Abstract
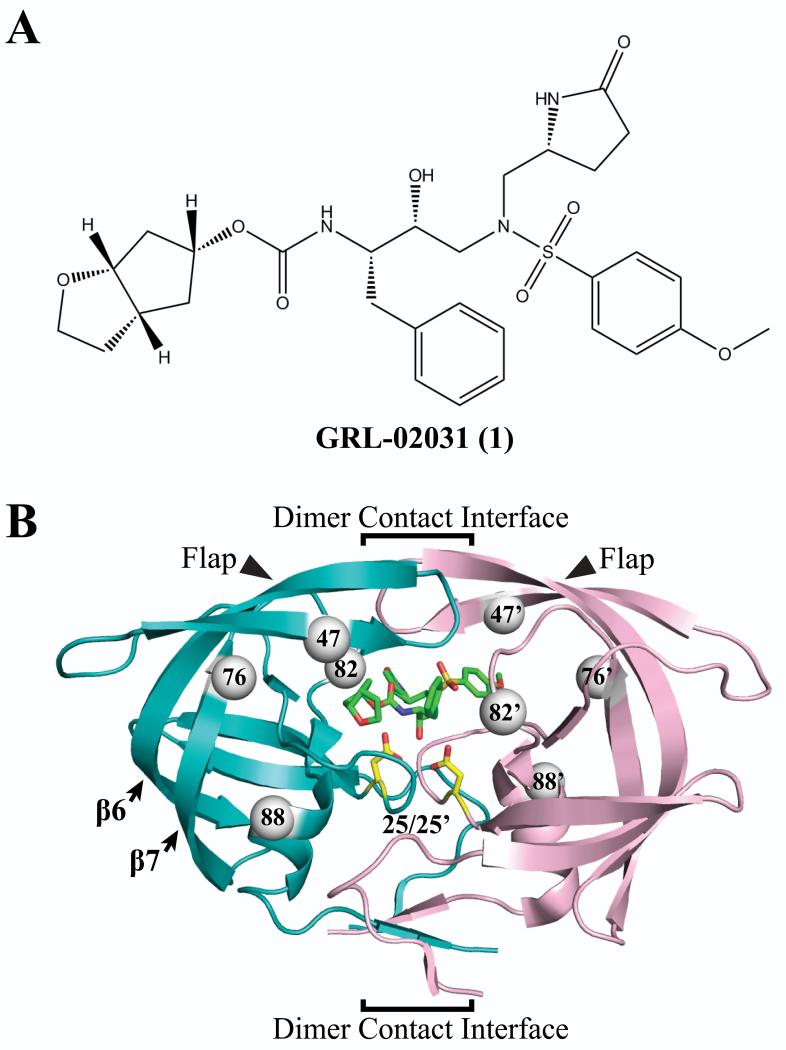
GRL-02031 (1) is an HIV-1 protease (PR) inhibitor containing a novel P1′ (R)-aminomethyl-2-pyrrolidinone group. Crystal structures at resolutions of 1.25 to 1.55 Å were analyzed for complexes of 1 with the PR containing major drug resistant mutations, PRI47V, PRL76V, PRV82A and PRN88D. Mutations of I47V and V82A alter residues in the inhibitor-binding site, while L76V and N88D are distal mutations having no direct contact with the inhibitor. Substitution of a smaller amino acid in PRI47V and PRL76V, and the altered charge of PRN88D are associated with significant local structural changes compared to the wild-type PRWT, while substitution of alanine in PRV82A increases the size of the S1′ subsite. The P1′ pyrrolidinone group of 1 accommodates to these local changes by assuming two different conformations. Overall, the conformation and interactions of 1 with PR mutants resemble those of PRWT with similar inhibition constants in good agreement with the antiviral potency on multidrug resistant HIV-1.
Keywords: aspartic protease, X-ray crystallography, molecular recognition, drug resistance, drug design
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) protease (PR) is a retroviral aspartyl protease with an essential role in the final step of viral maturation.1 The proteolytic cleavage of Gag and Gag-Pol polyproteins by PR releases the individual viral proteins2 and leads to morphological rearrangements3, which are required for the viral infectivity. Thus PR serves as an excellent target for antiretroviral drugs. Since 1995, nine protease inhibitors (PIs) have been approved by the Food and Drug Administration (FDA) for clinical use together with reverse transcriptase inhibitors and other antiviral agents, as part of the highly active antiretroviral therapy (HAART) regimens.4-6 Despite the success of HAART regimens, the treatments suffer from complications such as drug cytotoxicity, side effects and poor oral bioavailability, as well as drug resistance.7 Probably the most critical obstacle for long-term success of therapy is the emergence of multidrug-resistant HIV-1 variants due to the high drug selection pressure and extremely error-prone viral reverse transcriptase that lacks the proofreading step.8, 9
Among the clinical PIs, darunavir (DRV) exerts high antiviral activity against a wide spectrum of HIV-1 variants10, 11 with the enzyme inhibitory potency in the low picomolar range (Ki = 16 pM).12 DRV is a second generation PI that utilized the “backbone binding” strategy to maximize the interaction between inhibitor and PR backbone atoms.13 The recently described nonpeptidic PI GRL-02031 (1) (Figure 1A) based on the DRV scaffold retains potent activity against laboratory and primary HIV-1 strains.14 Compound 1 offers additional benefits over other clinical PIs with its low dose cytotoxicity (CC50 >100 μM) and high selectivity index (CC50/EC50 >3,600) against HIV-1LAI. Furthermore, compound 1 inhibited a wide spectrum of multidrug-resistant HIV-1 variants with less than 2-fold change in the EC50 relative to the effect on the wild-type HIV-1ERS104pre.15
Figure 1.
Protease inhibitor 1 and dimer structure of HIV-1 PRWT/1 (PDB ID 3H5B). (A) Chemical structure of compound 1. (B) The backbone of the two subunits is shown in teal and light pink ribbons, the inhibitor 1 is represented by green sticks, and the catalytic residues Asp25/25′ are represented by yellow sticks. The PR structure features of flaps, catalytic pocket, and dimer contact interface are indicated. The arrows indicate the flaps and secondary structure of 6th and 7th β-strands. The dimer interface indicated by the brackets extends from the interacting flap tips through the catalytic triplets to the 4-stranded β-sheet formed by both N- and C-termini. The gray spheres represent the location of mutated residues I47V, L76V, V82A and N88D in both subunits.
HIV-1 PR is a member of the aspartyl protease family comprising 99-residues in each monomer of the homodimer. Two conserved triplet residues (Asp25-Thr26-Gly27) interact closely in the center of the active site formed by two monomers.16 Important features of the PR dimer are: the catalytic pocket, the dimer interface and the mobile flaps. Inhibitors bind within a cavity between the catalytic triplets and the flaps (Figure 1B). In the presence of substrate or inhibitor, the flaps undergo conformation changes from an open state to a closed state.17, 18
HIV-1 develops resistance to PIs by selection of mutations in various regions of the PR. Resistance mutations have been observed in nearly forty different residues. Seventeen of the sites are considered major mutations providing high levels of resistance to one or more PIs.19 In this work, we have selected four of the major mutations that are involved in the cross-resistance of PIs. These mutations alter residues in distinct structural environments. Two of the mutations, I47V and V82A, are located inside the inhibitor binding site (Figure 1B). Residues Ile47 and Val82 contact the P2/P2′ and P1/P1′ groups of inhibitor 1, respectively.14, 15 These two residues are located in very different environments with Ile47 buried in a hydrophobic region of the catalytic pocket while Val82 is located on a surface loop of the protein. The other two residues, Leu76 and Asn88, are located in distal regions farther from the catalytic pocket and separated by greater than 5 Å from the inhibitor (Figure 1B). Residue 76 forms hydrophobic contacts with several residues in the hydrophobic core, as described previously.20 In contrast, residue 88 lies on the short helix near the protein surface.
Mutation of Ile47 shows a strong association with decreased susceptibility to amprenavir (APV), darunavir (DRV), lopinavir (LPV) and tipranavir (TPV).19, 21 Similarly, L76V mutation shows decreased susceptibility for APV, DRV and LPV. Interestingly, this mutation has an opposing effect on other drugs as it becomes hypersensitive to atazanavir (ATV), saquinavir (SQV) and TPV.22, 23 Mutations of V82A/T/F/S/L are very common in PI resistance and show reduced susceptibility to all the clinical PIs except DRV.19 Lastly, mutation of N88D alone is associated primarily with resistance to nelfinavir (NFV). However, N88D is common in association with D30N, which exerts a synergistic effect on resistance to NFV, and the double mutant shows 260-fold increased Ki value for NFV compared to wild-type.24-27 To analyze binding of compound 1 with various drug-resistant mutants, crystal structures were determined of its complexes with PRI47V, PRL76V, PRV82A and PRN88D. Analysis of these structures and the relative inhibition values will assist in the design of improved drugs for resistant HIV.
RESULTS
Crystallographic analysis of mutant/inhibitor complexes
Crystal structures of PRI47V, PRL76V, PRV82A and PRN88D drug-resistant mutants were determined with inhibitor 1 bound in the active site. All four structures were solved in the same space group P21212 as the PRWT/1 complex (PDB ID 3H5B)14 with X-ray data in the high resolution range of 1.25 to 1.55 Å. The data collection and refinement statistics are listed in Table 1. The asymmetric unit contains one homodimer PR with residues labeled 1-99 and 1′-99′. Superposition of wild-type and mutant dimers shows almost identical backbone conformations with a root mean square deviation (RMSD) of 0.12-0.19 Å over 198 Cα atoms. Two opposite orientations of inhibitor 1 were seen in the PRI47V and PRV82A mutants with occupancies of 0.7/0.3 and 0.65/0.35, respectively, comparable to the values of 0.8/0.2 in the PRWT complex. These alternate orientations of inhibitor related by 180° commonly occur due to the pseudosymmetry in the protease dimer. The conformation of the inhibitor is almost identical in all structures with pairwise RMSD of 0.1-0.3 Å for all non-hydrogen atoms; the largest difference is seen for the major orientation in PRI47V. PRL76V and PRN88D mutants showed a single orientation of the inhibitor. Furthermore, the P1′ group of 1 in PRI47V and PRL76V also shows two alternate conformations resembling the major orientation of PRWT/1. For PRI47V mutant, the alternate conformations of the P1′ group have 0.5/0.2 relative occupancy for the major orientation of 1, and a single conformation of P1′ group was refined for the minor inhibitor orientation. In the structure of PRL76V, the alternate conformations of P1′ group have 0.6/0.4 relative occupancy. In all four mutant structures, the inhibitor shows similar numbers of hydrogen bonds and hydrophobic interactions with the enzyme. Both major and minor orientations of inhibitor in the PRI47V and PRL76V structures have comparable interactions with the protease.
Table 1.
Data collection and refinement statistics for HIV-1 protease mutants in complex with 1
| PRI47V | PRL76V | PRV82A | PRN88D | |
|---|---|---|---|---|
| Crystal | ||||
| Space group | P21212 | P21212 | P21212 | P21212 |
| a, b, c (Å) | 58.20, 86.39, 45.95 | 57.82, 86.24, 46.01 | 58.64, 86.46, 45.44 | 58.23, 86.39, 46.07 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Data collection | ||||
| Resolution (Å) | 50-1.25 | 50-1.30 | 50-1.43 | 50-1.55 |
| Completeness (%) | 94.6 (63.7) | 98.1 (86.3) | 90.5 (57.0) | 99.9 (99.5) |
| Rsym (%) | 6.8 (32.6) | 9.4 (41.1) | 9.8 (45.0) | 7.5 (42.5) |
| <I/σ(I)> | 19.9 (2.4) | 14.3 (2.1) | 10.2 (2.0) | 23.1 (4.2) |
| Unique reflections | 61338 | 56338 | 39482 | 34554 |
| Redundancy | 6.1 (2.0) | 5.8 (2.7) | 12.1 (2.0) | 7.1 (6.3) |
| Refinement | ||||
| Resolution (Å) | 10-1.25 | 10-1.30 | 10-1.43 | 10-1.55 |
| Rwork (%) | 14.7 | 13.5 | 16.8 | 14.8 |
| Rfree (%) | 16.9 | 17.2 | 21.8 | 20.7 |
| RMSD bonds (Å) | 0.013 | 0.012 | 0.010 | 0.009 |
| RMSD angle dist. (Å) | 0.033 | 0.032 | 0.029 | 0.028 |
| Protein atoms | 1510 | 1510 | 1510 | 1512 |
| Solvent atoms | 205 | 225 | 129 | 183 |
| Total residues | 198 | 198 | 198 | 198 |
| Average B-factors (Å2) | ||||
| Main chain atoms | 13.4 | 12.5 | 17.7 | 16.1 |
| Side chain atoms | 18.7 | 19.3 | 24.8 | 23.5 |
| Inhibitor | 9.8 | 13.1 | 15.1 | 17.8 |
| Solvent | 24.4 | 23.8 | 24.8 | 24.7 |
| Ramachandran: | ||||
| Favored regions (%) | 100.0 | 100.0 | 100.0 | 100.0 |
| Allowed regions (%) | 100.0 | 100.0 | 100.0 | 100.0 |
| Relative Occup. of 1 | 0.70/0.30 | 1.00 | 0.65/0.35 | 1.00 |
Rsym = Σ|Iobs - Iavg|/ΣIavg; Rwork = Σ∥Fobs - Fcalc∥/ΣFobs; Rfree was calculated using 5% of data and the same sums. Parentheses denote the highest resolution bin.
Kinetic parameters of protease mutants with compound 1
The inhibition constants (Ki) for the wild-type and mutants, PRI47V, PRL76V, PRV82A and PRN88D, were determined using a p2/NC based fluorogenic hexapeptide substrate (Table 2). The compound was effective with all four mutants showing less than 2-fold changes in the inhibition constant. Furthermore, Koh et al. previously reported that 1 was a potent PI with comparable activity to DRV.15
Table 2.
Kinetic parameters of compound 1
The kinetic parameters of the mutants were similar to those of wild-type PR with less than 2-fold changes in the catalytic efficiency (kcat/Km). Interestingly, V82A and N88D mutations have opposite effects on the enzyme activity; PRV82A increases catalytic efficiency and PRN88D decreases catalytic efficiency. In our previous study, the catalytic efficiency of PRN88D had a 2.6 to 4.4-fold reduction over the wild-type value with the tested peptide substrates.28 In contrast, PRV82A showed differences in catalytic efficiency with two substrates based on the MA/CA or CA/p2 cleavage sites.29, 30 The contrasting observations are consistent with the location of the V82A mutation in the substrate binding site, while N88D is a distal mutation without direct effect on the substrate.
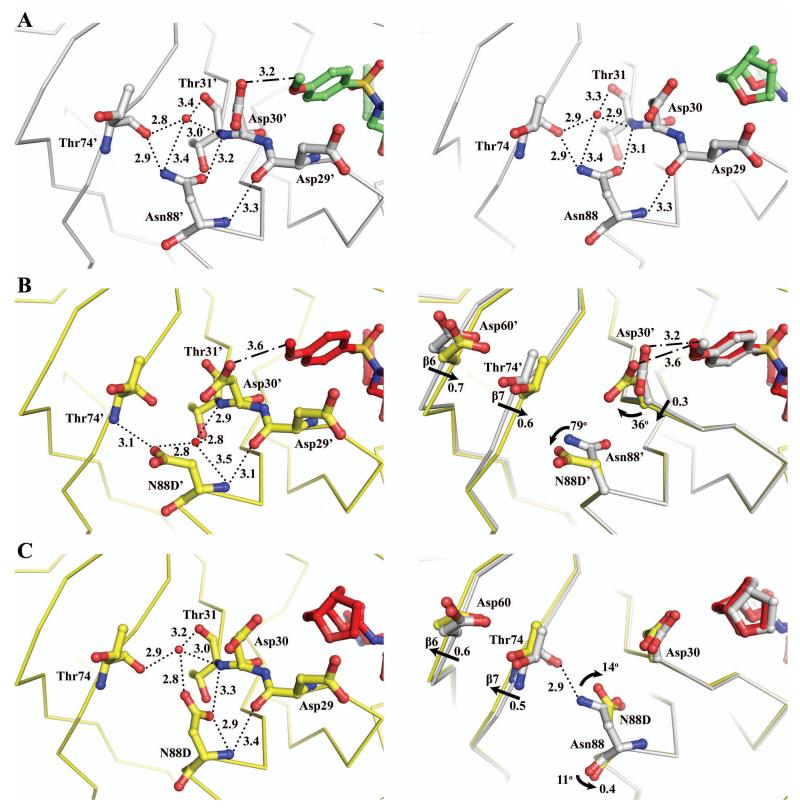
Influence of distal N88D and L76V mutations on the conformation of Asp30
Asn88 lies in the N-terminal helix near the protein surface in PRWT. Interestingly, the polar side chain of this residue is directed inward toward the 7th β-strand and loop between the 3rd and 4th β-strand rather than facing the protein surface. The side chain of Asn88 forms a network of several direct and water mediated hydrogen bonds with Asp30, Thr31 and Thr74 connecting to the inhibitor (Figure 2A). The carboxylate group of Asn88 interacts with the hydroxyl group and main chain amide of Thr31 with interatomic distances of 2.8 and 3.2 Å, respectively. The side chain amino group of Asn88 forms a hydrogen bond with the main chain carbonyl oxygen of Thr74 at 2.9 Å and a longer interaction with a water molecule at 3.4 Å. This water molecule is centered between Asp30, Thr31, Thr74 and Asn88 with hydrogen bonds to the carbonyl oxygen of Thr31 and Thr74, main chain amide of Thr31, and the side chain amide of Asn88 (Figure 2A). The mutation in the PRN88D/1 complex introduces a negatively charged side chain showing different conformations in the two subunits. These changes are accompanied by significant alteration in the hydrogen bonding network seen for Asn88 in PRWT. In monomer 1′-99′ of PRN88D, the mutated Asp88′ side chain has rotated 79° outward with a 1.9 Å relative translation at the Cγ position, and in the other monomer, the Asp88 side chain has rotated 14° inward with a 0.4 Å shift at Cγ position (Figure 2B and 2C).
Figure 2.
Comparison of interactions of Asn88 and Asp88 in PRN88D. PRWT/1 is colored gray and green, and PRN88D/1 is colored yellow and red. Hydrogen bond interactions are denoted by dotted lines with interatomic distances in Å. C-H…O interactions are denoted by dashed-dotted lines. Straight arrows denote a translational shift between the two Cα atoms. Curved arrows denote a rotation between side chains of the two structures. Red spheres represent water molecules. (A) Left panel, PRWT monomer 1′-99′; Right Panel, PRWT monomer 1-99. Both monomers 1-99 and 1′-99′ showed similar contacts for residue 88 with less than 0.1 Å difference in atomic position. (B) Left panel, PRN88D monomer 1′-99′. Right panel, superposition of PRWT and PRN88D monomer 1′-99′ illustrates a side chain rotation at Asp30′ and N88D′, and a backbone movement at Asp30′, Asp60′ and Thr74′ (6th and 7th β-strand). (C) Left panel, PRN88D monomer 1-99. Right panel, superposition of PRWT and PRN88D monomer 1-99 illustrates the inward rotation of the side chain N88D and backbone movement at Asp60 and Thr74.
The outward movement of the Asp88′ side chain in the mutant creates a cavity between Thr31′ and Thr74′, and as a result the hydrogen bond interactions with the carbonyl oxygen and side chain hydroxyl group of Thr31′ are abolished. Interestingly, the enzymatic activity is not altered significantly by the hydrogen bond disruption as a water molecule apparently assumes the role of the Asn88′ side chain. The newly created cavity is occupied by a water molecule in a new position that maintains hydrogen bond interactions with Thr31′, Thr74′ and Asp88′ (Figure 2A). Thus the cavity between the 7th β-strand and 3rd/4th β-strand loop generates a more compact structure with a 0.6-0.7 Å backbone shift in the 6th and 7th β-strands of monomer 1′-99′. In addition, the Asp30′ side chain has moved closer to Thr74′, which increases the C-H…O contact distance between the carboxylate group of Asp30′ and methyl group of inhibitor 1 by 0.4 Å (Figure 2B). In contrast, the inward movement of the Asp88 side chain in the other monomer breaks the hydrogen bond interactions with the main chain of Thr74 and shifts the backbone of 6th and 7th β-strand by 0.5-0.6 Å away from residue 88 (Figure 2C). Furthermore, the side chain movement of Asp88 has created an 11° rotation at the carbonyl oxygen to form a shorter hydrogen bond with the hydroxyl group of Thr91 of 2.9 Å in PRN88D relative to 3.2 Å in the PRWT complex. This inward movement of the side chain of Asp88 was also observed in the mutant structures of PRN88D with substrate analog peptides CA/p2 and p2/NC (PDB ID 1FG8 and 1FG6).31 In the PRWT structure, two alternative conformations were modeled for the pyrrolidinone ring of inhibitor 1, while in the PRN88D structure only a single inhibitor conformation was observed. Despite these differences, the protease interaction with inhibitor 1 was not disturbed by the N88D mutation.
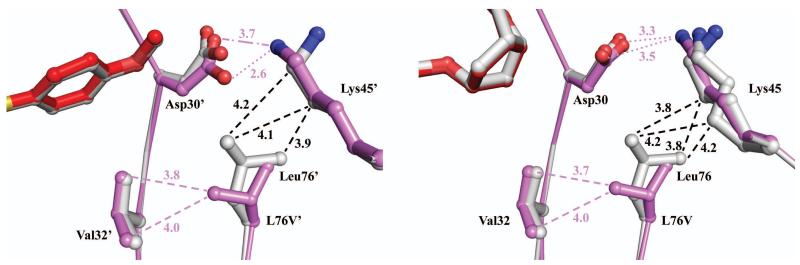
Leu76 is located in a hydrophobic environment toward the end of the 7th β-strand with no direct contact to the inhibitor (closest distance of >4.7 Å from P2/P2′ of inhibitor 1). Leu76 is positioned between Asp30 and Gln58 with its side chain directed toward Lys45 and Val56 of the flap. The Leu76 side chain forms multiple hydrophobic contacts with the side chains of Asp30, Val32, Lys45, Ile47, Val56, Gln58 and Thr74.20 The substitution of leucine to the shorter valine in the mutant eliminates several hydrophobic interactions and the side chain rotates by 180° to face Asp30. However, the mutant shows closer hydrophobic contacts between the side chains of the Val76 mutation and Val32 by up to 0.6 Å (Figure 3). Similar changes were also reported in the structures of PRL76V/DRV and PRL76V/SQV (PDB ID 3PWM and 3PWR).20 Aside from the changes in van der Waals interactions, the smaller size of the valine side chain allows movement of the side chains of Asp30/30′ and Lys45/45′ to form an ionic interaction with interatomic distances of 2.6-3.5 Å (Figure 3). A similar effect was seen in the structures of PRL76V/DRV and PRL76V/SQV, although there was a somewhat weaker ionic interaction (3.3-4.2 Å) between Asp30/30′ and Lys45/45′. Overall, the L76V and N88D mutations appear to disrupt the internal van der Waals contacts and hydrogen bonding network, which may alter the stability of the enzyme with minimal effect on the binding of inhibitors. In agreement with the loss of hydrophobic contacts, PRL76V exhibited increased sensitivity to urea and thermal denaturation as well as increased dimer dissociation relative to the PRWT.20
Figure 3.
Comparison of interactions of Leu76 and Val76. PRWT/1 is colored gray and PRL76V/1 is colored violet and red. Hydrogen bond interactions are denoted with dotted lines and van der Waals contacts are shown in dashed lines with interatomic distances in Å. Left panel, superposition of monomer 1′-99′ of PRWT and PRL76V. Right panel, superposition of monomer 1-99 of PRWT and PRL76V. Both panels illustrate the different side chain conformations of residue 76 and the ionic interaction of Asp30/Asp30′ and Lys45/Lys45′.
Effect of conservative mutation of I47V in the catalytic pocket
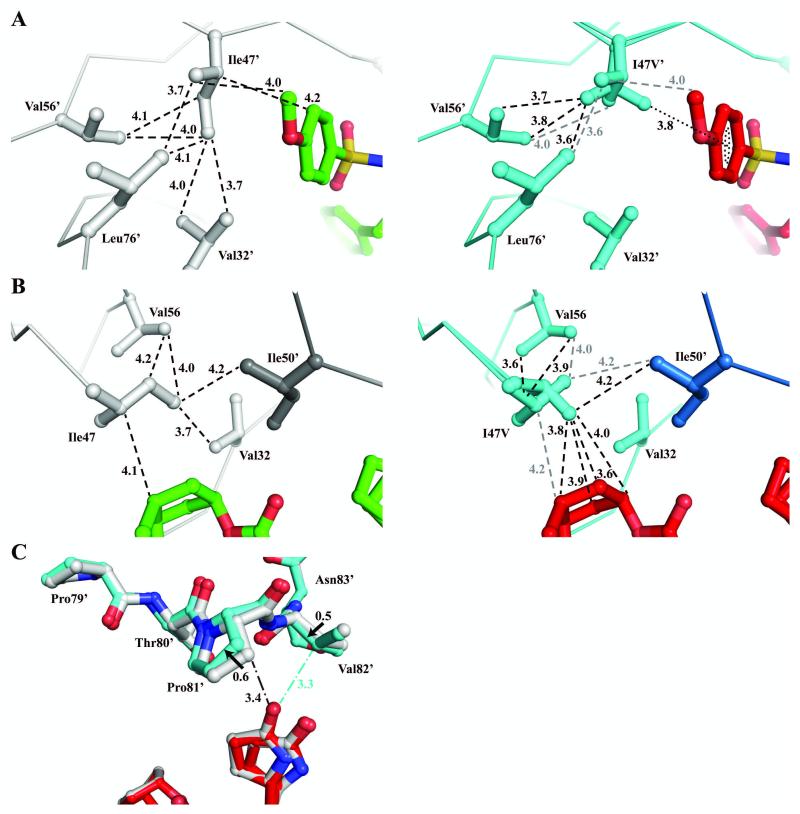
Residue Ile47 lies in the flap, which comprises two anti-parallel β-strands with most of the hydrophobic side chains extended into the catalytic pocket. Ile47 forms hydrophobic interactions with the side chains of Val32, Lys45, Ile54, Val56, Leu76 and Ile50′ from the other flap in the dimer. In addition, Ile47 forms van der Waals contacts with the p-methoxyphenyl and cyclopentanyltetrahydrofuran (Cp-THF) groups of inhibitor 1. Examination of various HIV protease complexes shows that the Ile47 side chain is maintained in a relatively fixed orientation with Cγ1 and Cγ2 positioned toward the side chain of Val56 and the Cδ methyl group extended toward the side chain of Val32 (Figure 4A).
Figure 4.
Comparison of interactions of Ile47 and Val47. PRWT/1 is colored in gray and green, and PRI47V/1 is colored in cyan and red. Van der Waals contacts are denoted by dashed lines and C-H…π interaction is indicated by dotted line with interatomic distances in Å. Only the major orientation of inhibitor is shown for clarity. C-H…O interactions are denoted by dash-dotted lines. (A) Left panel, PRWT monomer 1′-99′; Right panel, PRI47V monomer 1′-99′. (B) Left panel, PRWT monomer 1-99 and Ile50′ from other monomer shown in darker gray. Right panel, PRI47V monomer 1-99 and Ile50′ from other monomer are shown in lighter and darker blue, respectively. The grey dashed lines indicate interactions of the alternate conformation of Val47/47′ that is similar to Ile47 in PRWT. (C) Superposition of PRWT and PRI47V monomer 1′-99′ illustrates a movement in the 80’s loop and a shift of contacts between Pro81′, Val82′ and inhibitor 1. Solid arrows denote a shift between Cβ atoms of the two structures with distance in Å.
The drug-resistant mutations of Ile47 show very conservative changes with valine as the most frequent substitution. The loss of a methyl group from the isoleucine to valine substitution alters the van der Waals contacts and side chain conformation. In the PRI47V structure, the Val47 side chain has adopted two conformations with equal occupancy. One of the Val47 side chain conformations resembles the wild-type isoleucine with both methyl groups directed toward Val56. Despite this similarity, the mutated Val47 side chain has fewer van der Waals contacts with neighboring residues and abolished contacts with Val32. In the PRWT/1 complex, Ile47 and Ile47′ side chains have a total of 23 van der Waals contacts with the neighboring residues (11 contacts in S2 subsite and 12 contacts in S2′ subsite), whereas Val47 in the mutant has only 12 van der Waals contacts to the neighboring residues. In the other conformation, Val47 shows a significant movement at the main chain with both phi and psi angles rotated by 8-13° and the Cα is shifted by 0.5-0.6 Å. The side chain is rotated 142° and shifted by 0.8-1.0 Å at Cβ and the two methyl groups are shifted closer to Val56 and P2/P2′ of inhibitor. As a result, Val47 in the mutant has lost contacts with Lys45 and shows only 4-6 internal hydrophobic contacts with neighboring residues. Despite the loss of internal interactions, the new side chain position of the alternate conformation of Val47/47′ has gained hydrophobic contacts with the p-methoxyphenyl and Cp-THF of inhibitor 1 (Figure 4B).
Superposition of PRWT and PRI47V inhibitor complexes showed no significant change in the protein backbone with a RMSD of less than 0.2 Å. The only notable variation is seen at residues 79′-82′ of the 80’s loop with 0.6 Å shift at Cα of Pro81′. In the mutant structure, Pro81′ moves away from the oxygen of the pyrrolidinone ring by 0.5 Å and Val82′ moves 0.5 Å closer to the oxygen of the pyrrolidinone ring (Figure 4C). The pyrrolidinone ring has two alternate conformations with a 0.5/0.2 occupancy that favors interaction with the 80’s loop. The two different conformations of pyrrolidinone are also observed in the PRWT complex with a 0.4/0.4 occupancy. Overall, the inhibitor conformations and protease interactions are not significantly altered by the I47V mutation consistent with essentially identical inhibition of 0.4 nM (Table 2).
Effect of V82A mutation on inhibitor P1′ conformation
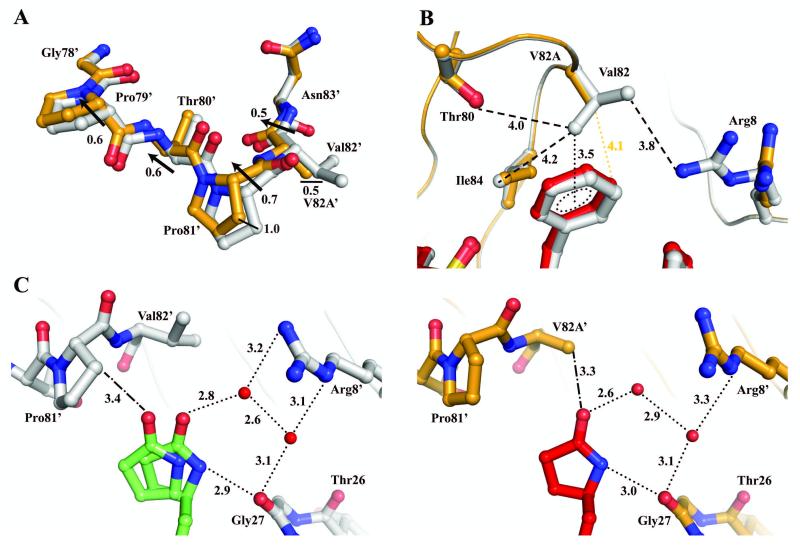
Mutations of Val82 are common in drug-resistant strains. Superposition of PRWT and PRV82A structures showed minimal changes in the backbone, except for the two surface loops formed by residues 16′-18′ and 79′-82′, which show a Cα deviation of 0.5-0.7 Å (Figure 5A). Residues 16′-18′ form the turn between the 2nd and 3rd β-strand at a region remote from the inhibitor binding site. Residues 79′-82′ are located on the loop between 7th and 8th β-strand and contribute direct contacts to the bound inhibitor. Residues from the 80’s loops and the flaps interact with the P1/P1′ groups of inhibitor 1. Val82 lies perpendicular to the P1 phenylalanine of inhibitor forming extensive C-H…π interactions (Figure 5B). In addition, the phenylalanine forms multiple van der Waals contacts with side chains of Asp25, Pro81, Ile84, Gly27′ and main chain of Gly48′, Gly49′, Ile50′ of the flap. The valine to alanine substitution changes the type of interaction from C-H...π to a single van der Waals contact at 4.1 Å from the P1 group.
Figure 5.
Comparison of interactions of Val82 and Ala82. PRWT/1 is colored in gray and green, and PRV82A/1 is colored in orange and red. Hydrogen bond and C-H…π interactions are denoted by dotted lines and van der Waals contacts are indicated by dashed lines with interatomic distances given in Å. C-H…O interactions are denoted by dashed-dotted lines. Black arrows denote a difference in Cα position between the two superimposed structures. Only the major orientation of inhibitor is shown for clarity. Red spheres represent water molecules. (A) Superposition of PRWT and PRV82A monomer 1′-99′ illustrates a movement in the 80’s loop. (B) PRWT/1 showing van der Waals contacts between Val82 and neighboring residues, and C-H…π interaction between Val82 and P1 group of inhibitor. PRV82A/1 loses van der Waals interactions and has increased distance from V82A to neighboring residues. (C) Left panel, PRWT/1 illustrates the alternative conformation of inhibitor P1′ group. Right panel, PRV82A/1 illustrates the single conformation of inhibitor P1′ group with hydrogen bond network to Gly27 and Arg8′.
In the wild-type S1′ subsite, the major orientation with 80% occupancy of inhibitor 1 shows two distinct conformations of the pyrrolidinone ring; one favors van der Waals interaction, and the other one favors hydrogen bond interaction with PRWT (Figure 5C). One conformation of the pyrrolidinone ring forms van der Waals contacts with Gly49 and Ile50 from the flap, Thr80′, Pro81′, Val82′and Ile84′ from the 80’s loop, and Gly27, Leu23′ and Asp25′ from the catalytic site. The alternative conformation has fewer van der Waals interactions and no contact with Pro81′ and Ile50. The loss of hydrophobic interactions is compensated by the two hydrogen bonds between the pyrrolidinone N15 and the main chain carbonyl oxygen of Gly27, and a water-mediated interaction between the O46 oxygen and Arg8′. In the mutant structure, only a single pyrrolidinone conformation is observed forming hydrogen bond interactions with the Gly27 and water molecules (Figure 5C). The valine to alanine substitution increases the flexibility of the 80’s loop so that the mutated side chain of Ala82′ no longer forms van der Waals contacts with Leu23′ and Ile84′ thus enlarging the S1′ pocket. This shift decreases hydrophobic interactions of the protease and P1′ pyrrolidinone with interatomic distances increased up to 0.7 Å. A similar movement in the 80’s loop was also observed in the PRV82A complexes with inhibitors DRV, IDV and SQV (PDB ID 2IDW, 1SDV and 2NMZ).30, 32, 33 Despite the variation in the P1′ conformation, the inhibition is not affected by the mutation as the pyrrolidinone ring maintains interactions with the mutant independent of changes at Pro81′ and Val82′/V82A′.
DISCUSSION AND CONCLUSIONS
Inhibitor 1 exerts potent activity against a wide spectrum of HIV-1 isolates and multidrug-resistant clinical strains.14, 15 Here, we present four high resolution crystal structures of 1 in complex with PRI47V, PRL76V, PRV82A and PRN88D bearing drug resistance mutations. All four single mutants shared comparable inhibition values as the wild-type enzyme with the largest difference of only 2-fold observed in the Ki of PRL76V and PRN88D. In both cases, the mutation increases the mobility of the Asp30 side chain, thus decreasing interactions with the inhibitor. The dynamic side chain position of Asp30 has been observed in various D30N27 single mutants and double mutants such as D30N/N88D and D30N/N88S34. This mutation has significantly reduced the effectiveness of the clinical PIs as observed from the kinetic studies.27 Moreover, D30N is one of the major mutations associated with NFV resistance.19, 35 Despite the variable position of the Asp30 side chain, the p-methoxysulfonamide in 1 maintains a hydrogen bond with the main chain amide of Asp30 and van der Waals contacts with side chain of Ala28 and Asp29. Although a mutation at residue 28 has been observed in the in vitro studies, mutations at Ala28 and Asp29 have not been identified in the clinical PI-resistance isolates.19, 36
The A28S mutation was identified in the UIC-94003 (also referred to as TMC-126)37 or GRL-139838 treated laboratory HIV-1 variants, suggesting an association between the presence of p-methoxysulfonamide in both compounds and the development of A28S resistance mutation. Interestingly, structural analysis and kinetic studies give a different result. Superposition of PRWT/1 and PRA28S/U-89360E (PDB ID 3H5B and 1AXA)39 suggests the A28S side chain may form multiple interactions with p-methoxysulfonamide and Cp-THF of inhibitor 1. Furthermore, kinetic studies showed the mutation of A28S significantly reduces the catalytic activity of the enzyme and viral infectivity, consistent with the absence of this mutation in clinical isolates.40, 41 A further study is needed to clarify the association of p-methoxysulfonamide and Ala28.
In the S2/S2′ subsite, Ile47 lies in the internal hydrophobic region with its side chain aligned parallel to the P2/P2′ group of inhibitor 1. In the mutant structure, I47V loses contacts with neighboring residues, which would potentially decrease the stability of the flaps as suggested by molecular dynamics simulations.42 In addition, the loss of the methyl group would likely affect the binding of substrates and other PIs. As observed in the PRWT/JE-2147 structure (PDB ID 1KZK)43, C5 of Ile47 forms multiple van der Waals contacts with the methylbenzamide group at P2′ position, and the isoleucine to valine mutation would reduce contacts with inhibitor JE-2147 as suggested by Yoshimaru.44 However, compound 1 remains a strong inhibitor of PRI47V. The Cp-THF in the P2 position forms multiple contacts with the flap residues Gly48 and Gly49, thus acting to stabilize the flap and S2 subsite. Notably, the PRI47V structure illustrates an important interaction between Val32 and Ile47.45 The Val32 side chain would potentially restrict the side chain conformations of Ile47 to maintain optimal van der Waals contacts among neighboring residues. This isoleucine-valine pair is retained in other lentivirus PR structures, although the residue types are reversed becoming Ile32 and Val47 in human immunodeficiency virus type 2 (HIV-2) PR, simian immunodeficiency virus (SIV) PR and many HIV-1 drug-resistant mutants.46
The S1/S1′ subsite is composed of the flap and 80s loop from each monomer. One unique feature is the flexibility of the 80s loop that accommodates different peptide or inhibitor groups at P1/P1′.47 A comparison of various inhibitor bound V82A mutant structures shows increased variability of the conformation of 80s loop, which suggested the Val82 side chain plays a role in stabilizing the loop. Val82 shows highly mobile Cγ1 and Cγ2 methyl groups in the electron density maps. The mobility of the Val82 side chain enables hydrophobic contacts with Arg8, Leu23 and/or Ile84, which cannot occur for the short alanine side chain in the PRV82A mutant. The diminished hydrophobic interactions of Ala82′ in the mutant shifts the 80’s loop farther away from the P1′ group of inhibitor 1. Interestingly, the rearrangement of the 80’s loop has a minimal effect on the inhibition. Instead of forming van der Waals contacts with Pro81′, the inhibitor 1 forms hydrogen bond interactions with Gly27 and Arg8′ side chain as described in the previous study of the wild-type complex14. The unusual ability of the P1′-pyrrolidinone ring to assume two alternate conformations has several advantages. First, the inhibitor is less dependent on the P1′ interaction with the 80s loop that is the location of several drug resistance mutations. Second, the inhibitor interactions with PR backbone atoms will be less affected by the high viral mutation rate. Lastly, Gly27 and Arg8′ provide a more stable inhibitor interaction as both residues are highly conserved and excluded from PI-resistant mutations seen in clinical isolates.
The new nonpeptidic PI 1 shows excellent nanomolar inhibition against four of the drug-resistant mutants with the Ki comparable to that of the wild-type enzyme. Moreover, it demonstrated potent antiviral activity against multidrug-resistant clinical variants of HIV-1.14, 15 The crystal structures of inhibitor 1 complexes with four of the major HIV-1 PR drug-resistant mutants, PRI47V, PRL76V, PRV82A and PRN88D, have provided new molecular insights for the interaction of the inhibitor and mutants. Importantly, this structural analysis demonstrates that the P1′ pyrrolidinone group is capable of adapting to fit the binding site of drug resistant mutants. Drug resistance mutations in PR often cause changes in the inhibitor binding site that weaken the PR-inhibitor interactions. One potential improvement to antiviral inhibitors might be the introduction of flexible groups, as observed for compound 1 where the alternate binding conformations of the P1′ pyrrolidinone ring within the S1′ subsite maintain strong inhibition of mutants. Moreover, flexible groups at the P2 or P2′ positions of the inhibitors would permit accommodation to mutations that alter the S2 and S2′ subsites. The flexible groups, however, might weaken the inhibition of wild-type PR. This information can improve the design of the next generation PIs and help to control the emerging multidrug-resistant HIV variants.
EXPERIMENTAL SECTION
General
Inhibitor 1 has shown analytical purity >99% by HPLC analysis.14 HPLC conditions: linear gradient from 25% acetonitrile and 75% water to 90% acetonitrile and 10% water over 20 min; flow rate 1.5 mL/min, XDB-C18 5 μM, 4.6 × 150 mm column, retention time 5.42 min. HRMS (m/z) calcd for C30H40N3O8S [M+H]+ 602.2536, found 602.2536.
Plasmid construction and protein purification
The HIV-1 PR (GenBank HIVHXB2CG) gene was cloned into pET11a expression vector (Novagen) and further optimized to restrict autoproteolysis (Q7K, L33I and L63I) and cysteine-thiol oxidation (C67A and C95A).1 HIV-1 PR point mutants (I47V, L76V, V82A and N88D) were constructed using optimized HIV-1 PR template with Quikchange site-directed mutagenesis kit (Stratagene). Plasmids were transformed into Escherichia coli BL21(DE3) for protein expression. The proteases were purified from inclusion bodies as described previously.48
Spectroscopic assay and determination of the inhibition constants
The kinetic parameters of HIV-1 PR were determined by monitoring the hydrolysis of the fluorescence substrate, Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 (where Abz is anthranilic acid, Nle is norleucine and p-nitro-Phe is p-Nitrophenylalanine), that is derived from the p2/NC cleavage site of the viral Gag-Pol polyprotein (Bachem Americas).49 The enzymatic assay was performed as follows; 10 μL protease to final concentration of 12-45 nM was pre-incubated with 98 μL of reaction buffer (0.1 M MES, pH 5.6, 0.4 M NaCl, 1 mM EDTA and 5% glycerol) and 2 μL of DMSO or inhibitor at 37 °C for 5 minutes. The reaction was initiated by addition of 90 μL anthranilyl to a final concentration of 6-96 μM and monitored over a period of 5 minutes using a BMG LABTECH POLARstar OPTIMA 96-well plate reader with excitation wavelength set at 340 nm and emission at 420 nm. The reactions were analyzed using FLUOstar OPTIMA 1.30-0 (BMG LABTECH). Values of Km and kcat were determined by fitting the data to the Michaelis-Menten equation using SigmaPlot 9.01 (Systat Software). Inhibitor Ki values were determined from the fluorescence assay of IC50 dose-response inhibition curve using the equation Ki = (IC50 − [E]/2)/(1 + [S]/Km), where [E] and [S] are protease and anthranilyl concentrations, respectively.48
Crystallographic analysis
PRI47V, PRL76V, PRV82A and PRN88D were co-crystallized with compound 1 by the hanging-drop vapor diffusion method at 23 °C. Protein (4.0-7.0 mg/mL) was pre-incubated with the inhibitor at 5-fold molar excess (1:5, protein-inhibitor ratio) for 30 minutes at 4 °C. Each crystallization drop contained equal volumes of protein and reservoir solution (0.1 M sodium acetate buffer, pH 4.2-5.4 and 0.6-1.5 M NaCl). The crystals grew within 1-3 days as bricks, plates or clusters. The cluster crystals were cut and separated into individual crystals before mounting. Prior to the diffraction experiment, crystals were soaked for 30 seconds in reservoir solution containing 30% (v/v) glycerol as cryoprotectant then flash-frozen in liquid nitrogen.
X-ray diffraction data were collected at the SER-CAT beamline of the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). All the experiments were done using a 100 K stream of gaseous nitrogen for crystal cooling. Redundant data sets were collected and processed with the HKL-2000 package.50 Initial phases for PRI47V, PRL76V, PRV82A and PRN88D were determined by molecular replacement using the CCP4 program suite.51 PRWT/DRV (PDB ID 2IEN)32 was used as the initial model for PRI47V and PRL76V. PRWT/1 (PDB ID 3H5B) was used as the initial model for PRV82A and PRN88D. Structural refinement was performed using SHELX52 with the molecular graphics program O53 and Coot54. Alternate conformations were modeled for the protease residues, inhibitor and solvent molecules based on the observed electron density maps. The relative occupancy was calculated during refinement. Anisotropic B-factors were refined for all atoms. Hydrogen atoms were added in the last round of refinement using SHELXL.52 The models were validated with SFCHECK51 and MolProbity55. The atomic coordinates and structure factors were deposited in the Protein Data Bank (PDB).56 Data collection and refinement statistics are listed in Table 1. Structural figures were generated using PyMOL program suite.57
ACKNOWLEDGEMENT
This work was supported in part by the Georgia State University Molecular Basis of Disease Program, the Georgia State University Research Program Enhancement award in Bioinformatics, the Georgia Research Alliance, and the National Institutes of Health grants GM062920 and GM053386. We thank Ting-Yi Chiu and Jing Wang for assisting with construction of PRI47V. We thank the staff at the SER6 CAT beamline at the Advanced Photon Source, Argonne National Laboratory, for assistance during X6 ray data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38.
ABBREVIATIONS
- APV
amprenavir
- ATV
atazanavir
- Cp6THF
cyclopentanyltetrahydrofuran
- DRV
darunavir
- FDA
Food and Drug Administration
- HAART
highly active antiretroviral therapy
- HIV-1
human immunodeficiency virus type 1
- HIV-2
human immunodeficiency virus type 2
- LPV
lopinavir
- NFV
nelfinavir
- PDB
Protein Data Bank
- PI
protease inhibitor
- PR
protease
- PRI47V
PR with I47V mutation
- PRL76V
PR with L76V mutation
- PRV82A
PR with V82A mutation
- PRN88D
PR with N88D mutation
- RMSD
root mean square deviation
- SIV
simian immunodeficiency virus
- SQV
saquinavir
- TPV
tipranavir
Footnotes
PROTEIN DATA BANK: Crystallographic data are available for inhibitor complexes with: PRI47V (PDB ID: 3VF5), PRL76V (PDB ID: 3VF7), PRV82A (PDB ID: 3VFA), and PRN88D (PDB ID: 3VFB).
REFERENCES
- 1.Louis JM, Clore GM, Gronenborn AM. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 1999;6:868–875. doi: 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 2.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RA, Scolnick EM, Sigal IS. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sepkowitz KA. AIDS66the first 20 years. N. Engl. J. Med. 2001;344:1764–1772. doi: 10.1056/NEJM200106073442306. [DOI] [PubMed] [Google Scholar]
- 5.Pomerantz RJ, Horn DL. Twenty years of therapy for HIV-1 infection. Nat. Med. (New York, NY, U.S.) 2003;9:867–873. doi: 10.1038/nm0703-867. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett JA, Fath MJ, Demasi R, Hermes A, Quinn J, Mondou E, Rousseau F. An updated systematic overview of triple combination therapy in antiretroviral-naive HIV-infected adults. AIDS. 2006;20:2051–2064. doi: 10.1097/01.aids.0000247578.08449.ff. [DOI] [PubMed] [Google Scholar]
- 7.Grabar S, Pradier C, Le Corfec E, Lancar R, Allavena C, Bentata M, Berlureau P, Dupont C, Fabbro-Peray P, Poizot-Martin I, Costagliola D. Factors associated with clinical and virological failure in patients receiving a triple therapy including a protease inhibitor. AIDS. 2000;14:141–149. doi: 10.1097/00002030-200001280-00009. [DOI] [PubMed] [Google Scholar]
- 8.Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954–958. doi: 10.1021/bi00119a002. [DOI] [PubMed] [Google Scholar]
- 9.Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J. Biol. Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 10.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Meyer S, Azijn H, Surleraux D, Jochmans D, Tahri A, Pauwels R, Wigerinck P, de Bethune MP. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 2005;49:2314–2321. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh AK, Kincaid JF, Cho W, Walters DE, Krishnan K, Hussain KA, Koo Y, Cho H, Rudall C, Holland L, Buthod J. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg. Med. Chem. Lett. 1998;8:687–690. doi: 10.1016/s0960-894x(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh AK, Dawson ZL, Mitsuya H. Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV. Bioorg. Med. Chem. 2007;15:7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh AK, Leshchenko-Yashchuk S, Anderson DD, Baldridge A, Noetzel M, Miller HB, Tie Y, Wang YF, Koh Y, Weber IT, Mitsuya H. Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1′-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2009;52:3902–3914. doi: 10.1021/jm900303m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koh Y, Das D, Leschenko S, Nakata H, Ogata-Aoki H, Amano M, Nakayama M, Ghosh AK, Mitsuya H. GRL-02031, a novel nonpeptidic protease inhibitor (PI) containing a stereochemically defined fused cyclopentanyltetrahydrofuran potent against multi-PI-resistant human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 2009;53:997–1006. doi: 10.1128/AAC.00689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wlodawer A, Gustchina A. Structural and biochemical studies of retroviral proteases. Biochim. Biophys. Acta. 2000;1477:16–34. doi: 10.1016/s0167-4838(99)00267-8. [DOI] [PubMed] [Google Scholar]
- 17.Weber IT, Agniswamy J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis JM, Ishima R, Torchia DA, Weber IT. HIV-1 protease: structure, dynamics, and inhibition. Adv. Pharmacol. 2007;55:261–298. doi: 10.1016/S1054-3589(07)55008-8. [DOI] [PubMed] [Google Scholar]
- 19.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, Schapiro JM, Richman DD. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 2010;18:156–163. [PubMed] [Google Scholar]
- 20.Louis JM, Zhang Y, Sayer JM, Wang YF, Harrison RW, Weber IT. The L7-V drug resistance mutation decreases the dimer stability and rate of autoprocessing of HIV-1 protease by reducing internal hydrophobic contacts. Biochemistry. 2011;50:4786–4795. doi: 10.1021/bi200033z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee SY, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young TP, Parkin NT, Stawiski E, Pilot-Matias T, Trinh R, Kempf DJ, Norton M. Prevalence, mutation patterns, and effects on protease inhibitor susceptibility of the L7-V mutation in HIV-1 protease. Antimicrob. Agents Chemother. 2010;54:4903–4906. doi: 10.1128/AAC.00906-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiesmann F, Vachta J, Ehret R, Walter H, Kaiser R, Sturmer M, Tappe A, Daumer M, Berg T, Naeth G, Braun P, Knechten H. The L7-V mutation in HIV-1 protease is potentially associated with hypersusceptibility to protease inhibitors Atazanavir and Saquinavir: is there a clinical advantage? AIDS Res. Ther. 2011;8:7. doi: 10.1186/1742-6405-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsuya Y, Winters MA, Fessel WJ, Rhee SY, Hurley L, Horberg M, Schiffer CA, Zolopa AR, Shafer RW. N88D facilitates the co-occurrence of D30N and L90M and the development of multidrug resistance in HIV type 1 protease following nelfinavir treatment failure. AIDS Res. Hum. Retroviruses. 2006;22:1300–1305. doi: 10.1089/aid.2006.22.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata J, Sugiura W, Ode H, Iwatani Y, Sato H, Tsang H, Matsuda M, Hasegawa N, Ren F, Tanaka H. Within-host co-evolution of Gag P453L and protease D30N/N88D demonstrates virological advantage in a highly protease inhibitor-exposed HIV-1 case. Antiviral Res. 2011;90:33–41. doi: 10.1016/j.antiviral.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Santos AF, Soares MA. The impact of the nelfinavir resistance-conferring mutation D30N on the susceptibility of HIV-1 subtype B to other protease inhibitors. Mem. Inst. Oswaldo Cruz. 2011;106:177–181. doi: 10.1590/s0074-02762011000200010. [DOI] [PubMed] [Google Scholar]
- 27.Kozisek M, Bray J, Rezacova P, Saskova K, Brynda J, Pokorna J, Mammano F, Rulisek L, Konvalinka J. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J. Mol. Biol. 2007;374:1005–1016. doi: 10.1016/j.jmb.2007.09.083. [DOI] [PubMed] [Google Scholar]
- 28.Mahalingam B, Louis JM, Reed CC, Adomat JM, Krouse J, Wang YF, Harrison RW, Weber IT. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 1999;263:238–245. doi: 10.1046/j.1432-1327.1999.00514.x. [DOI] [PubMed] [Google Scholar]
- 29.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J. Med. Chem. 2006;49:1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahalingam B, Wang YF, Boross PI, Tozser J, Louis JM, Harrison RW, Weber IT. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur. J. Biochem. 2004;271:1516–1524. doi: 10.1111/j.1432-1033.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins: Struct., Funct., Genet. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 32.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 33.Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins: Struct., Funct., Bioinf. 2007;67:232–242. doi: 10.1002/prot.21304. [DOI] [PubMed] [Google Scholar]
- 34.Bihani SC, Das A, Prashar V, Ferrer JL, Hosur MV. Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S. Biochem. Biophys. Res. Commun. 2009;389:295–300. doi: 10.1016/j.bbrc.2009.08.138. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Picado J, Savara AV, Sutton L, D’Aquila RT. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura K, Kato R, Kavlick MF, Nguyen A, Maroun V, Maeda K, Hussain KA, Ghosh AK, Gulnik SV, Erickson JW, Mitsuya H. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 2002;76:1349–1358. doi: 10.1128/JVI.76.3.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ide K, Aoki M, Amano M, Koh Y, Yedidi RS, Das D, Leschenko S, Chapsal B, Ghosh AK, Mitsuya H. Novel HIV-1 protease inhibitors (PIs) containing a bicyclic P2 functional moiety, tetrahydropyrano-tetrahydrofuran, that are potent against multi-PI-resistant HIV-1 variants. Antimicrob. Agents Chemother. 2011;55:1717–1727. doi: 10.1128/AAC.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong L, Hartsuck JA, Foundling S, Ermolieff J, Tang J. Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant. Protein Sci. 1998;7:300–305. doi: 10.1002/pro.5560070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ido E, Han HP, Kezdy FJ, Tang J. Kinetic studies of human immunodeficiency virus type 1 protease and its active-site hydrogen bond mutant A28S. J. Biol. Chem. 1991;266:24359–24366. [PubMed] [Google Scholar]
- 41.Rose JR, Babe LM, Craik CS. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bandyopadhyay P, Meher BR. Drug resistance of HIV-1 protease against JE-2147: I47V mutation investigated by molecular dynamics simulation. Chem. Biol. Drug Des. 2006;67:155–161. doi: 10.1111/j.1747-0285.2006.00348.x. [DOI] [PubMed] [Google Scholar]
- 43.Reiling KK, Endres NF, Dauber DS, Craik CS, Stroud RM. Anisotropic dynamics of the JE-2147-HIV protease complex: drug resistance and thermodynamic binding mode examined in a 1.09 A structure. Biochemistry. 2002;41:4582–4594. doi: 10.1021/bi011781z. [DOI] [PubMed] [Google Scholar]
- 44.Yoshimura K, Kato R, Yusa K, Kavlick MF, Maroun V, Nguyen A, Mimoto T, Ueno T, Shintani M, Falloon J, Masur H, Hayashi H, Erickson J, Mitsuya H. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8675–8680. doi: 10.1073/pnas.96.15.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao W, Everitt L, Manchester M, Loeb DD, Hutchison CA, 3rd, Swanstrom R. Sequence requirements of the HIV-1 protease flap region determined by saturation mutagenesis and kinetic analysis of flap mutants. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2243–2248. doi: 10.1073/pnas.94.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagossi P, Sperka T, Feher A, Kadas J, Zahuczky G, Miklossy G, Boross P, Tozser J. Amino acid preferences for a critical substrate binding subsite of retroviral proteases in type 1 cleavage sites. J. Virol. 2005;79:4213–4218. doi: 10.1128/JVI.79.7.4213-4218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munshi S, Chen Z, Yan Y, Li Y, Olsen DB, Schock HB, Galvin BB, Dorsey B, Kuo LC. An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2000;56:381–388. doi: 10.1107/s0907444900000469. [DOI] [PubMed] [Google Scholar]
- 48.Maibaum J, Rich DH. Inhibition of porcine pepsin by two substrate analogues containing statine. The effect of histidine at the P2 subsite on the inhibition of aspartic proteinases. J. Med. Chem. 1988;31:625–629. doi: 10.1021/jm00398a022. [DOI] [PubMed] [Google Scholar]
- 49.Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J. Mol. Biol. 2008;381:102–115. doi: 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW Jr., Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 51.Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Sheldrick GM. A short history of SHELX. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 53.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., Sect. A: Found. Crystallogr. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The PyMOL Molecular Graphics System, Version 1.3. LLC; Schrödinger: [Google Scholar]