Abstract
Five organisms having completely sequenced genomes and belonging to all major branches of green plants (Viridiplantae) were analyzed with respect to their content of P-type ATPases encoding genes. These were the chlorophytes Ostreococcus tauri and Chlamydomonas reinhardtii, and the streptophytes Physcomitrella patens (a non-vascular moss), Selaginella moellendorffii (a primitive vascular plant), and Arabidopsis thaliana (a model flowering plant). Each organism contained sequences for all five subfamilies of P-type ATPases. Whereas Na+ and H+ pumps seem to mutually exclude each other in flowering plants and animals, they co-exist in chlorophytes, which show representatives for two kinds of Na+ pumps (P2C and P2D ATPases) as well as a primitive H+-ATPase. Both Na+ and H+ pumps also co-exist in the moss P. patens, which has a P2D Na+-ATPase. In contrast to the primitive H+-ATPases in chlorophytes and P. patens, the H+-ATPases from vascular plants all have a large C-terminal regulatory domain as well as a conserved Arg in transmembrane segment 5 that is predicted to function as part of a backflow protection mechanism. Together these features are predicted to enable H+ pumps in vascular plants to create large electrochemical gradients that can be modulated in response to diverse physiological cues. The complete inventory of P-type ATPases in the major branches of Viridiplantae is an important starting point for elucidating the evolution in plants of these important pumps.
Keywords: evolution, P-type ATPases, plants, salt tolerance, Na+ pumps
Introduction
P-type ATPases are primary transporters energized by hydrolysis of ATP with a wide range of specificities for small cations and apparently also phospholipids (Møller et al., 1996; Palmgren and Harper, 1999). P-type ATPases are characterized by forming a phosphorylated intermediate (hence the name P-type), by being inhibited by vanadate, and by having a number of sequence motifs in common (Serrano, 1989; Axelsen and Palmgren, 1998). Plant P-type ATPases are characterized structurally by having a single catalytic subunit, 8–12 transmembrane segments, N and C termini exposed to the cytoplasm, and a large central cytoplasmic domain including the phosphorylation and ATP binding sites.
P-type ATPases constitute a large and indispensable family in most organisms. The P-type ATPase family can be divided into five major evolutionarily related subfamilies, P1–P5, which group in a phylogenetic tree according to the ions they transport (Axelsen and Palmgren, 1998). The P-type ATPases are involved in a wide range of fundamental cellular processes such as the efflux or organismal redistribution of micronutrients (P1B Zn2+- and Cu2+-ATPases), cellular signaling and Ca2+ compartmentalization (P2A and P2B Ca2+-ATPases), energizing the electrochemical gradient used as the driving force for the secondary transporters (P3A H+-ATPases in plants and fungi and P2C Na+/K+-ATPases in animals), and being involved in membrane vesicle budding (P4 ATPases). The function of P5 ATPases is not known but they have been implicated in vesicle budding from the endoplasmic reticulum (Poulsen et al., 2008a).
The bioenergetic systems of flowering plants and animals use different coupling ions (Skulachev, 1988; Rodríguez-Navarro, 2000). In animals, the potential energy that can be harvested to drive, e.g., nutrient transport across the plasma membrane derives from a Na+ gradient across this membrane. A Na+ pump in the plasma membrane is a very efficient system for extrusion of toxic Na+ (Gonzalez, 2011; Whittamore, 2012). A plasma membrane Na+ pump in the plasma membrane is also important for the extrusion of toxic Na+. Fish and invertebrates living in the salty oceans are dependent on such a pump for survival. For animals living in marine environments a Na+/K+ pumps (P2C ATPases) remain the sole system for formation of electrochemical ion gradients gradients in the plasma membrane (Morth et al., 2011).
In contrast, in plant and fungal cells, it is an electrochemical gradient of H+ that energizes the plasma membrane (Morth et al., 2011). In cells of flowering plants, the plasma membrane completely lacks Na+ pumps and depends entirely on plasma membrane H+ pumps for establishing a steep electrochemical ion gradient across the plasma membrane. In those flowering plants in which salt tolerance mechanisms have been investigated, strategies other than primary active extrusion of Na+ (i.e., efflux through a Na+ pump) appear to have evolved (Tester and Davenport, 2003; Flowers and Colmer, 2008). Unfortunately, most flowering plants remain very sensitive to saline environments and salinization of soils due to extensive irrigation is becoming an increasing problem for productivity of agriculture, especially in arid regions of the world (Tester and Davenport, 2003; Yamaguchi and Blumwald, 2005).
The apparent absence of Na+ pumps in flowering plants raises several questions. In ancestral plants containing both Na+ and H+ pumps, did both pumps help energize the plasma membrane, and did those membranes function with co-transport systems that could utilize both Na+ and H+ gradients? During the evolution of flowering plants, was there a physiological reason why plasma membranes might not function well with both Na+ and H+ pumps? Did the flowering plant lineage evolve from an ancestor living exclusively in a fresh water environment, or were Na+ pumps gradually lost at later points in evolution? Is it possible that Na+ pumps are still present in some flowering plants that have not yet been studied?
With the completion of genome sequences from all major branches of Viridiplantae, it is now possible to study the evolution of primary transport capabilities throughout the green plant lineage and to compare these transporters with those of protists, animals, and fungi. Our analysis of five selected plant genomes suggest that Na+ pumps coexisted with primitive plasma membrane H+ pumps in both early aquatic and terrestrial plants. However, at some point in the evolution of vascular plants, Na+ pumps appear to have been lost. The evolution of multicellular land plants also correlates with an expansion in number and potential regulatory features for H+ pumps, Ca2+ pumps, heavy metal pumps, and lipid flippases. In contrast, the P5 subfamily did not show an equivalent change, with only one or two isoforms present in all five plant lineages. This study provides an evolutionary framework for considering how P-type ATPases contribute to the biology of all Viridiplantae, from single celled algae to multicellular flowering plants.
Materials and Methods
The P-type ATPase sequences were identified by searching for sequences in UniProtKB (The UniProt Consortium, 2012) and in Phytozome1 from the relevant genomes that matched the PFAM profile PF00122 and in UniProtKB also the PROSITE pattern PS00154 (DKTG[T,S][L,I,V,M][T,I]; Axelsen and Palmgren, 1998). The PROSITE pattern is unique for P-type ATPases while the PFAM profile is more inclusive and also matches sequences that cannot function as active P-type ATPases as they lack the phosphorylated Asp residue that is present in the PROSITE pattern. Since the sequences in Phytozome do not include information about matches to PROSITE patterns, only PFAM was used to select sequences from this resource. The identified sequences were aligned and duplicates were removed. All genomes except Arabidopsis are still draft versions in the databases, so often the predicted P-type ATPases do not represent complete proteins. Furthermore, the versions identified in UniProtKB and in Phytozome were not 100% identical. When possible, the UniProtKB sequence was chosen.
The resulting dataset of 150 sequences were aligned using Muscle (Edgar, 2004). The resulting alignment of the 150 full-length sequences was used to construct a phylogenetic tree using the Neighbor Joining method as implemented in QuickTree (Howe et al., 2002). The standard parameters of Muscle and QuickTree were used. For visualization of the constructed tree we used Dendroscope (Huson et al., 2007). A phylogenetic tree for each subgroup (P1–P5) of the 150 sequences was constructed and visualized similarly. For each subgroup, the corresponding set of sequences was selected from the dataset of 150 sequences. The selected sequences and a few additional outlier sequences were aligned using Muscle and a phylogenetic tree was constructed using QuickTree. The trees for the individual subfamilies are all rooted with the sequence of the E. coli P1A ATPase KdpB (P03960) as outgroup. The 150 sequences as well as sequences for the outliers employed are available as Fasta files in Supplementary Information.
Results
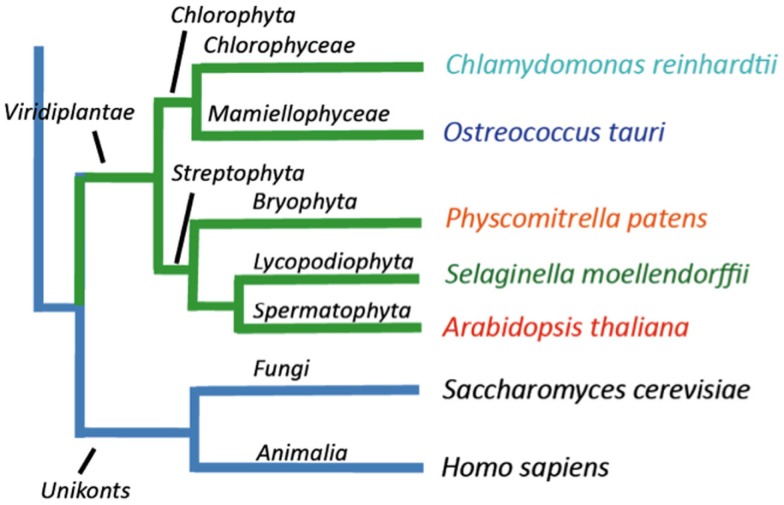
In this work we identified genes encoding P-type ATPases in the sequenced genomes from five representatives of the green plant lineage (Viridiplantae), which previously diverged from opisthokonts (animals, fungi, and Choanozoa; Yoon et al., 2004; Figure 1). The Chlorophytae (green algae, including Chlamydomonas and Ostreococcus) diverged from the Streptophytae (land plants and their close relatives; Figure 1) over a billion years ago. In this work, the genomes of two green algae were analyzed, namely those of Ostreococcus tauri (Derelle et al., 2006) and Chlamydomonas reinhardtii (Merchant et al., 2007), and three land plants, the moss Physcomitrella patens (Rensing et al., 2008), the primitive vascular plant Selaginella moellendorffii2 and the flowering plant Arabidopsis thaliana (The Arabidopsis Genome Initiative, 2000). The latter has previously been investigated for its content of P-type ATPases (Axelsen and Palmgren, 2001).
Figure 1.
Overview of the evolutionary relationship between the green plant genomes analyzed in this study.
Ostreococcus tauri is an extremely small (0.8 μm wide) unicellular green alga, which belongs to the Prasinophyceae, one of the most ancient groups within the lineage of green plants (Courties et al., 1998). This organism is a naked, non-flagellated cell possessing a single mitochondrion and a single chloroplast, and a common member of global oceanic picoplankton populations. C. reinhardtii is a much bigger (∼10 μm), likewise unicellular green alga. It lives in terrestrial soils and has multiple mitochondria, two anterior flagella for motility and mating, and a chloroplast (Rochaix, 1995). P. patens is a moss, a representative for primitive land plants without a vascular system. S. moellendorffii is a member of an ancient vascular plant lineage that first appears in the fossil record about 400 million years ago (Banks, 2009). A. thaliana was the first angiosperm to have its genome sequenced, and is a model plant for understanding the molecular biology of flowering plants.
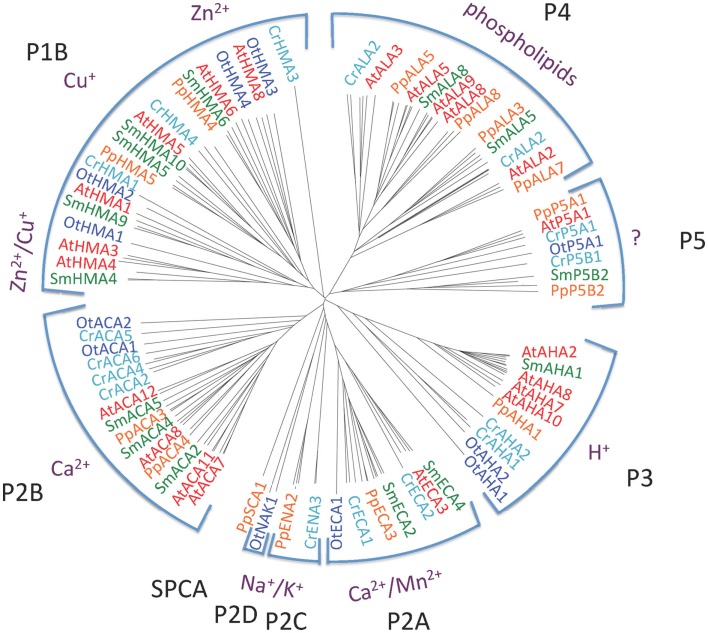
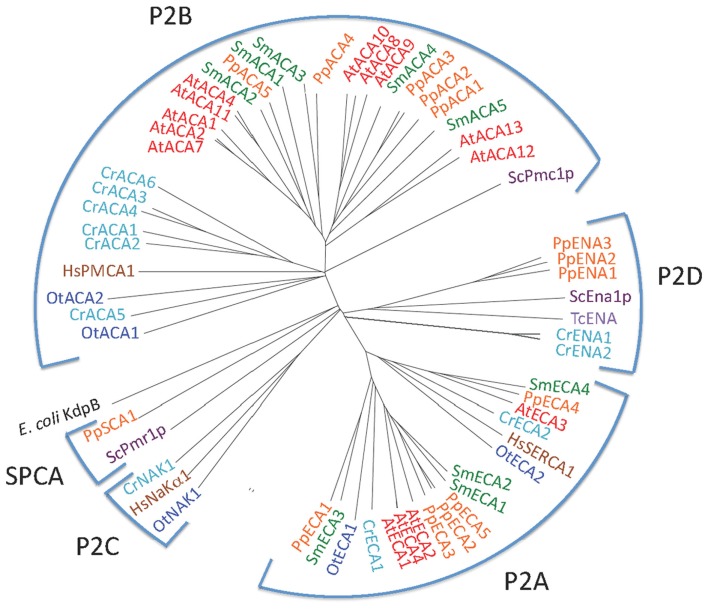
Figure 2 shows the phylogenetic relationship of all the 150 sequences listed in Table 2, which were evaluated in this study. They are distributed in all five major families of P-type ATPases with some noteworthy comparisons discussed below.
Figure 2.
Phylogenetic tree of P-type ATPases analyzed in this study. Not all branches are labeled. Accession numbers for sequences are given in Table 2.
Table 2.
List of P-type ATPases analyzed in this study.
| Class | Gene | aa | Accession # | Locus/ORF |
|---|---|---|---|---|
| OSTREOCOCCUS TAURI | ||||
| P1B | OtHMA1 | 1052 | Q01EZ3 | Ot02g01930 |
| P1B | OtHMA2 | 681 | Q00V551 | Ot15g01530 |
| P1B | OtHMA3 | 1359 | Q014R9 | Ot07g03370 |
| P1B | OtHMA4 | 861 | Q00YQ6 | Ot11g02480 |
| P1B | OtHMA5 | 925 | Q018N8 | Ot05g03820 |
| P2A | OtECA1 | 561 | Q01FX01 | Ot01g04600 |
| P2A | OtECA2 | 1013 | Q01C29 | Ot03g04740 |
| P2B | OtACA1 | 1062 | Q011R1 | Ot09g02100 |
| P2B | OtACA2 | 780 | Q00U351 | Ot16g02150 |
| P2C | OtNAK1 | 1172 | Q01GZ3 | Ot01g00900 |
| P3A | OtAHA1 | 1043 | Q00RY32 | Ot20g00330 |
| P3A | OtAHA2 | 879 | Q017J63 | Ot06g01560 |
| P4 | OtALA1 | 1258 | Q016N2 | Ot06g03680 |
| P5A | OtP5A1 | 1398 | Q00 × 11 | Ot13g01720 |
| CHLAMYDOMONAS REINHARDTII | ||||
| P1B | CrHMA1 | 1189 | A8J6D5 | CHLREDRAFT_195998 |
| P1B | CrHMA2/CTP2 | 1086 | A8IC93 | CHLREDRAFT_205938 |
| P1B | CrHMA3/CTP3 | 504 | A8ICL14 | CHLREDRAFT_165985 |
| P1B | CrHMA4/CTP1 | 1097 | A8JBB5 | CHLREDRAFT_206047 |
| P1B | CrHMA5 | 1041 | A8J829 | CHLREDRAFT_195962 |
| P2A | CrECA1 | 1069 | A8I542 | CHLREDRAFT_77047 |
| P2A | CrECA2 | 845 | g7623505,6 | Cre22.g762350 |
| P2B | CrACA1 | 1179 | A8IZL7 | CHLREDRAFT_196702 |
| P2B | CrACA2 | 1009 | A8IJV9 | CHLREDRAFT_128099 |
| P2B | CrACA3/FAB39 | 930 | A8J0V21 | CHLREDRAFT_118223 |
| P2B | CrACA4 | 1191 | A8IS11 | CHLREDRAFT_189266 |
| P2B | CrACA5 | 873 | A8JEM47 | CHLREDRAFT_179136 |
| P2B | CrACA6 | 1434 | A8HM60/g0363508 | CHLREDRAFT_116583/Cre01.g036350 |
| P2C | CrNAK1 | 1112 | A8HX15 | CHLREDRAFT_187139 |
| P2D | CrENA1 | 1004 | A8J4M6 | CHLREDRAFT_104709 |
| P2D | CrENA2 | 1001 | A8J4M4 | CHLREDRAFT_130931 |
| P3A | CrAHA1/CrPMH1 | 1081 | A8IFK0/Q9FNS3 | CHLREDRAFT_54949 |
| P3A | CrAHA2 | 802 | A8IFH01 | CHLREDRAFT_38137 |
| P3A | CrAHA3/CrPMA2 | 1081 | Q93Z22 | CHLREDRAFT_182602 |
| P3A | – | 1628 | g3169509 | Cre07.g316950 |
| P4 | CrALA1 | 1183 | A8IVJ3 | CHLREDRAFT_172401 |
| P4 | CrALA2 | 1300 | A8IVJ6 | CHLREDRAFT_190292 |
| P4 | CrALA3 | 1243 | A8J8G96 | CHLREDRAFT_193025 |
| P5A | CrP5A1 | 1168 | A8JI261 | CHLREDRAFT_60708 |
| P5B | CrP5B1 | 1308 | A8I9J21 | CHLREDRAFT_186680 |
| PHYSCOMITRELLA PATENS | ||||
| P1B | PpHMA1 | 902 | A9RNK6 | PHYPADRAFT_117222 |
| P1B | PpHMA2 | 743 | A9S8B91 | PHYPADRAFT_125638 |
| P1B | PpHMA3 | 1004 | A9T8Q3 | PHYPADRAFT_192723 |
| P1B | PpHMA4 | 841 | A9SUQ2 | PHYPADRAFT_215914 |
| P1B | PpHMA5 | 1009 | A9SME3 | PHYPADRAFT_81365 |
| P1B | PpHMA6 | 893 | A9TB46 | PHYPADRAFT_142921 |
| P1B | PpHMA7 | 743 | A9TQB5 | PHYPADRAFT_148958 |
| P2A | PpECA1 | 1060 | A9RZK8 | PHYPADRAFT_179791 |
| P2A | PpECA2 | 1039 | A9SH40 | PHYPADRAFT_184915 |
| P2A | PpECA3 | 1055 | A9SHQ4 | PHYPADRAFT_212461 |
| P2A | PpECA4 | 1000 | A9TIL4 | PHYPADRAFT_222630 |
| P2A | PpECA5 | 1037 | A9TK26 | PHYPADRAFT_223041 |
| P2A | PpSCA1 | 822 | A9T9F0 | PHYPADRAFT_220109 |
| P2B | PpACA1/PCA1 | 1098 | Q70TF0 | PHYPADRAFT_202276 |
| P2B | PpACA2/PCA2 | 1105 | Q70TF1 | PHYPADRAFT_224496 |
| P2B | PpACA3 | 1074 | A9SLT6 | PHYPADRAFT_230135 |
| P2B | PpACA4 | 948 | A9RXA710 | PHYPADRAFT_121055 |
| P2B | PpACA5 | 1035 | A9RZJ8 | PHYPADRAFT_121834 |
| P2D | PpENA1 | 967 | Q7XB51 | |
| P2D | PpENA2 | 1058 | Q7XB50 | |
| P2D | PpENA3 | 963 | C1L359 | PHYPADRAFT_112089 |
| P3A | PpAHA1 | 936 | A9U0N9 | PHYPADRAFT_153928 |
| P3A | PpAHA2 | 465 | A9TU441 | PHYPADRAFT_198308 |
| P4 | PpALA1 | 1219 | A9RVW0 | PHYPADRAFT_205967 |
| P4 | PpALA2 | 1062 | A9S030 | PHYPADRAFT_121975 |
| P4 | PpALA3 | 1229 | A9S076 | PHYPADRAFT_122321 |
| P4 | PpALA4 | 1194 | A9SKC3 | PHYPADRAFT_165384 |
| P4 | PpALA5 | 1251 | A9SY94 | PHYPADRAFT_189702 |
| P4 | PpALA6 | 1262 | A9T6U6 | PHYPADRAFT_88857 |
| P4 | PpALA7 | 1104 | A9T776 | PHYPADRAFT_168461 |
| P4 | PpALA8 | 1151 | A9TDQ8 | PHYPADRAFT_221270 |
| P5A | PpP5A1 | 1178 | A9RG70 | PHYPADRAFT_113743 |
| P5B | PpP5B1 | 435 | A9TN061 | PHYPADRAFT_147950 |
| P5B | PpP5B2 | 1290 | A9SGC2 | PHYPADRAFT_229474 |
| SELAGINELLA MOELLENDORFFII11 | ||||
| P1B | SmHMA1 | 819 | D8RBL1 | SELMODRAFT_89397 |
| P1B | SmHMA2 | 696 | D8QRZ31 | SELMODRAFT_60690 |
| P1B | SmHMA3 | 952 | D8SD62 | SELMODRAFT_114297 |
| P1B | SmHMA4 | 831 | D8SJM4 | SELMODRAFT_118425 |
| P1B | SmHMA5 | 1018 | D8SPX5 | SELMODRAFT_122320 |
| P1B | SmHMA6 | 910 | D8TEP8 | SELMODRAFT_138129 |
| P1B | SmHMA7 | 953 | D8RFP0 | SELMODRAFT_92276 |
| P1B | SmHMA8 | 790 | D8QYH61 | SELMODRAFT_60775 |
| P1B | SmHMA9 | 817 | D8R2L0 | SELMODRAFT_167322 |
| P1B | SmHMA10 | 960 | D8R2W8 | SELMODRAFT_84115 |
| P2A | SmECA1 | 1047 | D8RSK1 | SELMODRAFT_267711 |
| P2A | SmECA2 | 1042 | D8RUH8 | SELMODRAFT_102055 |
| P2A | SmECA3 | 1045 | D8SA88 | SELMODRAFT_112465 |
| P2A | SmECA4 | 1009 | D8SUV3 | SELMODRAFT_158488 |
| P2B | SmACA1 | 1014 | D8QTC2 | SELMODRAFT_437746 |
| P2B | SmACA2 | 1030 | D8QTC3 | SELMODRAFT_266601 |
| P2B | SmACA3 | 1068 | D8T1F8 | SELMODRAFT_129812 |
| P2B | SmACA4 | 1105 | D8S012 | SELMODRAFT_451372 |
| P2B | SmACA5 | 1069 | D8T4Q6 | SELMODRAFT_451597 |
| P3A | SmAHA1 | 875 | D8T8I0 | SELMODRAFT_430150 |
| P3A | SmAHA2 | 952 | D8QT851 | SELMODRAFT_76771 |
| P4 | SmALA1 | 1207 | D8QMQ3 | SELMODRAFT_164122 |
| P4 | SmALA2 | 1157 | D8R8G6 | SELMODRAFT_86830 |
| P4 | SmALA3 | 1109 | D8RG22 | SELMODRAFT_410847 |
| P4 | SmALA4 | 1009 | D8RKR6 | SELMODRAFT_95836 |
| P4 | SmALA5 | 1208 | D8S239 | SELMODRAFT_107016 |
| P4 | SmALA6 | 1221 | D8SBS1 | SELMODRAFT_113552 |
| P4 | SmALA7 | 1153 | D8SGU6 | SELMODRAFT_116847 |
| P4 | SmALA8 | 1184 | D8TF22 | SELMODRAFT_138337 |
| P5A | SmP5A1 | 1109 | D8TBK6 | SELMODRAFT_187116 |
| P5B | SmP5B1 | 1290 | D8SFE9 | SELMODRAFT_445079 |
| P5B | SmP5B2 | 1246 | D8R5P0 | SELMODRAFT_439526 |
| ARABIDOPSIS THALIANA | ||||
| P1B | AtHMA1 | 819 | Q9M3H5 | At4g37270 |
| P1B | AtHMA2 | 951 | Q9SZW4 | At4g30110 |
| P1B | AtHMA3 | 760 | P0CW78 | At4g30120 |
| P1B | AtHMA4 | 1172 | O64474 | At2g19110 |
| P1B | AtHMA5 | 995 | Q9SH30 | At1g63440 |
| P1B | AtHMA6/PAA1 | 949 | Q9SZC9 | At4g33520 |
| P1B | AtHMA7/RAN1 | 1001 | Q9S7J8 | At5g44790 |
| P1B | AtHMA8/PAA2 | 856 | Q9C594 | At5g21930 |
| P2A | AtECA1 | 1061 | P92939 | At1g07810 |
| P2A | AtECA2 | 1054 | O23087 | At4g00900 |
| P2A | AtECA3 | 998 | Q9SY55 | At1g10130 |
| P2A | AtECA4 | 1061 | Q9XES1 | At1g07670 |
| P2B | AtACA1 | 1020 | Q37145 | At1g27770 |
| P2B | AtACA2 | 1014 | O81108 | At4g37640 |
| P2B | AtACA4 | 1030 | O22218 | At2g41560 |
| P2B | AtACA7 | 1015 | O64806 | At2g22950 |
| P2B | AtACA8 | 1074 | Q9LF79 | At5g57110 |
| P2B | AtACA9 | 1086 | Q9LU41 | At3g21180 |
| P2B | AtACA10 | 1069 | Q9SZR1 | At4g29900 |
| P2B | AtACA11 | 1025 | Q9M2L4 | At3g57330 |
| P2B | AtACA12 | 1033 | Q9LY77 | At3g63380 |
| P2B | AtACA13 | 1017 | Q9LIK7 | At3g22910 |
| P2B | – | 1049 | F4KHQ212 | At5g53010 |
| P3A | AtAHA1 | 949 | P20649 | At2g18960 |
| P3A | AtAHA2 | 948 | P19456 | At4g30190 |
| P3A | AtAHA3 | 949 | P20431 | At5g57350 |
| P3A | AtAHA4 | 960 | Q9SU58 | At3g47950 |
| P3A | AtAHA5 | 949 | Q9SJB3 | At2g24520 |
| P3A | AtAHA6 | 949 | Q9SH76 | At2g07560 |
| P3A | AtAHA7 | 961 | Q9LY32 | At3g60330 |
| P3A | AtAHA8 | 948 | Q9M2A0 | At3g42640 |
| P3A | AtAHA9 | 954 | Q42556 | At1g80660 |
| P3A | AtAHA10 | 947 | Q43128 | At1g17260 |
| P3A | AtAHA11 | 956 | Q9LV11 | At5g62670 |
| P3A | – | 813 | Q9T0E013 | At4g11730 |
| P4 | AtALA1 | 1158 | P98204 | At5g04930 |
| P4 | AtALA2 | 1107 | P98205 | At5g44240 |
| P4 | AtALA3 | 1213 | Q9XIE6 | At1g59820 |
| P4 | AtALA4 | 1216 | Q9LNQ4 | At1g17500 |
| P4 | AtALA5 | 1228 | Q9SGG3 | At1g72700 |
| P4 | AtALA6 | 1240 | Q9SLK6 | At1g54280 |
| P4 | AtALA7 | 1247 | Q9LVK9 | At3g13900 |
| P4 | AtALA8 | 1189 | Q9LK90 | At3g27870 |
| P4 | AtALA9 | 1200 | Q9SX33 | At1g68710 |
| P4 | AtALA10 | 1202 | Q9LI83 | At3g25610 |
| P4 | AtALA11 | 1203 | Q9SAF5 | At1g13210 |
| P4 | AtALA12 | 1184 | P57792 | At1g26130 |
| P5A | AtP5A1 | 1179 | Q9LT02 | At5g23630 |
1Fragment.
2A possible chimera. The last 200 amino acid residues do not match AtAHA4. Similarity ends at the position equivalent to position 862 in AtAHA4.
3Possibly a fragment. Maybe 80 amino acid residues missing from C-terminus (when compared to AtAHA2). The similarity extends to position 864 in AtAHA2.
4A possible fragment or pseudogene. When compared to AtHMA8/PAA1, positions 523–827 are missing (this includes the motif DKTGT). The last 60 amino acid residues are also missing.
5200 amino acid residues from the C-terminal are missing.
6No corresponding UniProtKB entry.
7When compared to AtACA9, amino acid residues 1–170 are missing.
8Fragment with insert. Similar to AtACA10 (107–529 + 576–1046; the positions on C. reinhardtii 3–462 + 856–1347). The UniProtKB entry only covers part of the protein taken from Phytozome.
9Possibly a pseudogene or chimera. Partly covered by A8I6H0 (460–576) and A8I6H2 (1521–1622). Many regions of low complexity. Similar to AtAHA10 (58–209 + 584–974, AtAHA10 numbering).
10First 130 aa missing compared to AtACA8.
11Near identical copies of most S. moellendorffii genes are present in databases indicating co-sequencing of two related cultivars.
12This protein is not likely to function as an ATPase as several key features are missing including core sequence 2 (TGES− > TASD), core seq. 4 (PEGL− > PVGL, core seq. 5 (the entire intron including DKTGTLT is missing), core sequence 6 (TGDN− > TDND), and core sequence 7 (VVAVTGDGTNDAPAL− > IVAATGMGIHDPKTL). cDNA: AY078942 (613–1049, but different splice variant), CB264713 (772–980), this splice variant. Probably similar to PMAX.
13Probable pseudogene (Axelsen and Palmgren, 2001).
P1B ATPases: Heavy metal pumps
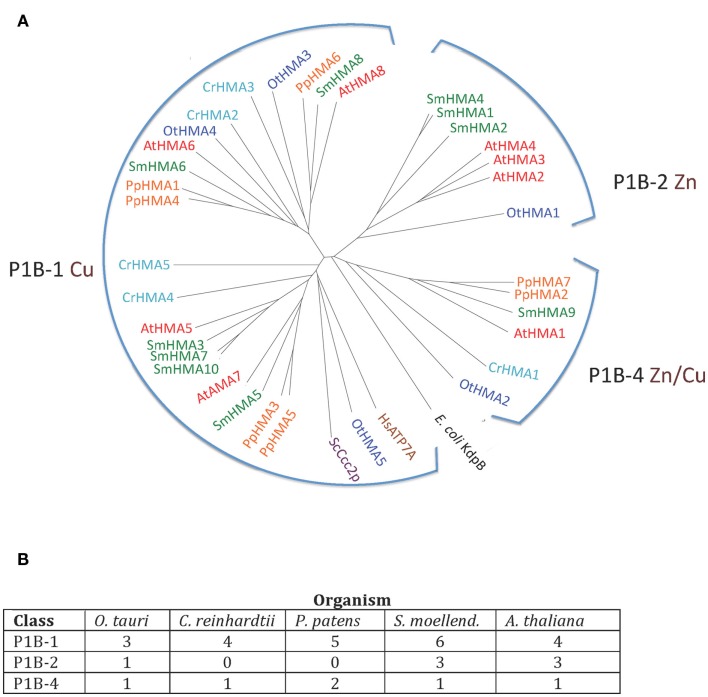
Heavy metal pumps (P1B ATPases) are found in all life forms including bacteria. In eukaryotes, these pumps are typically encoded by multigene families (Axelsen and Palmgren, 1998). The model dicotyledonous plant A. thaliana contains eight P1B ATPases, which can be divided into three groups according to conserved sequence motifs and their putative substrate specificity (Axelsen and Palmgren, 2001; Williams and Mills, 2005; Argüello et al., 2007). AtHMA5 to 8 belong to group P1B-1 and are predicted to transport Cu/Ag, while AtHMA2 to 4 belong to group P1B-2 predicted to transport Zn/Cd, and AtHMA1 belongs to group P1B-4 with a predicted broad substrate specificity (Zn/Cu/Co/Cd/Pb/Ca).
P1B ATPases were found in all organisms investigated in this study (Figure 3). The genome of C. reinhardtii encodes five P1B ATPases, four of which (CrHMA2-5) belong to the P1B-1 cluster. A representative protein in A. thaliana from this cluster is AtHMA8/PAA2. This protein localizes to the thylakoid membrane of chloroplasts and is required for Cu delivery during the biogenesis of plastocyanin (Abdel-Ghany et al., 2005).
Figure 3.
(A) Phylogenetic tree of P1B ATPases (heavy metal pumps) from Viridiplantae. Accession numbers for sequences are given in Table 2. For comparison, the following outliers were included: a bacterial P1A ATPase E. coli KdpB (P03960), and HsATP7A (Q04656), and ScCcc2p (P38995), Cu+ pumps from H. sapiens and S. cerevisiae, respectively. (B) Numbers of P1B ATPases by subgroups (Argüello et al., 2007) in the five plant genomes analyzed.
The only heavy metal ATPase in C. reinhardtii that is not a P1B-1 pump, is CrHMA1, which belongs to the P1B-4 cluster in which AtHMA1 is also found. AtHMA1 is localized to the chloroplast inner envelope membrane (Seigneurin-Berny et al., 2006; Kim et al., 2009). This ATPase has been reported to function as a transporter for Cu and Ca in addition to Zn/Cd/Co (Seigneurin-Berny et al., 2006; Moreno et al., 2008; Kim et al., 2009). All five organisms investigated in this study had one or two representatives of P1B-4 ATPases.
Zn transporting P1B ATPases are common in prokaryotes but have not been identified in fungi and animals. In O. tauri, which has one of the smallest known eukaryotic genomes, there are five P1B ATPases (∼30% of its P-type ATPase genes), which distribute among all three clusters of P1B ATPases, including a single P1B-2 pump. In our analysis, P1B-2 ATPases were found in O. tauri (OtHMA1), S. moellendorffii (SmHMA1, SmHMA2, and SmHMA4), and A. thaliana (AtHMA2, AtHMA3, and AtHMA4; Figures 3 and 4). P1B-2 ATPases could not be identified in C. reinhardtii and P. patens. Among the A. thaliana P1B-2 ATPases, AtHMA2, and AtHMA4 are localized to the plasma membrane. They show redundant function in cellular export of Zn and Cd into plant vascular tissues where they facilitate xylem loading and transport to the shoot (Hussain et al., 2004; Verret et al., 2004; Mills et al., 2003; Wong and Cobbett, 2009).
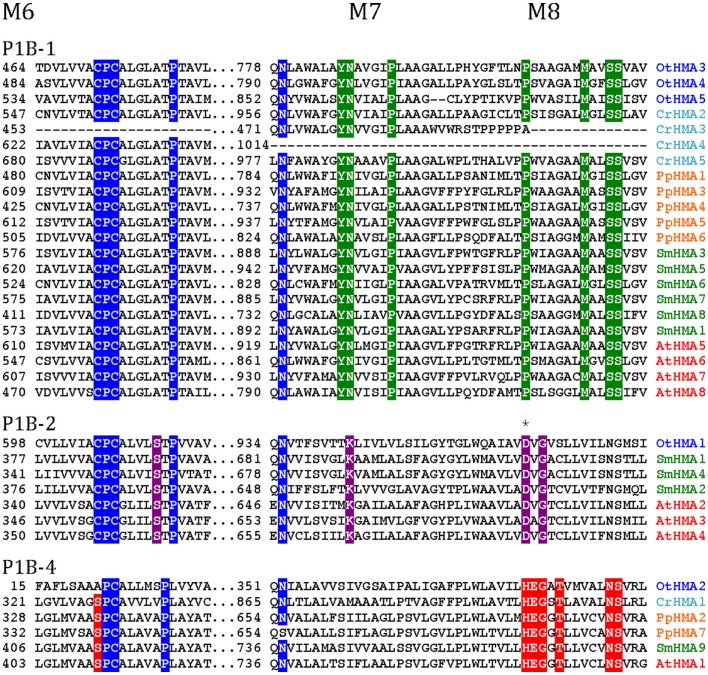
Figure 4.
Alignment of three transmembrane segments from Viridiplantae P1B ATPases analyzed in this study. Only the predicted transmembrane segments M6, M7, and M8 are shown. P1B-1: Cu+ transporting ATPases; P1B-2: Zn2+ transporting ATPase: P1B-4: Mixed specificity heavy metal pumps. The asterisk indicates an Asp residue (D) conserved in P1B-2 ATPases, which could be important for coordination of Zn2+ (Dutta et al., 2006).
As P1B-2 Zn ATPases are absent from fungi and animals, but common in bacteria, it seems likely that in plants these pumps have evolved from chloroplastic pumps. With the advent of vascular plants, it is likely that a subset of P1B-2 ATPases was targeted to the plasma membrane and acquired a new role in redistribution of Zn within the plant body.
P2A ATPases: ER-type Ca2+-ATPases
P2 ATPases form a large subfamily further divided into at least four clusters of pumps, two (P2A and P2B) having specificity for Ca2+ and two (P2C and P2D) for Na+ as the transported ligand (Figure 5).
Figure 5.
Phylogenetic tree of P2 ATPases (Ca2+ and Na+ pumps). Accession numbers for sequences are given in Table 2. For comparison, the following outliers were included: a bacterial P1A ATPase E. coli KdpB (P03960), the S. cerevisiae pumps ScPmr1p (P13586), ScPmc1p (P38929), and ScEna1p (P13857), the H. sapiens pumps HsPMCA1 (P20200), HsSERCA1 (O14983), and HsNaKα1 (P98194), and the Trypanosoma cruzi pump TcENA (Q76DT8).
P2A ATPases were identified in all plants studied here (Figure 5). A single pump from each organism (OtECA2, CrECA4, PpECA4, SmECA4, and AtECA3) form a distinct subset of ER-type Ca2+-ATPases (ECAs) closely related to the animal sarco-endoplasmic reticulum Ca2+ pump SERCA1 (Figure 5). In the flowering plant A. thaliana, AtECA3 is a Golgi-localized pump that can transport Ca2+ and Mn2+ (Li et al., 2008; Mills et al., 2008).
In a comparison among many eukaryotes, a subset of P2A Ca2+-ATPases form a distinct cluster and have been named secretory pathway Ca2+-ATPases (SPCAs; Wuytack et al., 2003). These pumps are identified in fungal and animal cells and are localized to the Golgi apparatus or other membranes of the secretory pathway. As evident from Figure 6, representatives of these pumps from fungi (S. cerevisiae and S. pombe) are characterized by having lost Ca2+ binding site 1.
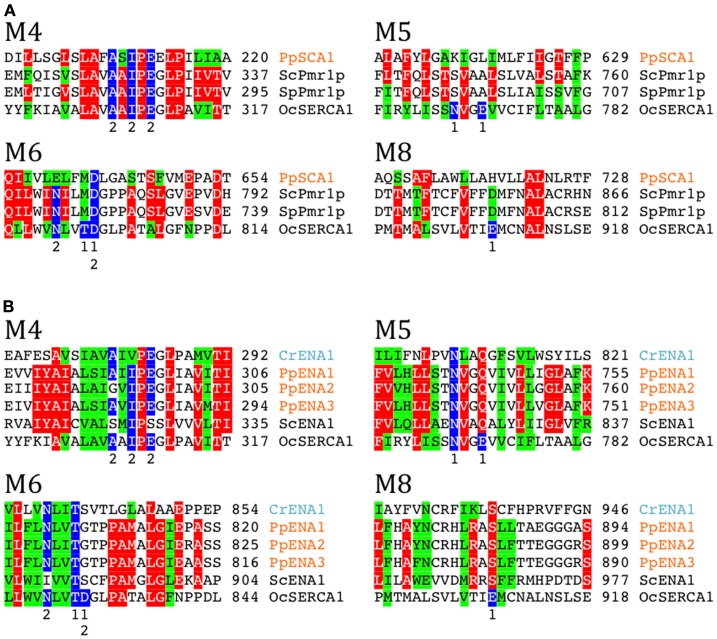
Figure 6.
Alignment of transmembrane segments showing differences in a potential cation binding site for selected Ca2+ and Na+ pumps. Secretory pathway Ca2+ATPases (SPCA pumps) and ENA Na+ pumps (P2D ATPases) are missing Ca2+ binding site 1 present in P2A pumps typified by rabbit OcSERCA1 (P04191). In P2A pumps a conserved Asp in M6 contributes to coordination of both Ca2+ ions. (A) Secretory pathway pumps (SPCAs) have a conserved Asp in M6. Fungal examples shown are S. cerevisiae Pmr1p (P13586) and S. pombe Pmr1p (O59868). (B) P2D ATPases (ENA pumps) do not have a conserved Asp in M6 like all other P2 ATPases. Residues contributing with Ca2+ coordinating oxygen molecules in rabbit SERCA1 (Toyoshima et al., 2000) are marked in blue. Only sequences including the predicted transmembrane segments M4, M5, M6, and M8 are shown.
Only a single likely SPCA protein was found in our analysis, namely PpSCA1. This protein clusters with other SPCAs and does not contain conserved residues expected for Ca2+ binding site 1 (Figure 6A). Among reference plant and animal genomes surveyed, PpSCA1 showed the greatest identity (30%) to the secretory pathway Ca2+-ATPase Pmr1p from S. cerevisiae (Rudolph et al., 1989; Antebi and Fink, 1992).
P2B ATPases: Autoinhibited Ca2+-ATPases
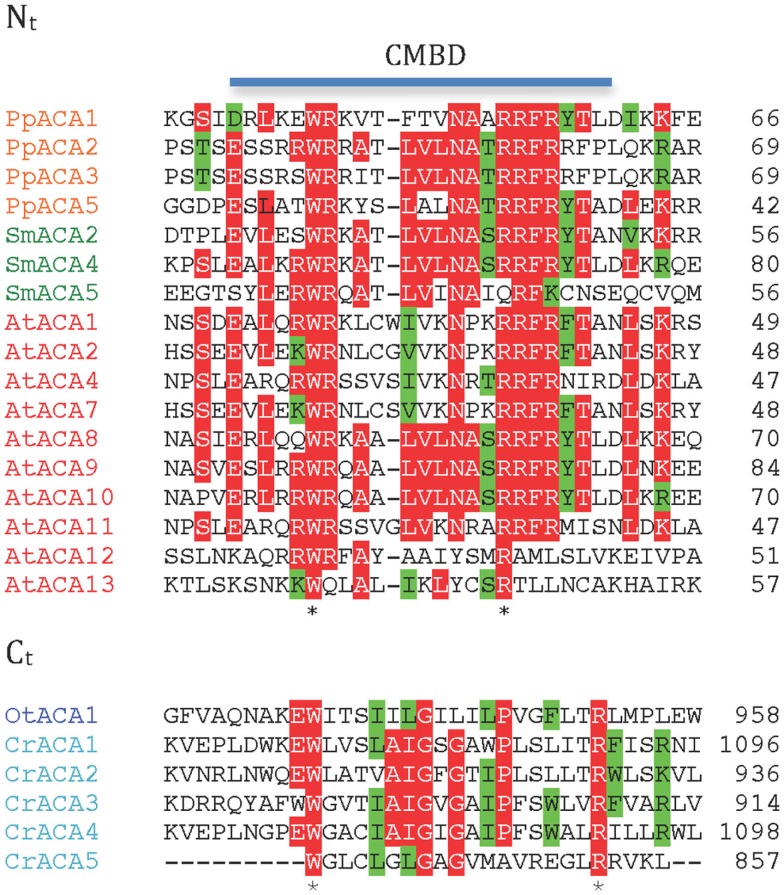
P2B ATPases are Ca2+ pumps that are activated by binding of calmodulin to autoinhibitory terminal domains. A marked difference between higher plant and animal P2B ATPases is that their calmodulin-binding domains (CMBDs) are situated in the N- and C-terminal domains, respectively (Sze et al., 2000). Autoinhibited Ca2+-ATPases (ACAs) from flowering plants have been shown to be activated by Ca2+ in the presence of calmodulin and are characterized by an N-terminally situated CMBD (Malmström et al., 1997; Harper et al., 1998; Curran et al., 2000; Hwang et al., 2000). The CMBD overlaps partially with an autoinhibitory pump sequence (Bækgaard et al., 2006) and it has been proposed that calmodulin, by binding to the CMBD, neutralizes the constraint set by the autoinhibitory sequence on the pump molecule (Bækgaard et al., 2006). Animal P2B ATPases are likewise activated by calmodulin, but in these the CMBD is situated in an extended C-terminal domain (James et al., 1988).
Putative CMBDs were identified in the N-terminal domains of most P2B ATPases of P. patens, S. moellendorffii, and A. thaliana (Figure 7). The proposed CMBDs in AtACA12 and AtACA13 are weakly defined. Importantly, an N-terminal CMBD could not be identified in P2B ATPases from the chlorophytes O. tauri and C. reinhardtii.
Figure 7.
Alignment of putative calmodulin-binding sites in the N- and C-terminal regions of ACA pumps. Nt, N-terminal domain; Ct, C-terminal domain.
As animal P2B ATPases are equipped with a C-terminally located CMBD, we analyzed all of the plant P2B ATPases for a putative regulatory domain in this location. CMBDs have very little similarity between calmodulin-binding proteins and can be difficult or impossible to predict with certainty although as a rule there is alternation between bulky aromatic and positively charged residues. No C-terminal extensions could be identified in Streptophyte P2B ATPases. In contrast, a conserved sequence with weak resemblance of a CMBD was identified in the C-terminal domain of P2B ATPases from the chlorophytes O. tauri and C. reinhardtii (Figure 7), which are missing a similar sequence from their N-terminal domain. The calmodulin-binding capacity of these putative CMBDs remains to be tested, but it is an attractive working hypothesis that swapping of the CMBD from the C-terminal to the N-terminal domain occurred at the split between Chlorophytae and Streptophytae.
P2C ATPases: Na+/K+-ATPases
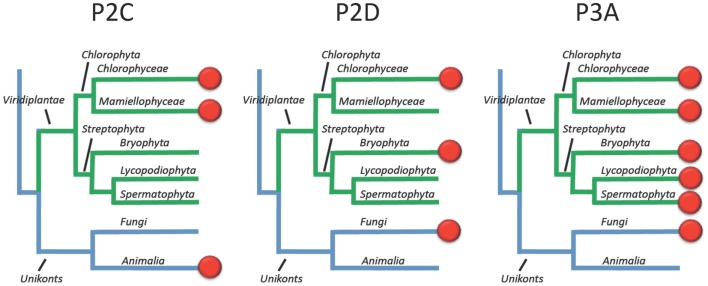
The Na+/K+-ATPase of animal cells was the first P-type ATPase to be discovered (Skou, 1957), but all subsequent attempts to identify a Na+ pump in vascular plants failed. Vascular plants tend to be very sensitive to elevated Na+ in the soil, which is probably due to the lack of an effective Na+ extrusion system such as a Na+/K+-ATPase. In animal cells, the plasma membrane is energized by the Na+/K+-ATPase whereas in a typical plant, the plasma membrane H+-ATPases (P3A ATPases) carry out this function. It has therefore been hypothesized that P2C Na+/K+-ATPases were lost at a branch point in the Streptophyta plant lineage, presumably in an organism that evolved in a fresh water environment and utilized a plasma membrane H+-ATPase to energize its plasma membrane (Palmgren, 2001).
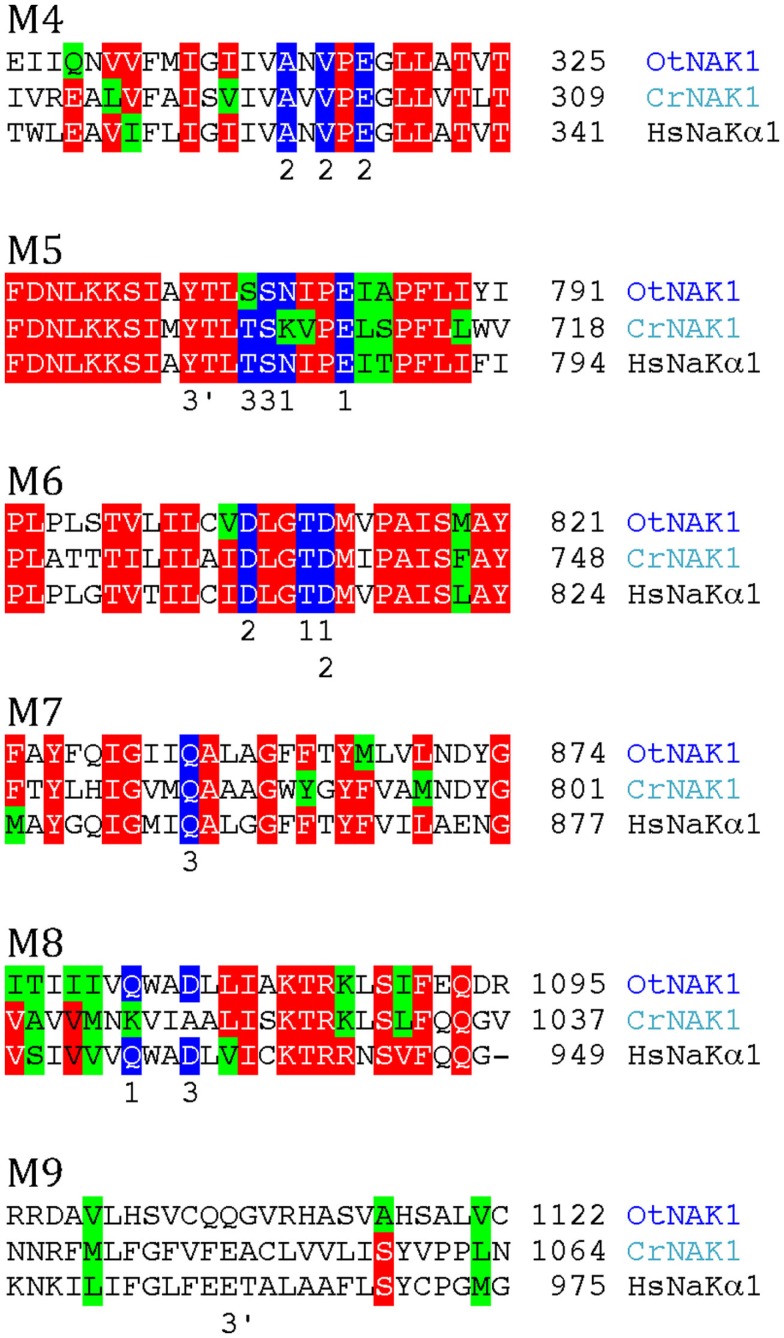
Biochemical evidence has pointed to the presence an electrogenic, vanadate-sensitive, ouabain-resistant Na+-ATPase in the plasma membrane of marine chlorophytes (Popova et al., 1999, 2005; Gimmler, 2000) and chlorophyte expressed sequence tags with similarity to Na+/K+-ATPase have been identified (Barrero-Gil et al., 2005). Both chlorophytes analyzed in this work contain sequences with strong similarity to an animal Na+/K+-ATPase (Figure 8). No Na+/K+-ATPase has been crystallized in a form with bound Na+, but a homology model has been built (Morth et al., 2011) based on the structure of a Na+/K+-ATPase with bound Rb+ (as a substitute for K+) and the structure of SERCA1 with bound Ca2+. According to this model, residues in several transmembrane segments of Na+/K+-ATPase contribute with binding ligands to Na+ (Figure 8). All these residues are conserved in OtNAK1 and CrNAK1, strongly suggesting that these ATPases operate as Na+ pumps. O. tauri is a chlorophyte that lives in oceans, where a Na+/K+-ATPase is of obvious benefit for extrusion of Na+ leaking in from sea water. However, it is peculiar that C. reinhardtii, a green alga of terrestrial soils, is also equipped with such a pump. This suggests that the presence of Na+/K+ pumps in green algae is a primitive character that was lost with the emergence of Streptophytae. This hypothesis is supported by the widespread presence of P2C pumps in other eukaryotes and archaea (Sáez et al., 2009).
Figure 8.
Alignment of predicted transmembrane segments of putative chlorophyte Na+/K+-ATPases (OtNAK1 and CrNAK1) with similar regions in the human Na+/K+-ATPase α1 subunit (ATP1A1; P05023). Residues that in Na+/K+-ATPase are likely to contribute with oxygen atoms for coordination of Na+ (Morth et al., 2007) are marked by blue. Numbers above these residues refer to which of the three Na+ sites (1–3) they contribute to. Residues contributing to an alternative Na+ site (site 3b) are marked 3′.
As mentioned above, the substrate specificity of PpSCA1 is uncertain, with features that are both consistent and contrary to speculations on the transport of either Na+ or Ca2+. This pump does have some similarity with chlorophyte Na+/K+-ATPases, but ligands contributing to Na+ Site 3 are not present and similarity to sites contributing to sites 1 and 2 are not absolute (Figure 6A). Biochemical and genetic experiments are needed to address the question of substrate specificity for PpSCA1.
P2D ATPases: Na+ or K+ pumps of mosses and fungi
P2D ATPases form a unique group of pumps so far only found in mosses, fungi, and protozoa and confer Na+ tolerance to organisms in which they are expressed (Rodríguez-Navarro and Benito, 2010). The moss P. patens encodes three P2D ATPases: PpENA1, PpENA2, and PpENA3. Related pumps are present in liverworts (Marchantiophyta), such as Marchantia polymorpha and Riccia fluitans, which are primitive non-vascular land plants related to mosses. A close fungal homolog to PpENA1 is from Neurospora crassa (Q9UUX8; 43% identity). When protein databases were searched for similar proteins outside plants and fungi, hits were only found in protist sequences.
In P2A ATPases, two Ca2+ sites are present: Site 1 and Site 2 (Toyoshima et al., 2000). In SPCAs (Figure 6A) and P2D ATPases (Figure 6B), Site 1 is missing. Thus, two negatively charged Glu residues in M6 and M8, which in SERCA1 contribute to Ca2+ coordination in Site 1, are absent in both secretory pathway pumps and P2D pumps. Notably, a negatively charged Asp in M6, which is conserved in all other P2-type ATPases, is replaced by a neutral residue in P2D pumps (Rodríguez-Navarro and Benito, 2010; Figure 6B). In the available structures of P2A, P2C, and P3A ATPases (Morth et al., 2007; Olesen et al., 2007; Pedersen et al., 2007), this Asp contributes to coordination of all transported cations including Ca2+, Na+, K+, and H+. The absence of the Asp in M6, in addition to other negatively charged amino acid residues in the membrane domain, therefore appears to be a hallmark of P2D ATPases. When the sequences of P-type ATPases retrieved in this study were analyzed in detail it appeared that two pumps from C. reinhardtii, here named CrENA1 and CrENA2, are likely to represent chlorophyte P2D ATPases as they lack a negatively charged Asp in M6 (Figure 6B). Although these branch out close to P2D pumps (Figure 5) they have less than 33% identity to these or any other plant pumps.
P3A ATPases: Autoinhibited H+-ATPases
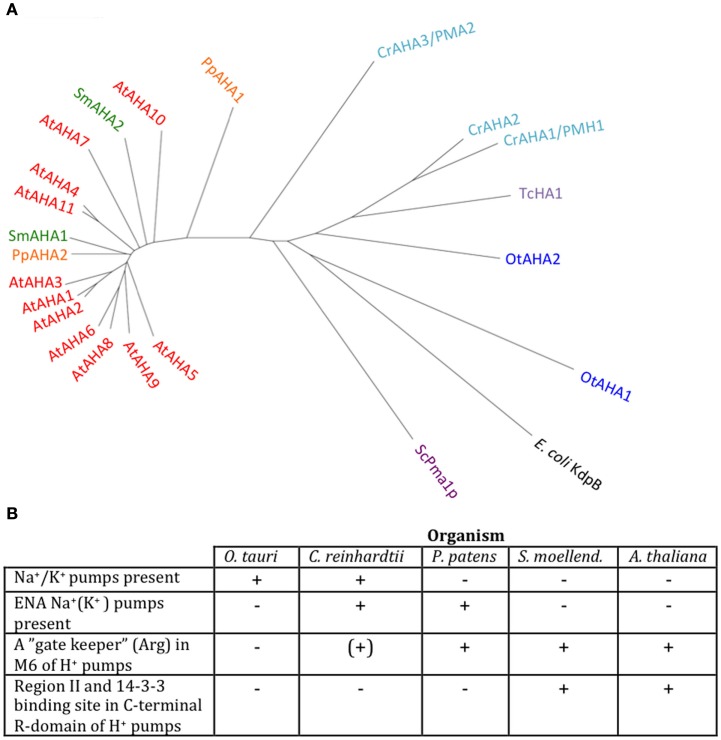
We identified P3A ATPases in all genomes of Viridiplantae analyzed in this work (Figure 9). P3A ATPases energize the plasma membrane of plants and fungi by establishing a large proton gradient and membrane potential (negative on the inside) across the plasma membrane (Palmgren, 2001). The potential energy stored in this gradient serves as a proton motive force that drives a large number of transport processes carried out by secondary active transporters and channel proteins. The plasma membrane proton pump isoform 2 from A. thaliana (autoinhibited H+-ATPase 2, AHA2) is expressed throughout the plant and, together with the closely related isoform AHA1, is essential for plant growth (Palmgren, 2001; Haruta et al., 2010). In this respect, they serve as functional analogs to the Na+/K+-ATPases of animal cells (Morth et al., 2011).
Figure 9.
Phylogenetic tree of P3 ATPases (H+-pumps) (A) and table summarizing conserved features (B). Accession numbers for sequences are given in Table 2. For comparison, the following outliers were included: a bacterial P1A ATPase E. coli KdpB (P03960), the S. cerevisiae pump ScPma1p (P05030), and the Trypanosoma cruzi pump TcHA1 (Q8T7V7).
In Chlamydomonas, two plasma membrane H+-ATPases have been described in the literature (Campbell et al., 2001). Closely related homologs to OtAHA2 are found in other green algae but not in streptophytes. Outside this group, OtAHA2 has highest similarity (42–44% identity) to plasma membrane H+-ATPases of protists that have been characterized biochemically as P-type H+-ATPases (Luo et al., 2002, 2006; Figure 9). OtAHA2 has lesser but marked similarity to AtAHA2 (38% identity). The most divergent P3A ATPase analyzed in this study is OtAHA1.
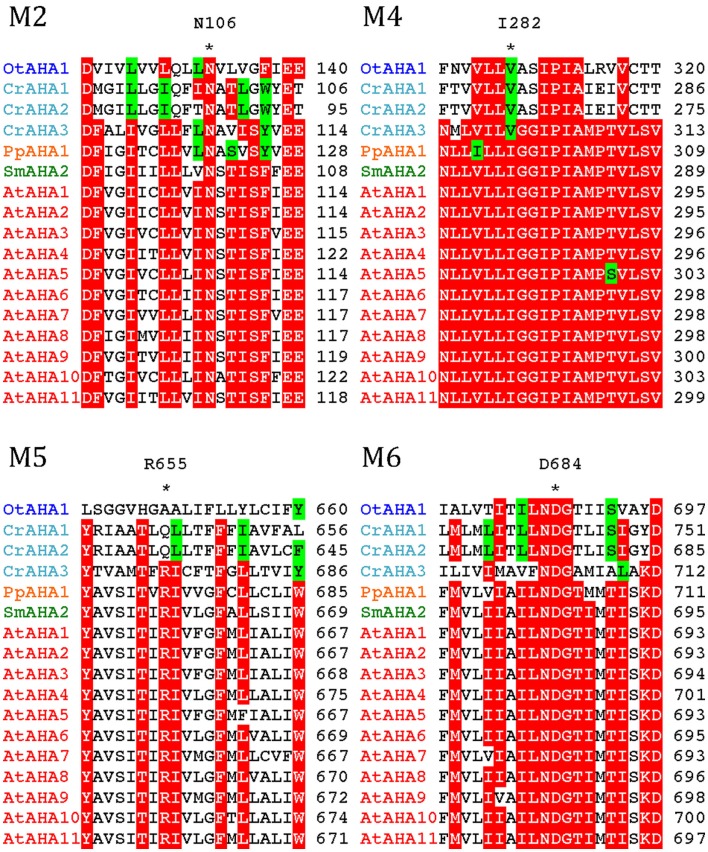
All P3A plasma membrane H+-ATPases have conserved residues that have been implicated as being important for H+ transport (Pedersen et al., 2007; Figure 10). These include the H+ acceptor/donor Asp684 (AtAHA2 numbering) in M6 and the proposed gate-keeper residue Asn106 in M2 (Pedersen et al., 2007; Buch-Pedersen et al., 2009). Arg655, which in AtAHA2 has been proposed to prevent backflow of H+ through the pump, a feature likely to be essential when electrochemical gradients get steep, is strictly conserved in all streptophyte pumps. Interestingly, this residue is absent in chlorophyte P3A ATPases except for CrAHA3 (Figure 10). CrAHA3 is the chlorophyte pump that shows the highest similarity to a streptophyte H+ pump (Figure 9).
Figure 10.
Alignment of predicted transmembrane segments in P3A H+-ATPases. Asterisks mark residues of potential importance for proton coordination and pumping based on evidence from mutagenesis and analysis of a crystal structure for AtAHA2 (Pedersen et al., 2007). The number of the amino acid residue in AtAHA2 is indicated above each asterisk. R655 in M5, which seems important for controlling backflow of H+ at high electrochemical gradients (Pedersen et al., 2007), is lacking in some chlorophyte H+-ATPases.
Notably, typical protists and chlorophytes are characterized by having both Na+ and H+ pumps. Protists are equipped with P2D Na+ pumps and chlorophytes with P2C Na+/K+ pumps (Table 1; Figures 8 and 9). As plasma membrane H+-ATPases in these organisms lack the residue corresponding to Arg655 (to block H+ backflow), this would suggest that in organisms with co-expression of electrogenic H+ and Na+ pumps, it is the role of Na+ ATPases to generate a plasma membrane electrochemical gradient, which in turn can be used as an energy source to drive a variety of cellular processes, such as secondary active transport. P3A ATPases in protists and chlorophytes might therefore have other roles than establishing electrochemical gradients, e.g., controlling intracellular pH.
Table 1.
Overview of number of pumps in different P-type ATPase subfamilies in Viridiplantae.
| Class | Organism |
Total | ||||
|---|---|---|---|---|---|---|
| O. tauri | C. reinhardtii | P. patens | S. moellendorffii | A. thaliana | ||
| P1B | 5 | 5 | 7 | 10 | 8 | 35 |
| P2A | 2 | 2 | 5 | 4 | 4 | 17 |
| P2B | 2 | 6 | 6 | 5 | 10 | 29 |
| P2C | 1 | 1 | 0 | 0 | 0 | 2 |
| P2D | 0 | 2 | 3 | 0 | 0 | 5 |
| P3A | 2 | 3 | 2 | 2 | 11 | 20 |
| P4 | 1 | 3 | 8 | 8 | 12 | 32 |
| P5A | 1 | 1 | 1 | 1 | 1 | 5 |
| P5B | 0 | 1 | 2 | 2 | 0 | 5 |
| Total | 14 | 24 | 34 | 32 | 46 | 150 |
Angiosperm plasma membrane H+-ATPases are regulated by an extended C-terminal domain that functions as a pump auto inhibitor (Palmgren et al., 1991). All residues in this domain of ∼100 residues have been mutagenized and two clusters of autoinhibitory sequences have been identified, Region I and Region II (Axelsen et al., 1999). Further, in the extreme C-terminal end, a 14-3-3 binding site has been identified. 14-3-3 binding results in pump activation, but in order for 14-3-3 binding to occur, the penultimate residue (a Thr or Ser) first has to become phosphorylated (Fuglsang et al., 1999; Svennelid et al., 1999; Maudoux et al., 2000).
When C-terminal sequences of putative P3A ATPases were analyzed, the complete set of regulatory sequences (Region I–II and the 14-3-3 binding site) could be identified in all AHAs of S. moellendorffii and A. thaliana (Figure 11). In S. moellendorffii the shorter C-terminal regions of CrAHA2 and PpAHA1, a stretch of residues with weak but notable similarity to Region I could be identified (Figure 11). In these pumps, sequences with similarity to Region II and the 14-3-3 binding site could not be observed. In Chlorophyte P3A ATPases resembling protist plasma membrane H+-ATPases (OtAHA1, CrAHA1, and CrAHA3) no sequences with similarity to any of these regions could be identified. The presence of a putative Region I in the C termini of CrAHA2 and PpAHA1 suggests that the basic regulatory apparatus of the higher plant C-terminus could have been present in the first green plants, but that more complex features (e.g., a 14-3-3 binding site) evolved latter in the evolution of vascular plants. In support of an early origin of Region I, the S. cerevisiae plasma membrane H+-ATPase Pma1p has a short autoinhibitory sequence in its C-terminal domain with weak similarity to Region I, which appears to be involved in regulation of pump efficiency (Portillo et al., 1989; Venema and Palmgren, 1995).
Figure 11.
Alignment of putative autoinhibitory regions in the C-terminal region of P3A H+-ATPases.
P4 ATPases: Putative lipid pumps
P4 ATPases in plants have been implicated in flipping phospholipids across biological membranes (Poulsen et al., 2008a). They are completely absent from eubacteria and archaebacteria, whereas in eukaryotes they are typically encoded for by multigene families (Axelsen and Palmgren, 1998).
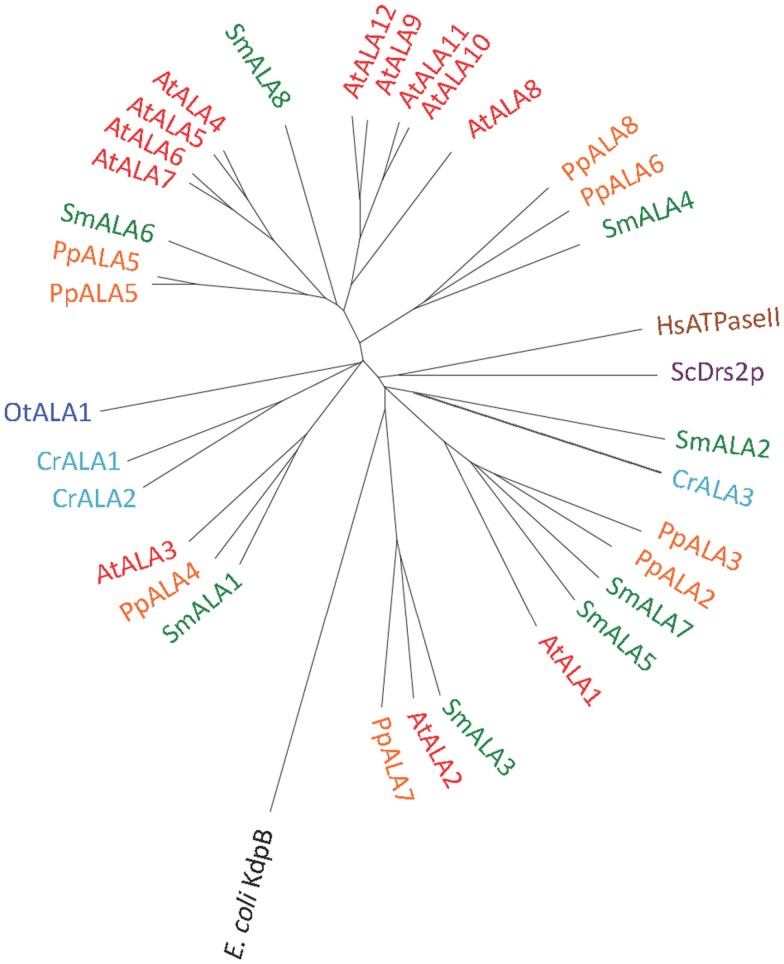
We identified P4 ATPases in all of the Viridiplantae investigated in this study (Figure 12). The chlorophyte O. tauri was the only organism with a single P4 ATPase. This solitary P4 ATPase groups in the phylogenetic tree in the same larger branch as AtALA3. AtALA3 activity is connected with transport of phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine in A. thaliana (Poulsen et al., 2008b). Further, it is a resident of the trans Golgi of root tip cells where it is connected to the generation of secretory vesicles leaving the Golgi apparatus (Poulsen et al., 2008b).
Figure 12.
Phylogenetic tree of P4 ATPases (lipid flippases). Accession numbers for sequences are given in Table 2. For comparison, the following outliers were included: A bacterial P1A ATPase E. coli KdpB (P03960), the S. cerevisiae pump ScDrs2p (P39524), and the H. sapiens pump HsATPaseII (ATP8A1; Q9Y2Q0).
P5 ATPases: Pumps with no assigned specificity
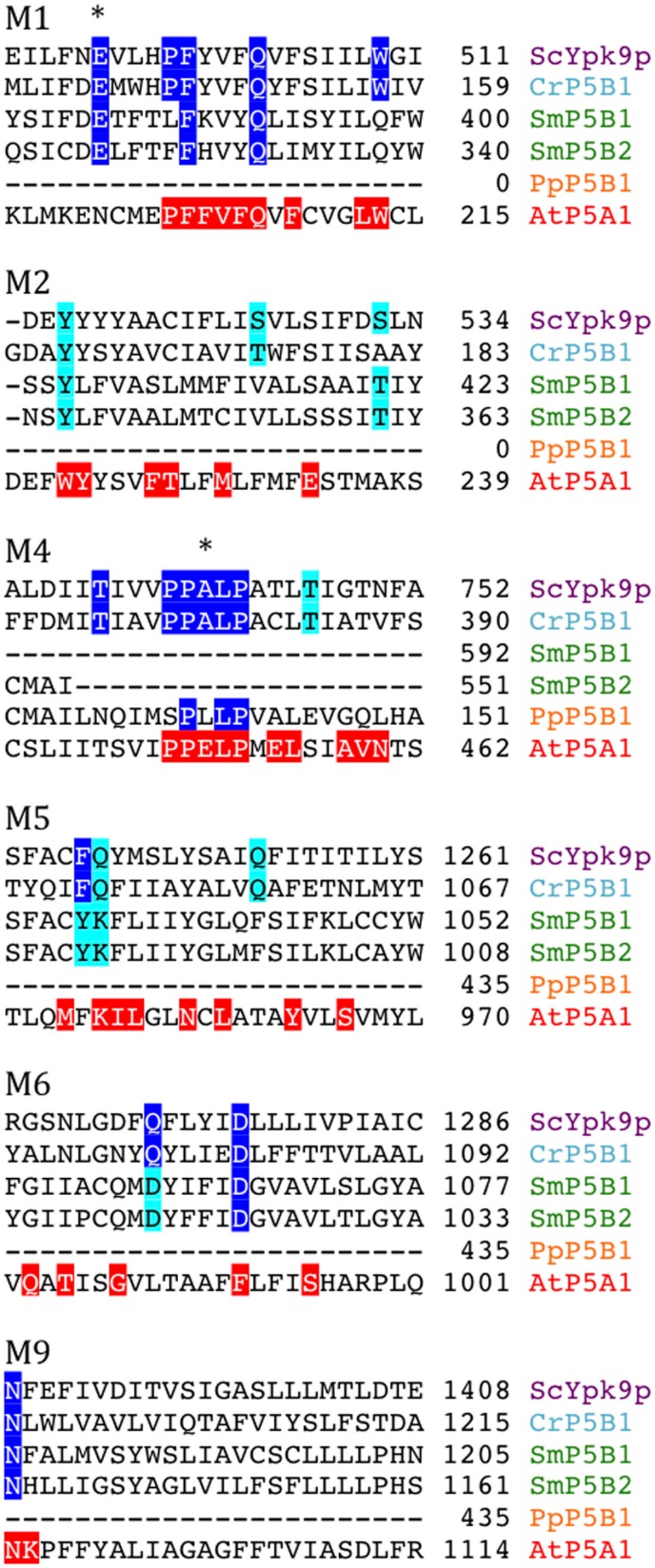
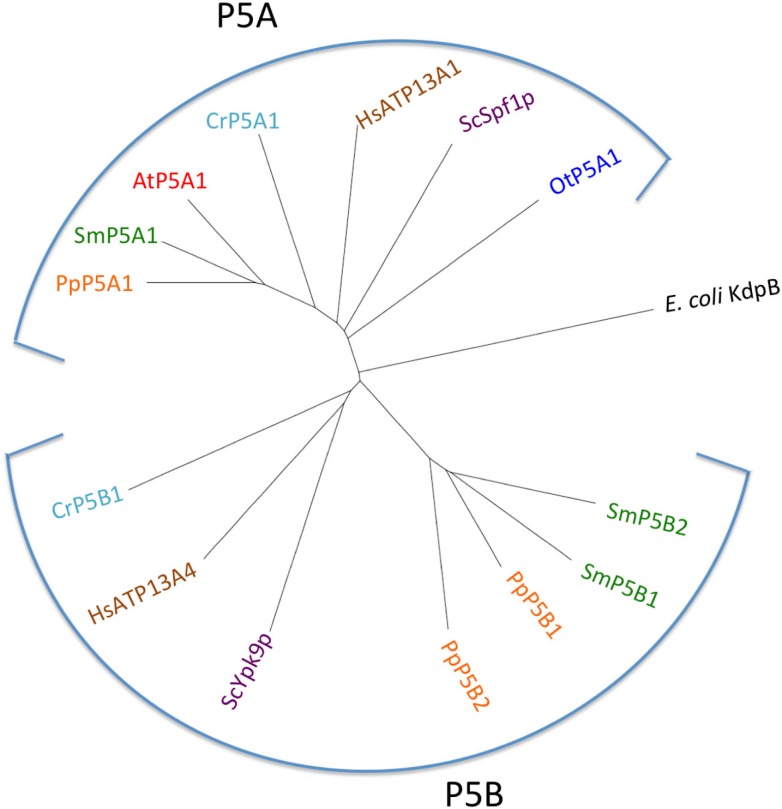
P5 ATPases constitute the least characterized group of P-type pumps and their transported ligand – if any – has not been identified. These pumps are absent from prokaryotes and are confined to eukaryotes where they reside in internal membrane systems (Møller et al., 2008). Based on sequence analysis, they are divided into two groups, P5A and P5B, each of which is predicted to transport different substrates based on differences in their transmembrane segments (Figure 14; Sørensen et al., 2010).
Figure 14.
Alignment of predicted transmembrane segments of putative P5B ATPases aligned with similar regions in the S. cerevisiae P5B ATPase ScYpk9p (Q12697). Residues conserved in all P5B ATPases (according to Sørensen et al., 2010) are marked in blue. Those that are highly conserved are marked in cyan. The P5A ATPase AtP5A/AtMIA is shown with residues conserved in P5A ATPases highlighted in red (Sørensen et al., 2010). Asterisks mark residues that are likely to play a role in ligand coordination.
P5A ATPases have been found in all eukaryotic genomes analyzed so far (Møller et al., 2008) and were identified in all Viridiplantae analyzed in this study (Figure 13). Only a single P5A ATPase could be identified in each organism.
Figure 13.
Phylogenetic tree of P5 ATPases (having unknown transport activity). Accession numbers for sequences are given in Table 2. For comparison, the following outliers were included: a bacterial P1A ATPase E. coli KdpB (P03960), the S. cerevisiae pumps ScSpf1p (P39986) and ScYpk9 (Q12697), and the H. sapiens pumps HsATP13A1 (Q6NT90) and HsATP13A4 (Q4VNC1).
P5B ATPase sequences have so far been identified in the genomes of all eukaryotes examined, except for two plant lineages. Their widespread distribution supports a model in which they arose at an early point in the evolution of eukaryotes (Sørensen et al., 2010). When the Viridiplantae genomes analyzed here were searched for P5B sequences, we could identify P5B ATPases in C. reinhardtii, P. patens, and S. moellendorffii, but not in O. tauri and A. thaliana (Figure 13). This suggests that loss of P5B ATPases occurred at least twice in the evolution of Viridiplantae.
Discussion
Evolution of plant P-type ATPases
This survey of P-type ATPases provides evidence that members of the green plant lineage require at least five different types of P-type ATPases. Each of the five reference genomes analyzed showed at least one representative from each of the five subgroups of P-type ATPases. However, within the five different subgroups there is still evidence for considerable evolution of biochemical functions (Figure 15). For example, within the P2-type pumps, there are at least four subdivisions that delineate two subfamilies each of Na+ and Ca2+ pumps (Figure 5).
Figure 15.
Overview of the evolution of Na+ and H+ transporting P-type ATPases in Viridiplantae. Most likely, the ancestor of green plants had two types of Na+ pumps (P2C and P2D) in addition to a plasma membrane H+ pump (P3A). In present day plants, the terrestrial green algae C. reinhardtii still has all three types of pumps whereas Na+ pumps have been lost completely in vascular plants (here represented by Lycopodiophyta and Spermatophyta).
In Arabidopsis there are 46 pumps, compared to 14 in O. tauri, 22 in C. reinhardtii, 33 in P. patens, and 32 in S. moellendorffii. This is consistent with a speculation that multicellular organisms require more pumps, presumably to provide specialized functions associated with more complex developmental programs or more variable environments. While loss of function phenotypes have been established for at least one member of each pump type in Arabidopsis, most of the 46 pumps remain uncharacterized at the genetic level.
The evolutionary diversity of P-type ATPases raises many questions to be explored. For example, why does O. tauri require only one lipid flippase (P4-type pump), whereas rice and Arabidopsis have 10 and 12 members, respectively (Baxter et al., 2003). What are the cellular functions of the lipid flippases? Are their biochemical functions limited to flipping lipids, or do they also help insert or remove lipids from membranes? At a structural level, how have the P4-pumps evolved from an ancestor that recognized simple cation substrates into having a dynamic interaction with lipids?
The relative expansion of the proton pump family (P3A) in flowering plants is also noteworthy, with 11 and 10 members in Arabidopsis and rice, respectively (Baxter et al., 2003), and only two in P. patens and S. moellendorffii. Do physiological complexities of flowering plants necessitate different isoform specific features?
In contrast, P5 pumps have only one or two representatives in all five reference organisms. The biochemical functions of these pumps are not clear, but their general importance to eukaryotes is supported by the presence of at least one representative in reference genomes from yeast to man.
The observation that all five reference genomes have two types of Ca2+ pumps, P2A and P2B suggests that both subgroups have conserved functions. A role in Ca2+ signaling has been proposed for P2B pumps, based on the presence of a regulatory domain that provides for activation of the pump by Ca2+/calmodulin. Genetic evidence supports a model in which regulation of pump activity could modulate the magnitude or duration of a Ca2+ signal (Qudeimat et al., 2008; Boursiac et al., 2010; Zhu et al., 2010; Spalding and Harper, 2011). For the P2A pumps, genetic evidence suggests that they function in the homeostasis of both Ca2+ and Mn2+ (Wu et al., 2002; Li et al., 2008; Mills et al., 2008). Delineating signaling and nutritional functions of these pumps is an important challenge for the future.
The evolution of plasma membrane H+ and Na+ pumps
The evolution of Na+ pumps (type 2C and 2D) is of interest from several perspectives. First, the relatively close relationship between Na+ and Ca2+ pumps raises an interesting evolutionary question of which came first? Since both Na+ and Ca2+ can be toxic within the cytosol, did both types of pumps arise early in evolution as a way to efflux toxic ions? Second, it appears that ancestral plants had both P2C and P2D ATPases. Both groups of pumps have remained in Chlorophyceae, represented here by the terrestrial green algae C. reinhardtii, but appear to have been lost in vascular plants (Figure 15).
An interesting question is why do the chlorophytes examined here have both H+ and Na+ pumps? For organisms such a C. reinhardtii that can live in fresh water, what is the evolutionary pressure for the retention of a Na+ pump? For organisms that live in saline environments, such as O. tauri, why retain a plasma membrane H+ pump? It seems reasonable to assume that Na+ pumps alone could control cytoplasmic Na+ levels and energize the plasma membrane for signaling and co-transport systems, as they do in typical animal cells. However, it is possible that the H+ pumps actually evolved in marine organisms not to energize the plasma membrane, but rather to control cytoplasmic pH.
Regardless of their evolutionary origins, the observation that both H+ and Na+ pumps co-exist in the plasma membrane of well studied chlorophytes raises a question about which ion is used to drive secondary active transport systems? Are there different sets of H+ or Na+ specific co-transporters, or can the cotransporters be driven by either H+ or Na+ gradients? In vascular plants, Na+ pumps appear to have been lost, leaving only plasma membrane H+-ATPases to drive secondary transport systems. As a consequence it is thought that most plasma membrane cotransporters in vascular plants are H+ coupled.
In the Arabidopsis pump AtAHA2, a conserved Arg in transmembrane segment M5 (Arg655) appears to be an important part of the H+ pumping apparatus (Pedersen et al., 2007). This residue has been proposed to serve as a built-in cation that allows for rapid transition of the pump from the E1P to the E2P conformational state and as a gate-keeper that prevents H+ from flowing backward (“backflow protection”; Pedersen et al., 2007; Buch-Pedersen et al., 2009). Further, in AHA2, a complex C-terminal regulatory domain with two autoinhibitory regions and a binding site for activating 14-3-3 protein is present (Axelsen et al., 1999). Backflow protection and multiple regulatory features are characteristic of all well studied H+ pumps in flowering plants. The observation of similar features associated with the S. moellendorffi pumps (Figure 11), suggests that H+ pumps with “advanced features” may be universal to all vascular plants.
In contrast, the plasma membrane H+ pumps in chlorophytes appear to be more primitive in at least two aspects (Figure 9B). First, both P3A pumps of O. taurii lack the conserved Arg of M5 and two out of the three P3A pumps in C. reinhardtii also lack this residue (Figure 10). Second, a complex C-terminal regulatory domain is absent from the chlorophyte pumps.
The presence of “more advanced” H+ pumps in vascular plants would suggest that these organisms have the capacity to utilize H+ pumps to energize their plasma membranes with large electrochemical gradients. In flowering plants, H+ pumps can create steep H+ gradients and membrane potentials that can exceed 200 mV (negative on the inside; Hirsch et al., 1998). In comparison, Na+/K+ pumps in animal cells typically only produce membrane potentials around 60 mV. It is tempting to hypothesize that vascular plants have evolved to rely on these “more advanced” pumps to create very large electrochemical gradients for special purposes, such as signaling or nutrient transport. However, during evolution, large H+-based electrochemical gradients may have been problematic in the presence of a Na+ pump. For example, a large membrane potential or pH gradient may have resulted in ancestral Na+ pumps mis-functioning as H+ or Na+ leaks. As a result, Na+ pumps may have been lost as vascular plants evolved to rely on their more advanced H+ pumps.
Frontiers in P-type ATPase research
The frontiers of P-type ATPase research can be divided into three areas. The first is to understand cellular and organismal functions of each pump. The second is to match biological functions with a structural understanding of how pumps transport specific substrates, and how their activities are modulated by signaling systems. The third is to explore ideas about how P-type ATPases can be altered or used to improve crop plants. For example, can Na+ pumps from chlorophytes be moved into crop plants to provide improved Na+ tolerance? Or would these pumps require re-engineering to prevent them from becoming H+ or Na+ leaks in cell types with large membrane potentials or H+ gradients. As these frontiers are considered, it is worth remembering that P-type ATPases have already proven themselves as flexible substrates for evolution, producing pumps with a wide diversity of functions, from pumping protons to flipping lipids.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00031/abstract
Acknowledgments
Funding of Kristian B. Axelsen by The Swiss Federal Government through the Federal Office of Education and Science; and by the European Union (SLING: Serving Life-science Information for the Next Generation [226073] is gratefully acknowledged.
Footnotes
References
- Abdel-Ghany S. E., Muller-Moule P., Niyogi K. K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251 10.1105/tpc.104.030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A., Fink G. R. (1992). The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3, 633–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello J. M., Eren E., González-Guerrero M. (2007). The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248 10.1007/s10534-006-9055-6 [DOI] [PubMed] [Google Scholar]
- Axelsen K. B., Palmgren M. G. (1998). Evolution of substrate specificities in the P-Type ATPase superfamily. J. Mol. Evol. 46, 84–101 10.1007/PL00006286 [DOI] [PubMed] [Google Scholar]
- Axelsen K. B., Palmgren M. G. (2001). Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 126, 696–706 10.1104/pp.126.2.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen K. B., Venema K., Jahn T., Baunsgaard L., Palmgren M. G. (1999). Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38, 7227–7234 10.1021/bi982482l [DOI] [PubMed] [Google Scholar]
- Bækgaard L., Luoni L., De Michelis M. I., Palmgren M. G. (2006). The plant plasma membrane Ca2+ pump ACA8 contains overlapping as well as physically separated autoinhibitory and calmodulin-binding domains. J. Biol. Chem. 281, 1058–1065 10.1074/jbc.M508299200 [DOI] [PubMed] [Google Scholar]
- Banks J. A. (2009). Selaginella and 400 million years of separation. Annu. Rev. Plant Biol. 60, 223–238 10.1146/annurev.arplant.59.032607.092851 [DOI] [PubMed] [Google Scholar]
- Barrero-Gil J., Garciadeblás B., Benito B. (2005). Sodium, potassium-ATPases in algae and oomycetes. J. Bioenerg. Biomembr. 37, 269–278 10.1007/s10863-005-6637-x [DOI] [PubMed] [Google Scholar]
- Baxter I., Tchieu J., Sussman M. R., Boutry M., Palmgren M. G., Gribskov M., Harper J. F., Axelsen K. B. (2003). Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 132, 618–628 10.1104/pp.103.021923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y., Lee S. M., Romanowsky S., Blank R., Sladek C., Chung W. S., Harper J. F. (2010). Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 154, 1158–1171 10.1104/pp.110.159038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch-Pedersen M. J., Pedersen B. P., Veierskov B., Nissen P., Palmgren M. G. (2009). Protons and how they are transported by proton pumps. Pflugers Arch. 457, 573–579 10.1007/s00424-008-0503-8 [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Coble A. J., Cohen L. D., Ch’ng T. H., Russo K. M., Long E. M., Armburst E. V. (2001). Identification and DNA sequence of a new H+-ATPase in the unicellular green alga Chlamydomonas reinhardtii (Chlorophyceae) J. Phycol. 37, 536–542 10.1046/j.1529-8817.2001.037004536.x [DOI] [Google Scholar]
- Courties C., Perasso R., Chrétiennot-Dinet M.-J., Gouy M., Guillou L., Troussellier M. (1998). Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Prasinophyceae). J. Phycol. 34, 844–849 10.1046/j.1529-8817.1998.340844.x [DOI] [Google Scholar]
- Curran A. C., Hwang I., Corbin J., Martinez S., Rayle D., Sze H., Harper J. F. (2000). Autoinhibition of a calmodulin-dependent calcium pump involves a structure in the stalk that connects the transmembrane domain to the ATPase catalytic domain. J. Biol. Chem. 275, 30301–30308 10.1074/jbc.M002047200 [DOI] [PubMed] [Google Scholar]
- Derelle E., Ferraz C., Rombauts S., Rouzé P., Worden A. Z., Robbens S., Partensky F., Degroeve S., Echeynié S., Cooke R., Saeys Y., Wuyts J., Jabbari K., Bowler C., Panaud O., Piégu B., Ball S. G., Ral J. P., Bouget F. Y., Piganeau G., De Baets B., Picard A., Delseny M., Demaille J., Van de Peer Y., Moreau H. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. U.S.A. 103, 11647–11652 10.1073/pnas.0604795103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S. J., Liu J., Hou Z., Mitra B. (2006). Conserved aspartic acid 714 in transmembrane segment 8 of the ZntA subgroup of P1B-type ATPases is a metal-binding residue. Biochemistry 45, 5923–5931 10.1021/bi0523456 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T. J., Colmer T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963 10.1111/j.1469-8137.2008.02531.x [DOI] [PubMed] [Google Scholar]
- Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999). Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780 10.1074/jbc.274.51.36774 [DOI] [PubMed] [Google Scholar]
- Gimmler H. (2000). Primary sodium plasma membrane ATPases in salt-tolerant algae: facts and fictions. J. Exp. Bot. 51, 1171–1178 10.1093/jexbot/51.348.1171 [DOI] [PubMed] [Google Scholar]
- Gonzalez R. J. (2011). The physiology of hyper-salinity tolerance in teleost fish: a review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. (in press). [DOI] [PubMed] [Google Scholar]
- Harper J. F., Hong B., Hwang I., Guo H. Q., Stoddard R., Huang J. F., Palmgren M. G., Sze H. (1998). A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J. Biol. Chem. 273, 1099–1106 10.1074/jbc.273.2.1099 [DOI] [PubMed] [Google Scholar]
- Haruta M., Burch H. L., Nelson R. B., Barrett-Wilt G., Kline K. G., Mohsin S. B., Young J. C., Otegui M. S., Sussman M. R. (2010). Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285, 17918–17929 10.1074/jbc.M110.101733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R. E., Lewis B. D., Spalding E. P., Sussman M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921 10.1126/science.280.5365.918 [DOI] [PubMed] [Google Scholar]
- Howe K., Bateman A., Durbin R. (2002). QuickTree: building huge neighbour-joining trees of protein sequences. Bioinformatics 18, 1546–1547 10.1093/bioinformatics/18.11.1546 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007). Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain D., Haydon M. J., Wang Y., Wong E., Sherson S. M., Young J., Camakaris J., Harper J. F., Cobbett C. S. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16, 1327–1339 10.1105/tpc.020487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Harper J. F., Liang F., Sze H. (2000). Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal autoinhibitory domain. Plant Physiol. 122, 157–168 10.1104/pp.122.1.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Maeda M., Fischer R., Verma A., Krebs J., Penniston J., Carafoli E. (1988). Identification and primary structure of a calmodulin binding domain of the Ca2+ pump of human erythrocytes. J. Biol. Chem. 263, 2905–2910 [PubMed] [Google Scholar]
- Kim Y. Y., Choi H., Segami S., Cho H. T., Martinoia E., Maeshima M., Lee Y. (2009). AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 58, 737–753 10.1111/j.1365-313X.2008.03770.x [DOI] [PubMed] [Google Scholar]
- Li X., Chanroj S., Wu Z., Romanowsky S. M., Harper J. F., Sze H. (2008). A distinct endosomal Ca2+/Mn2+ pump affects root growth through the secretory process. Plant Physiol. 147, 1675–1689 10.1104/pp.108.119909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Fang J., Docampo R. (2006). Molecular characterization of Trypanosoma brucei P-type H+-ATPases. J. Biol. Chem. 281, 21963–21973 10.1074/jbc.M601559200 [DOI] [PubMed] [Google Scholar]
- Luo S., Scott D. A., Docampo R. (2002). Trypanosoma cruzi H+-ATPase 1 (TcHA1) and 2 (TcHA2) genes complement yeast mutants defective in H+ pumps and encode plasma membrane P-type H+-ATPases with different enzymatic properties. J. Biol. Chem. 277, 44497–44506 10.1074/jbc.M111072200 [DOI] [PubMed] [Google Scholar]
- Malmström S., Askerlund P., Palmgren M. G. (1997). A calmodulin-stimulated Ca2+-ATPase from plant vacuolar membranes with a putative regulatory domain at its N-terminus. FEBS Lett. 400, 324–328 10.1016/S0014-5793(96)01448-2 [DOI] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275, 17762–17770 10.1074/jbc.M909690199 [DOI] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Maréchal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernández E., Fukuzawa H., González-Ballester D., González-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riaño-Pachón D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martínez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. F., Doherty M. L., López-Marqués R. L., Weimar T., Dupree P., Palmgren M. G., Pittman J. K., Williams L. E. (2008). ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 146, 116–128 10.1104/pp.107.110817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. F., Krijger G. C., Baccarini P. J., Hall J. L., Williams L. E. (2003). Functional expression of AtHMA4, a P1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J. 35, 164–176 10.1046/j.1365-313X.2003.01790.x [DOI] [PubMed] [Google Scholar]
- Møller A. B., Asp T., Holm P. B., Palmgren M. G. (2008). Phylogenetic analysis of P5 P-type ATPases, a eukaryotic lineage of secretory pathway pumps. Mol. Phylogenet. Evol. 46, 619–634 10.1016/j.ympev.2007.10.023 [DOI] [PubMed] [Google Scholar]
- Møller J. V., Juul B., le Maire M. (1996). Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta 1286, 1–51 [DOI] [PubMed] [Google Scholar]
- Moreno I., Norambuena L., Maturana D., Toro M., Vergara C., Orellana A., Zurita-Silva A., Ordenes V. R. (2008). AtHMA1 is a thapsigargin-sensitive Ca2+/heavy metal pump. J. Biol. Chem. 283, 9633–9641 10.1074/jbc.M800736200 [DOI] [PubMed] [Google Scholar]
- Morth J. P., Pedersen B. P., Buch-Pedersen M. J., Andersen J. P., Vilsen B., Palmgren M. G., Nissen P. (2011). A structural overview of the plasma membrane Na+,K+-ATPase and H+-ATPase ion pumps. Nat. Rev. Mol. Cell Biol. 12, 60–70 10.1038/nrm3031 [DOI] [PubMed] [Google Scholar]
- Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007). Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 10.1038/nature06419 [DOI] [PubMed] [Google Scholar]
- Olesen C., Picard M., Winther A. M., Gyrup C., Morth J. P., Oxvig C., Møller J. V., Nissen P. (2007). The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 10.1038/nature06418 [DOI] [PubMed] [Google Scholar]
- Palmgren M. G. (2001). Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845 10.1146/annurev.arplant.52.1.817 [DOI] [PubMed] [Google Scholar]
- Palmgren M. G., Harper J. F. (1999). Pumping with plant P-type ATPases. J. Exp. Bot. 50, 883–893 10.1093/jexbot/50.suppl_1.883 [DOI] [Google Scholar]
- Palmgren M. G., Sommarin M., Serrano R., Larsson C. (1991). Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266, 20470–20475 [PubMed] [Google Scholar]
- Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. (2007). Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 10.1038/nature06417 [DOI] [PubMed] [Google Scholar]
- Popova L., Balnokin Y., Dietz K. J., Gimmler H. (1999). Characterization of phosphorylated intermediates synthetized during the catalytic cycle of the sodium adenosine triphosphatase in the plasma membrane of the marine unicellular alga Tetraselmis (Platymonas) viridis. J. Plant Physiol. 155, 302–309 10.1016/S0176-1617(99)80109-6 [DOI] [Google Scholar]
- Popova L. G., Shumkova G. A., Andreev I. M., Balnokin Y. V. (2005). Functional identification of electrogenic Na+-translocating ATPase in the plasma membrane of the halotolerant microalga Dunaliella maritima. FEBS Lett. 579, 5002–5006 10.1016/j.febslet.2005.07.087 [DOI] [PubMed] [Google Scholar]
- Portillo F., de Larrinoa I. F., Serrano R. (1989). Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett. 247, 381–385 10.1016/0014-5793(89)81375-4 [DOI] [PubMed] [Google Scholar]
- Poulsen L. R., López-Marqués R. L., Palmgren M. G. (2008a). Flippases: still more questions than answers. Cell. Mol. Life Sci. 65, 3119–3125 10.1007/s00018-008-8341-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. R., López-Marqués R. L., McDowell S. C., Okkeri J., Licht D., Schulz A., Pomorski T., Harper J. F., Palmgren M. G. (2008b). The Arabidopsis P4-ATPase ALA3 localizes to the Golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 20, 658–676 10.1105/tpc.107.054767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qudeimat E., Faltusz A. M., Wheeler G., Lang D., Brownlee C., Reski R., Frank W. (2008). A PIIB-type Ca2+-ATPase is essential for stress adaptation in Physcomitrella patens. Proc. Natl. Acad. Sci. U.S.A. 105, 19555–19560 10.1073/pnas.0800864105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P. F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin-I T., Kuroki Y., Toyoda A., Suzuki Y., Hashimoto S., Yamaguchi K., Sugano S., Kohara Y., Fujiyama A., Anterola A., Aoki S., Ashton N., Barbazuk W. B., Barker E., Bennetzen J. L., Blankenship R., Cho S. H., Dutcher S. K., Estelle M., Fawcett J. A., Gundlach H., Hanada K., Heyl A., Hicks K. A., Hughes J., Lohr M., Mayer K., Melkozernov A., Murata T., Nelson D. R., Pils B., Prigge M., Reiss B., Renner T., Rombauts S., Rushton P. J., Sanderfoot A., Schween G., Shiu S. H., Stueber K., Theodoulou F. L., Tu H., Van de Peer Y., Verrier P. J., Waters E., Wood A., Yang L., Cove D., Cuming A. C., Hasebe M., Lucas S., Mishler B. D., Reski R., Grigoriev I. V., Quatrano R. S., Boore J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. (1995). Chlamydomonas reinhardtii as the photosynthetic yeast. Annu. Rev. Genet. 29, 209–230 10.1146/annurev.ge.29.120195.001233 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A., Benito B. (2010). Sodium or potassium efflux ATPase – a fungal, bryophyte, and protozoal ATPase. Biochim. Biophys. Acta 1798, 1841–1853 10.1016/j.bbamem.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469, 1–30 [DOI] [PubMed] [Google Scholar]
- Rudolph H. K., Antebi A., Fink G. R., Buckley C. M., Dorman T. E., LeVitre J., Davidow L. S., Mao J. I., Moir D. T. (1989). The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell 58, 133–145 10.1016/0092-8674(89)90410-8 [DOI] [PubMed] [Google Scholar]
- Sáez A. G., Lozano E., Zaldívar-Riverón A. (2009). Evolutionary history of Na,K-ATPases and their osmoregulatory role. Genetica 136, 479–490 10.1007/s10709-009-9356-0 [DOI] [PubMed] [Google Scholar]
- Seigneurin-Berny D., Gravot A., Auroy P., Mazard C., Kraut A., Finazzi G., Grunwald D., Rappaport F., Vavasseur A., Joyard J., Richaud P., Rolland N. (2006). HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 281, 2882–2892 10.1074/jbc.M508333200 [DOI] [PubMed] [Google Scholar]
- Serrano R. (1989). Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim. Biophys. Acta 947, 1–28 [DOI] [PubMed] [Google Scholar]
- Skou J. C. (1957). The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim. Biophys. Acta 23, 394–401 10.1016/0006-3002(57)90343-8 [DOI] [PubMed] [Google Scholar]
- Skulachev V. P. (1988). Membrane Bioenergetics. Berlin: Springer-Verlag [Google Scholar]
- Sørensen D. M., Buch-Pedersen M. J., Palmgren M. G. (2010). Structural divergence between the two subgroups of P5 ATPases. Biochim. Biophys. Acta 1797, 846–855 10.1016/j.bbabio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Spalding E. P., Harper J. F. (2011). The ins and outs of cellular Ca2+ transport. Curr. Opin. Plant Biol. 14, 715–720 10.1016/j.pbi.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F., Olsson A., Piotrowski M., Rosenquist M., Ottman C., Larsson C., Oecking C., Sommarin M. (1999). Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11, 2379–2392 10.1105/tpc.11.12.2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H., Liang F., Hwang I., Curran A. C., Harper J. F. (2000). Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 433–462 10.1146/annurev.arplant.51.1.433 [DOI] [PubMed] [Google Scholar]
- Tester M., Davenport R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91, 503–527 10.1093/aob/mcg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 10.1038/35048692 [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium (2012). Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000). Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Ǻ resolution. Nature 405, 647–655 10.1038/35015017 [DOI] [PubMed] [Google Scholar]
- Venema K., Palmgren M. G. (1995) Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J. Biol. Chem. 270, 19659–19667 10.1074/jbc.270.33.19659 [DOI] [PubMed] [Google Scholar]
- Verret F., Gravot A., Auroy P., Leonhardt N., David P., Nussaume L., Vavasseur A., Richaud P. (2004). Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 576, 306–312 10.1016/j.febslet.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Whittamore J. M. (2012). Osmoregulation and epithelial water transport: lessons from the intestine of marine teleost fish. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 182, 1–39 10.1007/s00360-011-0601-3 [DOI] [PubMed] [Google Scholar]
- Williams L. E., Mills R. F. (2005). P1B-ATPases – an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 10, 491–502 10.1016/j.tplants.2005.08.008 [DOI] [PubMed] [Google Scholar]
- Wong C. K., Cobbett C. S. (2009). HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 181, 71–78 10.1111/j.1469-8137.2008.02637.x [DOI] [PubMed] [Google Scholar]
- Wu Z., Liang F., Hong B., Young J. C., Sussman M. R., Harper J. F., Sze H. (2002). An endoplasmic reticulum-bound Ca2+/Mn2+ pump, ECA1, supports plant growth and confers tolerance to Mn2+ stress. Plant Physiol. 130, 128–137 10.1104/pp.004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuytack F., Raeymaekers L., Missiaen L. (2003). PMR1/SPCA Ca2+ pumps and the role of the Golgi apparatus as a Ca2+ store. Pflugers Arch. 446, 148–153 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Blumwald E. (2005). Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 10, 615–620 10.1016/j.tplants.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Hackett J. D., Ciniglia C., Pinto G., Bhattacharya D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 10.1093/molbev/msh075 [DOI] [PubMed] [Google Scholar]
- Zhu X., Caplan J., Mamillapalli P., Czymmek K., Dinesh-Kumar S. P. (2010). Function of endoplasmic reticulum calcium ATPase in innate immunity-mediated programmed cell death. EMBO J. 29, 1007–1018 10.1038/emboj.2009.402 [DOI] [PMC free article] [PubMed] [Google Scholar]