evolution of phosphorylation at these sites may be a more recent adaptation (Figure 4C).
Figure 4.
Conserved phosphorylation sites of interest. The conservation of observed phosphorylation sites is detailed by frequency plots. Red asterisks are placed above positions in which phosphorylated peptides were observed, but the specific residue phosphorylated was ambiguous. Black asterisks are place above unambiguous phosphorylation. A region near the catalytic sites has strongly conserved serines/threonines in the primary CesAs and the moss and lycophyte sequences (A). Serines/threonines observed to be phosphorylated in CesA3 are strongly conserved and aligned to a strongly conserved serine/threonine in CesA1 and the moss and lycophyte sequences (B). A phosphorylated region shows strong conservation within the CesA1 family is also found to have several phosphorylated serines/threonines in the CesA3 family, but only in eudicots (labeled 3D), possibly indicating phosphorylation here is a recent adaptation in eudicot CesA3s (C).
The second phosphorylated region occurs in the hypervariable region in the second cytoplasmic domain near the active sites (Figure 4A). Primary CesA clades 1, 3, and 6A, and also moss/lycophyte sequences show strong conservation of serine/threonines in this region, suggesting that phosphorylation at this position is an ancient regulatory event. These sites and their flanking residues represent the fourth class-specific region identified previously (Figure 2B), following the cysteine-rich region (Figure 3C). The secondary CesAs 4, 7, and 8 have lost the region corresponding to this site. Site-directed mutagenesis at this position has been shown to impair normal cellulose synthesis in CesA1 (Chen et al., 2010). The conservation of this position across the other primary CesAs suggests they may be similarly regulated.
Identification of a point mutation site which distinguishes CesA classes
A growing number of CesA mutants have been characterized in Arabidopsis. Characterized mutations provide an excellent opportunity to look for class-specific residues within the CesA family and the CesA superfamily as a whole. If a mutation at a particular residue in one of the CesA clades causes a phenotype, but the residue is not well conserved in other CesA clades, it is a good indication that the residue is involved in some class-specific function.
Mutations are also of relevance with respect to the CslD and CslF families, which share a common architecture of transmembrane, catalytic, and hypervariable regions. Since the similarity between CesA, CslD, and CslF is so high, any mutations known to impair function in CesAs, but are not conserved in the CslD or CslF families may provide valuable clues about the mechanisms for synthesis in these groups. To compare the CesAs to the CslDs and CslFs we acquired 62 CslD and 8 CslF sequences and aligned these with the 88 CesA sequences to create a superfamily alignment.
There are currently 18 characterized missense mutants in CesAs (Table 4). All characterized mutations except rsw5 occur in residues that are strongly conserved amongst CesAs. However, a number of the characterized mutants in the CesA clades occur at residues that are quite different in the CslD and CslF families. In some cases, particularly mutations in the C-terminal region, the positions of CesA mutations are absent from the alignment of CslD and CslF sequences. In other cases, such as in lew2-2, irx1-1, or bc11, the observed residue in CslD or CslF is often different from the conserved Arabidopsis residue and occasionally identical to the mutation that causes a phenotype in the CesAs. Understanding the nature of these mutations may prove useful to determining the mechanisms behind the function of CesAs, CslDs, and CslFs. In addition, a number of premature stop codons have been observed in mutant screens of CesAs. We have listed the ones we are aware of at the time of this writing (Table 5).
Table 4.
Observed point mutants.
| Mutant | CesA | Species | Mutation | Region | CesA conserved | CslD conserved | CslF conserved | Reference |
|---|---|---|---|---|---|---|---|---|
| rsw1-1 | 1 | A. thaliana | A549 → V | Catalytic | A or G | A | G | Baskin et al. (1992) |
| rsw1-2 | 1 | A. thaliana | G631 → S | Catalytic | G | G | G | Gillmor et al. (2002) |
| rsw1-45 | 1 | A. thaliana | E779 → K | Catalytic | E | E | E | Beeckman et al. (2002) |
| rsw1-20 | 1 | A. thaliana | D780 → N | Catalytic | D | D | D | Beeckman et al. (2002) |
| eli1-1 | 3 | A. thaliana | S301 → F | 2nd TM | S | S | mostly S; also M, T |
Cano-Delgado et al. (2000) |
| eli1-2 | 3 | A. thaliana | A522 → V | Catalytic | A | A | A | Cano-Delgado et al. (2000) |
| tha1** | 3 | A. thaliana | P578 → S | Catalytic | P | P | P | Daras et al. (2009) |
| cev1 | 3 | A. thaliana | G617 → E | Catalytic | G | G | G | Ellis et al. (2002) |
| ixr1-2* | 3 | A. thaliana | T942 → I | 3rd Outer | T | T | T | Scheible et al. (2001) |
| ixr1-1* | 3 | A. thaliana | G998 → D | 7th TM | G | G | G,M,L,A | Scheible et al. (2001) |
| rsw5 | 3 | A. thaliana | P1056 → S | C-terminus | P except in CesAs 1, 8 |
Absent | Absent | Wang et al. (2006) |
| bc11 | 4 | O. sativa | G858 → R | 5th TM | G | G | mostly G; also K, R, L |
Zhang et al. (2009) |
| ixr2-1* | 6 | A. thaliana | R1064 → W | C-terminus | R, K | Absent | Absent | Desprez et al. (2002) |
| irx1-1 | 7 | A. thaliana | G683 → N | Catalytic | G | N | Absent | Taylor et al. (2000) |
| fra5** | 7 | A. thaliana | P557 → T | Catalytic | P | P | P | Zhong et al. (2003) |
| mur10-2 | 7 | A. thaliana | H805 → Y | Catalytic | H | H | H | Bosca et al. (2006) |
| irx1-2 | 8 | A. thaliana | S679 → L | Catalytic | S | S | T, I | Taylor et al. (2000) |
| lew2-2 | 8 | A. thaliana | L791 → F | 2nd outer | L | L | P | Chen et al. (2005) |
A list of all missense mutations observed in the CesA family which have been categorized. Region indicates which part of the gene the mutation occurs in (TM is transmembrane domain, catalytic refers to the second cytoplasmic loop). Also listed are the conserved amino acids in the global alignment at the position of the mutation. Additionally, the dominant amino acids in the CslDs and CslFs at the same position are reported.
*These mutations confer resistance to the drug isoxaben but do not impair CesA function.
**These mutations are dominant-negative (and occur at the same position in the alignment).
Table 5.
Observed nonsense mutations.
| Mutant | CesA | Species | Mutation | Region | Reference |
|---|---|---|---|---|---|
| prc1-1 | 6 | A. thaliana | Y752 → Stop | Catalytic | Desnos et al. (1996) |
| prc1-4 | 6 | A. thaliana | W777 → Stop | Catalytic | Desnos et al. (1996) |
| mur10-1 | 7 | A. thaliana | W444 → Stop | Catalytic | Bosca et al. (2006) |
| irx3 | 7 | A. thaliana | W859 → Stop | 3rd Inner | Taylor et al. (1999) |
| lew2-1 | 8 | A. thaliana | W217 → Stop | Catalytic | Chen et al. (2005) |
A list of nonsense mutations observed in the CesA family. The classification of region is the same as in Table 4.
Identification of aromatic positions potentially important for catalysis
When an amino acid is absolutely conserved at a particular position it is often difficult to determine which of its chemical characteristics are essential to its function. Understanding the nature of an amino acid's contribution to protein function can be important for understanding the mechanism of the protein's action and being able to engineer the protein in the future. We screened for positions with strong conservation of an amino acid property, but without strong conservation of amino acid identity (Table 8). For most amino acid properties, it was impossible to suggest a function for the residues. However, we identified two positions with the unusually characteristic of conserving an aromatic property without conserving amino acid identity (Tables 6 and 7). Each of these aromatic residues is found in close proximity to one of the aspartate residues identified as important to catalysis in GT2 family glycosyltransferases (Figure 5). The corresponding position in the CslD and CslF clades also invariably contained an amino acid with an aromatic ring.
Table 6.
Amino acids observed at first identified aromatic residue.
| Clade | Phenylalanine (%) |
Histidine (%) |
Tyrosine (%) |
|---|---|---|---|
| CesA1 | 33 | 67 | 0 |
| CesA3 | 7 | 0 | 93 |
| CesA6 | 62 | 0 | 38 |
| CesA4 | 37.5 | 25 | 37.5 |
| CesA7 | 91 | 0 | 9 |
| CesA8 | 33 | 0 | 67 |
| Physcomitrella | 100 | 0 | 0CesA |
| Selaginella | 75 | 0 | 25CesA |
| CslDs | 0 | 100 | 0 |
| CslFs | 0 | 100 | 0 |
| Total | 35 | 39 | 26 |
A breakdown of the amino acid identity of the amino acids found at the first identified conserved aromatic property in CesAs, CslDs, and CslF by clade. In the CesAs, the identity of the amino acid found at this position does not show a clear class-specific pattern.
Table 7.
Amino acids observed at second identified aromatic residue.
| Clade | Phenylalanine (%) |
Tyrosine (%) |
|---|---|---|
| CesA1 | 0 | 100 |
| CesA3 | 53 | 47 |
| CesA6 | 0 | 100 |
| CesA4 | 100 | 0 |
| CesA7 | 100 | 0 |
| CesA8 | 17 | 83 |
| Physcomitrella CesA | 50 | 50 |
| Selaginella CesA | 50 | 50 |
| CslDs | 100 | 0 |
| CslFs | 100 | 0 |
| Total | 57 | 43 |
A breakdown of the amino acid identity of the amino acids found at the second identified conserved aromatic property in CesAs, CslDs, and CslF by clade. In the CesAs, the identity of the amino acid found at this position does not show a clear class-specific pattern.
Figure 5.
Residues with conserved aromatic property but not amino acid identity. The location of positions in the global alignment with strong aromatic property but not strong conservation of identity are marked by a orange hexagon on the schematic of the CesA protein. Shown in red are the conserved aspartate residues identified as important to catalysis in family 2 processive glycosyltransferases. The black Q marks the location of the QxxRW motif. Shown below this are frequency plots derived from the alignment of all CesA, CslD, and CslF sequences in this region. The amino acid with conserved aromatic property is labeled with an orange hexagon above it. The aspartate identified as important in the D,D,D,QxxRW motif is identified with a red asterisk.
Discussion
In this paper we have examined the sequences of the CesA family, identifying that the evolution of the family involved early duplication events giving rise to the six essential clades of CesAs found in seed plants. We have identified class-specific regions that may be responsible for the differences between these clades. We have determined which observed phosphorylation sites are strongly conserved and mapped these onto the class-specific regions identified. We have determined that only one of the observed CesA mutations may indicate functional differences between CesA clades, but that a number of mutant sites indicate possible differences between the CesAs and their closest related families: the CslDs and CslFs.
Our phylogeny indicates that the specialization of the CesA family occurred at some time between the evolution of the lycophytes and the evolution of the gymnosperms. Unfortunately, no fern CesA sequences are available to further narrow the exact time of the major duplications. Fern sequences would be particularly useful since there are some regions in which the available gymnosperm CesAs have characteristics which more closely resemble the ancestral moss sequences than the derived monocot and eudicot sequences. For example, the gymnosperm CesA3 lacks conservation around a number of phosphorylation sites strongly conserved in monocot and eudicot sequences, and the gymnosperm CesA3 contains an N-terminal region which closely resembles the region conserved across mosses, lycophytes, and CesAs 1, 6, and 7.
In every species in our analysis, the genes in each of the primary CesA clades (CesAs 1, 3, and 6) had more copies than the genes in the secondary CesA clades (CesAs 4, 7, and 8). Of the 78 CesA sequences in the seed plants, 53 were in the primary CesA clades while only 25 were in secondary ones. Among the primary CesAs, the CesA6 clade has undergone much more diversification and divergence than the other positions, and it is also the only clade to be further subdivided due to a duplication event likely occurring after the evolution of the eudicots. Unfortunately, Arabidopsis has lost its CesAs that correspond to the angiosperm CesA6B branch, retaining only eudicot-specific CesA6A members. One potential explanation is that the recent duplication of the Arabidopsis CesA6A family into CesAs 2, 5, 6, and 9 was either a cause or a result of the loss of its CesA6B gene. The greater diversification of the CesA6 position may suggest that this position is regulated to achieve subtler control of CesA complex.
We identified class-specific regions potentially responsible for the differences between the clades. Some of these represent regions found in the earliest CesA sequences that were subsequently lost in certain clades. The loss of the conserved N-terminal region in CesA clades 3, 4, and 8 may indicate that these CesAs rely on the presence of this region in the other CesA clades to provide whatever essential function it provides the CesA complex.
The phosphorylation sites in the catalytic domain represent the fourth class-specific region. This region is found in the primary CesAs and mosses, suggesting its role in regulating cellulose synthesis is ancient. However, the region is absent from all secondary CesAs. This could suggest that this region serves a purpose that is unnecessary in the secondary CesAs. For example, since cellulose synthesis in the secondary cell wall is a terminal event, CesAs might not need to be recycled from the membrane.
The class specific region in the C-terminus is particularly interesting because there is a mutant which suggests a possible function for the region. The proline to serine mutation in the CesA3 mutant rsw5 was less able to competitively access the complex (Wang et al., 2006). This proline is absolutely conserved in all sequences of CesAs 1, 3, 4, and 7, but is not conserved in CesAs 1 and 8. In addition, certain residues near this proline show greater similarity between the clades CesA3 and CesA7 and between the clades of CesA6 and CesA4. This is the only region observed which shows a stronger mapping between primary and secondary CesAs than within primary or secondary CesAs. This could suggest that this region is responsible for some aspect of assembly found in both primary and secondary complexes and could indicate a common stoichiometry or assembly between the primary and secondary CesAs that is at least partially mediated by the C-terminus. This could also explain why no complementing C-terminal fluorescent protein fusions have been reported in the literature.
Many of the phosphorylation events in the N-terminal region of the CesA1 and 3 are found in a similar pattern. With the exception of the S211 site, many CesA3 phosphorylation events are not found in the gymnosperm representatives, while others are only found in eudicots. This could suggest that phosphorylation in this region developed over time in the CesA3 family. Starting from the S211 site, additional sites emerged in early angiosperms and later in evolution further sites emerged in the early eudicots. This suggests that phosphorylation in this region may have emerged convergently in the CesA1 and CesA3 clades, indicating that this region is in an ideal position to control some aspect of CesA behavior. Although the sequences surrounding the phosphorylation sites differ between CesA1 and CesA3, the distribution of the phosphorylated serines and threonines follows a similar overall pattern, possibly suggesting that phosphorylation in these residues may trigger a similar conformational change.
All phosphorylation events observed in CesA6-like isoforms are not-conserved even within the closely related CesA6-like isoforms of Arabidopsis. If these sites are functionally relevant, this would indicate that regulation at these positions is a very recent innovation, and a potentially important way for plant cells to differentially control the CesA6-like isoforms.
The fact that with only one exception every documented mutation is strongly conserved across CesAs should be considered in the light of how these mutations were found. These mutations were characterized from mutagenic screens that looked for hypocotyl and root elongation defects. These screens may miss more subtle phenotypes. This could suggest that mutations to residues that would be class-specific have less severe effects than most mutations documented so far, perhaps reflecting partial redundancy of function in other isoforms over certain class-specific regions. A similar observation could be made about the fact that no missense mutations have been observed in CesA6-like genes, while multiple nonsense mutations have been reported. This may suggest that the partial redundancy at the CesA6 position protects the complex from disrupting point mutations as long as enough protein is present to maintain stiochiometric balance between CesA1, CesA3, and CesA6-like isoforms.
The observation of two positions with conserved aromatic property is interesting in the context of the activity of CesAs as glycosyltransferases. Many protein–carbohydrate interactions are mediated by the stacking effects of the electrons in an aromatic amino acid with those of the sugar ring. This has been documented in a variety of systems such as certain glycohydrolases and glycosyltransferases (Haga et al., 2003).
In conclusion, we have identified a number of CesA sequence properties that can aid in the understanding of how this essential gene family has evolved and specialized during the course of the evolution of the plant kingdom. In addition, this analysis provides a wealth of targets that can be probed in more detail through site-directed mutagenesis, domain swapping, or inter-species rescue experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Andrew Carroll is supported by the U.S. Department of Energy (grant no. DE-FG02-09ER16008). Chelsea Specht is supported by NSF IOS 085641 and the Hellman Family Faculty Fund.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/plant_genetics_and_genomics/abstract/9743
CesA_alignment.doc: The alignment of CesA sequences analyzed used in the analyses of the phylogeny, class-specific regions, phosphorylation, conservation of mutation sites, and unique residue requirements
Csl_alignment.doc: The alignment of the CesA, CslD, and CslF used in the analysis of the conservation of mutation sites.
References
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Institute. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- Baskin T. I., Betzner A. S., Hoggart R., Cork A., Williamson R. E. (1992). Root morphology mutants in Arabidopsis thaliana. Funct. Plant Biol. 19, 427–437 [Google Scholar]
- Beeckman T., Przemeck G. K. H., Stamatiou G., Lau R., Terryn N., De Rycke R., Inze D., Berleth T. (2002). Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol. 130, 1883–1893 10.1104/pp.102.010603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca S., Barton C. J., Taylor N. G., Ryden P., Neumetzler L., Pauly M., Roberts K., Seifert G. J. (2006). Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 142, 1353–1363 10.1104/pp.106.087700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling A. J., Brown R. M. (2008). The cytoplasmic domain of the cellulose-synthesizing complex in vascular plants. Protoplasma 233, 115–127 10.1007/s00709-008-0302-2 [DOI] [PubMed] [Google Scholar]
- Brown R. M., Montezinos D. (1976). Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 73, 143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R. A., Wilson S. M., Hrmova M., Harvey A. J., Shirley N. J., Medhurst A., Stone B. A., Newbigin E. J., Bacic A., Fincher G. B. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311, 1940–1942 10.1126/science.1122975 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A. I., Metzlaff K., Bevan M. W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127, 3395–3405 [DOI] [PubMed] [Google Scholar]
- Carpita N. C., McCann M. C. (2010). The maize mixed-linkage (1->3),(1->4)-β-D-glucan polysaccharide is synthesized at the Golgi membrane. Plant Physiol. 153, 1362–1371 10.1104/pp.110.156158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D. W., Somerville C. R. (2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 17188–17193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Hong X., Zhang H., Wang Y., Li X., Zhu J.-K., Gong Z. (2005). Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 43, 273–283 10.1111/j.1365-313X.2005.02452.x [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E. F., Bischoff V., Desprez T., Rolland A., Stierhof Y.-D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154 10.1105/tpc.108.065334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G., Rigas S., Penning B., Milioni D., McCann M. C., Carpita N. C., Fasseas C., Hatzopoulos P. (2009). The thanatos mutation in Arabidopsis thaliana cellulose synthase 3 (AtCesA3) has a dominant-negative effect on cellulose synthesis and plant growth. New Phytol. 184, 114–126 10.1111/j.1469-8137.2009.02960.x [DOI] [PubMed] [Google Scholar]
- Desnos T., Orbovic V., Bellini C., Kronenberger J., Caboche M., Traas J., Hofte H. (1996). Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsis seedlings. Development 122, 683–693 [DOI] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E. F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez T., Vernhettes S., Fagard M., Refrégier G., Desnos T., Aletti E., Py N., Pelletier S., Höfte H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128, 482–490 10.1104/pp.010822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin M. S., Kurek I., Jacob-Wilk D., Delmer D. P. (2002). Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol. 43, 1407–1420 10.1093/pcp/pcf164 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J. G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566 10.1105/tpc.002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor C. S., Poindexter P., Lorieau J., Palcic M. M., Somerville C. (2002). α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156, 1003–1013 10.1083/jcb.200111093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. A., Ricke D., Lan T. H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Hadley D., Hutchison D., Martin C., Katagiri F., Lange B. M., Moughamer T., Xia Y., Budworth P., Zhong J., Miguel T., Paszkowski U., Zhang S., Colbert M., Sun W. L., Chen L., Cooper B., Park S., Wood T. C., Mao L., Quail P., Wing R., Dean R., Yu Y., Zharkikh A., Shen R., Sahasrabudhe S., Thomas A., Cannings R., Gutin A., Pruss D., Reid J., Tavtigian S., Mitchell J., Eldredge G., Scholl T., Miller R. M., Bhatnagar S., Adey N., Rubano T., Tusneem N., Robinson R., Feldhaus J., Macalma T., Oliphant A., Briggs S. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 10.1126/science.1068275 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- Gupta S. K., Kececioglu J. D., Schäffer A. A. (1995). Improving the practical space and time efficiency of the shortest-paths approach to sum-of-pairs multiple sequence alignment. J. Comput. Biol. 2, 459–472 [DOI] [PubMed] [Google Scholar]
- Gutierrez R., Lindeboom J. J., Paredez A. R., Emons A. M. C., Ehrhardt D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806 [DOI] [PubMed] [Google Scholar]
- Haga K., Kanai R., Sakamoto O., Aoyagi M., Harata K., Yamane K. (2003). Effects of essential carbohydrate/aromatic stacking interaction with Tyr100 and Phe259 on substrate binding of cyclodextrin glycosyltransferase from Alkalophilic Bacillus sp. 1011. J. Biochem. 134, 881–891 10.1093/jb/mvg215 [DOI] [PubMed] [Google Scholar]
- Heazlewood J. L., Durek P., Hummel J., Selbig J., Weckwerth W., Walther D., Schulze W. X. (2008). PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 36, D1015-1021-D1015-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 89, 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C., Vezzi A., Legeai F., Hugueney P., Dasilva C., Horner D., Mica E., Jublot D., Poulain J., Bruyère C., Billault A., Segurens B., Gouyvenoux M., Ugarte E., Cattonaro F., Anthouard V., Vico V., Del Fabbro C., Alaux M., Di Gaspero G., Dumas V., Felice N., Paillard S., Juman I., Moroldo M., Scalabrin S., Canaguier A., Le Clainche I., Malacrida G., Durand E., Pesole G., Laucou V., Chatelet P., Merdinoglu D., Delledonne M., Pezzotti M., Lecharny A., Scarpelli C., Artiguenave F., Pè M. E., Valle G., Morgante M., Caboche M., Adam-Blondon A. F., Weissenbach J., Quétier F., Wincker P., French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., Hellsten U., Mitros T., Poliakov A., Schmutz J., Spannagl M., Tang H., Wang X., Wicker T., Bharti A. K., Chapman J., Feltus F. A., Gowik U., Grigoriev I. V., Lyons E., Maher C. A., Martis M., Narechania A., Otillar R. P., Penning B. W., Salamov A. A., Wang Y., Zhang L., Carpita N. C., Freeling M., Gingle A. R., Hash C. T., Keller B., Klein P., Kresovich S., McCann M. C., Ming R., Peterson D. G., Mehboob-ur-Rahman, Ware D., Westhoff P., Mayer K. F., Messing J., Rokhsar D. S. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Pear J. R., Kawagoe Y., Schreckengost W. E., Delmer D. P., Stalker D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 12637–12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B., Argos P. (1994). Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237, 182–192 [DOI] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C. R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper Z. A. (2008). Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 286–292 [DOI] [PubMed] [Google Scholar]
- Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P. F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin-I T., Kuroki Y., Toyoda A., Suzuki Y., Hashimoto S., Yamaguchi K., Sugano S., Kohara Y., Fujiyama A., Anterola A., Aoki S., Ashton N., Barbazuk W. B., Barker E., Bennetzen J. L., Blankenship R., Cho S. H., Dutcher S. K., Estelle M., Fawcett J. A., Gundlach H., Hanada K., Heyl A., Hicks K. A., Hughes J., Lohr M., Mayer K., Melkozernov A., Murata T., Nelson D. R., Pils B., Prigge M., Reiss B., Renner T., Rombauts S., Rushton P. J., Sanderfoot A., Schween G., Shiu S. H., Stueber K., Theodoulou F. L., Tu H., Van de Peer Y., Verrier P. J., Waters E., Wood A., Yang L., Cove D., Cuming A. C., Hasebe M., Lucas S., Mishler B. D., Reski R., Grigoriev I. V., Quatrano R. S., Boore J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Scheible W. R., Eshed R., Richmond T., Delmer D., Somerville C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. U.S.A. 98, 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T. A., Minx P., Reily A. D., Courtney L., Kruchowski S. S., Tomlinson C., Strong C., Delehaunty K., Fronick C., Courtney B., Rock S. M., Belter E., Du F., Kim K., Abbott R. M., Cotton M., Levy A., Marchetto P., Ochoa K., Jackson S. M., Gillam B., Chen W., Yan L., Higginbotham J., Cardenas M., Waligorski J., Applebaum E., Phelps L., Falcone J., Kanchi K., Thane T., Scimone A., Thane N., Henke J., Wang T., Ruppert J., Shah N., Rotter K., Hodges J., Ingenthron E., Cordes M., Kohlberg S., Sgro J., Delgado B., Mead K., Chinwalla A., Leonard S., Crouse K., Collura K., Kudrna D., Currie J., He R., Angelova A., Rajasekar S., Mueller T., Lomeli R., Scara G., Ko A., Delaney K., Wissotski M., Lopez G., Campos D., Braidotti M., Ashley E., Golser W., Kim H., Lee S., Lin J., Dujmic Z., Kim W., Talag J., Zuccolo A., Fan C., Sebastian A., Kramer M., Spiegel L., Nascimento L., Zutavern T., Miller B., Ambroise C., Muller S., Spooner W., Narechania A., Ren L., Wei S., Kumari S., Faga B., Levy M. J., McMahan L., Van Buren P., Vaughn M. W., Ying K., Yeh C. T., Emrich S. J., Jia Y., Kalyanaraman A., Hsia A. P., Barbazuk W. B., Baucom R. S., Brutnell T. P., Carpita N. C., Chaparro C., Chia J. M., Deragon J. M., Estill J. C., Fu Y., Jeddeloh J. A., Han Y., Lee H., Li P., Lisch D. R., Liu S., Liu Z., Nagel D. H., McCann M. C., SanMiguel P., Myers A. M., Nettleton D., Nguyen J., Penning B. W., Ponnala L., Schneider K. L., Schwartz D. C., Sharma A., Soderlund C., Springer N. M., Sun Q., Wang H., Waterman M., Westerman R., Wolfgruber T. K., Yang L., Yu Y., Zhang L., Zhou S., Zhu Q., Bennetzen J. L., Dawe R. K., Jiang J., Jiang N., Presting G. G., Wessler S. R., Aluru S., Martienssen R. A., Clifton S. W., McCombie W. R., Wing R. A., Wilson R. K. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Somerville C. (2006). Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 22, 53–78 [DOI] [PubMed] [Google Scholar]
- Stork J., Harris D., Griffiths J., Williams B., Beisson F., Li-Beisson Y., Mendu V., Haughn G., Debolt S. (2010). Cellulose Synthase9 serves a nonredundant role in secondary cell wall synthesis in Arabidopsis epidermal testa cells. Plant Physiol. 153, 580–589 10.1104/pp.110.154062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Murata K., Yamazaki M., Onosato K., Miyao A., Hirochika H. (2003). Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 133, 73–83 10.1104/pp.103.022442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. G. (2007). Identification of cellulose synthase AtCesA7 (IRX3) in vivo phosphorylation sites-a potential role in regulating protein degradation. Plant Mol. Biol. 64, 161–171 [DOI] [PubMed] [Google Scholar]
- Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. G., Laurie S., Turner S. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2540 10.1105/tpc.12.12.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor N. G., Scheible W. R., Cutler S., Somerville C. R., Turner S. R. (1999). The irregular xylem3 locus of arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780 10.1105/tpc.11.5.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Schein J., Sterck L., Aerts A., Bhalerao R. R., Bhalerao R. P., Blaudez D., Boerjan W., Brun A., Brunner A., Busov V., Campbell M., Carlson J., Chalot M., Chapman J., Chen G. L., Cooper D., Coutinho P. M., Couturier J., Covert S., Cronk Q., Cunningham R., Davis J., Degroeve S., Déjardin A., Depamphilis C., Detter J., Dirks B., Dubchak I., Duplessis S., Ehlting J., Ellis B., Gendler K., Goodstein D., Gribskov M., Grimwood J., Groover A., Gunter L., Hamberger B., Heinze B., Helariutta Y., Henrissat B., Holligan D., Holt R., Huang W., Islam-Faridi N., Jones S., Jones-Rhoades M., Jorgensen R., Joshi C., Kangasjärvi J., Karlsson J., Kelleher C., Kirkpatrick R., Kirst M., Kohler A., Kalluri U., Larimer F., Leebens-Mack J., Leplé J. C., Locascio P., Lou Y., Lucas S., Martin F., Montanini B., Napoli C., Nelson D. R., Nelson C., Nieminen K., Nilsson O., Pereda V., Peter G., Philippe R., Pilate G., Poliakov A., Razumovskaya J., Richardson P., Rinaldi C., Ritland K., Rouzé P., Ryaboy D., Schmutz J., Schrader J., Segerman B., Shin H., Siddiqui A., Sterky F., Terry A., Tsai C. J., Uberbacher E., Unneberg P., Vahala J., Wall K., Wessler S., Yang G., Yin T., Douglas C., Marra M., Sandberg G., Van de Peer Y., Rokhsar D. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Wang J., Howles P. A., Cork A. H., Birch R. J., Williamson R. E. (2006). Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol. 142, 685–695 10.1104/pp.106.084004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Guo K., Li Y., Tu Y., Hu H., Wang B., Cui X., Peng L. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282. 10.1186/1471-2229-10-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Huang J., Xu Y. (2009). The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9, 99–99 10.1186/1471-2229-9-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Deng L., Qian Q., Xiong G., Zeng D., Li R., Guo L., Li J., Zhou Y. (2009). A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol. Biol. 71, 509–524 [DOI] [PubMed] [Google Scholar]
- Zhong R., Morrison W. H., Freshour G. D., Hahn M. G., Ye Z. H. (2003). Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132, 786–795 10.1104/pp.102.019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CesA_alignment.doc: The alignment of CesA sequences analyzed used in the analyses of the phylogeny, class-specific regions, phosphorylation, conservation of mutation sites, and unique residue requirements
Csl_alignment.doc: The alignment of the CesA, CslD, and CslF used in the analysis of the conservation of mutation sites.