Abstract
Abscisic acid (ABA) and Gibberellic acid (GA) play key roles in regulating seed dormancy and germination. First, when examining germination of different rice cultivars, we found that their germination timing and dormancy status are rather distinct, coupled with different GA/ABA ratio. Second, we studied genomic sequences of ABA and GA dormancy- and germination-associated genes in rice and discovered single nucleotide polymorphisms and insertions/deletions (Indels) in both coding and regulatory sequences. We aligned all these variations to the genome assemblies of 9311 and PA64s and demonstrated their relevance to seed dormancy both quantitatively and qualitatively based on gene expression data. Third, we surveyed and compared differentially expressed genes in dry seeds between 9311 and PA64s to show that these differentially expressed genes may play roles in seed dormancy and germination.
Keywords: dormancy and germination, ABA, GA
Introduction
Seed dormancy has been defined as a temporary failure of seed germination under conditions that favor germination (Bewley, 1997). The initiation of dormancy can either occur in the dry state (after-ripening) or be triggered by imbibition under defined conditions in mature seeds (Gubler et al., 2005). Pre-harvest sprouting of cereal grain leads to reduced grain yield and poor quality products, resulting in economic losses of cultivars. Strong dormancy also prevents grain such as barley from being re-planted immediately after harvest, causing delayed or non-uniform germination. Both situations would result in poor crop establishment and grain performance.
Seed dormancy and germination are regulated by nearly all plant hormones. Several studies have shown that ethylene, auxin, and brassinosteroids promote the germination of dormant seeds, but it is now generally accepted that ABA and GA are the leading regulators (Koornneef et al., 2002; Gubler et al., 2005). ABA deficiency during seed development is associated with the absence of primary dormancy in mature seeds, whereas over-expression of ABA biosynthesis genes can increase seed ABA content and enhance seed dormancy or delay germination. ABA plays important roles in many cellular processes including seed development, dormancy, germination, vegetative growth, and environmental stress responses. ABA level can increase significantly during seed maturation and in response to environmental stresses. In addition to hormone content, the transition from the dormant to the non-dormant state of many seeds is characterized by a decrease in ABA sensitivity and an increase in GA sensitivity (Chiwocha et al., 2005).
Recently, the major enzymes involved in ABA and GA metabolism pathways have been identified. ABA metabolic genes have been identified such as ABA biosynthetic gene NCED that encodes a 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA catabolic gene CYP707A that encodes a ABA 8′-hydroxylase (CYP707A; Seo and Koshiba, 2002; Lefebvre et al., 2006; Okamoto et al., 2006; Yang and Guo, 2007). Genes associated with GA metabolism have also been identified such as GA biosynthetic gene GA20ox that encodes a GA 20-oxidase (GA20ox), GA3ox that encodes a GA 3-hydroxylase (GA3ox), and GA catabolic gene GA2ox that encodes a GA 2-oxidase (GA2ox; Yamauchi et al., 2004; Finch-Savage et al., 2007). However, no potential receptors that could perceive this increase in abscisic acid had been identified until recent reports of four abscisic acid binding proteins: the chloroplast protein magnesium protoporphyrin-IX chelatase H subunit (Shen et al., 2006), the membrane-associated protein G protein coupled receptor 2 (Liu et al., 2007; Verslues and Zhu, 2007), two GPCR-type G proteins (GTG1 and GTG2) (Ma et al., 2009; Pandey et al., 2009), and the PYR/PYL/RCAR family proteins (Ma et al., 2009; Park et al., 2009). Since there have been controversies over structural and functional classification of ABA receptors, we carried out analyses on the recently available recent receptor candidates in this study. The identification of GA receptors has also improved our understanding of GA function in relation to germination (Nakajima et al., 2006).
Rice is one of the most important cereal crops, providing food for billions of people. In addition, it is also a model plant with various cultivars and mutants for the study of heterosis and domestic process. 9311 is a cultivar of Oryza sativa L. ssp. indica – the major rice subspecies grown in Asia-Pacific regions. Pei-Ai 64s (PA64s) has a major background of indica and a minor background of japonica and javanica, and the first two are two other commonly cultivated subspecies in China. 9311 and PA64s as parental lines show significant difference in their phenotype of seed dormancy. Their F1 offspring, an elite super hybrid rice LYP9, also shows significant heterosis including weak dormancy like its paternal line 9311. Compared to its wild ancestor O. rufipogon, cultivated rice typically exhibits reduced dormancy (Veasey et al., 2004; Sweeney and Mccouch, 2007). Therefore, to elucidate the molecular mechanisms regulating rice seed dormancy and germination is of importance for both plant biology and crop development. Toward this end, we identified genes encoding ABA and GA metabolic enzymes and found that there were many single nucleotide polymorphisms (SNPs) and Indels in both protein-coding and regulatory regions between different rice cultivars. We also found differentially expressed transcription units in the seed of our rice model 9311 and PA64s. We demonstrate that seed dormancy state in rice may be influenced by both gene sequence variation as well as expression patterns of ABA and GA metabolic genes.
Materials and Methods
Plant materials
Mature seeds from fresh harvest of 9311, PA64s, IR24, SHOEMED, AZUCENA, DOURADO AGULHA, MAKALIOKA34, and IR36 were harvested at the maturing stage then dried and stored at room temperature. The seeds of 9311 and PA64s were provided by the Chinese National R&D Center on Hybrid Rice, Changsha, China. The seeds of IR24, SHOEMED, AZUCENA, DOURADO AGULHA, MAKALIOKA34, and IR36 were provided by the International Rice Research Institute (IRRI). Six months after harvesting the seeds were used for the experiments. Mature rice embryos were separated manually from their seeds using scalpels, and then stored in −80°C immediately before RNA extraction.
Germination experiments
For the germination experiment, we started with 50 seeds in a Petri dish with multiple layers of wet filter papers (sterile double-distilled water) at 37°C. The germination rate was calculated daily based on radical emergence. Experiments were performed in triplicate for each cultivar examined.
Determination of ABA and GA levels
Mature rice embryos were homogenized in 80% (v/v) acetone. After adding internal standards 13C-abscisic acid (ABA) and 3H-labled GA3, the homogenate was shaken for 10 h on ice in darkness and then centrifuged at 2,000g for 30 min. The precipitate was then re-extracted, and the combined supernatant was evaporated to remove residual acetone. After a series of organic extractions, the extracts were purified through C18 column. ABA was methylated with diazomethane, whereas GA3 were trimethylsilylated with BSTFA at 100°C for 60 min. Gas chromatography–electron impact ionization mass spectrometry was carried out to determine ABA and GA concentrations. The following mass-to-charge ratio peaks were used for quantification: for ABA, 192 (labeled) and 190 (endogenous); and for GA3, 506 (labeled) and 504 (endogenous). ABA and GA concentrations of 9311 and PA64s from each individual assay of the triplicate are provided in Data Sheet S1 in Supplementary Material.
Sequence similarity analysis and alignment of amino acid sequences
We acquired sequences of seed dormancy- and germination-associated genes from Arabidopsis and rice databases (Goff et al., 2002; Yu et al., 2002). We analyzed ABA metabolism-related genes from rice Nipponbare including OsNCED1 (AY838897), OsNECD2 (AY838898), OsNCED3 (AY838899), OsNCED4 (AY838900), OsNCED5 (AY838901), OsCYP707A5 (AB277270), OsCYP707A6 (NM_001068556), OsCYP707A7 (NM_001069901), and OsGPCR (CM000147), and homologs from Arabidopsis including AtNCED1 (AT3G63520), AtNCED2 (AT4G18350), AtNCED3 (AT3G14440), AtNCED4 (AT4G19170), AtNCED5 (AT1G30100), AtNCED6 (AT3G24220), AtNCED9 (AT1G78390), AtCYP707A1 (AT4G19230), AtCYP707A2 (AT2G29090), AtCYP707A3 (AT5G45340), AtCYP707A4 (AT3G19270), and AtGCR2 (AT1G52920) (Note: not all NCED enzymes are involved in ABA biosynthesis in Arabidopsis). GA metabolism-related genes from rice are OsGA20ox1 (AC096690), OsGA20ox2 (NM_001051549), OsGA20ox3 (AP005840), OsGA20ox4 (NM_001062119), OsGA3ox1 (NM_001048721), OsGA3ox2 (AC144738), OsGA2ox1 (AC119288), OsGA2ox2 (NM_001048899), OsGA2ox3 (NM_001050827), OsGA2ox4 (AC132485), OsGA2ox5 (NM_001062846), and OsGID1 (AB211399), and homologs from Arabidopsis are AtGA20ox1 (At4g25420), AtGA20ox2 (At5g51810), AtGA20ox3 (At5g07200), AtGA3ox1 (At1g15550), AtGA3ox2 (At1g80340), AtGA3ox3 (At4g21690), AtGA2ox1 (AT1G78440), AtGA2ox2 (AT1G30040), AtGA2ox3 (AT2G34555), and AtGID1 (AT3G05120). Alignment of sequences and phylogenetic analysis were carried out by using ClustalW with default parameters and MEGA4.1 with neighbor-joining method (Tamura et al., 2007). All sequences of 9311, PA64s, and Nipponbare used in this study are provided in Data Sheet S2 in Supplementary Material. For each gene pair, we calculated the number of non-synonymous substitutions per non-synonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) using the maximum-likelihood method (Goldman and Yang, 1994). Ka/Ks = 1 indicates neutral evolution, where the number of non-synonymous changes at each possible non-synonymous site is the same as the number of synonymous changes per synonymous site. Ka/Ks < 1 suggests purifying selection, where selection generally eliminates deleterious mutations; Ka/Ks > 1 indicates positive selection, where selection bring more amino acid changes.
Template preparation and real-time PCR validation
Total RNA was purified from each sample using Trizol (Invitrogen) according to the manufacturer's instructions. Eight microgram total RNA of each sample was used for first-strand cDNA synthesis in 25 μl reaction containing 5 μl 5 × RT buffer, 2.5 μl 10 mM dNTP, 50 ng random primer, 50 ng oligoDT(15), 2.5 μl RNase inhibitor (20 U/μl), 4 μl reverse transcriptase (50 U/μl; Invitrogen), 2.5 μl DTT. Reverse transcription was performed at 42°C for 60 min with a final denaturation at 70°C for 15 min.
Specific primers for Real-time PCR in the experiments are listed as follows:
OsNCED1 (upper: 5′-TGGAGCACATGGAGCTAGT-3′, lower: 5′-CCGAAGTAGCCGTACCTG-3′); OsNCED2 (upper: 5′-GCTTGACTTTGATCTCGTC-3′, lower: 5′-TCTCCGGTGTCCTCTTC-3′); OsNCED3 (upper: 5′-CCACCATGATCCACGACT-3′, lower: 5′-AGAGGTGGAAGCAGAAGC-3′); OsNCED4 (upper: 5′-GCTTGACTTTGATCTCGTC-3′, lower: 5′-AGAGGTGGAAGCAGAAGC-3′); OsNCED5 (upper: 5′-CATCTTCAACGAGTCGGA-3′, lower: 5′-GTAGGCGTACCTCGTCTTC-3′); OsCYP707A5 (upper : 5′-AGATTGCCAAGGAGAAAGA-3′, lower: 5′-TGAAGGTGAAGGAGAGGATG-3′); OsCYP707A6 (upper: 5′-AGACGAGGAGCATGACACT-3′, lower: 5′-CGGGTTGTGATGGATGTT-3′); OsCYP707A7 (upper: 5′-GCTCACCTTCTTCCTCAAC-3′, lower: 5′-GCCATTCCTTTGCTTGAT-3′); OsGA20ox1 (upper: 5′-CATGCGCCTCAACTACTAC-3′, lower: 5′-GAGCGCCATGAAGGTGT-3′); OsGA20ox2 (upper: 5′-TCATGCGGTGCAACTACTA-3′, lower: 5′-GTGTCGCCGATGTTGAT-3′); OsGA20ox3 (upper: 5′-GCTCACCTTCTTCCTCAAC-3′, lower: 5′-GCCATTCCTTTGCTTGA-3′); OsGA20ox4 (upper: 5′-CGTCGCCGATTACTTCTC-3′, lower: 5′-TAGTAGTTGCACCGCATGA-3′); OsGA3ox1 (upper: 5′-CGACGAGTTGCTGAGGTT-3′, lower: 5′-AGGTGAAGAAGCCCGAGT-3′); OsGA3ox2 (upper: 5′-ATGCCCTACTTCCTCGGT-3′, lower: 5′-CTTGTCCTCTTCCTTCGCTA-3′); OsGA2ox1 (upper: 5′-TCAATGTTGGTGATGTCCTC-3′, lower: 5′-GCTGGCTGTGATTGTCTCT-3′); OsGA2ox2 (upper: 5′-AGATCATCTCCGTGCTCC-3′, lower: 5′-CTCGACTTCTCGCTGTTCA-3′); OsGA2ox3 (upper: 5′-CGACTCCTTCTTCGTCAAC-3′, lower: 5′-TGCAATCCTCTGTGCTAAC-3′); OsGA2xo4 (upper: 5′-GCCAACAAGCTCAAGTC-3′, lower: 5′-GAGAGGTAGGCAGCCTTC-3′); OsGA2ox5 (upper: 5′-CATTTCTGGTAACTAAAGGC-3′, lower: 5′-GAGCAGCACATAATCACAT-3′).
Real-time PCR was performed with Quant SYBR Green PCR kit (BIO-RAD). The data were quantitated by referencing to a PCR product of house-keeping gene of GAPDH under the same reaction condition (94°C for 4 min, then 40 cycles of 94°C for 15 s, 60°C for 20 s, and 68°C for 30 s). Melting curves for each PCR reaction were carefully monitored to avoid non-specific amplifications. Gene expression quantification was performed with the comparative Ct formula normalized with the expression of GADPH.
Statistic analysis
We used the unpaired Student's t-test to analyze expression levels of ABA and GA, transcripts of ABA- and GA- related genes in dry seeds when comparing the two groups of 9311 and PA64s, and Ka/Ks ratios. Statistical significance (p < 0.01) was determined based on the t-test in GraphPad Prism 5. Wilcoxon rank sum test was performed to compare SNPs and Indels between coding regions and regulatory regions, also between paralogs and orthologs.
Results
Germination test and ABA/GA concentration measurement of rice seeds from various cultivars
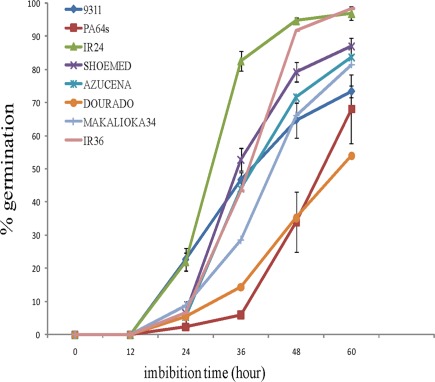
We initiated our study by carrying out a germination experiment using seeds from various cultivars (9311, PA64s, IR24, SHOEMED, AZUCENA, DOURADO AGULHA, MAKALIOKA34, and IR36) with diverse genetic backgrounds (Figure 1). Although all seeds started to germinate after 1 day rehydration, we observed significant differences among the cultivars. For instance, IR24 showed the highest germination rate (82%) and about half of the seeds from 9311, SHOEMED, IR36, and AZUCENA germinated within 36-h imbibition. In contrast, only 29% of MAKALIOKA34, 14% of DOURADO, and less than 10% of PA64s seeds germinated at this time period.
Figure 1.
Germination test for rice cultivars 9311, PA64s, IR24, SHOEMED, AZUCENA, DOURADOAGULHA, MAKALIOKA34, and IR36 (after imbibition). Measurements are averaged over three replicates with error bars representing SE.
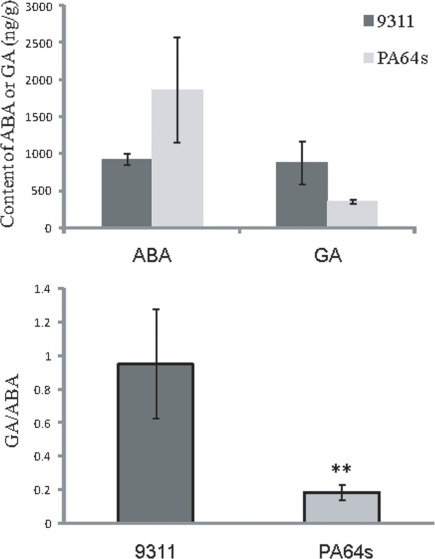
Since rice germination rate is highly associated with the ABA/GA ratio (Nonogaki et al., 2010), we quantified ABA and GA in 9311 and PA64s dry seeds (see Materials and Methods). The endogenous ABA level of 9311 was obviously lower than those of PA64s and the GA/ABA ratio was much higher in 9311 than in PA64s, which is consistent with our experimental results that PA64s exhibited stronger dormancy than 9311 (Figure 2).
Figure 2.
Quantification of endogenous ABA and GA levels in dry seeds of 9311 and PA64s. Measurements are averaged over three replicates with error bars representing SE. The GA/ABA ratio is significantly different in 9311 and PA64s (t-test: **p < 0.01).
Identification of rice ABA and GA metabolic genes
Because ABA and GA metabolic genes and their receptors are not well-annotated in the rice genome assemblies, we extracted well-annotated Arabidopsis genes from the public databases and searched for orthologs and paralogs from the rice databases1. To confirm the predicted protein sequences, we used PFAM database2 to identify conserved domains and found that rice has five NCED, three CYP707A, four GA20ox, two GA3ox, and five GA2ox gene family members. They are distributed on chromosome 1 (OsGA20ox2, OsGA3ox2, OsGA2ox2, and OsGA2ox3), chromosome 2 (OsNCED1 and OsCYP707A5), chromosome 3 (OsNCED3 and OsGA20ox1), chromosome 5 (OsGA20ox4, OsGA3ox1, OsGA2ox1, OsGA2ox4, and OsGA2ox5), chromosome 7 (OsNCED4), chromosome 8 (OsCYP707A6), chromosome 9 (OsCYP707A7), and chromosome 12 (OsNCED2, OsNCED5, and OsGA20ox3). Our annotations were further confirmed in GBROWSE3 (Table 1).
Table 1.
ABA and GA metabolism-related genes in Nipponbare.
| Number | Gene name | Entry name | DNA sequence length (bp) | Annotation | Chromosome |
|---|---|---|---|---|---|
| ABA BIOSYNTHESIS | |||||
| 1 | OsNCED1 | AY838897 | 1917 | 9-cis-epoxycarotenoid dioxygenase 1 | II |
| 2 | OsNCED2 | AY838898 | 1731 | 9-cis-epoxycarotenoid dioxygenase 2 | XII |
| 3 | OsNCED3 | AY838899 | 1827 | 9-cis-epoxycarotenoid dioxygenase 3 | III |
| 4 | OsNCED4 | AY838900 | 1749 | 9-cis-epoxycarotenoid dioxygenase 4 | VII |
| 5 | OsNCED5 | AY838901 | 1842 | 9-cis-epoxycarotenoid dioxygenase 5 | XII |
| ABA CATABOLISM | |||||
| 6 | OsCYP707A5 | AB277270 | 1416 | ABA 8′-hydroxylase 1 | II |
| 7 | OsCYP707A6 | NM_001068556 | 1521 | Cytochrome P450 family protein | VIII |
| 8 | OsCYP707A7 | NM_001069901 | 1503 | Cytochrome P450 family protein | IX |
| GA BIOSYNTHESIS | |||||
| 9 | OsGA20ox1 | AC096690 | 1113 | Putative gibberellin 20-oxidase | III |
| 10 | OsGA20ox2 | NM_001051549 | 1170 | Gibberellin 20-oxidase | I |
| 11 | OsGA20ox3 | AP005840 | 1104 | Putative gibberellin 20-oxidase | XII |
| 12 | OsGA20ox4 | NM_001062119 | 1332 | Gibberellin 20-oxidase | V |
| 13 | OsGA3ox1 | NM_001048721 | 1113 | GA 3 beta-hydroxylase | V |
| 14 | OsGA3ox2 | AC144738 | 1155 | Putative gibberellin 3 beta-hydroxylase | I |
| GA CATABOLISM | |||||
| 15 | OsGA2ox1 | AC119288 | 1149 | Gibberellin 2-oxidase | V |
| 16 | OsGA2ox2 | NM_001048899.1 | 1008 | Gibberellin 2-oxidase | I |
| 17 | OsGA2ox3 | NM_001050827 | 984 | Gibberellin 2-oxidase | I |
| 18 | OsGA2ox4 | AC132485 | 729 | Putative gibberellin 2-oxidase | V |
| 19 | OsGA2ox5 | NM_001062846 | 1062 | Gibberellin 2-oxidase | V |
| ABA AND GA PERCEPTION | |||||
| 20 | OsGPCR | CM000147 | 1386 | Lanthionine synthetase C-like | X |
| 21 | OsGID1 | AB211399 | 1065 | Gibberellin insensitive dwarf 1 | V |
Phylogenic analysis of ABA and GA metabolic genes
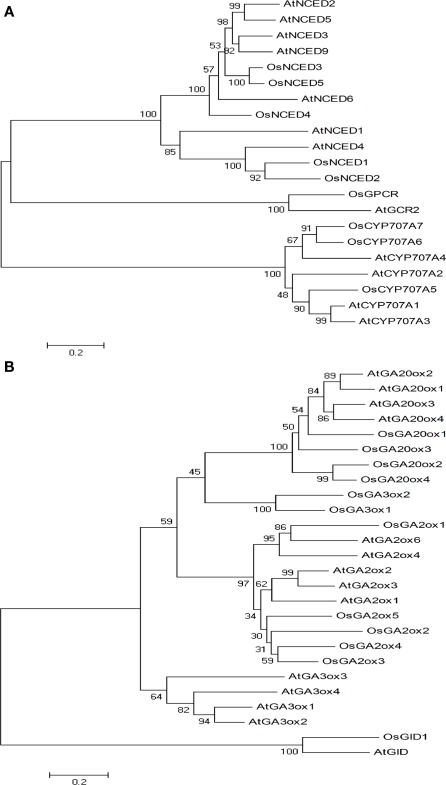
We identified five NCED genes in rice and nine in Arabidopsis (Schwartz et al., 2003; Tan et al., 2003; Kushiro et al., 2004). OsNCED2 and OsNCED5 are both located on chromosome 12 and OsNCED1, OsNCED3, and OsNCED4 are located on chromosomes 2, 3, and 7, respectively. The amino acid sequence of Arabidopsis AtNCED4 showed higher similarities with OsNCED1 and OsNCED2 (51–57% identities) but relatively lower identities with OsNCED3 to OsNCED5 (ranging between 33 and 36%). Phylogenetic analysis (Figure 3A) revealed that OsNCED3 and OsNCED5, which were more similar to each other (80% identity) than any other reported NCED proteins from Arabidopsis, were also closely related to AtNCED2, AtNCED3, AtNCED5, AtNCED6, and AtNCED9 (59–63% identities).
Figure 3.
Neighbor-joining trees for ABA (A) and GA (B) metabolic genes were constructed by using MEGA4. 1 and generated based on full-length amino acid sequences from rice and Arabidopsis.
Since Arabidopsis CYP707A has four paralogs (AtCYP707A1, AtCYP707A2, AtCYP707A3, and AtCYP707A4), we tried really hard to find each of their orthologs in rice. However, we only identified three rice CYP707A genes; one of them is identical to what was reported previously (Saika et al., 2007). OsCYP707A5, OsCYP707A6, and OsCYP707A7 reside on different chromosomes: 2, 8, and 9, respectively. We observed independent gene duplication events in both rice and Arabidopsis (Figure 3A).
As to GA3ox in rice, we identified only two previously reported genes (Itoh et al., 2001), matching to four GA3ox orthologs in Arabidopsis (Hedden et al., 2001). OsGA3ox1 and OsGA3ox2 are located on chromosomes 5 and 1, respectively. The amino acid sequence of OsGA3ox1 and OsGA3ox2 displayed about 37% similarities to Arabidopsis GA3ox proteins. Phylogenetic analysis (Figure 3B) revealed that GA3ox genes duplicated separately after the monocot and dicot divide.
We identified four OsGA20ox genes; two were previously reported in rice (Sasaki et al., 2002) and five were reported in Arabidopsis (Hedden et al., 2001). In rice, OsGA20ox1, OsGA20ox2, OsGA20ox3, and OsGA20ox4 are located on chromosomes 3, 1, 12, and 5, respectively. The amino acid sequence of OsGA20ox1 has the highest homology with Arabidopsis GA20ox proteins (52–57% identities). In comparison, OsGA20ox2 and OsGA20ox4 have 46–51% identities, respectively. Phylogenetic analysis (Figure 3B) revealed that GA20ox proteins were subdivided into two groups. OsGA20ox2 and OsGA20ox4 were more closely related (64% identity) than the other OsGA20ox proteins, and they formed one separate branch, but then OsGA20ox1 and OsGA20ox3 were grouped with AtGA20ox1, AtGA20ox2, and AtGA20ox3 to form another.
We identified five GA2ox genes in rice as compared to seven previously reported Arabidopsis counterparts (Schomburg et al., 2003); four (OsGA2ox1, OsGA2ox2, OsGA2ox3, and OsGA2ox4) of them were reported previously (Sakamoto et al., 2001; Sakai et al., 2003). OsGA2ox1, OsGA2ox4, and OsGA2ox5 are located on chromosome 5, and OsGA2ox2 and OsGA2ox3 are on chromosome 1. Although some OsGA2ox genes are localized on the same chromosomes, they are well separated with large phylogenetic distance that suggests early duplication events. OsGA2ox3 showed highest similarity to Arabidopsis GA2ox (49–51% identities), whereas the other OsGA2ox showed identities between 35 and 48% with the Arabidopsis counterparts. Phylogenetic analysis (Figure 3B) revealed that OsGA2ox1 has lower identities with those of Arabidopsis counterparts, constituting a separate clade.
Comparative analysis of ABA and GA metabolic genes among rice cultivars
Using a threshold of nucleotide identity over 95% and coverage over 80%, we defined orthologous gene pairs in 9311, PA64s, and Nipponbare (also including a 3-kb upstream sequence containing promoters and UTRs) for comparative analysis (Table 2). Although 9311, PA64s, and Nipponbare share a common ancestor of O. sativa, there are many sequence variations between orthologs that may be a result of artificial selection. In general, there are significantly more SNPs and Indels in coding regions between paralogs than between orthologs (Wilcoxon rank sum test: p < 0.01). Compared to coding regions, regulatory regions have significantly higher numbers of substitutions and Indels (Wilcoxon rank sum test: p < 0.01), which may also collectively contribute to the differential expression of these genes.
Table 2.
Single nucleotide polymorphisms and Indels among 9311, PA64s, and Nipponbare.
| Gene name | Genes on chromosomea | Regulation region | ||||
|---|---|---|---|---|---|---|
| 9311 | PA64s | SNP no.b | SNP ratec | Indel no.d | Indel lengthe | |
| OsNCED1 | Chr02_3173 | Chr02_4135 | 43|33|10 | 0.0141|0.0108|0.0033 | 12|7|6 | 116|96|20 |
| OsNCED2 | Chr12_1047 | Chr12_0855 | 31|139|158 | 0.0102|0.0425|0.0478 | 8|8|16 | 96|540|614 |
| OsNCED3 | Chr03_3073 | Chr03_2198 | NA|NA|NA | NA|NA|NA | NA|NA|NA | NA|NA|NA |
| OsNCED4 | Chr07_0366 | Chr07_0350 | 100|174|228 | 0.033|0.0577|0.0757 | 3|16|9 | 60|30|22 |
| OsNCED5 | Chr12_2021 | Chr12_1628 | 53|125|125 | 0.0173|0.0399|0.0406 | 9|10|12 | 116|264|164 |
| OsCYP707A5 | Chr02_3170 | Chr02_4132 | 7|17|11 | 0.0023|0.0054|0.0035 | 3|3|2 | 2|262|260 |
| OsCYP707A6 | Chr08_2087 | Chr08_1729 | 37|38|3 | 0.012|0.012|0.001 | 16|13|7 | 32|58|56 |
| OsCYP707A7 | Chr09_1329 | Chr09_1219 | 262|262|2 | 0.084|0.0845|0.0007 | 25|25|0 | 204|204|0 |
| OsGA20ox1 | Chr03_4546 | Chr03_3632 | 9|10|1 | 0.003|0.0033|0.0003 | 6|4|3 | 80|36|44 |
| OsGA20ox2 | Chr01_4645 | Chr01_4410 | 34|NA|NA | 0.011|NA|NA | 7|NA|NA | 188|NA|NA |
| OsGA20ox3 | Chr12_1260 | Chr12_1026 | 0|0|0 | 0|0|0 | 0|0|2 | 0|0|2 |
| OsGA20ox4 | Chr05_1936 | Chr05_1801 | 45|45|0 | 0.0147|0.0147|0 | 9|9|0 | 102|102|0 |
| OsGA3ox1 | Chr05_0563 | Chr05_0539 | 28|28|0 | 0.009|0.009|0 | 18|18|0 | 242|242|0 |
| OsGA3ox2 | Chr01_0611 | Chr01_0665 | 40|40|0 | 0.0133|0.0133|0 | 6|6|0 | 12|12|0 |
| OsGA2ox1 | Chr05_0449 | Chr05_0429 | 5|NA|NA | 0.0016|NA|NA | 6|NA|NA | 10|NA|NA |
| OsGA2ox2 | Chr01_0837 | Chr01_0926 | 19|14|7 | 0.0063|0.0047|0.0023 | 11|6|8 | 28|18|12 |
| OsGA2ox3 | Chr01_3778 | Chr01_3512 | 36|39|3 | 0.0119|0.0129|0.001 | 7|7|0 | 42|42|0 |
| OsGA2ox4 | Chr05_2528 | Chr05_2378 | 370|18|348 | 0.117|0.0059|0.1103 | 69|10|67 | 306|76|306 |
| OsGA2ox5 | Chr05_2857 | Chr05_2689 | 36|33|7 | 0.012|0.011|0.0023 | 6|5|4 | 10|6|8 |
| OsGPCR | Chr10_1648 | Chr10_1340 | NA|25|NA | 0.5375|0.0079|0.5209 | 80|11|95 | 184|340|228 |
| OsGID1 | Chr05_1857 | Chr05_1721 | NA|NA|NA | NA|NA|NA | NA|NA|NA | NA|NA|NA |
| Gene name | Genes on chromosomea | Coding region | ||||
|---|---|---|---|---|---|---|
| 9311 | PA64s | SNP no.b | SNP ratec | Indel no.d | Indel lengthe | |
| OsNCED1 | Chr02_3173 | Chr02_4135 | 5|5|0 | 0.0026|0.0026|0 | 1|4|1 | 48|239|189 |
| OsNCED2 | Chr12_1047 | Chr12_0855 | 10|0|9 | 0.0057|0|0.0051 | 0|0|0 | 0|0|0 |
| OsNCED3 | Chr03_3073 | Chr03_2198 | 8|16|8 | 0.0044|0.0088|0.004 | 0|2|2 | 0|129|129 |
| OsNCED4 | Chr07_0366 | Chr07_0350 | 1|9|8 | 0.0006|0.0051|0.004 | 1|0|1 | 177|0|177 |
| OsNCED5 | Chr12_2021 | Chr12_1628 | 1|4|1 | 0.0007|0.0022|0.001 | 0|0|0 | 0|0|0 |
| OsCYP707A5 | Chr02_3170 | Chr02_4132 | 1|2|0 | 0.0007|0.0014|0 | 0|2|2 | 0|306|306 |
| OsCYP707A6 | Chr08_2087 | Chr08_1729 | 1|6|0 | 0.0007|0.0036|0 | 0|2|0 | 0|159|0 |
| OsCYP707A7 | Chr09_1329 | Chr09_1219 | 1|4|0 | 0.0007|0.0027|0 | 0|0|0 | 0|0|0 |
| OsGA20ox1 | Chr03_4546 | Chr03_3632 | 2|2|0 | 0.0018|0.0018|0 | 0|0|0 | 0|0|0 |
| OsGA20ox2 | Chr01_4645 | Chr01_4410 | 2|2|0 | 0.0017|0.0017|0 | 2|2|2 | 126|381|411 |
| OsGA20ox3 | Chr12_1260 | Chr12_1026 | 0|0|0 | 0|0|0 | 0|0|0 | 0|0|0 |
| OsGA20ox4 | Chr05_1936 | Chr05_1801 | 41|41|0 | 0.031|0.031|0 | 2|1|1 | 540|369|171 |
| OsGA3ox1 | Chr05_0563 | Chr05_0539 | 3|5|2 | 0.0026|0.0043|0.017 | 2|6|4 | 18|84|66 |
| OsGA3ox2 | Chr01_0611 | Chr01_0665 | 3|3|0 | 0.0027|0.0027|0 | 1|1|0 | 9|9|0 |
| OsGA2ox1 | Chr05_0449 | Chr05_0429 | 3|2|2 | 0.00247|0.0017|0.002 | 1|1|1 | 63|471|534 |
| OsGA2ox2 | Chr01_0837 | Chr01_0926 | 81|5|57 | 0.08|0.0049|0.057 | 4|1|5 | 276|3|279 |
| OsGA2ox3 | Chr01_3778 | Chr01_3512 | 1|1|0 | 0.001|0.001|0 | 0|0|0 | 0|0|0 |
| OsGA2ox4 | Chr05_2528 | Chr05_2378 | 3|32|8 | 0.0028|0.0318|0.011 | 1|10|2 | 336|276|62 |
| OsGA2ox5 | Chr05_2857 | Chr05_2689 | 59|6|65 | 0.0527|0.0056|0.061 | 7|2|9 | 60|95|154 |
| OsGPCR | Chr10_1648 | Chr10_1340 | 10|40|38 | 0.0072|0.0288|0.027 | 2|5|4 | 597|388|553 |
| OsGID1 | Chr05_1857 | Chr05_1721 | 20|20|18 | 0.0119|0.0019|0.017 | 5|0|5 | 708|0|708 |
aMatched genes of 9311 and PA64s with chromosome and gene ID information.
bNumber of SNPs between the rice cultivars. | Number of SNPs between PA64s and Nipponbare | Number of SNPs between 9311 and PA64s. NA strands for poor similarity.
cRate of SNPs between 9311 and Nipponbare | Rate of SNPs between PA64s and Nipponbare | Rate of SNPs between 9311 and PA64s.
dNumber of Indels between 9311 and Nipponbare | Number of Indels between PA64s and Nipponbare | Number of Indels between 9311 and PA64s.
eTotal length (bp) of Indels between 9311 and Nipponbare | Total length (bp) of Indels between PA64s and Nipponbare | Total length (bp) of Indels between 9311 and PA64s.
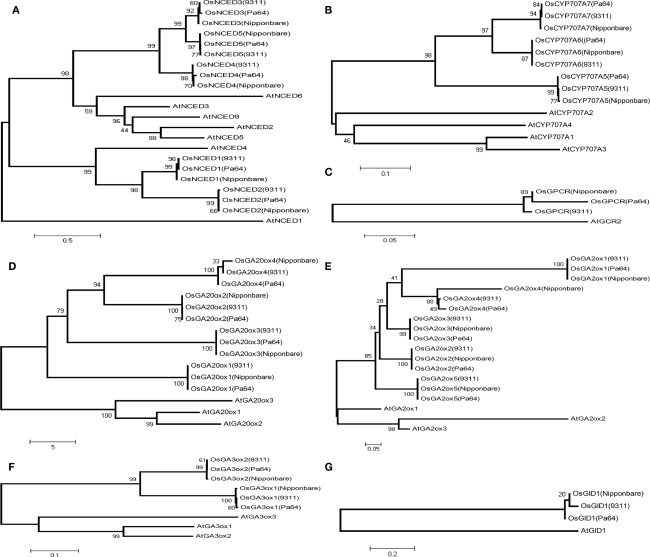
Based on the degree of sequence variations, we can readily identify the gene family members and their relationship with regard to obvious inheritances from their ancestor subspecies, indica or japonica. For instance, in NCED family, three genes (OsNCED1, OsNCED3, and OsNCED5) of PA64s and 9311 were inherited from the same indica ancestor as shown in Figure 4, which is consistent with lower variation between 9311 and PA64s (Table 2). In another gene family, OsCYP707A6 and OsCYP707A7 of PA64s were from indica (Figure 4B). In GA20ox family, PA64s inherited OsGA20ox2 from indica, whereas other genes were not obviously separated into indica or japonica (Figure 4D). In GA3ox family, both OsGA3ox1 and OsGA3ox2 were from indica background, and in GA2ox family, PA64s inherited OsGA2ox4 from its indica background (Figures 4E and 4F). ABA receptor (OsGPCR) of PA64s was most likely from japonica or javanica rather than from indica background. Both ABA and GA receptors were found with large amount of variations between 9311 and PA64s due to frequent hybridization with complex genetic background during rice domestication and breeding. We concluded that the dormancy trait of PA64s was mainly inherited from indica, though it also maintained a partial background of japonica and javanica. The mixed background may contribute to its significant difference in germination when compared with 9311.
Figure 4.
Neighbor-joining phylogenetic analysis of (A) NCED, (B) CYP707A, (C) ABA receptor, (D) GA20ox, (E) GA2ox, (F) GA3ox, and (G) GA receptor among 9311, PA64s, and Nipponbare.
Sequence and functional analysis of ABA and GA metabolic genes of 9311, PA64s, and Nipponbare
We essentially identified two classes of differences among the three cultivars: SNPs and Indels (Table 2). Some genes are obviously more conserved (by and large due to selection) than others, such as OsGA3ox2 and OsGA2ox3, who have no Indel or SNP discovered in either 9311 or PA64s (Table 2). One possible reason is that these genes play major functional roles and therefore are strongly selected. Another explanation is that these genes were recently duplicated and therefore less mutation was accumulated. Other genes in 9311, PA64s, and Nipponbare appear high variable, such as OsGA20ox4 (41 SNPs and 2 Indels with a total length of 540 bp) and OsGA2ox5 (65 SNPs and 9 Indels with a total length of 154 bp) (Table 2). One possible reason for the variability is that they are relative older or less conserved due to weaker selection.
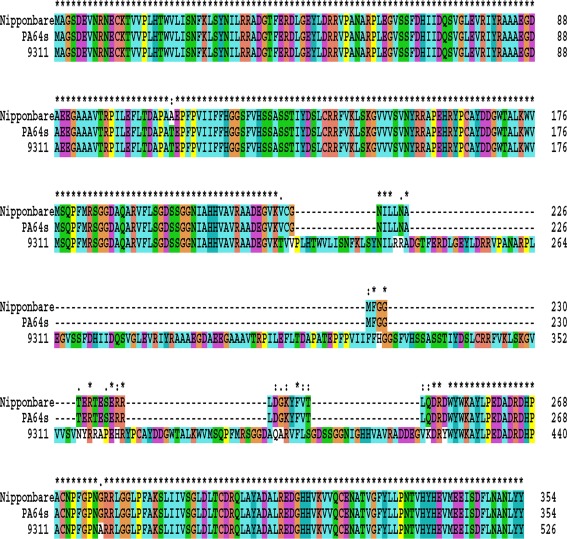
We have further scrutinized some SNPs and Indels of protein domains of the related genes to look for potential changes in protein structure and function. Ninety-seven candidate SNPs and 618 candidate Indels involving amino acid changes were found to lie in domains of RPE membrane protein, cytochrome P450, 2OG-Fe(II) oxygenase superfamily, lanthionine synthetase C-like protein, and alpha/beta hydrolase fold, as well as other residues associated with specific functions predicted by pfam (Table 3). We found that 2OG-Fe(II) oxygenase of OsGA3ox family is highly conserved but alpha/beta hydrolase fold of OsGID1 has 24 SNPs and 172 amino acid Indels (Figure 5). Compared to the sequences of PA64s and Nipponbare, 9311 has a large insertion in the alpha/beta hydrolase fold (Ollis et al., 1992), and this insertion may lead to enhanced sensitivity to GA. Sequence polymorphisms in the functional domains that differ between the three subspecies may be one of the direct causes for functional variation, although further functional tests are required to confirm their precise roles.
Table 3.
Candidate SNPs and Indels identified in conserved regions as indicated by Pfam.
| Gene | Changea | Positionb | Indel length (aa) | Domain/family | Domain/family position |
|---|---|---|---|---|---|
| OsNCED1 | E/K | 448 | 63 | RPE (retinal pigment epithelial) membrane protein | 131–630 |
| G/R | 465 | ||||
| OsNCED2 | I/L | 188 | 0 | RPE membrane protein | 68–569 |
| V/A | 199 | ||||
| R/Q | 359 | ||||
| OsNCED3 | R/A | 238 | 43 | RPE membrane protein | 108–601 |
| A/G | 240 | ||||
| C/W | 241 | ||||
| G/R | 242 | ||||
| D/E | 245 | ||||
| N/Y | 254 | ||||
| L/F | 257 | ||||
| V/I | 258 | ||||
| D/G | 282 | ||||
| C/R | 492 | ||||
| OsNCED4 | P/S | 87 | 59 | RPE membrane protein | 76–575 |
| V/L | 109 | ||||
| G/A | 116 | ||||
| V/L | 125 | ||||
| A/T | 131 | ||||
| G/S | 549 | ||||
| OsNCED5 | A/V | 260 | 0 | RPE membrane protein | 110–606 |
| OsCYP707A5 | A/T | 237 | 59 | Cytochrome P450 | 40–432 |
| OsCYP707A6 | NA | NA | 4 | Cytochrome P450 | 52–489 |
| OsCYP707A7 | N/H | 69 | 0 | Cytochrome P450 | 45–474 |
| OsGA20ox1 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 208–307 |
| OsGA20ox2 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 225–324 |
| OsGA20ox3 | NA | NA | 9 | 2OG-Fe(II) oxygenase superfamily | 199–304 |
| S/V | 304 | ||||
| C/S | 312 | ||||
| S/A | 313 | ||||
| I/T | 315 | ||||
| OsGA20ox4 | R/L | 316 | 49 | 2OG-Fe(II) oxygenase superfamily | 234–369 |
| L/V | 318 | ||||
| L/F | 320 | ||||
| T/R | 321 | ||||
| T/Q | 331 | ||||
| Q/S | 347 | ||||
| OsGA3ox1 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 226–327 |
| OsGA3ox2 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 212–304 |
| OsGA2ox1 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 190–321 |
| OsGA2ox2 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 179–287 |
| OsGA2ox3 | NA | NA | 0 | 2OG-Fe(II) oxygenase superfamily | 174–278 |
| OsGA2ox4 | 25 | / | 113 | 2OG-Fe(II) oxygenase superfamily | 113–177 |
| OsGA2ox5 | F/P | 206 | 0 | 2OG-Fe(II) oxygenase superfamily | 179–292 |
| G/R | 207 | ||||
| OsGPCR | 18 | / | 47 | Lanthionine synthetase C-like protein | 120–461 |
| OsGID1 | 24 | / | 172 | alpha/beta hydrolase fold | 116–330 |
aAmino acid changes in functional regions are indicated as well as Indels numbers.
bThe position of amino acid changes in functional regions. NA stands for no change. Positions of three proteins, OsGA2ox4, OsGPCR, and OsGID1 were not listed due to a high number of changes.
/ represents that there are too many SNPs to list in the Table.
Figure 5.
Amino acid sequence alignment of GA receptors among 9311, PA64s, and Nipponbare. Positions in the sequence where all three proteins are the same or physiochemically similar are marked with an asterisk above.
Different functional constraints on ABA and GA metabolic genes between 9311, PA64s, and Nipponbare
Following a gene duplication event, the two duplicates may be subjected to different selective constraints and even new functions. To investigate whether or not different rice cultivars have undergone different types of artificial selection, we identified reciprocal best matches – orthologs – among ABA and GA metabolic genes from Arabidopsis and rice. We calculated non-synonymous and synonymous substitution rate (Ka and Ks) for each pair (Figure 6). As indicated by Ka/Ks < 1, all of pairwise comparisons appear to undergo purifying selection. Interestingly, the Ka/Ks ratio of OsGA20ox2 is significantly different from other members of OsGA20ox family (t-test: p < 0.01), indicating that it may evolve faster than others. This difference may be either a result of positive selection or relaxed purifying selection. Our results suggest that genes with multiple copies tend to evolve in different patterns, consistent with the view that one of the duplicates may undergo purifying selection after gene duplication while the other may enjoy more relaxed selective pressure. Furthermore, we also found that some of these genes (such as OsNCED1 and OsGA3ox2) had different Ka/Ks values between different rice cultivars.
Figure 6.
Ka/Ks ratios of ABA and GA metabolic genes for different rice cultivars.
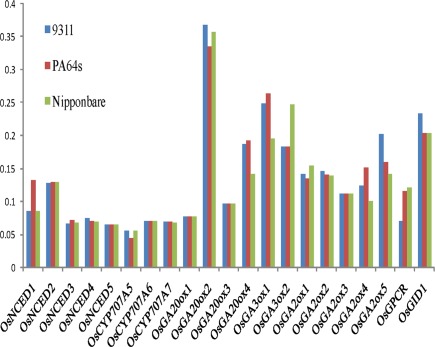
Expression analysis of NCED, CYP707a, GA20ox, GA3ox, and GA2ox genes in 9311 and PA64s
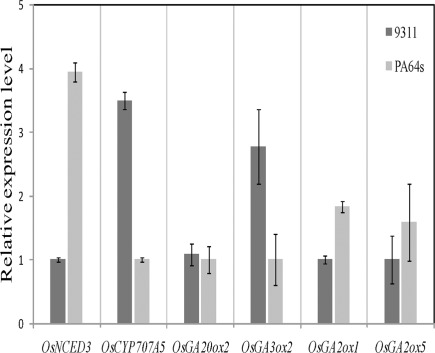
In imbibing seeds, ABA and GA contents are regulated by ABA biosynthetic enzyme NCED, ABA catabolic enzyme CYP707A, GA biosynthetic enzymes GA20ox and GA3ox, and GA catabolic enzyme GA2ox. We detected expression levels of these genes which are important for the regulation of ABA and GA contents (Xiong and Zhu, 2003; Welsch et al., 2008). We also performed real-time PCR to confirm the expression levels of 19 genes in 9311 and PA64s, including 6 genes (OsNCED3, OsCYP707A5, OsGA20ox2, OsGA3ox2, OsGA2ox1, and OsGA2ox5) high-abundance and 13 (OsNCED1, OsNCED2, OsNCED4, OsNCED5, OsCYP707A6, OsCYP707A7, OsGA20ox1, OsGA20ox4, OsGA20ox3, OsGA3ox1, OsGA2ox2, OsGA2ox3, OsGA2ox4) low-abundance transcripts. Our results showed that the expression of ABA and GA metabolic genes was relative low and that there were significant differences between 9311 and PA64s (Figure 7). For instance, the expression of ABA synthetic gene OsNCED3 was 3.9-folds higher (t-test: p < 0.01) and catabolic gene OsCYP707A5 was 3.5-folds lower (t-test: p < 0.01) in PA64s than in 9311; this result implied a lower ABA level in 9311 (Barrero et al., 2006). The GA synthetic gene OsGA3ox2 was expressed at a much higher level (2.8-folds) in 9311 than in PA64s. In contrast, both of GA catabolic genes OsGA2ox1 and OsGA2ox5 were expressed at a much higher level in PA64s than in 9311; this result suggested higher GA level or less dormant seeds in 9311. Indeed, a correlation between the expression level of GA metabolism genes and the content of active GA has been reported (Busov et al., 2003; Oh et al., 2006). The differential expression among gene family members also confirmed the complexity of the regulatory network, also footnoted by higher variations in regulatory regions.
Figure 7.
Real-time PCR based analyses of OsNCED3, OsCYP707A5, OsGA20ox2, OsGA3ox2, OsGA2ox1, and OsGA2ox5 discovered in 9311 and PA64s seeds. Transcripts were normalized with GAPDH as an internal control. Measurements were averaged over three replicates with error bars representing SE.
Discussion
Studying the plasticity of dormancy and germination is of necessity in plant biology and crop breeding (Roberts, 1961). We engaged our study on seed germination and discovered that dormancy states are different among rice cultivars, consistent with previous publications (Seshu and Dadlani, 1991; Gu et al., 2004, 2006; Veasey et al., 2004). Aligning sequences in coding and regulatory regions, we observed large amount of Indels and SNPs in genes that are related to regulate the ABA and GA metabolic pathways. For instance, two Indels of 306 bp in the coding sequence and two Indels of 260 bp in the upstream regulatory region of OsCYP707A5 between 9311 and PA64s may affect the function of OsCYP707A5 and the binding of its regulatory transcriptional factors. In addition, we compared genes differentially expressed in dry seed between 9311 and PA64s, and hypothesized that different cellular concentrations of ABA and GA may reflect the genetic variations among these genes, especially those related to the ABA and GA pathways (Xiong and Zhu, 2003; Welsch et al., 2008).
As rice cultivars have been under strong artificial selection during domestication and the loss of seed dormancy is one of the most important traits in the domestication syndrome, dormancy- and germination-associated genes are absolutely the targets of selection. Thus, we further calculated Ka/Ks values for the ABA and GA metabolism-related genes and found that all were lower than 0.5; the result suggests that they are most likely undergoing purifying selection despite the fact that most of them belong to multi-gene families. Compared to other OsNCED family members, OsNCED3, due to their high expression levels, may play a major role in controlling dormancy and germination (Lefebvre et al., 2006; Hwang et al., 2010), which is consistent with that the ectopic expression of OsNCED3 in Arabidopsis leads to a delay in seed germination (Hwang et al., 2010). Expression analysis of CYP707A genes suggests that OsCYP707A5 may play a key role in seed dormancy and germination (Kushiro et al., 2004; Millar et al., 2006; Okamoto et al., 2006). One SNP and several Indels with a total length of 59 amino acids were identified in functional regions of OsCYP707A5, indicating its potential involvement in regulating ABA concentration. In OsGA20ox and OsGA3ox families, OsGA20ox2 and OsGA3ox2 may play major roles in the regulation of GA synthesis (Calvo et al., 2004); both are highly conserved as no SNP/Indel was found in their functional regions. Therefore, their differential expression may control the cellular concentration of GA (Mitchum et al., 2006; Rieu et al., 2008). By the same token, OsGA2ox1 and OsGA2ox5 of OsGA2ox family may play major roles in regulating GA catabolism (Mitchum et al., 2006) so do OsGPCR and OsGID1 (Aleman et al., 2008; Hirano et al., 2008; Murase et al., 2008).
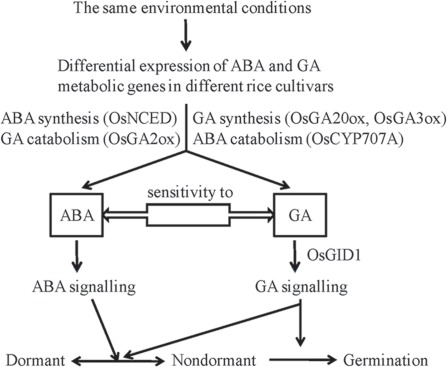
A balanced ABA/GA ratio governed by dynamics of hormone synthesis and catabolism regulates dormancy and germination through changing seed sensitivity to the external environment (Cadman et al., 2006; Seo et al., 2006). We propose a model extending the ABA/GA balance theory to address changes in gene function and expression (Figure 8). According to this model, different rice cultivars exhibit different dormancy states due to differential expression of ABA and GA metabolic genes. Environmental conditions, such as high temperature and light (Toh et al., 2008), can enhance expression of specific ABA biosynthesis genes (OsNCED family) and GA catabolic genes (OsGA2ox family), resulting in seed dormancy through induction of ABA dominance. Low temperature (Finch-Savage et al., 2007) can also enhance expression of specific ABA catabolic genes (OsCYP707A family) and GA biosynthesis genes (OsGA20ox and OsGA3ox families). In order to further illustrate molecular mechanisms of embryonic dormancy in rice, additional studies should be carried out, focusing on comparative sequence analysis and expression profiling of dormancy-associated genes among large samples of rice cultivars to construct gene regulation networks covering all the stages of seed dormancy and germination.
Figure 8.
A schematic model for the regulation of dormancy and germination by ABA and GA in response to the environment in rice seeds. Under the same environmental conditions, different rice cultivars exhibit different dormancy states due to their differential expression of ABA and GA metabolic genes. Environmental conditions such as higher temperature enhance expression of specific ABA biosynthesis genes (OsNCED family) and GA catabolic genes (OsGA2ox family), which result in dormancy by inducing dominance of ABA. Moreover, whether a seed remains dormancy or germinates may depend on a balanced ABA and GA metabolism influenced by genetic and epigenetic mechanisms, a collective outcome determined by sequence and expression variations of ABA and GA metabolic genes and the function of ABA/GA receptors.
Conclusion
In this study, we identified rice ABA and GA metabolism-related genes based on comparative and phylogenetic analyses among rice cultivars. We found that there are many SNPs and Indels in the coding and regulatory sequences of these genes due to their redundancy and diverse genetic background. We further traced the variations on the genome sequences of 9311 and PA64s and showed that sequence variations may lead to functional variations and variable seed dormancy states regulated by ABA and GA. In addition, we also surveyed and compared differentially expressed genes between 9311 and PA64s and demonstrated that differential expression of related genes may also play roles in the variable dormancy and germination states in rice. Although precise correlation between sequence variations and dormancy states need further experimental verification, sequence and expression analyses of ABA and GA metabolism-related genes among rice cultivars pave a way to study molecular mechanism of seed dormancy and germination.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Genetics_and_Genomics/10.3389/fpls.2011.00017/abstract
ABA and GA concentrations of 9311 and PA64s from each individual assay of the triplicate.
DNA sequences of 9311, PA64s, and Nipponbare used in this study.
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (30221004) awarded to Weiwei Wang and from the National Basic Research Program (973 Program; 2011CB944100 and 2011CB944101) and National Natural Science Foundation of China (90919024) awarded to Jun Yu.
Footnotes
References
- Aleman L., Kitamura J., Abdel-Mageed H., Lee J., Sun Y., Nakajima M., Ueguchi-Tanaka M., Matsuoka M., Allen R. D. (2008). Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol. 68, 1–16 [DOI] [PubMed] [Google Scholar]
- Barrero J. M., Rodriguez P. L., Quesada V., Piqueras P., Ponce M. R., Micol J. L. (2006). Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 29, 2000–2008 10.1111/j.1365-3040.2006.01576.x [DOI] [PubMed] [Google Scholar]
- Bewley J. D. (1997). Seed germination and dormancy. Plant Cell 9, 1055–1066 10.1105/tpc.9.7.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V. B., Meilan R., Pearce D. W., Ma C., Rood S. B., Strauss S. H. (2003). Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from poplar that regulates tree stature. Plant Physiol. 132, 1283–1291 10.1104/pp.103.020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman C. S., Toorop P. E., Hilhorst H. W., Finch-Savage W. E. (2006). Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J. 46, 805–822 10.1111/j.1365-313X.2006.02738.x [DOI] [PubMed] [Google Scholar]
- Calvo A. P., Nicolas C., Nicolas G., Rodriguez D. (2004). Evidence of a cross-talk regulation of a GA 20-oxidase (FsGA20ox1) by gibberellins and ethylene during the breaking of dormancy in Fagus sylvatica seeds. Physiol. Plant 120, 623–630 10.1111/j.0031-9317.2004.0270.x [DOI] [PubMed] [Google Scholar]
- Chiwocha S. D., Cutler A. J., Abrams S. R., Ambrose S. J., Yang J., Ross A. R., Kermode A. R. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42, 35–48 10.1111/j.1365-313X.2005.02359.x [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Cadman C. S., Toorop P. E., Lynn J. R., Hilhorst H. W. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51, 60–78 10.1111/j.1365-313X.2007.03118.x [DOI] [PubMed] [Google Scholar]
- Goff S. A., Ricke D., Lan T. H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Hadley D., Hutchison D., Martin C., Katagiri F., Lange B. M., Moughamer T., Xia Y., Budworth P., Zhong J., Miguel T., Paszkowski U., Zhang S., Colbert M., Sun W. L., Chen L., Cooper B., Park S., Wood T. C., Mao L., Quail P., Wing R., Dean R., Yu Y., Zharkikh A., Shen R., Sahasrabudhe S., Thomas A., Cannings R., Gutin A., Pruss D., Reid J., Tavtigian S., Mitchell J., Eldredge G., Scholl T., Miller R. M., Bhatnagar S., Adey N., Rubano T., Tusneem N., Robinson R., Feldhaus J., Macalma T., Oliphant A., Briggs S. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 10.1126/science.1068275 [DOI] [PubMed] [Google Scholar]
- Goldman N., Yang Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736 [DOI] [PubMed] [Google Scholar]
- Gu X. Y., Kianian S. F., Foley M. E. (2004). Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa). Genetics 166, 1503–1516 10.1534/genetics.166.3.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Y., Kianian S. F., Foley M. E. (2006). Dormancy genes from weedy rice respond divergently to seed development environments. Genetics 172, 1199–1211 10.1534/genetics.105.049155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Millar A. A., Jacobsen J. V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8, 183–187 [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A. L., Rojas M. C., Carrera E., Tudzynski B. (2001). Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20, 319–331 10.1007/s003440010037 [DOI] [PubMed] [Google Scholar]
- Hirano K., Ueguchi-Tanaka M., Matsuoka M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13, 192–199 10.1016/j.tplants.2008.02.005 [DOI] [PubMed] [Google Scholar]
- Hwang S.-G., Chen H.-C., Huang W.-Y., Chu Y.-C., Shii C.-T., Cheng W.-H. (2010). Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 178, 12–22 10.1016/j.plantsci.2009.09.014 [DOI] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sentoku N., Kitano H., Matsuoka M., Kobayashi M. (2001). Cloning and functional analysis of two gibberellin 3 beta hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. U.S.A. 98, 8909–8914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36 [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656 10.1038/sj.emboj.7600121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319 10.1111/j.1365-313X.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Liu X., Yue Y., Li B., Nie Y., Li W., Wu W. H., Ma L. (2007). A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315, 1712–1716 10.1126/science.1135882 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- Millar A. A., Jacobsen J. V., Ross J. J., Helliwell C. A., Poole A. T., Scofield G., Reid J. B., Gubler F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J. 45, 942–954 10.1111/j.1365-313X.2006.02659.x [DOI] [PubMed] [Google Scholar]
- Mitchum M. G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T. P. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45, 804–818 10.1111/j.1365-313X.2005.02642.x [DOI] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T. P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 10.1038/nature07519 [DOI] [PubMed] [Google Scholar]
- Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., Maeda T., Matsuoka M., Yamaguchi I. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 10.1111/j.1365-313X.2006.02748.x [DOI] [PubMed] [Google Scholar]
- Nonogaki H., Bassel G. W., Bewley J. D. (2010). Germination – still a mystery. Plant Sci. 179, 574–581 10.1016/j.plantsci.2010.02.010 [DOI] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W. I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139 10.1111/j.1365-313X.2006.02773.x [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141, 97–107 10.1104/pp.106.079475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J., Sussman J. L., Verschueren K. H. G., Goldman A. (1992). The alpha/beta hydrolase fold. Protein Eng. 5, 197–211 10.1093/protein/5.3.197 [DOI] [PubMed] [Google Scholar]
- Pandey S., Nelson D. C., Assmann S. M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136–148 10.1016/j.cell.2008.12.026 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., Mccourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Ruiz-Rivero O., Fernandez-Garcia N., Griffiths J., Powers S. J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S. G., Phillips A. L., Hedden P. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53, 488–504 10.1111/j.1365-313X.2007.03356.x [DOI] [PubMed] [Google Scholar]
- Roberts E. H. (1961). Dormancy of rice seed. I. The distribution of dormancy periods. J. Exp. Bot. 12, 319–329 [Google Scholar]
- Saika H., Okamoto M., Miyoshi K., Kushiro T., Shinoda S., Jikumaru Y., Fujimoto M., Arikawa T., Takahashi H., Ando M., Arimura S., Miyao A., Hirochika H., Kamiya Y., Tsutsumi N., Nambara E., Nakazono M. (2007). Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 48, 287–298 10.1093/pcp/pcm003 [DOI] [PubMed] [Google Scholar]
- Sakai M., Sakamoto T., Saito T., Matsuoka M., Tanaka H., Kobayashi M. (2003). Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J. Plant Res. 116, 161–164 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayano T., Tanaka H., Iwahori S., Matsuoka M. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125, 1508–1516 10.1104/pp.125.3.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G. S., Kitano H., Matsuoka M. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice. Nature 416, 701–702 10.1038/416701a [DOI] [PubMed] [Google Scholar]
- Schomburg F. M., Bizzell C. M., Lee D. J., Zeevaart J. A., Amasino R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163 10.1105/tpc.005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. H., Qin X., Zeevaart J. A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601 10.1104/pp.102.017921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., North H., Marion-Poll A., Sun T. P., Koshiba T., Kamiya Y., Yamaguchi S., Nambara E. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366 10.1111/j.1365-313X.2006.02881.x [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48 10.1016/S1360-1385(01)02187-2 [DOI] [PubMed] [Google Scholar]
- Seshu D. V., Dadlani M. (1991). Mechanism of seed dormancy in rice. Seed Sci. Res. 1, 187–194 [Google Scholar]
- Shen Y. Y., Wang X. F., Wu F. Q., Du S. Y., Cao Z., Shang Y., Wang X. L., Peng C. C., Yu X. C., Zhu S. Y., Fan R. C., Xu Y. H., Zhang D. P. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826 10.1038/nature05176 [DOI] [PubMed] [Google Scholar]
- Sweeney M., Mccouch S. (2007). The complex history of the domestication of rice. Ann. Bot. 100, 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tan B. C., Joseph L. M., Deng W. T., Liu L., Li Q. B., Cline K., Mccarty D. R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35, 44–56 10.1046/j.1365-313X.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Toh S., Imamura A., Watanabe A., Nakabayashi K., Okamoto M., Jikumaru Y., Hanada A., Aso Y., Ishiyama K., Tamura N., Iuchi S., Kobayashi M., Yamaguchi S., Kamiya Y., Nambara E., Kawakami N. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385 10.1104/pp.107.113738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veasey E. A., Karasawa M. G., Santos P. P., Rosa M. S., Mamani E., Oliveira G. C. (2004). Variation in the loss of seed dormancy during after-ripening of wild and cultivated rice species. Ann. Bot. 94, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P. E., Zhu J. K. (2007). New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 10, 447–452 [DOI] [PubMed] [Google Scholar]
- Welsch R., Wust F., Bar C., Al-Babili S., Beyer P. (2008). A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 147, 367–380 10.1104/pp.108.117028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Zhu J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36 10.1104/pp.103.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16, 367–378 10.1105/tpc.018143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Guo Z. (2007). Cloning of a 9-cis-epoxycarotenoid dioxygenase gene (SgNCED1) from Stylosanthes guianensis and its expression in response to abiotic stresses. Plant Cell Rep. 26, 1383–1390 10.1007/s00299-007-0325-8 [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G. K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Cao M., Liu J., Sun J., Tang J., Chen Y., Huang X., Lin W., Ye C., Tong W., Cong L., Geng J., Han Y., Li L., Li W., Hu G., Huang X., Li W., Li J., Liu Z., Li L., Liu J., Qi Q., Liu J., Li L., Li T., Wang X., Lu H., Wu T., Zhu M., Ni P., Han H., Dong W., Ren X., Feng X., Cui P., Li X., Wang H., Xu X., Zhai W., Xu Z., Zhang J., He S., Zhang J., Xu J., Zhang K., Zheng X., Dong J., Zeng W., Tao L., Ye J., Tan J., Ren X., Chen X., He J., Liu D., Tian W., Tian C., Xia H., Bao Q., Li G., Gao H., Cao T., Wang J., Zhao W., Li P., Chen W., Wang X., Zhang Y., Hu J., Wang J., Liu S., Yang J., Zhang G., Xiong Y., Li Z., Mao L., Zhou C., Zhu Z., Chen R., Hao B., Zheng W., Chen S., Guo W., Li G., Liu S., Tao M., Wang J., Zhu L., Yuan L., Yang H. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92 10.1126/science.1068037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ABA and GA concentrations of 9311 and PA64s from each individual assay of the triplicate.
DNA sequences of 9311, PA64s, and Nipponbare used in this study.