Abstract
In angiosperms, sucrose uptake transporters (SUTs) have important functions especially in vascular tissue. Here we explore the evolutionary origins of SUTs by analysis of angiosperm SUTs and homologous transporters in a vascular early land plant, Selaginella moellendorffii, and a non-vascular plant, the bryophyte Physcomitrella patens, the charophyte algae Chlorokybus atmosphyticus, several red algae and fission yeast, Schizosaccharomyces pombe. Plant SUTs cluster into three types by phylogenetic analysis. Previous studies using angiosperms had shown that types I and II are localized to the plasma membrane while type III SUTs are associated with vacuolar membrane. SUT homologs were not found in the chlorophyte algae Chlamydomonas reinhardtii and Volvox carterii. However, the characean algae Chlorokybus atmosphyticus contains a SUT homolog (CaSUT1) and phylogenetic analysis indicated that it is basal to all other streptophyte SUTs analyzed. SUTs are present in both red algae and S. pombe but they are less related to plant SUTs than CaSUT1. Both Selaginella and Physcomitrella encode type II and III SUTs suggesting that both plasma membrane and vacuolar sucrose transporter activities were present in early land plants. It is likely that SUT transporters are important for scavenging sucrose from the environment and intracellular compartments in charophyte and non-vascular plants. Type I SUTs were only found in eudicots and we conclude that they evolved from type III SUTs, possibly through loss of a vacuolar targeting sequence. Eudicots utilize type I SUTs for phloem (vascular tissue) loading while monocots use type II SUTs for phloem loading. We show that HvSUT1 from barley, a type II SUT, reverted the growth defect of the Arabidopsis atsuc2 (type I) mutant. This indicates that type I and II SUTs evolved similar (and interchangeable) phloem loading transporter capabilities independently.
Keywords: sucrose transporter, SUT, phylogeny, evolution
Introduction
In angiosperms, H+-coupled sucrose-uptake transporters (SUTs) are involved in the long-distance transport of sucrose. They function to load sucrose into the phloem (vascular tissue) and in uptake of sucrose into sink tissues such as seeds and flowers. The physiological functions of SUTs have been reviewed recently (Braun and Slewinski, 2009; Kuhn and Grof, 2010; Ayre, 2011). In this paper we focus on the phylogenetic relationship between SUTs in photosynthetic organisms from algae to angiosperms. SUTs are members of the glycoside–pentoside–hexuronide (GPH): cation symporter family which is distantly related to the major facilitator superfamily (Chang et al., 2004). Transporters homologous to SUTs are found in bacteria, fungi, and animals. SUT function in angiosperms predominates in the phloem, and yet SUTs clearly existed prior to evolution of phloem tissue. So it is interesting to investigate SUT sequences in more simple non-vascular land plants and algae to understand the origins of angiosperm SUTs. Analysis of the structure/function of more divergent homologs may also help us understand the SUT transport mechanism.
The SUT homolog SpSUT1 from Schizosaccharomyces pombe is a proton-coupled α-glucoside symporter that has a higher affinity for maltose than sucrose (Reinders and Ward, 2001). SUT homologs in animals, including humans, are associated with melanosomes and mutations in the respective genes generally cause hypopigmentation. MATP (Harada et al., 2001) in humans is encoded by AIM1 or SLC45A2 and mutations result in oculocutaneous albinism type 4 (OCA4; Inagaki et al., 2006). In horse, MATP is associated with cream coat color (Mariat et al., 2003). Similarly, in mouse a SUT homolog is encoded by the underwhite (uw) gene (Newton et al., 2001; Costin et al., 2003), mutations in the AIM1 gene in medaka fish reduce melanin content (Fukamachi et al., 2001) and in birds, plumage color is controlled by alleles of the gene encoding MATP (Gunnarsson et al., 2007). The only animal SUT homolog for which transport activity has been reported is SCRT from Drosophila, SCRT is able to transport sucrose and it is localized to subcellular vesicles that resemble melanosomes (Meyer et al., 2011).
In plants, the first SUT was cloned using yeast functional expression (Riesmeier et al., 1992). SUTs are encoded by small gene families in all flowering plants and phylogenetic analysis shows the presence of three groups of SUTs called type I, II, and III (Aoki et al., 2003). Interestingly, type I SUTs are only found in eudicot species. Type I SUTs are necessary for essential functions in eudicots such as phloem loading (Riesmeier et al., 1994; Gottwald et al., 2000) and normal pollen function (Sivitz et al., 2008). All land plant species contain type II and III SUTs. Monocot species utilize type II SUTs for phloem loading (Slewinski et al., 2009). This indicates that evolution of type I SUTs coincided with monocot and eudicot divergence. Type III SUTs were first cloned from Arabidopsis, potato and tomato and characterized as H+-coupled symporters (Weise et al., 2000). Type III SUTs are localized at the vacuolar membrane (Endler et al., 2006; Reinders et al., 2008) and function in sucrose-uptake into the cytoplasm (Reinders et al., 2008; Schulz et al., 2011).
Advances in genome sequencing allow us for the first time to investigate the origins of angiosperm SUTs. Complete genome sequence is available for representative bryophyte (Physcomitrella patens), lycophyte (Selaginella moellendorffii), and chlorophytes (Chlamydomonas reinhardtii and Volvox carterii). In addition, partial sequence is available for the red algae Galdieria sulphuraria and Cyanidioschyzon merolae and EST sequence is available for several charophyte algae (Timme and Delwiche, 2010). The main questions that we can address by phylogenetic analysis are whether type I SUTs were derived from type II or type III SUTs and whether both type II and III SUTs were represented in the earliest land plants and algae.
Materials and Methods
SUT protein sequences
All SUT protein sequences were obtained from the following species in which genome sequence is available: the eudicot Arabidopsis thaliana, the monocot rice (Oryza sativa), the lycophyte Selaginella moellendorffii, and the bryophyte Physcomitrella patens using BLAST searches on the Phytozome website1. The same database was searched for SUT protein sequences from the chlorophytes Chlamydomonas reinhardtii and Volvox carterii. Dr. Charles F. Delwiche and Mr. James Thierer, University of Maryland, provided support by searching their algal sequence database (Timme and Delwiche, 2010) for SUT homologs in the charophytes Chlorokybus atmosphyticus, Klebsormidium flaccidum, Spirogyra pratensis, Coleochaete sp., Chaetosphaeridium globosum, Penium marinum, and Nitella hyalina. In addition, the genome sequence of the red algae Galdieria sulphuraria2 (Barbier et al., 2005) and Cyanidioschyzon merolae3 were searched for the presence of SUTs. Sequences of Galdieria sulphuraria SUTs were provided by Dr. Andreas P. M. Weber, University of Düsseldorf.
Phylogenetic analysis
Multiple protein sequence alignments were generated with Clustal X (Larkin et al., 2007). The variable length N- and C-terminal regions of the alignment were removed. Percent protein sequence identity is presented, based on the trimmed alignment, as average for each cluster (±SD). Sequences with greater than 90% overall sequence identity were not included in the phylogenetic analysis. Phylogenetic analysis was performed through the iPlant Collaborative website4. Maximum likelihood analysis was done using PhyML 3.0 with 100 bootstrap replicates (Guindon and Gascuel, 2003; Guindon et al., 2010). Trees were visualized using the FigTree program5.
Complementation of the Arabidopsis atsuc2-1 mutant
Constructs for plant transformation contained the AtSUC2 (At1g22710) promoter, coding region of either AtSUC2 or HvSUT1 (CAJ20123.1) cDNAs and the AtSUC2 3′UTR. The AtSUC2 promoter (2 kb) was amplified using the primers 5′ggggacaactttgtatagaaaagttgtaccagatttcggtaaatt and 5′ggggactgcttttttgtacaaacttgaagaaagtaagaaaaaaaagaaatt and cloned into the pDONR P4-P1R vector (Invitrogen) using BP clonase II. The AtSUC2 ORF was amplified using 5′caccggtttgtcaaatatggtcagccatcc and 5′atgaaatcccatagtagctttgaag. The HvSUT1 ORF was amplified using 5′caccggtttgtcaaatatggcgcgcggcggcgg and 5′tcagtgaccgccgccgctgac. The two ORFs were cloned into pENTR/D/TOPO (Invitrogen). The AtSUC2 3′UTR (500 bp) was amplified using 5′ggggacagctttcttgtacaaagtggattgaattttagcagtggt and 5′ggggacaactttgtataataaagttgaattaactaaaatagataa and cloned into pDONR P2R–P3 (Invitrogen). Constructs were assembled into the pB7m34GW binary vector (Karimi et al., 2005) by directional multi-fragment recombination cloning using LR Clonase Plus (Invitrogen). Agrobacterium tumefaciens strain C58C1 containing these plasmids was used to transform heterozygous atsuc2-1 Arabidopsis (WS ecotype) plants (Gottwald et al., 2000) by the floral dipping method (Clough and Bent, 1998). Basta-resistant transformed plants were selected on soil. Homozygous atsuc2-1 mutants were identified by PCR.
Results
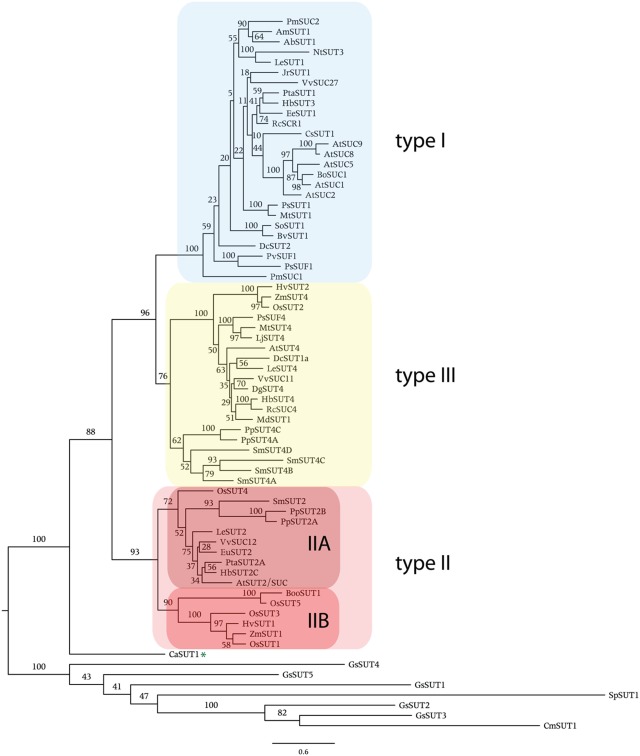
Phylogenetic analysis shows that angiosperm SUTs form three main groups (Figure 1). Here, we follow the nomenclature of Aoki et al. (2003) and name these groups type I, II, and III. All SUTs encoded by the eudicot Arabidopsis thaliana (seven sequences), the monocot Oryza sativa (five), the basal non-vascular moss Physcomitrella patens (four), the vascular non-seed spikemoss Selaginella moellendorffii (five), and the yeast Schizosaccharomyces pombe (one) were included as representatives of those groups where full genome sequence is available. In addition, SUTs that have been functionally characterized were included. In Figure 1 the commonly used abbreviated names for the transporter genes are listed. In Table 1 the protein accession number, gene name, protein length, and species are presented and sorted by phylogenetic group. Sequences from single-celled red algae Cyanidioschyzon merolae and Galdieria sulphuraria that are homologous to SUTs but did not cluster with SUTs encoded by land plants are present in a separate group in Figure 1 and Table 1. Additionally, the fungal sequence SpSUT1 from Schizosaccharomyces pombe is homologous (Reinders and Ward, 2001) but did not cluster with plant SUTs. The genome sequence from two chlorophyte green algae, Chlamydomonas reinhardtii and Volvox carteri is available, however no SUT sequences were identified in these chlorophytes. A single charophyte algal sequence from Chlorokybus atmosphyticus (CaSUT1) is present just basal to the plant SUT sequences but does not cluster with type I, II, or II (marked with an asterisk in Figure 1).
Figure 1.
Phylogenetic analysis of plant sucrose transporters and homologs. Protein alignment was done using Clustal X. Sequences with greater than 90% identity were not used in construction of the tree (they are shown in Table 1). The variable length N- and C-terminal regions were trimmed from the alignment. The maximum likelihood tree was generated using PhyML 3.0. Numbers indicate percent of 100 bootstrap analyses. Asterisk indicates the single charophyte SUT sequence, CaSUT1, from Chlorokybus atmosphyticus.
Table 1.
Sucrose transporter homologs.
| Type | Organism | Common name | Gene | Prot ID | Length (aa) | Reference |
|---|---|---|---|---|---|---|
| I | Alonsoa meridionalis | AmSUT1 | AAF04295 | 502 | Knop et al. (2001) | |
| I | Arabidopsis thaliana | Thale cress | AtSUC1 (At1g71880) | CAA53147 | 513 | Sauer and Stolz (1994) |
| I | Arabidopsis thaliana | Thale cress | AtSUC2 (At1g22710) | CAA53150 | 512 | Sauer and Stolz (1994) |
| I | Arabidopsis thaliana | Thale cress | AtSUC5 (At1g71890) | AAG52226 | 512 | Theologis et al. (2000) |
| I | Arabidopsis thaliana | Thale cress | AtSUC8 (At2g14670) | AAC69375 | 492 | Lin et al. (1999) |
| I | Arabidopsis thaliana | Thale cress | AtSUC9 (At5g06170) | BAB09682 | 491 | Tabata et al. (2000) |
| I | Asarina barclaiana (Maurandya barclaiana) | Twining snapdragon | AsSUT1 | AAF04294 | 510 | Knop et al. (2001) |
| I | Beta vulgaris | Sugar beet | BvSUT1 | CAA58730 | 523 | Vaughn et al. (2002) |
| I | Brassica oleracea | Broccoli | BoSUC1 | AAL58071 | 513 | Gapper et al. (2005) |
| I | Citrus sinensis | Sweet orange | CsSUT1 | AAM29150 | 528 | Li et al. (2003) |
| I | Daucus carota | Carrot | DcSUT2 | CAA76369 | 515 | Shakya and Sturm (1998) |
| I | Euphorbia esula | Leafy spurge | EeSUT1 | AAF65765 | 530 | |
| I | Hevea brasiliensis | Para rubber tree | HbSUT3/HbSUT1A | ABK60190 | 535 | Tang et al. (2010) |
| I | Juglans regia | English walnut | JrSUT1 | AAU11810 | 516 | Decourteix et al. (2006) |
| I | Solanum lycopersicum (Lycopersicon esculentum) | Tomato | LeSUT1 | CAA57726 | 512 | Barker et al. (2000) |
| I | Medicago truncatula | Barrel medic | MtSUT1 | TC175182, TC184317* | 525 | http://compbio.dfci.harvard.edu/tgi/ |
| I | Nicotiana tabacum | Common tobacco | NtSUT3 | AAD34610 | 521 | Lemoine et al. (1999) |
| I | Phaseolus vulgaris | Common bean | PvSUF1 | ABB30165 | 509 | Zhou et al. (2007) |
| I | Pisum sativum | Pea | PsSUT1 | AAD41024 | 524 | Tegeder et al. (1999) |
| I | Pisum sativum | Pea | PsSUF1 | ABB30163 | 511 | Zhou et al. (2007) |
| I | Plantago major | Common plantain | PmSUC1 | CAA59113 | 503 | Gahrtz et al. (1996) |
| I | Plantago major | Common plantain | PmSUC2 | CAA53390 | 510 | Gahrtz et al. (1996) |
| I | Populus trichocarpa | Black poplar | PtaSUT1/PtSUT1.2 | 18221401† | 535 | Tuskan et al. (2006) |
| I | Ricinus communis | Castor bean | RcSCR1 | CAA83436 | 533 | Weig and Komor (1996) |
| I | Spinacia oleracea | Spinach | SoSUT1 | CAA47604 | 526 | Riesmeier et al. (1992) |
| I | Vitis vinifera | Grape | VvSUC27 | AAF08331 | 505 | Davies et al. (1999) |
| IIA | Arabidopsis thaliana | Thale cress | AtSUT2/AtSUC3 (At2g02860) | CAB92307 | 595 | Meyer et al. (2000), Schulze et al. (2000) |
| IIA | Eucommia ulmoides | Gutta-percha tree | EuSUT2 | AAX49396 | 604 | Pang et al. (2008) |
| IIA | Hevea brasiliensis | Para rubber tree | HbSUT2C/HbSUT2A | CAM33449 | 539 | Dusotoit-Coucaud et al. (2009) |
| IIA | Oryza sativa japonica | Rice | OsSUT4 (Os02g58080) | BAC67164 | 595 | Aoki et al. (2003) |
| IIA | Physcomitrella patens | PpSUT2A | 18051919† | 635 | Rensing et al. (2008) | |
| IIA | Physcomitrella patens | PpSUT2B | 18064412† | 557 | Rensing et al. (2008) | |
| IIA | Populus trichocarpa | Black poplar | PtaSUT2A | 18241865† | 602 | Tuskan et al. (2006) |
| IIA | Selaginella moellendorffii | SmSUT2 | 15412113† | 521 | Banks et al. (2011) | |
| IIA | Solanum lycopersicum (Lycopersicon esculentum) | Tomato | LeSUT2 | AAG12987 | 605 | Barker et al. (2000) |
| IIA | Vitis vinifera | Grape | VvSUC12 | AAF08330 | 612 | Davies et al. (1999) |
| IIB | Bambusa oldhamii (Dendrocalamopsis oldhamii) | Bamboo | BooSUT1 | AAY43226 | 525 | |
| IIB | Hordeum vulgare | Barley | HvSUT1 | CAB75882 CAJ20123 | 523 | Weschke et al. (2000), Sivitz et al. (2005) |
| IIB | Oryza sativa japonica | Rice | OsSUT1 (Os03g07480) | BAA24071 | 537 | Hirose et al. (1997) |
| IIB | Oryza sativa japonica | Rice | OsSUT3 (Os10g26740) | BAB68368 | 506 | Aoki et al. (2003) |
| IIB | Oryza sativa japonica | Rice | OsSUT5 (Os02g36700) | BAC67165 | 535 | Aoki et al. (2003) |
| IIB | Saccharum hybrid cultivar | Sugarcane | ShSUT1# | AAV41028 | 517 | Rae et al. (2005) |
| IIB | Zea mays | Corn | ZmSUT1 | BAA83501 | 521 | Aoki et al. (1999) |
| III | Arabidopsis thaliana | Thale cress | AtSUT4 (At1g09960) | AAL59915 | 510 | Weise et al. (2000) |
| III | Datisca glomerata | Durango root | DgSUT4 | CAG70682 | 498 | Schubert et al. (2010) |
| III | Daucus carota | Carrot | DcSUT1a | CAA76367 | 501 | Shakya and Sturm (1998) |
| III | Hevea brasiliensis | Para rubber tree | HbSUT4/HbSUT4A | ABK60191 | 498 | Tang et al. (2010) |
| III | Hordeum vulgare | Barley | HvSUT2 | CAB75881 | 506 | Weschke et al. (2000) |
| III | Lotus japonicus | LjSUT4 | CAD61275 | 511 | Flemetakis et al. (2003) | |
| III | Malus x domestica | Apple | MdSUT1 | AAR17700 | 499 | Fan et al. (2009) |
| III | Medicago truncatula | Barrel medic | MtSUT4 | 17466537† | 504 | |
| III | Oryza sativa japonica | Rice | OsSUT2 (Os12g44380) | BAC67163 | 501 | Aoki et al. (2003) |
| III | Physcomitrella patens | PpSUT4A | 18040351† | 532 | Rensing et al. (2008) | |
| III | Physcomitrella patens | PpSUT4B# | 18037160† | 500 | Rensing et al. (2008) | |
| III | Physcomitrella patens | PpSUT4C | 18053343† | 524 | Rensing et al. (2008) | |
| III | Pisum sativum | Pea | PsSUF4 | ABB30162 | 507 | Zhou et al. (2007) |
| III | Ricinus communis | Castor bean | RcSUC4 | AAU21439 | 509 | |
| III | Selaginella moellendorffii | SmSUT4A | 15419655† | 514 | Banks et al. (2011) | |
| III | Selaginella moellendorffii | SmSUT4B | 15407332† | 492 | Banks et al. (2011) | |
| III | Selaginella moellendorffii | SmSUT4C | 15417411† | 493 | Banks et al. (2011) | |
| III | Selaginella moellendorffii | SmSUT4D | 15402611† | 531 | Banks et al. (2011) | |
| III | Solanum lycopersicum (Lycopersicon esculentum) | Tomato | LeSUT4 | AAG09270 | 501 | Weise et al. (2000) |
| III | Vitis vinifera | Grape | VvSUC11 | AAF08329 | 501 | Davies et al. (1999) |
| III | Zea mays | Corn | ZmSUT4 | AAT35810 | 501 | |
| Chlorokybus atmosphyticus | Soil alga | CaSUT1 | ||||
| Cyanidioschyzon merolae | CmSUT1 | CMO328C‡ | 502 | Matsuzaki et al. (2004) | ||
| Galdieria sulphuraria | GsSUT1 | Gs18190§ | 471 | Weber et al. (2004), Barbier et al. (2005) | ||
| Galdieria sulphuraria | GsSUT2 | Gs34550§ | 546 | Weber et al. (2004), Barbier et al. (2005) | ||
| Galdieria sulphuraria | GsSUT3 | Gs56570§ | 430 | Weber et al. (2004), Barbier et al. (2005) | ||
| Galdieria sulphuraria | GsSUT4 | Gs29860§ | 526 | Weber et al. (2004), Barbier et al. (2005) | ||
| Galdieria sulphuraria | GsSUT5 | Gs08920§ | 638 | Weber et al. (2004), Barbier et al. (2005) | ||
| Schizosaccharomyces pombe | Fission yeast | SpSUT1 | NP594387 | 553 | Reinders and Ward (2001) |
*sequence from DFCI (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb = medicago).
†sequence from Phytozome v7.0 (http://www.phytozome.net/).
#not included in the phylogenetic analysis (>90% identical to another SUT).
‡sequence from Cyanidioschyzonmerolae genome project (http://merolae.biol.s.u-tokyo.ac.jp/).
§sequence from Galdieriasulphuraria genome project (http://genomics.msu.edu/galdieria/index.html).
Type I SUTs
Type I SUTs were only found in eudicots. The 26 type I SUTs analyzed here cluster into a single group with an average of 69% (±5%) identity. The lack of type I SUTs in non-vascular land plants (Physcomitrella), vascular non-seed land plants (Selaginella), and the monocot branch of angiosperms indicates that development of type I SUTs occurred after divergence of monocots and eudicots, around 150 MYR ago (Laroche et al., 1995). Genome sequence representing early diverging eudicots such as Papaver sp. (poppy) and Ranunculus sp. (buttercup; Soltis et al., 2003) would be useful to more clearly determine the origins of type I SUTs. It is interesting that type I SUT genes were amplified in Arabidopsis and acquired specialized functions. Arabidopsis thaliana has five genes in this group (Figure 1; Table 1) and an additional two that have been identified as pseudogenes (Sauer et al., 2004) that were not included in the analysis.
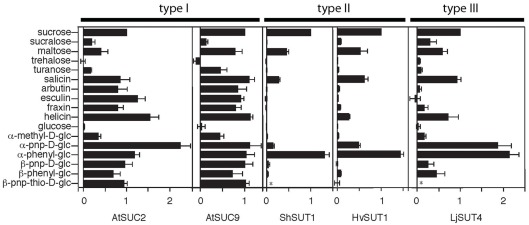
In Arabidopsis thaliana, type I SUTs display specialization in both expression and transport function. AtSUC2 is necessary for loading sucrose into the phloem (Gottwald et al., 2000). It has a Km (affinity) for sucrose of 1.4 mM (Chandran et al., 2003) and a wide substrate specificity for α and β glucosides that is shared with other type I SUTs (Figure 2; Chandran et al., 2003). AtSUC1 transport activity is very similar to AtSUC2 but its expression pattern is quite different. AtSUC1 is expressed in trichomes, pollen and roots (Sivitz et al., 2007). AtSUC1 is necessary for normal pollen function (Sivitz et al., 2008). Expression of AtSUC1 in the phloem, under control of the AtSUC2 promoter, has been shown to revert the growth defects of atsuc2 mutants (Wippel and Sauer, 2011). There are also examples of type I SUTs with modified transport activity. AtSUC9 has a much higher affinity for sucrose compared to other type I SUTs (66 μM; Sivitz et al., 2007) while the substrate specificity is typical of other type I SUTs (Figure 2; Sivitz et al., 2007).
Figure 2.
Substrate specificities of type I (AtSUC2, AtSUC9), type II (ShSUT1, HvSUT1), and type III (LjSUT4) plant sucrose transporters. Transport activity was assayed by expression in Xenopus oocytes and two-electrode voltage clamping. Oocytes were bathed in sodium Ringer solution containing substrates at concentrations between 0.5 and 25 mM (depending on the transporter affinity and substrate solubility). All currents were normalized to sucrose-dependent currents and are presented as mean ± SE with at least three oocytes per mean. *Indicates substrate not tested. Modified with permission from Chandran et al. (2003), Sivitz et al. (2005, 2007), Reinders et al. (2006, 2008).
Type II SUTs
Type II SUT sequences were identified in eudicots, monocots, non-vascular land plants (Physcomitrella), and vascular non-seed land plants (Selaginella). A total of 16 SUT sequences clustered in the type II group with an average of 62% (±9%) identity. The type II group was divided into two subgroups IIA and IIB. These two subgroups were identified previously (Braun and Slewinski, 2009). There is also a structural difference between type IIA and IIB SUTs. Type IIA proteins have a longer central cytoplasmic loop compared to type IIB SUTs. This is reflected in the average length of proteins in type IIA of 587 amino acids (aa) compared to 523 aa in type IIB (Table 1). Each angiosperm genome appears to have one gene in the IIA subgroup. Sequences from Physcomitrella (two) and Selaginella (one) are also included in the IIA subgroup. PpSUT2A and B from Physcomitrella and SmSUT2 contain longer central loops with conserved sequence characteristic of angiosperm type IIA transporters. Overall, this indicates that a type IIA transporter with a longer central loop was an ancestral form of the type II SUTs found in angiosperms.
The type IIB subgroup is monocot specific, rice encodes three type IIB transporters. This group contains the monocot phloem loading SUTs. ZmSUT1 has been shown to be expressed in vascular tissue and to function in phloem loading (Slewinski et al., 2009). Similar to the amplification of type I SUTs in Arabidopsis, type IIB SUTs appear to have been amplified in rice. Transport activities of OsSUT1 and OsSUT5 were analyzed by expression in oocytes and electrophysiology. OsSUT5 was found to have a higher affinity for sucrose (2.3 mM) compared to OsSUT1 (7.5 mM) and the activity of OsSUT5 was found to be less pH dependent (Sun et al., 2010).
It is interesting to note that monocots and eudicots utilize different SUTs to load sucrose into the phloem. Differences in substrate specificity between type I SUTs such as AtSUC2 that transport sucrose into the phloem in eudicots and type II SUTs such as HvSUT1 that performs the same function in monocots have been identified (Chandran et al., 2003; Sivitz et al., 2005, 2007; Reinders et al., 2006, 2008; Sun et al., 2008). Figure 2 shows a summary of substrate specificity results for five sucrose transporters. AtSUC2 and AtSUC9 are both type I sucrose transporters and although AtSUC9 has approximately a 20-fold lower K0.5 for sucrose (Sivitz et al., 2007) compared to AtSUC2, they have almost identical substrate specificities. These type I SUTs transport the plant β-glucosides salicin, arbutin, esculin, fraxin, and helicin. Notably, arbutin, esculin, and fraxin are not transported by the type II transporters ShSUT1 and HvSUT1 (Figure 2). Synthetic β phenyl glucosides are also transported by type I and not by type II SUTs (Figure 2).
The differences in substrate specificity between type I and type II SUTs might suggest that the specificity of phloem loading in eudicots is different from that in monocots. It is possible that type I SUTs load other glucosides, in addition to sucrose, into the phloem. To begin to address this question we used either AtSUC2 or HvSUT1 to complement the Arabidopsis atsuc2-1 mutant (Gottwald et al., 2000). The homozygous atsuc2-1 mutant has greatly reduced growth and accumulates starch in source leaves due to its reduced ability for phloem loading (Figure 3A). By comparison, growth of the atsuc2-1 heterozygous plants is indistinguishable from wild-type (Figures 3A,B). As expected, the atsuc2-1 mutant growth phenotype was complemented by expression of the AtSUC2 gene. Expression of the HvSUT1 coding region driven by the AtSUC2 promoter also resulted in growth that was indistinguishable from wild-type (Figure 3B). The type II SUT HvSUT1 appears to revert the growth reduction caused by the loss of AtSUC2 in Arabidopsis. This indicates that differences in substrate specificity between type I and II SUTs might not reflect a significant difference in physiological function, although this result is preliminary. Further work is necessary to determine if HvSUT1 fully complements under different growth and stress conditions.
Figure 3.
Complementation of suc2-1 sucrose transporter mutant. (A) Arabidopsis plants heterozygous for the suc2-1 insertion left, plants homozygous for the suc2-1 insertion right. (B) Both AtSUC2 and HvSUT1 complemented the growth defect of the homozygous suc2-1 mutant. The WS wild-type (left) is shown for comparison. All plants shown in (A) and (B) are 8 weeks old.
Finally, the grouping of moss type II SUTs can give us a few more clues about the evolution and function of these ancestral type SUTs. The type II moss and spikemoss sequences cluster with type IIA and contain longer central loops. Both Physcomitrella and Selaginella lack type I and type IIB SUTs. If early vascular plants such as Selaginella have SUTs that function in phloem loading, those transporters are likely to be type IIA such as SmSUT2 and are different from those used by monocots and eudicots. Also, type IIA SUTs in angiosperms do not compensate for loss of the main phloem loading SUT as evidenced by mutant phenotypes of atsuc2 (Gottwald et al., 2000) and zmsut1 (Slewinski et al., 2009) mutants.
Type III SUTs
The first type III SUTs were isolated from Arabidopsis, tomato, potato, and barley and named AtSUT4, LeSUT4, StSUT4, and HvSUT2, respectively (Weise et al., 2000; Weschke et al., 2000). AtSUT4 from Arabidopsis and HvSUT2 from barley (Endler et al., 2006), LjSUT4 from Lotus japonicus (Reinders et al., 2008), and OsSUT2 from rice (Eom et al., 2011) were demonstrated to localize to the vacuole membrane. Twenty type III SUT sequences were included in this study (Figure 1; Table 1) and these have an average of 65% (±8%) identity. Each angiosperm genome appears to contain a single type III SUT gene. Both Selaginella and Physcomitrella contain multiple type III SUT genes. No type III SUT homologs have been identified in green algae.
Transport activity has been characterized in detail for type III SUT LjSUT4 (Reinders et al., 2008). The substrate specificity of LjSUT4 is intermediate between type I and II SUTs (Figure 2). Like other type III SUTs (Weise et al., 2000; Weschke et al., 2000) LjSUT4 functions as a H+-coupled sucrose-uptake transporter. This indicates that its physiological function in the vacuolar membrane is sucrose-uptake into the cytoplasm from the vacuolar lumen. This activity for AtSUT4 has been demonstrated in Arabidopsis vacuoles (Schulz et al., 2011).
SUTs in Chlorokybus atmosphyticus, Galdieria sulphuraria, Cyanidioschyzon merolae, and Schizosaccharomyces pombe
No SUT sequences were found in chlorophytes Chlamydomonas reinhardtii and Volvox carteri. Charophyte green algae are considered to represent ancestors of land plants. A single SUT sequence was found in the charophyte Chlorokybus atmosphyticus (CaSUT1). It did not cluster with type I, II, or III SUTs from land plants but appears to be basal to these clades (Figure 1). Since a complete genome sequence of a charophyte is not yet available it remains to be determined whether additional SUTs are present in charophyte genomes. The central loop of CaSUT1 is not extended as in type IIA SUTs. Also, the N-terminal sequence for CaSUT1 is not available so we could not determine if the putative vacuole targeting sequence is present (see Discussion).
Galdieria sulphuraria and Cyanidioschyzon merolae are closely related, unicellular red microalgae. While G. sulphuraria can grow on 27 different sugars and sugar alcohols (Gross and Schnarrenberger, 1995), C. merolae can not grow heterotrophically (Matsuzaki et al., 2004). Five SUT homologs were identified in G. sulphuraria (GsSUT1-5) and one, CmSUT1, was identified in the C. merolae genome (Figure 1; Table 1). This is consistent with the larger number of genes encoding transporters and enzymes involved in carbohydrate metabolism identified in G. sulphuraria compared to C. merolae (Barbier et al., 2005).
Discussion
The origin of plant SUTs in charophyte algae
SUTs function as H+-coupled cellular sucrose uptake transporters. In angiosperms, type I and II SUTs are localized to the plasma membrane while type III SUTs are localized to the vacuole membrane. They are important for the long-distance transport of sucrose in apoplastic phloem loaders (requiring transmembrane transport). Another important function for SUTs in angiosperms is in sucrose-uptake into sinks that are symplastically isolated such as seeds and pollen. The availability of bryophyte (non-vascular), lycophyte (early vascular), and algal genome sequences allows us to begin to analyze the origins of SUTs in land plants. The presence of CaSUT1 in the charophyte alga Chlorokybus atmosphyticus as well as the absence of SUTs in chlorophyte algae (Chlamydomonas reinhardtii and Volvox carterii) is consistent with the hypothesis that charophyte algae are ancestral to land plants (McCourt et al., 2004).
The physiological function of SUT homologs in Chlorokybus, which exists as small clusters of cells and in the unicellular red algae Galdieria and Cyanidioschyzon is currently unknown but will depend on their membrane localization. They are likely to function as H+-coupled symporters for glucoside uptake into the cytosol whether they are localized to the plasma membrane or an internal membrane. Interestingly, Cyanidioschyzon lacks a central vacuole (Barbier et al., 2005), so it is more likely that CmSUT1 is a plasma membrane transporter. Bryophytes lack true vascular tissue yet Physcomitrella contains both type IIA and type III SUTs. In angiosperms, type IIA SUTs are localized to the plasma membrane (Barker et al., 2000; Meyer et al., 2000) while type III SUTs are vacuolar (Endler et al., 2006; Reinders et al., 2008). Therefore, it is likely that Physcomitrella contains both plasma membrane and vacuolar SUTs but this will need to be determined experimentally. Long-distance transport of photosynthate in mosses involves leptoid cells and the mechanism appears to be symplasmic, involving plasmodesmata not transmembrane transport (Raven, 2003). Therefore, if SUTs are localized to the plasma membrane in bryophytes their function is not in phloem loading but may be involved in recovery of sucrose that is released to the apoplast. Although leptoid cells evolved independently of phloem, many groups of angiosperms that utilize a similar passive mechanism for phloem loading (Rennie and Turgeon, 2009) also encode SUTs. The function of type III SUTs in bryophytes is likely to be the same as in angiosperms. Sucrose is transiently stored in the vacuole in angiosperms and type III SUTs function in the vacuole membrane to return sucrose from the vacuole lumen to the cytoplasm (Reinders et al., 2008; Schulz et al., 2011). The more recent development of type I SUTs in eudicots and type IIB SUTs in monocots is likely to be linked to the evolution of active phloem loading requiring energy and transmembrane transport.
Putative vacuolar targeting motif in type III SUTs
Recently, a dileucine-like motif (LXXXLL) in the N-terminal cytoplasmic domain of the Arabidopsis monosaccharide transporter ESL1was shown to be necessary for localization of the transporter to the vacuole membrane (Yamada et al., 2010). Dileucine-like motifs are recognized by a clathrin-associated, heterotetrameric adaptor protein (AP-3) complex and function in sorting of vacuole membrane proteins in yeast (Vowels and Payne, 1998). Similar dileucine motifs contain an acidic residue spaced several residues prior to the leucine pair with a consensus of DXXLL or [DE]XXXL[LI] (Braulke and Bonifacino, 2009). The AP-3 complex has been shown to be necessary for normal vacuole function in Arabidopsis (Zwiewka et al., 2011). An LXXLL motif is found in the cytoplasmic N-terminus of type III SUTs (Figure 4) but is lacking in type I and II SUTs. All of the angiosperm type III SUTs contain a perfect LXXLL motif with the exception of AtSUT4 that has the sequence KRVLL (Figure 4). AtSUT4 has been demonstrated to localize to the vacuole membrane (Endler et al., 2006) so it is likely that the first leucine of the motif is not strictly required. Recently, localization of AtSUT4 to the vacuole membrane in Arabidopsis was shown to be dependent on AP-3 (Wolfenstetter et al., 2012). None of the Physcomitrella or Selaginella type III SUTs contain a complete LXXLL motif and it is unknown whether they localize to the vacuole membrane.
Figure 4.
Putative dileucine-like vacuolar targeting sequence in type III SUTs. A part of the multiple protein alignment of sucrose transporters is shown. All type III SUTs and selected type I and II SUTs are shown for comparison. Numbers indicate the amino acid positions for each protein. Amino acid positions that conform to the dileucine-like motif LXXLL are shown in bold.
The origin of type I SUTs
Type I SUTs are localized to the plasma membrane in eudicots. Based on phylogeny (Figure 1) and substrate specificity (Figure 2) they are more similar to type III SUTs than to type II SUTs. Since type III SUTs are present in bryophytes and lycophytes, we suggest that type I SUTs are derived from vacuolar-type III SUTs. This would likely involve mutation of the vacuolar targeting information resulting in localization to the plasma membrane, the default targeting pathway for membrane proteins in plants. We hypothesize that the LXXLL motif found in type III SUTs serves as the vacuolar targeting domain but this needs to be tested directly.
Conclusion
Angiosperm SUTs clustered into three groups, type I, II, and III. Type I SUTs, only found in eudicots appear to have evolved from vacuolar-type III SUTs which were found in all land plants from bryophytes to angiosperms. Type II SUTs were divided into an ancestral form, type IIA, that exist in all land plants and have an extended central loop. Type IIB SUTs only exist in monocots and include the phloem loading transporters in those species. Here we identify an algal SUT (CaSUT1) from the charophyte Chlorokybus atmosphyticus. Based on phylogenetic analysis, CaSUT1 appears basal to three types of land plant SUTs and this is consistent with the hypothesis that charophytes are ancestral to land plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Charles F. Delwiche and Mr. James Thierer, Cell Biology and Molecular Genetics, University of Maryland for providing sequences from charophyte algae prior to publication. We thank Dr. Andreas Weber, University of Düsseldorf for providing sequences of SUT homologs from Galdieria sulphuraria. This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy grant DE-FG02-10ER15886 (John M. Ward).
Footnotes
References
- Aoki N., Hirose T., Scofield G. N., Whitfeld P. R., Furbank R. T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232 10.1093/pcp/pcg030 [DOI] [PubMed] [Google Scholar]
- Aoki N., Hirose T., Takahashi S., Ono K., Ishimaru K., Ohsugi R. (1999). Molecular cloning and expression analysis of a gene for a sucrose transporter in maize (Zea mays L.). Plant Cell Physiol. 40, 1072–1078 [DOI] [PubMed] [Google Scholar]
- Ayre B. G. (2011). Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 4, 377–394 10.1093/mp/ssr014 [DOI] [PubMed] [Google Scholar]
- Banks J. A., Nishiyama T., Hasebe M., Bowman J. L., Gribskov M., Depamphilis C., Albert V. A., Aono N., Aoyama T., Ambrose B. A., Ashton N. W., Axtell M. J., Barker E., Barker M. S., Bennetzen J. L., Bonawitz N. D., Chapple C., Cheng C., Correa L. G., Dacre M., Debarry J., Dreyer I., Elias M., Engstrom E. M., Estelle M., Feng L., Finet C., Floyd S. K., Frommer W. B., Fujita T., Gramzow L., Gutensohn M., Harholt J., Hattori M., Heyl A., Hirai T., Hiwatashi Y., Ishikawa M., Iwata M., Karol K. G., Koehler B., Kolukisaoglu U., Kubo M., Kurata T., Lalonde S., Li K., Li Y., Litt A., Lyons E., Manning G., Maruyama T., Michael T. P., Mikami K., Miyazaki S., Morinaga S., Murata T., Mueller-Roeber B., Nelson D. R., Obara M., Oguri Y., Olmstead R. G., Onodera N., Petersen B. L., Pils B., Prigge M., Rensing S. A., Riano-Pachon D. M., Roberts A. W., Sato Y., Scheller H. V., Schulz B., Schulz C., Shakirov E. V., Shibagaki N., Shinohara N., Shippen D. E., Sorensen I., Sotooka R., Sugimoto N., Sugita M., Sumikawa N., Tanurdzic M., Theissen G., Ulvskov P., Wakazuki S., Weng J. K., Willats W. W., Wipf D., Wolf P. G., Yang L., Zimmer A. D., Zhu Q., Mitros T., Hellsten U., Loque D., Otillar R., Salamov A., Schmutz J., Shapiro H., Lindquist E., Lucas S., Rokhsar D., Grigoriev I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier G., Oesterhelt C., Larson M. D., Halgren R. G., Wilkerson C., Garavito R. M., Benning C., Weber A. P. (2005). Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 137, 460–474 10.1104/pp.104.051169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L., Kuhn C., Weise A., Schulz A., Gebhardt C., Hirner B., Hellmann H., Schulze W., Ward J. M., Frommer W. B. (2000). SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12, 1153–1164 10.2307/3871262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J. S. (2009). Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793, 605–614 10.1016/j.bbamcr.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Braun D. M., Slewinski T. L. (2009). Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol. 149, 71–81 10.1104/pp.108.129049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D., Reinders A., Ward J. M. (2003). Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J. Biol. Chem. 278, 44320–44325 10.1074/jbc.M308490200 [DOI] [PubMed] [Google Scholar]
- Chang A. B., Lin R., Keith Studley W., Tran C. V., Saier M. H., Jr. (2004). Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 21, 171–181 10.1080/09687680410001720830 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Costin G. E., Valencia J. C., Vieira W. D., Lamoreux M. L., Hearing V. J. (2003). Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J. Cell Sci. 116, 3203–3212 10.1242/jcs.00598 [DOI] [PubMed] [Google Scholar]
- Davies C., Wolf T., Robinson S. P. (1999). Three putative sucrose transporters are differentially expressed in grapevine tissues. Plant Sci. 147, 93–100 10.1016/S0168-9452(99)00059-X [DOI] [Google Scholar]
- Decourteix M., Alves G., Brunel N., Ameglio T., Guillio A., Lemoine R., Petel G., Sakr S. (2006). JrSUT1, a putative xylem sucrose transporter, could mediate sucrose influx into xylem parenchyma cells and be up-regulated by freeze-thaw cycles over the autumn-winter period in walnut tree (Juglans regia L.). Plant Cell Environ. 29, 36–47 10.1111/j.1365-3040.2005.01398.x [DOI] [PubMed] [Google Scholar]
- Dusotoit-Coucaud A., Brunel N., Kongsawadworakul P., Viboonjun U., Lacointe A., Julien J. L., Chrestin H., Sakr S. (2009). Sucrose importation into laticifers of Hevea brasiliensis, in relation to ethylene stimulation of latex production. Ann. Bot. 104, 635–647 10.1093/aob/mcp150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A., Meyer S., Schelbert S., Schneider T., Weschke W., Peters S. W., Keller F., Baginsky S., Martinoia E., Schmidt U. G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141, 196–207 10.1104/pp.106.079533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom J. S., Cho J. I., Reinders A., Lee S.W., Yoo Y., Tuan P.Q., Choi S. B., Bang G., Park Y. I., Cho M. H., Bhoo S. H., An G., Hahn T. R., Ward J. M., Jeon J. S. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119 10.1104/pp.111.176982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R. C., Peng C. C., Xu Y. H., Wang X. F., Li Y., Shang Y., Du S. Y., Zhao R., Zhang X. Y., Zhang L. Y., Zhang D. P. (2009). Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 150, 1880–1901 10.1104/pp.109.141374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemetakis E., Dimou M., Cotzur D., Efrose R. C., Aivalakis G., Colebatch G., Udvardi M., Katinakis P. (2003). A sucrose transporter, LjSUT4, is up-regulated during Lotus japonicus nodule development. J. Exp. Bot. 54, 1789–1791 10.1093/jxb/erg179 [DOI] [PubMed] [Google Scholar]
- Fukamachi S., Shimada A., Shima A. (2001). Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat. Genet. 28, 381–385 10.1038/ng584 [DOI] [PubMed] [Google Scholar]
- Gahrtz M., Schmelzer E., Stolz J., Sauer N. (1996). Expression of the PmSUC1 sucrose carrier gene from Plantago major L. is induced during seed development. Plant J. 9, 93–100 10.1046/j.1365-313X.1996.09010093.x [DOI] [PubMed] [Google Scholar]
- Gapper N. E., Coupe S. A., McKenzie M. J., Sinclair B. K., Lill R. E., Jameson P. E. (2005). Regulation of harvest-induced senescence in broccoli (Brassica oleracea var. italica) by cytokinin, ethylene, and sucrose. J. Plant Growth Regul. 24, 153–165 10.1007/s00344-005-0028-8 [DOI] [Google Scholar]
- Gottwald J. R., Krysan P. J., Young J. C., Evert R. F., Sussman M. R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U.S.A. 97, 13979–13984 10.1073/pnas.250473797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross W., Schnarrenberger C. (1995). Heterotrophic growth of 2 strains of the acido-thermophilic red alga Galdieria sulphuraria. Plant Cell Physiol. 36, 633–638 [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Gunnarsson U., Hellstrom A. R., Tixier-Boichard M., Minvielle F., Bed’hom B., Ito S., Jensen P., Rattink A., Vereijken A., Andersson L. (2007). Mutations in SLC45A2 cause plumage color variation in chicken and Japanese quail. Genetics 175, 867–877 10.1534/genetics.106.063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M., Li Y. F., El-Gamil M., Rosenberg S. A., Robbins P. F. (2001). Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res. 61, 1089–1094 [PubMed] [Google Scholar]
- Hirose T., Imaizumi N., Scofield G. N., Furbank R. T., Ohsugi R. (1997). cDNA cloning and tissue specific expression of a gene for sucrose transporter from rice (Oryza sativa L.). Plant Cell Physiol. 38, 1389–1396 [DOI] [PubMed] [Google Scholar]
- Inagaki K., Suzuki T., Ito S., Suzuki N., Adachi K., Okuyama T., Nakata Y., Shimizu H., Matsuura H., Oono T., Iwamatsu H., Kono M., Tomita Y. (2006). Oculocutaneous albinism type 4: six novel mutations in the membrane-associated transporter protein gene and their phenotypes. Pigment Cell Res. 19, 451–453 10.1111/j.1600-0749.2006.00332.x [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10, 103–105 10.1016/j.tplants.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Knop C., Voitsekhovskaja O., Lohaus G. (2001). Sucrose transporters in two members of the Scrophulariaceae with different types of transport sugar. Planta 213, 80–91 10.1007/s004250000465 [DOI] [PubMed] [Google Scholar]
- Kuhn C., Grof C. P. (2010). Sucrose transporters of higher plants. Curr. Opin. Plant Biol. 13, 288–298 10.1016/j.pbi.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Laroche J., Li P., Bousquet J. (1995). Mitochondrial DNA and monocot-dicot divergence time. Mol. Biol. Evol. 12, 1151–1156 [Google Scholar]
- Lemoine R., Burkle L., Barker L., Sakr S., Kuhn C., Regnacq M., Gaillard C., Delrot S., Frommer W. B. (1999). Identification of a pollen-specific sucrose transporter-like protein NtSUT3 from tobacco. FEBS Lett. 454, 325–330 10.1016/S0014-5793(99)00843-1 [DOI] [PubMed] [Google Scholar]
- Li C. Y., Weiss D., Goldschmidt E. E. (2003). Effects of carbohydrate starvation on gene expression in citrus root. Planta 217, 11–20 [DOI] [PubMed] [Google Scholar]
- Lin X., Kaul S., Rounsley S., Shea T. P., Benito M. I., Town C. D., Fujii C. Y., Mason T., Bowman C. L., Barnstead M., Feldblyum T. V., Buell C. R., Ketchum K. A., Lee J., Ronning C. M., Koo H. L., Moffat K. S., Cronin L. A., Shen M., Pai G., Van Aken S., Umayam L., Tallon L. J., Gill J. E., Adams M. D., Carrera A. J., Creasy T. H., Goodman H. M., Somerville C. R., Copenhaver G. P., Preuss D., Nierman W. C., White O., Eisen J. A., Salzberg S. L., Fraser C. M., Venter J. C. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768 10.1038/45471 [DOI] [PubMed] [Google Scholar]
- Mariat D., Taourit S., Guerin G. (2003). A mutation in the MATP gene causes the cream coat colour in the horse. Genet. Sel. Evol. 35, 119–133 10.1186/1297-9686-35-1-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Misumi O., Shin I. T., Maruyama S., Takahara M., Miyagishima S. Y., Mori T., Nishida K., Yagisawa F., Yoshida Y., Nishimura Y., Nakao S., Kobayashi T., Momoyama Y., Higashiyama T., Minoda A., Sano M., Nomoto H., Oishi K., Hayashi H., Ohta F., Nishizaka S., Haga S., Miura S., Morishita T., Kabeya Y., Terasawa K., Suzuki Y., Ishii Y., Asakawa S., Takano H., Ohta N., Kuroiwa H., Tanaka K., Shimizu N., Sugano S., Sato N., Nozaki H., Ogasawara N., Kohara Y., Kuroiwa T. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 10.1038/nature02398 [DOI] [PubMed] [Google Scholar]
- McCourt R. M., Delwiche C. F., Karol K. G. (2004). Charophyte algae and land plant origins. Trends Ecol. Evol. (Amst.) 19, 661–666 10.1016/j.tree.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Meyer H., Vitavska O., Wieczorek H. (2011). Identification of an animal sucrose transporter. J. Cell Sci. 124, 1984–1991 10.1242/jcs.077883 [DOI] [PubMed] [Google Scholar]
- Meyer S., Melzer M., Truernit E., Hummer C., Besenbeck R., Stadler R., Sauer N. (2000). AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J. 24, 869–882 10.1046/j.1365-313x.2000.00934.x [DOI] [PubMed] [Google Scholar]
- Newton J. M., Cohen-Barak O., Hagiwara N., Gardner J. M., Davisson M. T., King R. A., Brilliant M. H. (2001). Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am. J. Hum. Genet. 69, 981–988 10.1086/324340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Zhang J., Cao J., Yin S. Y., He X. Q., Cui K. M. (2008). Phloem transdifferentiation from immature xylem cells during bark regeneration after girdling in Eucommia ulmoides Oliv. J. Exp. Bot. 59, 1341–1351 10.1093/jxb/ern041 [DOI] [PubMed] [Google Scholar]
- Rae A. L., Perroux J. M., Grof C. P. (2005). Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem: a potential role for the ShSUT1 sucrose transporter. Planta 220, 817–825 10.1007/s00425-004-1399-y [DOI] [PubMed] [Google Scholar]
- Raven J. A. (2003). Long-distance transport in non-vascular plants. Plant Cell Environ. 26, 73–85 10.1046/j.1365-3040.2003.00920.x [DOI] [Google Scholar]
- Reinders A., Sivitz A. B., Hsi A., Grof C. P., Perroux J. M., Ward J. M. (2006). Sugarcane ShSUT1: analysis of sucrose transport activity and inhibition by sucralose. Plant Cell Environ. 29, 1871–1880 10.1111/j.1365-3040.2006.01563.x [DOI] [PubMed] [Google Scholar]
- Reinders A., Sivitz A. B., Starker C. G., Gantt J. S., Ward J. M. (2008). Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol. Biol. 68, 289–299 10.1007/s11103-008-9370-0 [DOI] [PubMed] [Google Scholar]
- Reinders A., Ward J. M. (2001). Functional characterization of the alpha-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol. Microbiol. 39, 445–454 10.1046/j.1365-2958.2001.02237.x [DOI] [PubMed] [Google Scholar]
- Rennie E. A., Turgeon R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. U.S.A. 106, 14162–14167 10.1073/pnas.0902279106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A., Shapiro H., Nishiyama T., Perroud P. F., Lindquist E. A., Kamisugi Y., Tanahashi T., Sakakibara K., Fujita T., Oishi K., Shin I. T., Kuroki Y., Toyoda A., Suzuki Y., Hashimoto S., Yamaguchi K., Sugano S., Kohara Y., Fujiyama A., Anterola A., Aoki S., Ashton N., Barbazuk W. B., Barker E., Bennetzen J. L., Blankenship R., Cho S. H., Dutcher S. K., Estelle M., Fawcett J. A., Gundlach H., Hanada K., Heyl A., Hicks K. A., Hughes J., Lohr M., Mayer K., Melkozernov A., Murata T., Nelson D. R., Pils B., Prigge M., Reiss B., Renner T., Rombauts S., Rushton P. J., Sanderfoot A., Schween G., Shiu S. H., Stueber K., Theodoulou F. L., Tu H., Van De Peer Y., Verrier P. J., Waters E., Wood A., Yang L., Cove D., Cuming A. C., Hasebe M., Lucas S., Mishler B. D., Reski R., Grigoriev I. V., Quatrano R. S., Boore J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Riesmeier J. W., Willmitzer L., Frommer W. B. (1992). Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 11, 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier J. W., Willmitzer L., Frommer W. B. (1994). Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N., Ludwig A., Knoblauch A., Rothe P., Gahrtz M., Klebl F. (2004). AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 40, 120–130 10.1111/j.1365-313X.2004.02248.x [DOI] [PubMed] [Google Scholar]
- Sauer N., Stolz J. (1994). SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 6, 67–77 10.1046/j.1365-313X.1994.6010067.x [DOI] [PubMed] [Google Scholar]
- Schubert M., Melnikova A. N., Mesecke N., Zubkova E. K., Fortte R., Batashev D. R., Barth I., Sauer N., Gamalei Y. V., Mamushina N. S., Tietze L. F., Voitsekhovskaja O. V., Pawlowski K. (2010). Two novel disaccharides, rutinose and methylrutinose, are involved in carbon metabolism in Datisca glomerata. Planta 231, 507–521 10.1007/s00425-009-1085-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Beyhl D., Marten I., Wormit A., Neuhaus E., Poschet G., Buttner M., Schneider S., Sauer N., Hedrich R. (2011). Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136 10.1111/j.1365-313X.2011.04672.x [DOI] [PubMed] [Google Scholar]
- Schulze W., Weise A., Frommer W. B., Ward J. M. (2000). Function of the cytosolic N-terminus of sucrose transporter AtSUT2 in substrate affinity. FEBS Lett. 485, 189–194 10.1016/S0014-5793(00)02180-3 [DOI] [PubMed] [Google Scholar]
- Shakya R., Sturm A. (1998). Characterization of source- and sink-specific sucrose/H+ symporters from carrot. Plant Physiol. 118, 1473–1480 10.1104/pp.118.4.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A. B., Reinders A., Johnson M. E., Krentz A. D., Grof C. P., Perroux J. M., Ward J. M. (2007). Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 143, 188–198 10.1104/pp.106.089003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A. B., Reinders A., Ward J. M. (2005). Analysis of the transport activity of barley sucrose transporter HvSUT1. Plant Cell Physiol. 46, 1666–1673 10.1093/pcp/pci182 [DOI] [PubMed] [Google Scholar]
- Sivitz A. B., Reinders A., Ward J. M. (2008). Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 147, 92–100 10.1104/pp.108.118992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T. L., Meeley R., Braun D. M. (2009). Sucrose transporter 1 functions in phloem loading in maize leaves. J. Exp. Bot. 60, 881–892 10.1093/jxb/ern335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Senters A. E., Zanis M. J., Kim S., Thompson J. D., Soltis P. S., Ronse De Craene L. P., Endress P. K., Farris J. S. (2003). Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. Am. J. Bot. 90, 461–470 10.3732/ajb.90.11.1596 [DOI] [PubMed] [Google Scholar]
- Sun A. J., Xu H. L., Gong W. K., Zhai H. L., Meng K., Wang Y. Q., Wei X. L., Xiao G. F., Zhu Z. (2008). Cloning and expression analysis of rice sucrose transporter genes OsSUT2M and OsSUT5Z. J. Integr. Plant Biol. 50, 62–75 10.1111/j.1744-7909.2007.00627.x [DOI] [PubMed] [Google Scholar]
- Sun Y., Reinders A., Lafleur K. R., Mori T., Ward J. M. (2010). Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol. 51, 114–122 10.1093/pcp/pcq056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Kaneko T., Nakamura Y., Kotani H., Kato T., Asamizu E., Miyajima N., Sasamoto S., Kimura T., Hosouchi T., Kawashima K., Kohara M., Matsumoto M., Matsuno A., Muraki A., Nakayama S., Nakazaki N., Naruo K., Okumura S., Shinpo S., Takeuchi C., Wada T., Watanabe A., Yamada M., Yasuda M., Sato S., De La Bastide M., Huang E., Spiegel L., Gnoj L., O’Shaughnessy A., Preston R., Habermann K., Murray J., Johnson D., Rohlfing T., Nelson J., Stoneking T., Pepin K., Spieth J., Sekhon M., Armstrong J., Becker M., Belter E., Cordum H., Cordes M., Courtney L., Courtney W., Dante M., Du H., Edwards J., Fryman J., Haakensen B., Lamar E., Latreille P., Leonard S., Meyer R., Mulvaney E., Ozersky P., Riley A., Strowmatt C., Wagner-McPherson C., Wollam A., Yoakum M., Bell M., Dedhia N., Parnell L., Shah R., Rodriguez M., See L. H., Vil D., Baker J., Kirchoff K., Toth K., King L., Bahret A., Miller B., Marra M., Martienssen R., McCombie W. R., Wilson R. K., Murphy G., Bancroft I., Volckaert G., Wambutt R., Dusterhoft A., Stiekema W., Pohl T., Entian K. D., Terryn N., Hartley N., Bent E., Johnson S., Langham S. A., McCullagh B., Robben J., Grymonprez B., Zimmermann W., Ramsperger U., Wedler H., Balke K., Wedler E., Peters S., van Staveren M., Dirkse W., Mooijman P., Lankhorst R. K., Weitzenegger T., Bothe G., Rose M., Hauf J., Berneiser S., Hempel S., Feldpausch M., Lamberth S., Villarroel R., Gielen J., Ardiles W., Bents O., Lemcke K., Kolesov G., Mayer K., Rudd S., Schoof H., Schueller C., Zaccaria P., Mewes H. W., Bevan M., Fransz P., Kazusa DNA Research Institute, Cold Spring Harbor and Washington University in St Louis Sequencing Consortium and European Union Arabidopsis Genome Sequencing Consortium (2000). Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826 10.1038/35048507 [DOI] [PubMed] [Google Scholar]
- Tang C., Huang D., Yang J., Liu S., Sakr S., Li H., Zhou Y., Qin Y. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ. 33, 1708–1720 10.1111/j.1365-3040.2010.02175.x [DOI] [PubMed] [Google Scholar]
- Tegeder M., Wang X. D., Frommer W. B., Offler C. E., Patrick J. W. (1999). Sucrose transport into developing seeds of Pisum sativum L. Plant J. 18, 151–161 10.1046/j.1365-313X.1999.00439.x [DOI] [PubMed] [Google Scholar]
- Theologis A., Ecker J. R., Palm C. J., Federspiel N. A., Kaul S., White O., Alonso J., Altafi H., Araujo R., Bowman C. L., Brooks S. Y., Buehler E., Chan A., Chao Q., Chen H., Cheuk R. F., Chin C. W., Chung M. K., Conn L., Conway A. B., Conway A. R., Creasy T. H., Dewar K., Dunn P., Etgu P., Feldblyum T. V., Feng J., Fong B., Fujii C. Y., Gill J. E., Goldsmith A. D., Haas B., Hansen N. F., Hughes B., Huizar L., Hunter J. L., Jenkins J., Johnson-Hopson C., Khan S., Khaykin E., Kim C. J., Koo H. L., Kremenetskaia I., Kurtz D. B., Kwan A., Lam B., Langin-Hooper S., Lee A., Lee J. M., Lenz C. A., Li J. H., Li Y., Lin X., Liu S. X., Liu Z. A., Luros J. S., Maiti R., Marziali A., Militscher J., Miranda M., Nguyen M., Nierman W. C., Osborne B. I., Pai G., Peterson J., Pham P. K., Rizzo M., Rooney T., Rowley D., Sakano H., Salzberg S. L., Schwartz J. R., Shinn P., Southwick A. M., Sun H., Tallon L. J., Tambunga G., Toriumi M. J., Town C. D., Utterback T., Van Aken S., Vaysberg M., Vysotskaia V. S., Walker M., Wu D., Yu G., Fraser C. M., Venter J. C., Davis R. W. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816–820 10.1038/35048500 [DOI] [PubMed] [Google Scholar]
- Timme R. E., Delwiche C. F. (2010). Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 10, 96. 10.1186/1471-2229-10-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Schein J., Sterck L., Aerts A., Bhalerao R. R., Bhalerao R. P., Blaudez D., Boerjan W., Brun A., Brunner A., Busov V., Campbell M., Carlson J., Chalot M., Chapman J., Chen G. L., Cooper D., Coutinho P. M., Couturier J., Covert S., Cronk Q., Cunningham R., Davis J., Degroeve S., Dejardin A., Depamphilis C., Detter J., Dirks B., Dubchak I., Duplessis S., Ehlting J., Ellis B., Gendler K., Goodstein D., Gribskov M., Grimwood J., Groover A., Gunter L., Hamberger B., Heinze B., Helariutta Y., Henrissat B., Holligan D., Holt R., Huang W., Islam-Faridi N., Jones S., Jones-Rhoades M., Jorgensen R., Joshi C., Kangasjarvi J., Karlsson J., Kelleher C., Kirkpatrick R., Kirst M., Kohler A., Kalluri U., Larimer F., Leebens-Mack J., Leple J. C., Locascio P., Lou Y., Lucas S., Martin F., Montanini B., Napoli C., Nelson D. R., Nelson C., Nieminen K., Nilsson O., Pereda V., Peter G., Philippe R., Pilate G., Poliakov A., Razumovskaya J., Richardson P., Rinaldi C., Ritland K., Rouze P., Ryaboy D., Schmutz J., Schrader J., Segerman B., Shin H., Siddiqui A., Sterky F., Terry A., Tsai C. J., Uberbacher E., Unneberg P., Vahala J., Wall K., Wessler S., Yang G., Yin T., Douglas C., Marra M., Sandberg G., Van de Peer Y., Rokhsar D. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Vaughn M. W., Harrington G. N., Bush D. R. (2002). Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc. Natl. Acad. Sci. U.S.A. 99, 10876–10880 10.1073/pnas.172198599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels J. J., Payne G. S. (1998). A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J. 17, 2482–2493 10.1093/emboj/17.9.2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P., Oesterhelt C., Gross W., Brautigam A., Imboden L. A., Krassovskaya I., Linka N., Truchina J., Schneidereit J., Voll H., Voll L. M., Zimmermann M., Jamai A., Riekhof W. R., Yu B., Garavito R. M., Benning C. (2004). EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol. Biol. 55, 17–32 10.1007/s11103-004-0376-y [DOI] [PubMed] [Google Scholar]
- Weig A., Komor E. (1996). An active sucrose carrier (Scr1) that is predominantly expressed in the seedling of Ricinus communis L. J. Plant Physiol. 147, 685–690 10.1016/S0176-1617(11)81478-1 [DOI] [Google Scholar]
- Weise A., Barker L., Kuhn C., Lalonde S., Buschmann H., Frommer W. B., Ward J. M. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12, 1345–1355 10.2307/3871134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W., Panitz R., Sauer N., Wang Q., Neubohn B., Weber H., Wobus U. (2000). Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J. 21, 455–467 10.1046/j.1365-313x.2000.00695.x [DOI] [PubMed] [Google Scholar]
- Wippel K., Sauer N. (2011). Arabidopsis SUC1 loads the phloem in suc2 mutants when expressed from the SUC2 promoter. J. Exp. Bot. 63, 669–679 10.1093/jxb/err255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenstetter S., Wirsching P., Dotzauer D., Schneider S., Sauer N. (2012). Routes to the tonoplast: the sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Osakabe Y., Mizoi J., Nakashima K., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. J. Biol. Chem. 285, 1138–1146 10.1074/jbc.M109.054288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Qu H., Dibley K. E., Offler C. E., Patrick J. W. (2007). A suite of sucrose transporters expressed in coats of developing legume seeds includes novel pH-independent facilitators. Plant J. 49, 750–764 10.1111/j.1365-313X.2006.03000.x [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Moller B., Hwang I., Feraru M. I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21, 1711–1722 10.1038/cr.2011.99 [DOI] [PMC free article] [PubMed] [Google Scholar]