Abstract
Beyond their metabolic roles, sugars can also act as messengers in signal transduction. Trehalose, a sugar found in many species of plants and animals, is a non-reducing disaccharide composed of two glucose moieties. Its synthesis in plants is a two-step process, involving the production of trehalose-6-phosphate (T6P) catalyzed by trehalose-6-phosphate synthase (TPS) and its consecutive dephosphorylation to trehalose, catalyzed by trehalose-6-phosphate phosphatase (TPP). T6P has recently emerged as an important signaling metabolite, regulating carbon assimilation and sugar status in plants. In addition, T6P has also been demonstrated to play an essential role in plant development. This review recapitulates the recent advances we have made in understanding the role of T6P in coordinating diverse metabolic and developmental processes.
Keywords: trehalose, trehalose-6-phosphate, TPS, TPP, development
Introduction
Plants convert assimilated carbon into a range of monosaccharide sugars, many of which serve as precursors for the synthesis of more complex oligo- and polysaccharides. These complex carbohydrates fulfill very diverse functions in plants and are, for example, used as long-term energy storage or constitute important structural components of plant cells.
In addition to their metabolic and structural roles, carbohydrates can also influence plant growth and development by acting as elicitors or growth regulators (Creelman and Mullet, 1997). The regulation of plant development by carbohydrates has attracted a lot of interest over the last few years and several signaling pathways that integrate sugar responses have been identified. One such pathway involves the synthesis, perception, and response to the non-reducing disaccharide trehalose and its precursor trehalose-6-phosphate (T6P). Trehalose is widely distributed in actinomycetes, fungi, insects, and other invertebrates (Veluthambi et al., 1981; Elbein et al., 2003), where it reduces protein aggregation during stress conditions (Jain and Roy, 2010). However, the majority of land plants contain only low quantities of trehalose, making its role in stress protection seem unlikely (Zentella et al., 1999; Avonce et al., 2006; Fernandez et al., 2010). Exceptions to this rule are “resurrection” plants, in which trehalose is found at higher quantities and has an apparent function in protecting the plants against abiotic stresses by preventing the denaturation of cellular proteins (Márquez-Escalante et al., 2006).
The biosynthesis of trehalose in plants involves the generation of trehalose-6-phosphate (T6P) from glucose-6-phosphate and UDP-glucose by trehalose-6-phosphate synthase (TPS), and the subsequent dephosphorylation of T6P to trehalose by trehalose-6-phosphate phosphatase (TPP; Cabib and Leloir, 1958). The A. thaliana genome contains 11 TPS (AtTPS1–11) genes and 10 TPP (AtTPPA–J) genes (Table 1; Leyman et al., 2001). The AtTPS proteins carry both TPS- and TPP-like domains and have been classified into two distinct subfamilies based on their homology with the yeast (Saccharomyces cerevisiae) ScTPS. AtTPS1 to AtTPS4, which constitute the class I subfamily of TPSs, show the highest overall similarity to ScTPS1 and in them, the TPP-like domain is only weakly conserved. AtTPS1 differs from the other class I TPSs in that it contains an auto-inhibitory N-terminal extension that restricts its activity in vivo (Van Dijck et al., 2002). The class II subfamily TPSs (AtTPS5 to AtTPS11) display more similarity to ScTPS2 and contain conserved TPP motifs.
Table 1.
Genes involved in trehalose metabolism in Arabidopsis thaliana.
| Class | Gene name | AGI number |
|---|---|---|
| Class I TPSs | TPS1 | At1g78580 |
| TPS2 | At1g16980 | |
| TPS3 | At1g17000 | |
| TPS4 | At4g27550 | |
| Class II TPSs | TPS5 | At4g17770 |
| TPS6 | At1g68020 | |
| TPS7 | At1g06410 | |
| TPS8 | At1g70290 | |
| TPS9 | At1g23870 | |
| TPS10 | At1g60140 | |
| TPS11 | At2g18700 | |
| Class I TPPs | TPP1/TPPC | At1g22210 |
| TPP2/TPPD | At1g35910 | |
| TPP3/TPPB | At1g78090 | |
| TPP4/TPPE | At2g22190 | |
| TPP5/TPPF | At4g12430 | |
| TPP6/TPPG | At4g22590 | |
| TPP7/TPPH | At4g39770 | |
| Class II TPPs | TPP8/TPPI | At5g10100 |
| TPP9/TPPA | At5g51460 | |
| TPP10/TPPJ | At5g65140 | |
| Trehalase | TRE1 | At4g24040 |
Among the Arabidopsis TPSs, only TPS1 shows a demonstrable TPS activity (Blázquez et al., 1998). However, two active isoforms of TPS1, OsTPS1a, and OsTPS1b, have been identified in rice (Zang et al., 2011). Other TPSs including class II TPSs lack both TPS and TPP activities (Vogel et al., 2001; Harthill et al., 2006; Ramon et al., 2009). AtTPS1 expression has been detected by qRT-PCR in flower buds, ripening siliques, small rosette leaves, and in embryos using a TPS1-GUS reporter (van Dijken et al., 2004; Gómez et al., 2010). However, since this reporter mostly comprises the first intron, which is located in the 5′ untranslated region of TPS1, these results need to be verified. Similar to TPS1, class II TPSs are expressed widely throughout the plant (Paul et al., 2008; Ramon et al., 2009). In contrast, the other class I TPS genes, AtTPS2–4, have been reported to be specifically expressed in siliques and seeds (Paul et al., 2008).
All TPPs lack the N-terminal TPS-like domain found in the TPS proteins and contain only the conserved TPP domain, which shows significant similarity to the highly conserved phosphatase box in the C-terminal part of ScTPS2 (Lunn, 2007b). Out of the 10 TPPs in Arabidopsis, AtTPPA and AtTPPB genes have been shown to complement yeast TPP mutant (tps2) and thus encode active TPP enzymes (Vogel et al., 1998). Whether the other TPPs are enzymatically active, remains to be determined. Outside Arabidopsis, TPP2a from rice (Habibur Rahman Pramanik and Imai, 2005; Shima et al., 2007) and RAMOSA3 of maize (Satoh-Nagasawa et al., 2006) have been shown to encode active TPPs. In contrast to the TPS and TPP gene families, trehalase, which cleaves trehalose into two glucose molecules, appears to be encoded by a single gene (AtTRE1; Leyman et al., 2001; Lunn, 2007b).
T6P in Plant Carbohydrate Metabolism
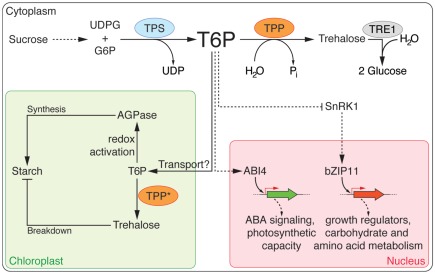
Sugars, including trehalose and, more importantly, T6P have been proposed to act as signaling molecules that modulate many important metabolic and developmental processes in plants (reviewed in Paul et al., 2008) and there is growing evidence that T6P plays a central role in regulating carbohydrate metabolism (Figure 1).
Figure 1.
Trehalose biosynthesis and its role in carbohydrate metabolism. Trehalose-6-phosphate (T6P) is synthesized from UDP-glucose (UDPG) and glucose-6-P (G6P) by the activity of trehalose-6-phosphate synthase (TPS) and subsequently converted to trehalose by trehalose-6-phosphate phosphatase (TPP). trehalase1 (TRE1) hydrolyzes trehalose into two molecules of glucose. T6P plays a central role in regulating sugar metabolism in plants. The precursors of T6P are derived from the sucrose metabolism. It has been suggested that T6P is transported by an unknown mechanism into plastids where it induces starch synthesis via thioredoxin-mediated activation of AGPase. T6P might be converted into trehalose, which has been shown to regulate starch breakdown in plastids. Several TPPs (marked with an asterisk) have been predicted to localize to plastids, but this still needs to be confirmed experimentally. SnRK1, which represses plant growth, is inhibited by T6P. A regulatory loop, which involves T6P, SnRK1, and bZIP11, and that is thought to control sucrose availability and utilization, has been proposed.
More specifically, T6P has been shown to regulate sucrose utilization in plants. Transgenic Arabidopsis lines expressing E. coli TPS (otsA) or TPP (otsB) genes displayed differences in T6P accumulation and responded differently to exogenous sucrose. In general, the ability of these plants to utilize sucrose, increased with rising concentration of T6P (Schluepmann et al., 2004). Conversely it was found that the amount of T6P strongly corresponds to sucrose availability in Arabidopsis wild-type plants. Lunn et al. (2006) showed that sucrose feeding rapidly induced T6P in carbon-starved seedlings. The rapid increase of T6P in response to exogenous sucrose may be due to an increase in the amount of available glucose-6-phosphate (G6P) and UDP-glucose (UDPG), which have been shown to be important in determining plant growth and biomass accumulation (Meyer et al., 2007). Thus T6P indirectly reflects sucrose concentrations and has now been widely accepted as an indicator of sucrose status in plants (Lunn et al., 2006; reviewed in Paul et al., 2008).
The importance of T6P was also made apparent in Arabidopsis and tobacco plants over-expressing E. coli TPS (otsA) and E. coli TPP (otsB; Schluepmann et al., 2003; Pellny et al., 2004). Interestingly, the tobacco plants over-expressing otsA exhibited large increase in photosynthetic capacity per unit leaf area and dry weight. This boost in photosynthetic capacity was shown to be in part as a result of increased expression of ABI4, a known regulator of RuBisCo genes (Acevedo-Hernández et al., 2005). An opposite effect was observed in E. coli TPP (otsB) over-expressing plants (Pellny et al., 2004). The observed changes in the photosynthetic capacity in plants with altered T6P levels reflect the combined effects of T6P on leaf development, in particular on cell division and cell wall biosynthesis, and a more direct effect on the regulation of factors involved in regulating photosynthesis such as ABI4. However, the details of how T6P imparts such a dramatic regulation in photosynthesis are not yet fully understood.
Trehalose-6-phosphate has also been shown to regulate starch metabolism in plants. In Arabidopsis, exogenous application of trehalose induces accumulation of starch by increasing the activity of ADP-glucose pyrophosphorylase (AGPase), a major enzyme controlling starch synthesis (Wingler et al., 2000). It has also been shown that T6P, synthesized by TPS1, which has been predicted to be mainly localized in the cytoplasm, promotes AGPase activation in chloroplasts via a thioredoxin-mediated redox reaction (Kolbe et al., 2005). Rising sucrose levels, which are accompanied by an increase in the level of T6P, also resulted in increased starch synthesis via AGPase activation (Lunn et al., 2006). As AGPase is localized in chloroplasts, T6P mediated AGPase activation suggests a link between cytosolic carbon status and plastidic starch storage. Accumulation of sucrose in the leaf has been shown to increase the T6P content, leading to activation of AGPase in the chloroplasts, without apparent change in the rate of photosynthesis. In this way, T6P might act as a signal between the cytosol and the chloroplast (Lunn, 2007a). Whether T6P is transported into the chloroplast remains to be determined.
Apart from its role in regulating starch metabolism, T6P has recently been shown to inhibit the sucrose non-fermenting 1-related protein kinase1 (SnRK1) complex of the SNF-1-related group of protein kinases in Arabidopsis (Zhang et al., 2009), which acts as a metabolite sensor to constantly adapt metabolism to the supply and demand of energy (Polge and Thomas, 2007). More recently, inhibition of SnRK1 by T6P has also been demonstrated in a transgenic sugarcane suspension cell line expressing a vacuole-targeted sucrose isomerase (Wu and Birch, 2010), in wheat grain extracts (Martínez-Barajas et al., 2011), and in potato tubers (Debast et al., 2011). SnRK1 is a hetero-trimeric protein complex composed of an AKIN10 or AKIN11 catalytic α-subunit, β-, and γ-subunits, which together form the active kinase complex (Polge and Thomas, 2007). Over-expression of SnRK1, promotes plant survival under low light and starvation conditions, in additions to altering inflorescence development and delaying senescence. In contrast, akin10 akin11 virus-induced gene silencing double mutant plants showed growth arrest coupled with premature senescence. The AKIN10/AKIN11 signaling cascade seems to be crucial for plant survival under stress, e.g., darkness and sugar deprivation (Baena-González et al., 2007). Microarray analysis showed that most of the up-regulated genes in E. coli TPS (otsA) over-expressing plants were down-regulated in AKIN10 over-expressors (Zhang et al., 2009). In addition, SnRK1 has also been implicated in sugar and ABA signaling pathways in an independent study (Jossier et al., 2009). Importantly, the effects on SnRK1 activity seem to be specific to T6P, as none of the other tested sugars or sugar phosphates inhibited SnRK1 (Zhang et al., 2009). Taken together, these experiments provide strong evidence that T6P inhibits SnRK1 to activate biosynthetic processes in growing tissues. These results suggest a regulatory loop in which sucrose-induced T6P inhibits SnRK1 when sucrose is plentiful. When the sucrose content decreases, with T6P decreasing as well, SnRK1 is released from repression, which leads to the induction of genes involved in photosynthesis related processes, so that more carbon is made available to the growing cells, to replenish a carbon deficit (Delatte et al., 2011).
More recently the Arabidopsis basic region leucine zipper transcription factor 11 (bZIP11) has been implicated in T6P signaling. In particular, over-expression of bZIP11, which is repressed by sucrose through a translational inhibition mechanism (Hanson et al., 2008), has been shown to result in reduced T6P levels (Ma et al., 2011). bZIP11 over-expressing plants also exhibited resistance to exogenously applied trehalose and accumulation of T6P, with concomitant reduction of SnRK1 activity (Delatte et al., 2011). The authors propose that bZIP11 acts as a powerful regulator of carbohydrate metabolism and is part of a growth regulatory network that includes T6P and the SnRK1 (Delatte et al., 2011).
The importance of T6P in metabolic processes has also been recently demonstrated in potato, where T6P has been shown to influence tuber growth (Debast et al., 2011). Transgenic potato lines over-expressing E.coli TPS (otsA) displayed reduced starch content, decreased amounts of ATP coupled with an increased respiration rate (indicating high metabolic activity), and delayed sprouting. Conversely, lines that over-expressed the E. coli TPP gene otsB, displayed significantly reduced T6P levels, and accumulated soluble carbohydrates, hexose-phosphates, and ATP. These plants no longer displayed any changes in starch content or early sprouting (Debast et al., 2011). Taken together, these results indicate that T6P functions as a key regulator of plant growth in response to environmental stimuli by regulating the central carbon metabolism.
T6P in Plant Development
Apart from its role in regulating carbohydrate metabolism, T6P has also been shown to be important for normal plant development. This was first noticed in plants over-expressing trehalose biosynthesis genes, which displayed a wide range of developmental defects. In addition, tps1 loss of function mutations in Arabidopsis demonstrated that TPS1 was required for successful embryo maturation. Homozygous tps1 mutant embryos were shown to initially develop normally, but further development was found to be progressively retarded, and eventually stalled completely at torpedo stage, when cell expansion and storage reserve accumulation occurs (Eastmond et al., 2002). Later studies revealed that cell cycle activity was perturbed in maturing mutant embryos and that the cell walls of these embryos were thicker than those of wild-type embryos (Gómez et al., 2006). Taken together these results indicate that T6P plays an important role in orchestrating cell cycle activity and cell wall biosynthesis with cellular metabolism during embryo development (Gómez et al., 2006).
The function of TPS1 in regulating plant development is, however, not limited to embryogenesis. When embryo-lethal tps1 mutant plants were rescued by dexamethasone-inducible transient expression of AtTPS1 during embryo maturation, the resulting plants showed phenotypic abnormalities throughout vegetative growth and floral transition (van Dijken et al., 2004). In particular, the rescued plants exhibited delayed germination, slow development, perturbed root growth, and a stunted stature. In addition, the plants displayed reduced apical dominance and aerial rosettes even under long day conditions. The abnormal growth patterns in rescued tps1 mutants, which can be almost recovered by inducing expression of TPS1, clearly show the role of T6P in plant vegetative growth and development in Arabidopsis. In a more recent study, expression of TPS1 from the seed-specific ABI3 promoter was used to rescue tps1 embryo-lethal phenotype (Gómez et al., 2010). The rescued plants displayed a severe growth arrest, accumulated soluble sugars and starch and resulted in up-regulation of several genes involved in ABA signaling. In addition, non-embryo-lethal weak alleles of tps1 isolated from a TILLING population showed reduced growth and delayed flowering when compared to wild-type plants. These plants were also found to be ABA hypersensitive, and accordingly displayed reduced stomatal aperture size (Gómez et al., 2010). Taken together, these results suggest that T6P might act through a mechanism involving ABA and sugar metabolism to coordinate embryo maturation and vegetative growth.
Later in development, TPS1 is required for the timely induction of flowering. This was first demonstrated in tps1 mutant plants that had been rescued through embryogenesis by means of dexamethasone-inducible TPS1 expression (van Dijken et al., 2004). Interestingly, these plants completely failed to flower, unless TPS1 expression was induced (Figure 2). Delayed flowering has also been observed in non-embryo-lethal weak alleles of tps1 isolated from a TILLING population (Gómez et al., 2010). Induction of flowering is known to be associated with starch mobilization, a transient increase in leaf carbohydrates and finally the export of sugars to the shoot apical meristem, suggesting that phloem-mobile carbohydrates are a critical factor in controlling the transition to flowering (Corbesier et al., 1998, 2002). The finding that exogenous supply of sucrose is sufficient to promote morphogenesis and flowering in Arabidopsis in the dark further emphasizes the importance of carbohydrates in flowering (Roldán et al., 1999). In addition, supplementation with 1% exogenous sucrose was shown to rescue the late flowering phenotype of several flowering time mutants (Roldán et al., 1999). The molecular mechanisms by which sugars control the transition to flowering are largely unknown. However, it seems likely that T6P as a signal of sucrose status plays an important role in floral transition.
Figure 2.
Plants defective in trehalose-6-phosphate synthesis are late flowering. Depicted are wild-type (Col-0) plants and homozygous tps1 mutants that carry a chemically inducible TPS1 rescue construct (GVG::TPS1), which flower late when compared to wild-type control. Plants were grown under long day photoperiod at 23°C and the images were taken on 20 and 50 days after sowing, for wild-type and homozygous tps1 mutant, respectively.
TPS1 is not the only Arabidopsis TPS gene for which a function in plant development has been reported. The cell shape phenotype-1 (csp-1) mutant, which has been mapped to a point mutation in AtTPS6, a class II TPS gene, displayed an absence of lobes in pavement cells, reduced trichome branching, altered leaf serration and stem branching, and increased stomatal density (Chary et al., 2008). However, since none of these alterations except for the cell shape defects could be observed in a TPS6 T-DNA insertion line (Chary et al., 2008), these findings need to be confirmed.
Finally, T6P metabolism has also been implicated in regulating inflorescence architecture or branching. The evidence for this largely comes from the ramosa3 (ra3) mutant in maize. In ra3 plants the inflorescence branching pattern is disturbed. The tassel or the male inflorescence of wild-type maize plants has elongated branches at the base and also possesses a central spike consisting of short branches with spikelet pairs. In contrast, the laterally placed ears or the female inflorescences have short branches, which aids in better packaging of seeds. However, the tassels of ra3 plants have additional long branches and the ears produce abnormally shaped long branches at their bases. The RA3 gene, which is expressed in distinct domains delineating axillary inflorescence meristems, has been shown to encode a TPP (Satoh-Nagasawa et al., 2006). Genetic and molecular data indicate that RA3 regulates inflorescence branching by modification of sugar signals possibly coordinated by T6P. RA3 is a functional TPP enzyme which acts upstream of RA1 transcription factor in the RAMOSA pathway, thereby regulating inflorescence development (Satoh-Nagasawa et al., 2006). This study nicely indicates that T6P metabolism can influence a specific developmental pathway in plants such as inflorescence architecture.
Conclusion and Perspective
The last few years have seen considerable progress in our understanding of T6P signaling as a central player in coordinating metabolism with growth and development. However, compared to other plant signaling pathways, our knowledge of T6P signaling is still relatively sparse. One reason for this has been the intricate and tight connection between T6P signaling and other developmental signaling pathways. Furthermore, uncovering the exact functions of T6P with respect to specific physiological processes has proven difficult, as it is present in plant cells only in low amounts. Another obstacle has been that T6P is essential for plant development, making the genetic analysis of the T6P signaling pathway rather difficult. However, lately most of these technical hurdles have been overcome and we can now expect new approaches to unravel the remaining mysteries behind T6P as a central regulator of plant growth and development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the DFG grants SCHM1560/6-1 within the framework of the Arabidopsis Functional Genomics Network (AFGN) and SCHM1560/8-1.
References
- Acevedo-Hernández G. J., León P., Herrera-Estrella L. R. (2005). Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J. 43, 506–519 10.1111/j.1365-313X.2005.02468.x [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G. (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol 6, 109. 10.1186/1471-2148-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J. M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 10.1038/nature06069 [DOI] [PubMed] [Google Scholar]
- Blázquez M. A., Santos E., Flores C. L., Martínez-Zapater J. M., Salinas J., Gancedo C. (1998). Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J. 13, 685–689 10.1046/j.1365-313X.1998.00063.x [DOI] [PubMed] [Google Scholar]
- Cabib E., Leloir L. F. (1958). The biosynthesis of trehalose Phosphate. J. Biol. Chem. 231, 259–275 [PubMed] [Google Scholar]
- Chary S. N., Hicks G. R., Choi Y. G., Carter D., Raikhel N. V. (2008). Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 146, 97–107 10.1104/pp.107.107441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Bernier G., Périlleux C. (2002). C: N ratio increases in the phloem sap during floral transition of the long-day plants sinapis alba and Arabidopsis thaliana. Plant Cell Physiol. 43, 684–688 10.1093/pcp/pcf071 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Lejeune P., Bernier G. (1998). The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta 206, 131–137 10.1007/s004250050383 [DOI] [PubMed] [Google Scholar]
- Creelman R. A., Mullet J. E. (1997). Oligosaccharins, brassinolides, and jasmonates: nontraditional regulators of plant growth, development, and gene expression. Plant Cell Online 9, 1211–1223 10.1105/tpc.9.7.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debast S., Nunes-Nesi A., Hajirezaei M.-R., Hofmann J., Sonnewald U., Fernie A. R., Börnke F. (2011). Altering trehalose-6-phosphate content in transgenic potato tubers affects tuber growth and alters responsiveness to hormones during sprouting. Plant Physiol. 156, 1754–1771 10.1104/pp.111.179903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T. L., Sedijani P., Kondou Y., Matsui M., de Jong G. J., Somsen G. W., Wiese-Klinkenberg A., Primavesi L. F., Paul M. J., Schluepmann H. (2011). Growth arrest by trehalose-6-phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol. 157, 160–174 10.1104/pp.111.180422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., van Dijken A. J. H., Spielman M., Kerr A., Tissier A. F., Dickinson H. G., Jones J. D. G., Smeekens S. C., Graham I. A. (2002). Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 29, 225–235 10.1046/j.1365-313x.2002.01220.x [DOI] [PubMed] [Google Scholar]
- Elbein A. D., Pan Y. T., Pastuszak I., Carroll D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17R–27R 10.1093/glycob/cwg047 [DOI] [PubMed] [Google Scholar]
- Fernandez O., Béthencourt L., Quero A., Sangwan R. S., Clément C. (2010). Trehalose and plant stress responses: friend or foe? Trends Plant Sci. 15, 409–417 10.1016/j.tplants.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Gómez L. D., Baud S., Gilday A., Li Y., Graham I. A. (2006). Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J. 46, 69–84 10.1111/j.1365-313X.2006.02662.x [DOI] [PubMed] [Google Scholar]
- Gómez L. D., Gilday A., Feil R., Lunn J. E., Graham I. A. (2010). AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J. 64, 1–13 [DOI] [PubMed] [Google Scholar]
- Habibur Rahman Pramanik M., Imai R. (2005). Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol. Biol. 58, 751–762 10.1007/s11103-005-7404-4 [DOI] [PubMed] [Google Scholar]
- Hanson J., Hanssen M., Wiese A., Hendriks M. M. W. B., Smeekens S. (2008). The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and proline dehydrogenase2. Plant J. 53, 935–949 10.1111/j.1365-313X.2007.03385.x [DOI] [PubMed] [Google Scholar]
- Harthill J. E., Meek S. E. M., Morrice N., Peggie M. W., Borch J., Wong B. H. C., MacKintosh C. (2006). Phosphorylation and 14-3-3 binding of Arabidopsis trehalose-phosphate synthase 5 in response to 2-deoxyglucose. Plant J. 47, 211–223 10.1111/j.1365-313X.2006.02780.x [DOI] [PubMed] [Google Scholar]
- Jain N. K., Roy I. (2010). Trehalose and protein stability. Curr. Protoc. Protein Sci. Chapter 4, Unit 4.9. [DOI] [PubMed] [Google Scholar]
- Jossier M., Bouly J.-P., Meimoun P., Arjmand A., Lessard P., Hawley S., Grahame Hardie D., Thomas M. (2009). SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59, 316–328 10.1111/j.1365-313X.2009.03871.x [DOI] [PubMed] [Google Scholar]
- Kolbe A., Tiessen A., Schluepmann H., Paul M., Ulrich S., Geigenberger P. (2005). Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. U.S.A. 102, 11118–11123 10.1073/pnas.0503410102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B., Van Dijck P., Thevelein J. M. (2001). An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 6, 510–513 10.1016/S1360-1385(01)02125-2 [DOI] [PubMed] [Google Scholar]
- Lunn J. E. (2007a). Compartmentation in plant metabolism. J. Exp. Bot. 58, 35–47 10.1093/jxb/erl134 [DOI] [PubMed] [Google Scholar]
- Lunn J. E. (2007b). Gene families and evolution of trehalose metabolism in plants. Funct. Plant Biol. 34, 550–563 10.1071/FP06315 [DOI] [PubMed] [Google Scholar]
- Lunn J. E., Feil R., Hendriks J. H. M., Gibon Y., Morcuende R., Osuna D., Scheible W.-R., Carillo P., Hajirezaei M.-R., Stitt M. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397, 139. 10.1042/BJ20060083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Hanssen M., Lundgren K., Hernández L., Delatte T., Ehlert A., Liu C.-M., Schluepmann H., Dröge-Laser W., Moritz T., Smeekens S., Hanson J. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191, 733–745 10.1111/j.1469-8137.2011.03735.x [DOI] [PubMed] [Google Scholar]
- Márquez-Escalante J. A., Figueroa-Soto C. G., Valenzuela-Soto E. M. (2006). Isolation and partial characterization of trehalose 6-phosphate synthase aggregates from Selaginella lepidophylla plants. Biochimie 88, 1505–1510 10.1016/j.biochi.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Martínez-Barajas E., Delatte T., Schluepmann H., de Jong G. J., Somsen G. W., Nunes C., Primavesi L. F., Coello P., Mitchell R. A. C., Paul M. J. (2011). Wheat grain development is characterized by remarkable trehalose 6-phosphate accumulation pregrain filling: tissue distribution and relationship to SNF1-related protein kinase1 activity. Plant Physiol. 156, 373–381 10.1104/pp.111.174524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. C., Steinfath M., Lisec J., Becher M., Witucka-Wall H., Törjék O., Fiehn O., Eckardt Ä., Willmitzer L., Selbig J., Altmann T. (2007). The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 4759–4764 10.1073/pnas.0610704104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59, 417–441 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Pellny T. K., Ghannoum O., Conroy J. P., Schluepmann H., Smeekens S., Andralojc J., Krause K. P., Goddijn O., Paul M. J. (2004). Genetic modification of photosynthesis with E. coli genes for trehalose synthesis. Plant Biotechnol. J. 2, 71–82 10.1111/j.1467-7652.2004.00053.x [DOI] [PubMed] [Google Scholar]
- Polge C., Thomas M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12, 20–28 10.1016/j.tplants.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Ramon M., De Smet I., Vandesteene L., Naudts M., Leyman B., Van Dijck P., Rolland F., Beeckman T., Thevelein J. M. (2009). Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 32, 1015–1032 10.1111/j.1365-3040.2009.01985.x [DOI] [PubMed] [Google Scholar]
- Roldán M., Gómez-Mena C., Ruiz-García L., Salinas J., Martínez-Zapater J. M. (1999). Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 20, 581–590 10.1046/j.1365-313X.1999.00632.x [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N., Nagasawa N., Malcomber S., Sakai H., Jackson D. (2006). A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441, 227–230 10.1038/nature04725 [DOI] [PubMed] [Google Scholar]
- Schluepmann H., Pellny T., van Dijken A., Smeekens S., Paul M. (2003). Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 6849–6854 10.1073/pnas.1132018100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H., van Dijken A., Aghdasi M., Wobbes B., Paul M., Smeekens S. (2004). Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135, 879–890 10.1104/pp.104.039503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S., Matsui H., Tahara S., Imai R. (2007). Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J. 274, 1192–1201 10.1111/j.1742-4658.2007.05658.x [DOI] [PubMed] [Google Scholar]
- Van Dijck P., Mascorro-Gallardo J. O., De Bus M., Royackers K., Iturriaga G., Thevelein J. M. (2002). Truncation of Arabidopsis thaliana and Selaginella lepidophylla trehalose-6-phosphate synthase unlocks high catalytic activity and supports high trehalose levels on expression in yeast. Biochem. J. 366, 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken A. J., Schluepmann H., Smeekens S. C. M. (2004). Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol. 135, 969–977 10.1104/pp.104.039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthambi K., Mahadevan S., Maheshwari R. (1981). Trehalose toxicity in cuscuta reflexa: correlation with low trehalase activity. Plant Physiol. 68, 1369–1374 10.1104/pp.68.6.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G., Aeschbacher R. A., Müller J., Boller T., Wiemken A. (1998). Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J. 13, 673–683 10.1046/j.1365-313X.1998.00064.x [DOI] [PubMed] [Google Scholar]
- Vogel G., Fiehn O., Jean-Richard-dit-Bressel L., Boller T., Wiemken A., Aeschbacher R. A., Wingler A. (2001). Trehalose metabolism in Arabidopsis: occurrence of trehalose and molecular cloning and characterization of trehalose-6-phosphate synthase homologues. J. Exp. Bot. 52, 1817–1826 10.1093/jexbot/52.362.1817 [DOI] [PubMed] [Google Scholar]
- Wingler A., Fritzius T., Wiemken A., Boller T., Aeschbacher R. A. (2000). Trehalose induces the ADP-glucose pyrophosphorylase gene, ApL3, and starch synthesis in Arabidopsis. Plant Physiol. 124, 105–114 10.1104/pp.124.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Birch R. G. (2010). Physiological basis for enhanced sucrose accumulation in an engineered sugarcane cell line. Funct. Plant Biol. 37, 1161–1174 10.1071/FP10055 [DOI] [Google Scholar]
- Zang B., Li H., Li W., Deng X. W., Wang X. (2011). Analysis of trehalose-6-phosphate synthase (TPS) gene family suggests the formation of TPS complexes in rice. Plant Mol. Biol. 76, 507–522 10.1007/s11103-011-9781-1 [DOI] [PubMed] [Google Scholar]
- Zentella R., Mascorro-Gallardo J. O., Van Dijck P., Folch-Mallol J., Bonini B., Van Vaeck C., Gaxiola R., Covarrubias A. A., Nieto-Sotelo J., Thevelein J. M., Iturriaga G. (1999). A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol. 119, 1473–1482 10.1104/pp.119.4.1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Primavesi L. F., Jhurreea D., Andralojc P. J., Mitchell R. A. C., Powers S. J., Schluepmann H., Delatte T., Wingler A., Paul M. J. (2009). Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149, 1860–1871 10.1104/pp.108.133934 [DOI] [PMC free article] [PubMed] [Google Scholar]