Abstract
Plant–microbe interactions are ubiquitous. Plants are threatened by pathogens, but they are even more commonly engaged in neutral or mutualistic interactions with microbes: belowground microbial plant associates are mycorrhizal fungi, Rhizobia, and plant-growth promoting rhizosphere bacteria, aboveground plant parts are colonized by internally living bacteria and fungi (endophytes) and by microbes in the phyllosphere (epiphytes). We emphasize here that a completely microbe-free plant is an exotic exception rather than the biologically relevant rule. The complex interplay of such microbial communities with the host–plant affects multiple vital parameters such as plant nutrition, growth rate, resistance to biotic and abiotic stressors, and plant survival and distribution. The mechanisms involved reach from direct ones such as nutrient acquisition, the production of plant hormones, or direct antibiosis, to indirect ones that are mediated by effects on host resistance genes or via interactions at higher trophic levels. Plant-associated microbes are heterotrophic and cause costs to their host plant, whereas the benefits depend on the current environment. Thus, the outcome of the interaction for the plant host is highly context dependent. We argue that considering the microbe-free plant as the “normal” or control stage significantly impairs research into important phenomena such as (1) phenotypic and epigenetic plasticity, (2) the “normal” ecological outcome of a given interaction, and (3) the evolution of plants. For the future, we suggest cultivation-independent screening methods using direct PCR from plant tissue of more than one fungal and bacterial gene to collect data on the true microbial diversity in wild plants. The patterns found could be correlated to host species and environmental conditions, in order to formulate testable hypotheses on the biological roles of plant endophytes in nature. Experimental approaches should compare different host–endophyte combinations under various relevant environmental conditions and study at the genetic, epigenetic, transcriptional, and physiological level the parameters that cause the interaction to shift along the mutualism–parasitism continuum.
Keywords: clavicipitaceous endophyte, extended phenotype, mycorrhiza, non-clavicipitaceous endophyte, pathogen, plant-associated microbe, plant–microbe interaction, rhizobia
Introduction
Plants are full of microbes. The phenotype of most plants in nature is the product of the concerted and highly co-regulated expression of both plant and microbial genes. In fact, significant parts of the plant phenotype represent the “extended phenotype” sensu Dawkins (1999) of one or several microorganism(s). Mutualistic plant–microbe interactions were originally considered as interesting, but specific, cases, which are of relevance only for some selected species or families. Likewise, most plant-infecting microorganisms were considered pathogens with detrimental effects on plant development, particularly in crop plants and much less so in wild plants under natural conditions. However, the presence of microorganisms in and on plants must be considered to be the rule, rather than the exception. Examples comprise pathogens, mutualistic fungal endophytes, mycorrhiza, nodulating and other plant-associated N-fixing bacteria, as well as microorganisms in the rhizosphere and phyllosphere (see Glossary). Many of these microorganisms develop within living plant tissue and therefore represent endophytes (see below).
Fossil evidence indicates the existence of arbuscular mycorrhizal fungi (AMF) on the earliest land plants, as structures that appear to be identical with modern mycorrhiza have been detected in fossils from the early Devonian (>400 million years ago: Remy et al., 1994) and from the Ordovician (ca. 460 million years ago: Redecker et al., 2000). Mycorrhiza are likely to have played a crucial role in the evolution of land plants (Brundrett, 2002; Bonfante and Genre, 2008), especially as it has been demonstrated that simulated CO2 rich-environments (similar to conditions under the Paleozoic) amplified net benefits for plants associated to AMF (Humphreys et al., 2010) in terms of carbon gain and improved growth. Considering the fact that nowadays more than 80% of all terrestrial plants are associated with mycorrhizal fungi it is tempting to state provocatively that an un-mycorrhized plant does (almost) not exist, neither actually nor in the history of land plants. This observation, together with studies that found leaf endophytes in all plant species that have been investigated so far (Arnold et al., 2003; Schulz and Boyle, 2005; Albrectsen et al., 2010; Gilbert et al., 2010), leads us to formulate the central message that we aim to emphasize here:
A plant that is completely free of microorganisms represents an exotic exception, rather than the – biologically relevant – rule
Endophytes have been defined by Wilson (1995) as “fungi that are present in host plant tissues, during at least part of their life cycle, without causing visible symptoms.” An increasing number of research articles published over the last 10 years demonstrated, however, that this definition suffers from three major drawbacks. First, microorganisms that develop within living plant tissue without causing visible symptoms are not restricted to fungi but also comprise bacteria and viruses. Second, the net effects of infection on the host are highly conditional and can shift from mutualism to antagonism for virtually any type of plant–microbe interaction (see The Conditional Outcomes of Plant–Microbe Interactions). Third, although classical endophytes admittedly do not cause visible disease symptoms, they certainly affect the phenotype of a plant: an endophyte-free plant looks and functions differently from an endophyte-infected one under most natural growing conditions (Figure 1).
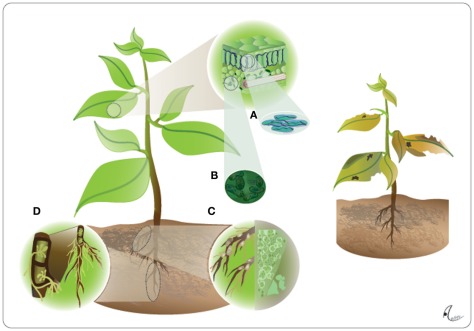
Figure 1.
The plant as the extended phenotype of endophytes. Endophytes including fungal leaf endophytes (A), bacterial leaf endophytes (B), Rhizobia (C), and arbuscular mycorrhizal fungi (D), can infect all organs of a plant, which in their presence exhibits what we usually know as its normal phenotype (left panel). These endophytes contribute so strongly to vitally important traits of the plant that a completely endophyte-free plant would suffer from chlorosis and lowered photosynthetic rates due to nutritional shortcomings, lowered resistance to abiotic stress and lowered resistance to both pathogens and herbivores and, in consequence, lowered competitive abilities and lower survival rates in nature (right panel).
Therefore, in the present article we use the term “endophyte” to denominate all microorganisms that develop within living plant tissue as the etymology of the word suggests. Realizing that it is impossible to assess the net balance of a certain host–microbe interaction for all possible sets of environmental conditions, we suggest that endophytes whose presence under the average conditions causes negative effects on host fitness should be termed “pathogens,” whereas endophytes with generally neutral or positive effects on host fitness represent the “classical” endophytes. As we explain below, these two terms must be considered operational terms that represent the extremes of a continuum of possible outcomes, rather than clearly separated groups of taxonomically defined organisms.
Although non-pathogenic endophytes cause no disease symptoms, they influence the phenotype and epigenome of their host plant (Gilbert et al., 2010). It is their omnipresence rather than the lack of visible effects, which led earlier researchers to the conclusion that endophytes do not cause visible symptoms (Wilson, 1995). The extended phenotype of the endophytes is easily overseen because it represents an integrative, ubiquitous part of the plant phenotype. Actually, Rosenberg et al. (2007) suggested the term “Holobiont” to denominate the eukaryotic host and its full symbiont population. Classical research takes the microbe-free plant as the control (and somehow “normal”) stage, whereas the plant that is interacting with a microbe is considered as the “treatment” stage. This position is dangerous.
First, many endophytic fungi and at least some endophytic bacteria are transmitted via the seeds, that is, vertically to the own offspring (Schardl et al., 2004; Ferreira et al., 2008; Rudgers et al., 2009; Johnston-Monje and Raizada, 2011), and many others have the ability to colonize plant tissues once the plant exists (during germination as well as at later developmental stages), which is referred as horizontal transmission. Thus, conclusions drawn on the assumption that all control plants are free of microbes because they have not been experimentally infected, are consequently overestimating the contribution of the plant genotype to the observed phenomenon. Second, given all the fitness-relevant plant traits affected by endophytic microbes it appears to be likely that a microbe-free plant would hardly be capable of surviving under natural conditions (Figure 1).
In this article, we aim at presenting a short and necessarily incomplete overview on plant phenotypic traits that are altered in the presence of endophytes, predominantly fungal ones as they have been more frequently investigated, and discuss how the above-described research strategy severely inhibits our progress in at least three disciplines: (1) the study of phenotypic plasticity, because many plastic responses of plants to changing environmental conditions are under control of microbial genes interacting with plant genes, (2) plant ecology, because the net fitness effects of multiple abiotic and biotic environmental factors are strongly altered when plants are colonized by endophytes, and (3) the study of plant evolution, because plant genomes are likely to co-evolve with microbial genomes, particularly those of mycorrhiza and vertically transmitted endophytes (see Glossary). We finish our article with some concrete suggestions on how to use currently available techniques to answer the most pertinent questions: “who is there?”, “who is where?”, and “who does what?”, that is, how does the colonization by endophytes affect plant ecology and physiology, plant phenotypic plasticity, and – in the long run – plant evolution?
Major Endophyte Groups and Their Positive Effects on Host Plants
Multiple reviews have been published on every group of microorganisms that are regularly associated with plants. We refer to the reviews on fungal endophytes (Muller and Krauss, 2005; Schulz and Boyle, 2005; Hartley and Gange, 2009; Rodriguez et al., 2009; Gao et al., 2010; Saikkonen et al., 2010; Eaton et al., 2011), fungal entomopathogens (Vega et al., 2009), bacterial endosymbionts (Weyens et al., 2009), mycorrhiza (Smith and Read, 2008; Bonfante, 2010; Koltai and Kapulnik, 2010; Ercolin and Reinhardt, 2011), and Rhizobia (Kiers and Denison, 2008; Oldroyd and Downie, 2008; Markmann and Parniske, 2009; Brevin, 2010). Here, we only provide some short paragraphs mainly aimed at defining the various terms and biological groups and highlighting the most important resistance effects that form the focus of the present article.
Groups of endophytes
Mycorrhiza
Mycorrhizal fungi form virtually ubiquitous associations with plant roots. Soil resources are accessed by fungi and exchanged for plant-derived assimilates (Smith and Read, 2008; Gehring and Bennett, 2009). Among the six major classes of mycorrhizas, ectomycorrhizas (EM), and arbuscular mycorrhizas (AM) have received most attention. EM occur in most woody plants and comprise a multitude of fungal species, whereas AM are formed by mosses, ferns, gymnosperms, and angiosperms with fungi of the division Glomeromycota (Bonfante and Genre, 2008; Smith and Read, 2008; Gehring and Bennett, 2009; Ercolin and Reinhardt, 2011). Anatomically, AMF represent “semi-endophytes” because one part of the fungus enters the intercellular space and forms haustoria that enter the roots cells without penetrating the plasma membrane, whereas the other part forms a dense mycelium that spreads into the surrounding soil. In general, the large and very fine mycelium dramatically increases the overall surface of the AMF-root complex and thereby improves nutrient uptake, particularly of phosphorous, from the soil (Bonfante, 2010; Koltai and Kapulnik, 2010). The level of host specificity of mycorrhizal fungi is low, as one mycelium can infect numerous plants that might belong to different species (Ercolin and Reinhardt, 2011). Interestingly, some AMF host in their cytoplasm endobacteria called bacteria-like-organisms (BLOs) related to the Mollicutes (Naumann et al., 2010), whereas in the Gigasporaceae family another endosymbiont named Candidatus Glomeribacter gigasporarum has been confirmed (Bianciotto et al., 2003), extending the symbiosis to a third partner. Besides their effects on nutrient uptake, mycorrhiza also have multiple effects on plant resistance both in the infected roots and in the aboveground compartment (Gehring and Bennett, 2009; Hartley and Gange, 2009; Van Dam and Heil, 2011). Because they connect different individual plants, AMF might even represent a medium for “plant-plant signaling” (Song et al., 2010).
N-fixing, nodulating bacteria – the diazotrophs
The second classical and highly investigated textbook example of a plant–microbe mutualism is formed by plants of the family Fabaceae (“Legumes”) and bacteria (Rhizobium and related genera, all belonging to the order Rhizobiales) that infect plant roots where they induce typical morphological structures called “nodules”(Kiers and Denison, 2008; Oldroyd and Downie, 2008; Markmann and Parniske, 2009). Within these nodules, the formerly mobile bacteria convert themselves into immobile bacteroids that exclusively live on plant-derived assimilates and in turn provide the plant with nitrogen that has been fixed from the atmosphere (Oldroyd and Downie, 2008; Brevin, 2010).
Type I or clavicipitaceous fungal endophytes
The first fungal endophytes were discovered in certain cultivars of tall fescue (Lolium arundinaceum) and of related grasses (mainly of the genera Festuca and Lolium) that cause livestock disorders. In these grasses, asexual, alkaloid-producing fungi of the genus Neotyphodium could be identified as the causal agent of their negative effects on mammal and insect herbivores (Clay, 1990). These “Type I fungal endophytes” sensu Yuan et al. (2010) are a taxonomically well defined group of the family Clavicipitaceae that are vertically transmitted via the seeds of their hosts and therefore cause systemic infection of certain grasses. Due to the production of alkaloids, these endophytes directly contribute to the resistance of their host plants to herbivores, but numerous studies have also reported effects on drought and flooding tolerance, biomass production and – in consequence – the competitive ability of the host grass (see Lehtonen et al., 2005 and Schulz and Boyle, 2005 for references).
Type II fungal endophytes
Type II fungal endophytes sensu Yuan et al. (2010) called class 3 endophytes by Rodriguez et al. (2009) are non-systemic, horizontally transmitted fungi that are just now being discovered. Numerous studies reported surprisingly high taxa numbers and infection rates by non-clavicipitaceous fungal endophytes. However, species saturation curves usually demonstrate that these studies were unlikely to discover all endophytes present in the investigated host plants (Arnold et al., 2000; Frohlich et al., 2000; Higgins et al., 2011). Thus, we must assume that we significantly underestimate the diversity of type II fungal endophytes and, hence, their potential importance in natural ecosystems. Type II fungal endophytes comprise species from both Ascomycota and Basidiomycota, although Ascomycota are usually dominating (Fisher, 1992; Frohlich et al., 2000; Arnold and Herre, 2003; Albrectsen et al., 2010).
These endophytes have been isolated from all plant organs, are usually found in 80–100% of the investigated samples, and multiple taxa can commonly be isolated from a single host species, or even a single surface-sterilized and symptomless leaf. For example, fungal species representing 18 taxonomic orders were reported from switchgrass (Panicum virgatum L.; Ghimire et al., 2011), 33 taxa of endophytic fungi were discovered in leaves of European aspen (Populus tremula; Albrectsen et al., 2010), 60 taxa were reported from leaves and twigs of Quercus ilex (Fisher et al., 1994), 58 operational taxonomic units were isolated from leaves of wild rubber trees (Hevea brasiliensis) in Peru (Gazis and Chaverri, 2010), more than 100 morphospecies of endophytes were found to be associated with tropical palms (Frohlich et al., 2000), and 418 fungal morphospecies could be isolated from 83 leaves from different tropical tree species (Arnold et al., 2000). Interestingly, type II endophytes can co-occur with type I endophytes in the same host species (Ghimire et al., 2011) although other authors found only numerous type II endophytes, but no clavicipitaceous species, in tropical grasses (Higgins et al., 2011). Information on the co-occurrence of endophytic fungi in the same host is just emerging. In Hevea spp., ca. 60–200 species of fungal endophytes were found in wild trees and 50–150 in planted ones, respectively, depending on sample size (P. Chaverri, pers. comm). On average 5.73 ± 1.94 species were found to co-occur in leaf disks of ca. 3 cm2. The most common genera/groups in wild rubber trees were Trichoderma, Penicillium, Paecilomyces, Pestalotiopsis, and Basidiomycota, whereas Trichoderma, Pestalotiopsis, Colletotrichum, Xylariales, and Basidiomycota were most common in planted trees. The most common combinations were Pestalotiopsis + Trichoderma (present in ca. 75% of the trees sampled), followed by Colletotrichum + Phomopsis (50%), Pestalotiopsis + Penicillium (45%), Pestalotiopsis + Colletotrichum (30%), and Pestalotiopsis + Trichoderma + Penicillium (20%). Pestalotiopsis or Trichoderma alone were only found in ca. 10–15% of the trees sampled, whereas Fusarium was always found associated with either Trichoderma or Penicillium (all data: pers. comm. by P. Chaverri and R. O. Gazis). Many more datasets of this type will be required to obtain insights into the general association patterns among endophytic fungi and to formulate testable hypotheses concerning the underlying mechanisms.
Bacterial endophytes
Bacterial endophytes represent a taxonomically and functionally highly diverse group that include non-nodulating nitrogen-fixing bacteria that infect shoots as well as other bacteria that exert multiple beneficial effects on the metabolism of their host plant (Weyens et al., 2009). Together with rhizosphere bacteria, these bacteria form the large functional group of “plant-growth promoting bacteria” (PGPB). Their growth promoting effects result from multiple mechanisms such as (1) the direct acquisition of mineral nutrients (via N-fixation, the solubilization of inorganic phosphates, the production of siderophores that facilitate iron uptake etc.), (2) the production of plant hormones, (3) the induction of resistance-related plant genes, and (4) direct antibiosis against pathogens (Weyens et al., 2009). Taxa that are frequently reported in this context belong to the genera Pseudomonas (in particular multiple strains of Pseudomonas fluorescens), Bradyrhizobium, Azorhizobium, Azospirillum, and Bacillus (commonly Bacillus thuringiensis). However, screening studies usually searched specifically for certain functional groups or were optimized for specific bacterial taxa. We therefore must assume that the true biodiversity of endophytic bacteria that infect wild plants in nature is as severely underestimated as is the biodiversity of type II fungal endophytes.
Positive effects on plant resistance to biotic stress
Microbial endophytes affect the physiology and ecology of their host plants in multiple ways. The underlying mechanisms can significantly differ among endophytes that belong to the same taxonomic group, whereas taxonomically different endophytes can act through similar or even identical molecular mechanisms. Therefore, we structure our – again necessarily highly incomplete – overview on beneficial effects of endophyte colonization of plants according to the major phenotypic outcomes, rather than on taxonomically defined groups of endophytes.
Resistance to pathogens
An increased resistance to pathogens appears to be a common consequence of plant infection or association with PGPB (Pieterse et al., 2001; Pieterse and Dicke, 2007) but is also commonly reported for plants that are infected with Rhizobia or AMF (Borowicz, 2001; Pozo and Azcón-Aguilar, 2007; Hartley and Gange, 2009; Pineda et al., 2010) and – less commonly – for plants that harbor fungal endophytes in their leaves (Weyens et al., 2009; Gao et al., 2010; Ownley et al., 2010). Although being phenotypically often similar, the enhanced resistance can be achieved by numerous direct and indirect mechanisms.
Direct effects are particularly likely to exist when the resistance effect is locally restricted to the infected tissue. For example, Theobroma cacao leaves that were infected with type I fungal endophytes showed a locally enhanced resistance to pathogens only in the infected areas, and direct in vitro confrontation assays demonstrated strong antagonisms between certain strains of endophytes and pathogens (Arnold et al., 2003). Direct effects of endophytes on the phenotypic resistance of their host can result from the production of antibiotics, of extracellular lytic enzymes such as chitinases, cellulases, proteases, and β-1,3-glucanases, of secondary compounds, when endophytes act as mycoparasites and directly parasitize plant pathogenic fungi, or when non-symptomatic endophytes outcompete the pathogens (Weyens et al., 2009; Gao et al., 2010; Ownley et al., 2010). Most of these traits have likely evolved in the context of antagonistic interactions among fungi. However, they will benefit any plant host that is infected by a competitively dominant fungus with low virulence. Belowground microorganisms are also very commonly reported to locally enhance the resistance of the roots (Borowicz, 2001; Pozo and Azcón-Aguilar, 2007; Elsen et al., 2008; Pineda et al., 2010), which makes it likely that similar mechanisms are involved. However, the molecular mechanism(s) that lead(s) to the enhanced resistance phenotype remain to be unraveled for most cases.
As a second mechanism, the infection of plants with endophytes that enhance the nutritional status of their host (MF, Rhizobia, and other PGPB) usually decreases the plant-internal competition among different traits for limited nutrients (Herms and Mattson, 1992). This effect alone might allow for systemically enhanced resistance levels (Van Dam and Heil, 2011). As a consequence, mycorrhiza can improve resistance to leaf pathogens (Fritz et al., 2006; Pozo and Azcón-Aguilar, 2007; Alejo-Iturvide et al., 2008) and also plants whose roots are infected with Rhizobia commonly show enhanced resistance levels in their aboveground parts (for a short overview on ecologically relevant resistance-enhancing effects of mycorrhiza and nodulation on aboveground communities see Van Dam and Heil, 2011).
A further group of mechanisms by which endophytes affect the resistance of their host to pathogens should be termed “indirect” mechanisms, because they act via interactions with the host’s own resistance system or via effects at higher trophic levels. Rhizobia and mycorrhizal or leaf endophytic fungi are well-known to induce host resistance responses. It appears to be likely that a significant part of the resistance effects to pathogens is caused by the enhanced expression of host resistance genes in endophyte-infected plants (Mithöfer, 2002; Pozo and Azcón-Aguilar, 2007; Ownley et al., 2010). For example, AMF colonization of roots commonly enhances resistance to necrotrophic leaf pathogens, whereas the resistance to biotrophic pathogens is often impaired, with the most likely reason being AMF-induced shifts between the different signaling pathways that control plant resistance to these functional groups of pathogens (Pozo and Azcón-Aguilar, 2007). Similarly, many endophytic fungi induce important resistance-related genes of their host (Ownley et al., 2010) and also most studies on PGPB that infect the aerial parts of plants report an induction or priming of resistance-related genes (Pieterse et al., 2001; Pineda et al., 2010), as it is well-known for PGPB in the rhizosphere (van Wees, 1999; Verhagen et al., 2004). For example, treating harvested papaya fruits with the endophytic bacterium Pseudomonas putida MGY2 enhanced their resistance to anthracnose caused by Colletotrichum gloeosporioides (Shi et al., 2011), whereas a Pseudomonas strain enhanced resistance of potato to soft rot disease caused by the necrotrophic pathogen Pectobacterium atrospeticum (Pavlo et al., 2011). In the first case, the authors suggested effects of endophyte treatment on ethylene production and reported an enhanced gene expression (and also activity at the enzymatic level) of phenylalanine ammonia-lyase, catalase, and peroxidase. In the second case, an induction of both basal resistance and of induced resistance dependent on SA- and JA/ethylene-signaling was found. We propose that due to the systemic nature of most resistance responses, it does not matter too much whether the resistance-inducing agent is infecting belowground or aboveground tissues. Most importantly, all these phenomena are mediated by effects of the endophytes on host gene expression patterns and therefore represent the “extended phenotype” of the microorganisms.
An even “more indirect” mechanism has been reported by Lehtonen et al. (2006), who found lower virus infection rates in Neotyphodium-infected meadow ryegrass (Lolium pratense). In this case, the underlying mechanism was a lowered preference of aphids for endophyte-infected plants. Because these aphids represent the most important vector for the virus, their behavior dramatically affected virus transmission rate in nature (Lehtonen et al., 2006). This example nicely demonstrates the complexity of interactions at multiple trophic levels and – in consequence – the complexity of the “extended phenotype” of endophytes which cause “symptomless” infections of their host plants.
Resistance to herbivores
As explained for pathogen resistance, the enhanced resistance of host plants to herbivores can also result from direct and indirect mechanisms. In fact, indirect mechanisms are likely to cause a large part of the enhanced resistance to herbivores that is caused by endophytes. Although one might expect a positive effect due to the enhanced nutrient contents of mycorrhized plants, the performance of herbivorous insects on AMF-inoculated plants is often lower than on un-mycorrhized plants (Hartley and Gange, 2009; Kempel et al., 2010; Pineda et al., 2010). Similarly, the colonization of roots by Rhizobia can increase resistance in the aboveground parts of soybean to aphids (Dean et al., 2009). For mycorrhizal fungi, effects in both directions have been reported, that is, aboveground herbivory can affect the mycorrhization of plant roots and mycorrhization can affect the aboveground resistance and tolerance to herbivory (Gehring and Bennett, 2009). Effects were, however, quite mixed, and future studies will need to consider detailed combinations of plant species with fungal species and the community context in order to identify general patterns (Gehring and Bennett, 2009).
Fungal endophytes are likely to enhance the resistance to herbivores of their host plant via four distinct mechanisms: (1) via the production of alkaloids and other toxic compounds, (2) due to their function as entomopathogens, (3) via the induction or priming of resistance-related genes, and (4) by enhancing the genetic and biochemical diversity of their host. These modes of action are not necessarily exclusive, as, for example, entomopathogenic fungal endophytes can also associate with the rhizosphere, act as antagonists of plant pathogens and might possibly even function as plant-growth promoting agents (Vega et al., 2009).
Type I endophytes produce alkaloids and thereby can directly intoxicate herbivores (see above). A recent meta-analysis revealed that type I endophytes in general slightly enhance host resistance to insects, although this result might be biased due to the concentration of the experimental effort on few agronomically important model systems (Saikkonen et al., 2010). For type II endophytes, by contrast, the situation appears to be less clear. Beyond doubt, in vitro cultures of many class II endophytic fungi produce compounds with antibacterial, fungicidal, and herbicidal effects (see Schulz and Boyle, 2005; Gao et al., 2010; Ownley et al., 2010 for references) and these compounds could exert a direct resistance effect. Class II endophytes, however, reach in general much lower densities in their host than do class I endophytes, and we are not aware of any empirical study on the direct resistance-enhancing effect in planta of the secondary compounds that are produced by class II fungal endophytes. In European aspen (P. tremula), however, the phenotypic resistance to herbivores of different clones was positively correlated to their load of endophytic fungi (Albrectsen et al., 2010), which makes a direct effect that is mediated by some fungal metabolites likely.
Second, several plant endophytic fungi also live as entomopathogens and thereby exert a direct negative effect on herbivores (Vega et al., 2009; Ownley et al., 2010). Interestingly, the life style as an insect parasite might be the ancestral state of many plant endophytic fungi, including type I endophytes (Rodriguez et al., 2009). In fact, parasitic fungi can often infect a wide spectrum of hosts including plants, animals, and other fungi. For example, the ubiquitous fungus Beauveria bassiana (Ascomycota) has been found as an endophyte in banana, corn, cotton, potato, tomato, common cocklebur, opium poppy, coffee, and cocoa (for references see Gurulingappa et al., 2010) but is also known to occur naturally in hundreds of insect hosts (Vega et al., 2009; Ownley et al., 2010). Care must be taken before we consider B. bassiana as some kind of “mega-generalist,” because it likely represents a species complex (H. E. Roy, pers. comm). In a recent study, Ormond et al. (2010) reported a strong assortment of specific genotypes of B. bassiana with specific hosts and other environmental conditions. Specificity at the level of genetically defined strains might be, thus, much higher than suggested by the above-cited lists of host species. Still, the fungi beyond doubt can switch from plant to insect hosts and vice-versa. Because the effects on the various hosts can be highly different, a surprisingly high number of fungi cause no symptoms in plants but act as severe pathogens when infecting an insect host (Ownley et al., 2010). A likely scenario appears to be that the fungus uses the plant as vector, that is, to enhance its transmission rate among different insect hosts. Knowledge on the ecology of these entomopathogens is limited (Rodriguez et al., 2009; Vega et al., 2009; Roy et al., 2010) and it therefore remains to be proven whether the negative effects on insect hosts can positively feed back to plant fitness. However, “biocontrol” entomopathogens are commonly isolated from symptomless plant tissues (Ownley et al., 2010) and beyond doubt can kill their insect hosts, which makes it likely that they exhibit a positive effects at least at the level of plant populations.
Whereas the direct effect of entomopathogenic fungi on herbivores happens outside the plant, a third, indirect mechanism results from effects of endophytes on the expression levels of host resistance genes. An infection with the root endophytic fungus Acremonium strictum enhanced resistance of broad bean (Vicia faba) to the generalist herbivore (Helicoverpa armigera). Because the fungus could not be re-isolated from the leaves, the authors suggested that it caused a systemic resistance effect and that the anti-herbivore effect did not depend on a direct contact between fungus and herbivore (Jaber and Vidal, 2010). It appears to be likely that in most of these cases the enhanced resistance results, at least in part, from changed expression patterns of resistance genes of the host plant (Ownley et al., 2010).
Finally, the presence of endophytes enhances the genetic and, consequently, biochemical diversity of the leaf tissue, which should represent per se a beneficial effect in evolutionary terms, because this increased diversity can make the adaptation of herbivores to certain plants more difficult (Schulz and Boyle, 2005; Albrectsen et al., 2010).
The Conditional Outcomes of Plant–Microbe Interactions
Conditional outcomes: an introduction
Based on studies that were conducted with model plants under laboratory or agronomic field conditions, we usually conclude that plant-infecting microorganisms can be classified as either pathogens or mutualists. Specific strains of microorganisms evidently behave as pathogens of certain hosts under virtually all environmental conditions, whereas Rhizobia and mycorrhizal fungi usually improve the nutritional status of their host plants and might enhance their resistance to biotic or abiotic stress. Fungal endophytes and PGPB commonly enhance host resistance to herbivores and, in specific cases, to abiotic stress.
However, the situation is far more complex. It now appears to be generally accepted that the interactions of plants with Rhizobia and fungal endophytes (both leaf endophytes and mycorrhiza) shift along a continuum from antagonism to mutualism (Francis and Read, 1995; Johnson et al., 1997; Denison and Kiers, 2004; Schulz and Boyle, 2005; Paszkowski, 2006; Hoeksema et al., 2010; Yuan et al., 2010). For example, mycorrhization either enhanced or decreased the resistance of the aboveground parts of plants to insect herbivores (Gehring and Bennett, 2009). The situation is particularly difficult for the fungal endophytes that usually do not cause visible disease symptoms (Francis and Read, 1995; Muller and Krauss, 2005; Saikkonen et al., 2006). In a common garden experiment with several tall fescue accessions, any putative effects of infection by Neotyphodium on the abundance and diversity of herbivores were overridden by the influences of environmental conditions and plant genotype (Vesterlund et al., 2011). In another experiment, infection of Arizona fescue by Neotyphodium generally decreased host growth rates and even host fitness in terms of number, mass, and germination rate of seeds (Faeth and Sullivan, 2003). Similarly, Faeth and Shochat (2010) reported an increased, rather than decreased, abundance of herbivores on Neotyphodium-infested Festuca arizonica plants, and Jani et al. (2010) reported increasing abundances and diversity of herbivores on native grasses with increasing contents of endophyte-produced alkaloids. These observations led several authors to question the general role of fungal endophytes as mutualists (Faeth, 2002; Faeth and Fagan, 2002), consider the positive reports on endophyte-mediated resistance to herbivores as unrepresentative (Saikkonen et al., 2006) or call the endophyte-grass mutualism “usually parasitic” (Faeth and Sullivan, 2003). What are the reasons for these conditional outcomes of seemingly mutualistic symbiotic interactions, why are these interactions still so common in nature, which data are required to predict the outcome of a certain interaction under natural conditions, and what does this conditionality mean in the context of our present considerations?
Conditional outcomes: examples
Although admittedly not the focus of the present article, the net effects of phloem feeders on the fitness of their host plant can help us to understand the context dependency of mutualisms. Aphids and other insects feed on phloem sap and thereby deprive the plant of highly valuable resources. Still, a recent meta-analysis (Romero and Koricheva, 2011) demonstrated on average net positive effects on plants of the presence of sap sucking herbivores. How can herbivores benefit their host plant? Aphids and other phloem feeders are commonly attended by ants, which can exert a significant “indirect” defensive effect by enhancing the predation pressure on most herbivores (Heil, 2008). In consequence, the presence of phloem feeders can positively affect plant fitness in the presence of other, more detrimental, herbivores (Moreira and del Claro, 2005; Styrsky and Eubanks, 2007).
What does this story tell us in the context of plant–microbe mutualisms? The message is simple: whether a given interaction between two species has a positive or negative net effect on either of the partners depends on the current biotic and abiotic environment. The outcome can therefore only be defined for a given set of environmental conditions. In fact, classical pathogens can positively affect their host plant under certain circumstances, and cases of supposedly mutualistic endophytes that harm their host or that are counterselected under specific growing conditions in fact appear to be common.
As an example of the first possibility, a normally detrimental virus infection can benefit the host plant when inducing the emission of volatiles with a repellent effect on herbivores. This case has been reported for white clover (Trifolium repens) plants infected with White Clover Mosaic Virus, which induced certain volatiles that repelled females of fungus gnats (Bradysia sp.; Van Moelken, 2009). Logically, this effect will benefit virus-infected plants under intense pressure by this particular herbivore, but not when the herbivore is absent. Even infection of plants with strains of Colletotrichum, a genus comprising “classical” pathogens, can be mutualistic in plants that were not known to be host plants (Redman et al., 2001). We are not aware of further studies reporting a positive effect of a plant pathogen but actually suggest that “conditionally beneficial pathogens” might be common, for the following reason. Most, if not all, biotrophic pathogens induce a systemic acquired resistance of their host plant against future pathogen infection. As described above, many of the beneficial effects of non-pathogenic endophytes are caused by an enhancement of the host’s resistance status as well. In the presence of a second, more virulent or lethal pathogen, a previous local infection with a less virulent pathogen is likely to have a positive net effect on plant fitness, due to its resistance-enhancing effect.
More commonly, however, researchers report negative net effects of supposedly mutualistic symbionts or find that endophytes are counterselected in certain environments. For example, Argentinian populations of Bromus setifolius exhibited higher (>80% of plant sampled) rates of infection with the fungus Neotyphodium tembladerae in regions where herbivore pressure exerted by leaf-cutting ants was high; by contrast, infections rates below 20% were observed in regions where this herbivore species is rare (White et al., 2001 cited in Rodriguez et al., 2009). Apparently, infection by endophytes can be rapidly counterselected in environments where it is not likely to benefit host fitness.
Negative effects of mycorrhization might arise from a general shift in plant allocation patters toward growth, at the expense of defense (Barazani et al., 2005). A recent study demonstrated positive effects of fungicide treatment on reproductive success of Polemonium viscosum and suggested high costs of mycorrhization as a likely reason (Becklin et al., 2011). Unfortunately, potential effects on other fungi were not considered and more data will be required to understand effects of mycorrhization, or infection with leaf endophytes, on the production of flower rewards and, ultimately, plant reproductive success. In this context, also the type of the enemy matters with which an endophyte-infected plant is interacting. Belowground infection with microbial mutualists usually enhances the resistance to soil-borne pathogens and to necrotrophic leaf pathogens (Pozo and Azcón-Aguilar, 2007; Van Dam and Heil, 2011). By contrast, mycorrhized plants commonly suffer from an enhanced susceptibility to biotrophic pathogens and aphids, and occasionally also to leaf chewers (Pozo and Azcón-Aguilar, 2007; Hartley and Gange, 2009; see also Figure 1 in Van Dam and Heil, 2011). In the case of leaf chewers, the degree of specialization of the herbivore also matters, as mycorrhization commonly enhances the resistance of the host to generalists while increasing its susceptibility to specialists (Koricheva et al., 2009). At least the effect of endophyte-produced alkaloids can scale up to higher trophic levels and for example negatively affect the natural enemies of the herbivores (Faeth and Shochat, 2010; Jani et al., 2010), thereby reducing the potential for indirect plant defense via tritrophic interactions (Heil, 2008). Even the direct defense of the plant can be impaired, as demonstrated by the observation that mycorrhization of Plantago lanceolata roots was negatively correlated with the concentration in roots of aucubin, a defensive iridoid glycoside (De Deyn et al., 2009).
A second defensive strategy besides resistance is tolerance, which means a reduced negative effect of a given damage level due to an enhanced capacity of regrowth after defoliation (Fornoni, 2011). Mycorrhization reduced regrowth after defoliation (De Deyn et al., 2009) and similar observations led Garrido et al. (2010a,b) to speculate that colonization of plant roots with mycorrhizal fungi could “constrain the evolution of plant tolerance to herbivory.” Similarly, Neotyphodium infection of grasses can negatively affect their recovery rate after clipping (Saari et al., 2010), which indicates that the same trade-off might also apply to endophyte infection. Other studies, however, found positive effects of mycorrhization on plant regrowth capacity after defoliation (Kula et al., 2005). In a comparison of three different mycorrhizal fungi, the fungal species mattered, as different fungi did, or did not, cause an enhanced tolerance of P. lanceolata to caterpillar feeding (Bennett and Bever, 2007). Depending on the actual composition of the taxa involved, the colonization by a mycorrhizal or endophytic fungus on the same plant genotype can, thus, be positive, neutral, or negative.
In interaction with the presence or absence of certain herbivores, abiotic factors such as soil nutrients and light can be crucial in determining the net effects of plant infection by microorganisms. For example, light intensity determined the pathogenicity of the fungal endophyte Diplodia mutila in the tropical palm Iriartea deltoidea (Álvarez-Loayza et al., 2011). The authors observed that the fungus causes increasingly intense disease symptoms with increasing light intensity, whereas it can protect seedlings and adult palms from stem and seed borers when acting as a mutualist under understory (i.e., low-light) conditions. Whereas the detailed molecular mechanisms and the ecological and evolutionary consequences of this phenomenon remain to be studied (Álvarez-Loayza et al., 2011), it is common knowledge that nutritional mutualisms can benefit plants only under limiting conditions. The energetically expensive bacterial fixation of elementary nitrogen pays off for the plants only when supply of mineral nitrogen is limited. Likewise, net effects of mycorrhization are strongly dependent on soil nutrient availability (Hoeksema et al., 2010, and references therein). However, even the resistance effects that are mediated by endophytes are triggered by abiotic conditions, particularly nutrient supply. For example, plants of meadow fescue, L. pratense, supported significantly lower numbers of cherry aphid (Rhopalosiphum padi) when being infected with Neotyphodium under high nutrient conditions, whereas this resistance effect was strongly impaired under low nutrient conditions (Lehtonen et al., 2005). In the end, even completely mutualistic endophytes obtain resources from their hosts and therefore cause metabolic costs, which must be outweighed by the benefits of their presence in order to cause a net mutualistic interaction.
Finally, rather than negatively affecting the interaction of plants with other beneficial organisms, beneficial microorganisms can themselves be harmed by such interactions. For example, the systemic induction of pathogen resistance in leaves can impair the capacity of plant roots to establish their interaction with Rhizobia or AMF (see de Román et al., 2011 and references therein) and leaf infestation with whitefly (Bemisia tabaci) affected the composition of the bacterial rhizosphere with likely negative effects on PGPB (Yang et al., 2011). In summary, the net outcome of most, if not all plant–endophyte interactions is highly conditional and depends on the detailed biotic and abiotic environment as much as on the specific genotypes of both endophyte and plant host (Figure 2).
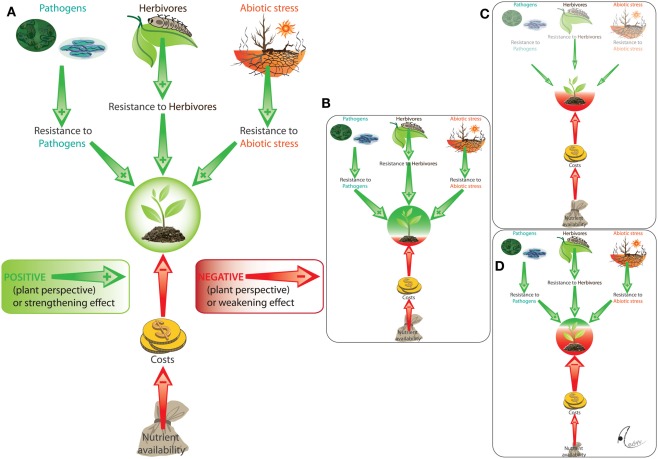
Figure 2.
Conditional outcomes of plant–microbe interactions. We illustrate here the multiple factors that mediate the outcome of the presence of a resistance-enhancing endophyte (A). Resistance against biotic stressors (pathogens and herbivores) and abiotic stressors (e.g., heat) has positive effects (from the perspective of the plant: green arrows), whereas the costs that are caused by the maintenance of the resistance-enhancing endophyte represent a negative effect (red arrows). Due to this interplay of positive and negative effects, the same interaction results mutualistic in presence of those stressors to which the endophyte provides resistance (B) whereas, by contrast, the net outcome becomes negative (antagonism) either in the absence of stressors [(C) lack of benefits] or when resource limitation enhances the relative costs of maintaining the endophyte (D).
Causes and consequences
Why are the effects of plant–microbe interactions so variable? Two non-exclusive frameworks have been proposed to understand the conditional outcomes for the infection of pants with fungal endophytes, but they are likely to generally apply to all plant–endophyte interactions: the virulence-resistance balance (Schulz and Boyle, 2005) and cost–benefit relations (Faeth, 2002).
Balanced antagonism: the molecular perspective
First, at the molecular level, all endophytes require at least some virulence to infect their host while the host requires some resistance traits to control their density (Schulz and Boyle, 2005; Eaton et al., 2011; Zamioudis and Pieterse, 2011). The partial suppression of the host resistance phenomenon represents a crucial step not only during the infection of plants with fungal endophytes but also in the nodulation or mycorrhization of plant roots (Garcia-Garrido and Ocampo, 2002; Mithöfer, 2002; Paszkowski, 2006; Liu et al., 2007; Pozo and Azcón-Aguilar, 2007; Zamioudis and Pieterse, 2011). Maintaining the delicate balance between host resistance and endophyte virulence represents a key factor that decides whether the endophyte can infect the plant at all, and whether it then causes visible disease symptoms (Schulz and Boyle, 2005; Paszkowski, 2006). For example, clavicipitaceous endophytes become pathogenic when developing in grass hosts that lack a component of the inducible NADPH oxidase pathway, because the host looses control over their growth rate (Eaton et al., 2011). Besides genetic and other endogenous factors, the balance depends also on exogenous (that is, environmental) factors.
The importance of induced plant resistance traits for root symbioses is supported by the observation that induced leaf resistance to pathogens can impair the capacity of roots to become colonized by AMF (de Román et al., 2011) and that also natural herbivory has commonly negative effects on mycorrhization levels, whereas experiments using clipping treatments mostly failed to confirm this effect (Gehring and Bennett, 2009). The explanation presented for the last phenomenon was that a low intensity of herbivory changes the amounts or quality of root exudates, which in turn has positive effects on mycorrhizal fungi and, likely, nutrient acquisition required for regrowth, whereas high levels of leaf damage reduce plant assimilation rates and thus the potential for carbon allocation to symbionts (Gehring and Bennett, 2009). An alternative explanation would be, however, that herbivory enhances JA-dependent resistance traits and, due to the general trade-off between JA-signaling and SA-signaling, impairs the resistance to microbial infections, thus causing shifts in the resistance–virulence balance that at least transiently cause higher colonization rates. Clipping treatments, by contrast, remove leaf area without liberating the elicitors that are required to mount an adequate resistance response and therefore only reduce leaf area, without inducing any resistance responses.
Host ontogeny is of crucial importance, because non-symptomatic endophytes can become pathogenic as soon as the increasing leaf age or abiotic stress weaken the resistance level of their hosts (Schulz and Boyle, 2005). Finally, many endophytes that did not cause visible disease symptoms under the specific study conditions belong to genera that also comprise important pathogens. For example, fungi that were isolated from surface-sterilized symptom-free leaves were determined or annotated as belonging to the genera Alternaria (Fisher et al., 1994; Albrectsen et al., 2010; Ghimire et al., 2011), Colletotrichum (Fisher et al., 1994; Frohlich et al., 2000; Ghimire et al., 2011; Higgins et al., 2011), Fusarium (Fisher et al., 1994; Gazis and Chaverri, 2010; Ghimire et al., 2011), Phomopsis (Fisher et al., 1994; Frohlich et al., 2000), or Verticillium (Frohlich et al., 2000). Few, if any, mutations in certain effector molecules or changes in host resistance status appear, therefore, to be sufficient to convert an asymptomatic endophyte into a highly virulent pathogen or vice-versa (Freeman and Rodriguez, 1993), as it is also being discussed for host–plant shifts of pathogens (Schulze-Lefert and Panstruga, 2011).
Cost–benefit relations: the level of the entire organism
At the level of the entire organism, using a general cost–benefit framework can help to understand the context dependency of plant–microbe interactions (Figure 2). Cost–benefit frameworks have already been suggested by for the specific case of AM fungi (Johnson et al., 1997; Gange and Ayres, 1999; Vannette and Hunter, 2011) and of class I fungal endophytes (Faeth, 2002) and have been well developed for plant defense traits (Herms and Mattson, 1992; Heil and Baldwin, 2002). In fact, however, essentially ALL plant traits (including traits that are encoded by plant genes and traits that represent the extended phenotype of associated microorganisms) come with certain costs and provide the plant with certain benefits.
On the one hand, costs can be direct ones and then usually result from the allocation of limited resources to trait A, which then cannot be allocated also to a trait B that is competing for the same resources. This situation is expressed in the “growth-differentiation balance” hypothesis (Herms and Mattson, 1992): plants cannot grow rapidly and at the same time effectively defend the newly developed tissue, because every atom of carbon or nitrogen that is invested into the synthesis of defensive secondary compounds has to be withdrawn from primary metabolism. Because endophytes compete with primary metabolism for certain nutrients, their presence can incur a considerable cost in terms of reduced growth rates under limiting sol nutrient condition (Cheplick et al., 1989). Costs of a given trait can, however, also occur as ecological costs (Heil, 2002), which result when the expression of a trait negatively affects other interactions of the host with its biotic or abiotic environment. For example, actively N-fixing nodules can attract root-feeding weevil larvae and in the presence of these herbivores cause negative net effects on plant growth and fitness (Johnson et al., 2005). The presence of Rhizobia can even significantly affect the community of aboveground herbivores and predators, which benefits or harms the plant, depending on the detailed shifts in the concrete insect communities (Katayama et al., 2011).
On the other hand, the benefits of a certain trait (or interaction) can also be direct or ecological ones. Rhizobia and mycorrhizal fungi provide their host plant with valuable nutrients, which clearly represents a direct benefit. Still, the relative contribution of this benefit to plant fitness depends on the environment, that is, the concentration of mineral nutrients in the soil. Plant roots in fact stop the allocation of assimilates toward their nodules or do not establish the mutualism under high soil nitrogen conditions. The benefits that type I fungal endophytes provide to their host plant are, by contrast, essentially ecological ones: these endophytes enhance the resistance of their host to herbivores (Clay, 1990, and above). In the absence of sensitive herbivores, this potential benefit cannot be realized and the net effect of infection is inevitably negative, because the (allocation) costs of nourishing the endophyte are not counterbalanced by any positive (ecological) effect.
We present here a graphic model to understand these complex interactions (Figure 3) and suggest that the net outcome of a certain interaction should be determined – for a given set of environmental conditions –as follows:
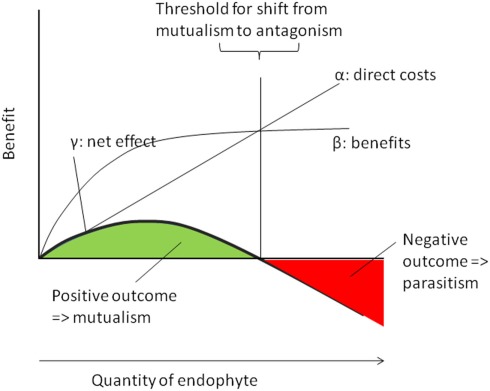
Figure 3.
Dose–response relations in plant–endophyte interactions. Endophytes cause direct metabolic costs that should be directly correlated to their abundance within the host (α) whereas their benefits are likely to follow a saturation curve (β). Therefore, the resulting net benefit (γ) follows an optimum curve and is predicted to reach positive values at low and intermediate densities (green area => mutualistic densities of the endophyte) but negative values (red area => antagonistic densities) at high (antagonistic) infection densities.
Terming an interaction “mutualism,” “commensalism,” or “parasitism” should be based on its average net outcome, considering the most common environments under which the interacting organisms occur naturally. We suggest consider only ecological benefits, but not ecological costs, in this context. Ecological benefits represent an important part of the overall benefits that are provided by microorganisms to their host plant. In the case of resistance-enhancing endophytes, all benefits provided are ecological ones. By contrast, ecological costs must be considered more cautiously, because most traits of an organism eventually attract predators or parasites and then impair rather than improve fitness. For example, fruits, pollen, and nectar are eventually consumed by animals that do not provide the expected service (dispersal and pollination). However, these traits in principle serve the reproduction of the plant. Thus, it would not be useful to consider, e.g., pollen grains as plant parasites under certain conditions, although their production eventually impairs plant fitness due to the losses to non-pollinating insects.
Consequences: predicting patterns in conditional outcomes
Host–endophyte interactions are highly variable and their net effects are strongly context dependent. Most importantly, however, we have seen above how important plant traits are strongly affected by the presence of endophytes in the plant. Due to the significant endophyte–environment interactions, we must assume that a large part of our current picture on general plant ecological traits is heavily biased or, at best, incomplete. Most studies on endophytes have been realized under controlled conditions, which often were not representative of the “normal” situation (Saikkonen et al., 2006). By contrast, most field ecological studies that were conducted in the wild did not even consider, let alone control for, the percentage of the plant population under investigation that was infected by endophytic microorganisms. Predictions on general or typical outcomes of endophyte–plant interactions under natural conditions are, therefore, still hardly possible in most contexts. A general pattern that we would expect is, however, that generalist herbivores are more commonly affected than specialists by any infection of the plant with a microorganism that interferes with the resistance of the host plant (Koricheva et al., 2009). Considering the fact that specialists are better adapted to the particular biochemistry and characteristic defenses of their host plant, any endophyte that indirectly affects the resistance of its host (that is, via an enhancement of the host’s own resistance) is likely to accelerate the normal effects of resistance: decreased performance of generalists but no or even positive effects on adapted specialists. Even endophytes that directly affect resistance via the synthesis of alkaloids or other toxic compounds might, however, attract specialists to their host plant, or at least affect specialists less strongly than generalists: due to the common and predictable presence of these endophytes in certain host species, co-evolutionary responses on the side of specialist enemies are likely. As far as we are aware, these patterns are exactly what researchers find (Koricheva et al., 1998, 2009; Hartley and Gange, 2009; Currie et al., 2011).
Similarly, the allocation of assimilates and resources toward the endophyte appears an inevitable consequence of their presence. Therefore, the presence of an endophyte is likely to cause a stronger negative effect on plant-growth rates under limiting nutrient conditions (Cheplick et al., 1989). We also predict that these costs will be proportional to the endophyte density reached in the plant, being generally higher for type I than for type II fungal endophytes. However, we are not aware of a study that has tested this prediction.
Areas for Future Research
Search for them
We need more field research in order to broaden our knowledge on the microorganisms that are common in nature, that affect the ecological interactions of plants with their environment and that, thereby, drive plant evolution. For horizontally transmitted, non-systemic endophytic fungi and bacteria in particular, we lack the most basic knowledge: “who is there?”, “who is where?”, and “who is common and/or ecologically important?”. Even these basic questions remain largely unanswered for most groups of endophytes, because research effort has mainly concentrated on the vertically transmitted clavicipitaceous fungi of grasses (Saikkonen et al., 2006; Saikkonen, 2007). Various studies have screened for fungal endophytes, but they used different methods and their results are therefore hard to compare, particularly with respect to quantitative questions. Second, factors such as host ontogeny, season, leaf age, microclimatic conditions, and the distance from other plants (that can serve as a source for infection with horizontally transmitted endophytes) affect the fungal (and likely also bacterial) population of a certain plant organ (Arnold and Herre, 2003; Schulz and Boyle, 2005; Ormond et al., 2010). As exemplified by the effect of light intensity of the pathogenicity of a fungal endophyte (Álvarez-Loayza et al., 2011), these factors can be subject to complex interactions: infection by D. mutila is likely to benefit seedlings and saplings in the understory but may harm the larger plants that are exposed to full light conditions. Third, species saturation curves usually demonstrate that the conducted studies were unlikely to discover the total endophyte diversity present (Arnold et al., 2000; Frohlich et al., 2000; Higgins et al., 2011). Finally, screenings for endophytic bacteria are usually restricted to taxa with a known plant-growth promoting effect.
As a first step into the future, we suggest to apply broad screening techniques to search for endophytic microorganisms in natural species and plant communities. Screening can be realized both empirically and in silico. Screening attempts that include an initial cultivation step have been applied successfully and revealed an as of yet unknown fungal and bacterial diversity in various host–plant species (Albrectsen et al., 2010; Gazis and Chaverri, 2010; Ormond et al., 2010; Ghimire et al., 2011; Higgins et al., 2011). However, the cultivation step is likely to filter against a significant part of the microbial flora. Culture-independent techniques such as the direct PCR of microbial DNA extracted from plant tissue (Higgins et al., 2011) and subsequent mass sequencing is therefore likely to improve our knowledge on the as yet overlooked microbial diversity within plants. Molecular, culture-independent methods to study microbial biodiversity of natural systems rely either on the extraction of DNA, cloning and sequencing of small DNA fragments (shot gun cloning – metagenomics) or in the a priori amplification of defined genes using polymerase chain reaction. Some of the genes that are commonly used as molecular markers are the ribosomal genes (16S and 18S/ITS), RNA polymerase B (rpoB), methane monooxygenase (pmoA), nitrogenase (nifH), nitrite reductase (nirS/nirK), among others. Later, such amplified products can be directly cloned and sequenced, or submitted to several fingerprinting methods such as: amplified ribosomal DNA restriction analysis or restriction fragment length polymorphism (RFLP); automated ribosomal intergenic spacer analysis (ARISA); terminal RFLP (T-RFLP); denaturing gradient gel electrophoresis (DGGE) or temperature gradient gel (TGGE); single strand conformation polymorphism (SSCP); and denaturing high-performance liquid chromatography (DHPLC). Some reviews about the advantages and disadvantages of each of these methods for community profiling have been published elsewhere (Binladen et al., 2007; Nocker et al., 2007; Huys et al., 2008; Reeder and Knight, 2010). It is important to note that although great progress has been achieved and massive sequencing technologies are becoming more feasible, addressing microbial communities, and microbial diversity in natural environments is still a challenge (Bent and Forney, 2008). Moreover, microbial communities seems to be greatly diverse within a single host organism (Arnold et al., 2000; Frohlich et al., 2000; Schoettner et al., 2009; Higgins et al., 2011) and even tripartite symbioses formed in a “babushka-like” style by a plant, an endophytic fungus, and an endo-fungal bacterium or virus have been reported (Partida-Martinez and Hertweck, 2005; Márquez et al., 2007; Hoffman and Arnold, 2010; Naumann et al., 2010) and might significantly contribute to the as of yet unknown microbial diversity in natural ecosystems.
Where exactly are all these microbes localized? Endophytic microbes that live within the plant tissue can be visualized, for example, by in situ hybridization and microscopy with selective stains (Singh et al., 2004). Additionally, in cases where the microbial cells can be transformed with plasmids coding for fluorescent proteins such as GFP, the transformed cells could be followed during the interaction with their host (Partida-Martinez et al., 2007). Studying the molecular mechanisms via which plants interact with microbes will rely on the combination of multiple molecular, genomic, and biochemical methods, depending on the culturability of the microbial endophytes, the feasibility of genetic transformation of both plant and endophyte, and the ability to create asymbiotic hosts plants. Finally, we should apply Koch’s postulates to investigate the phenotypic effects of endophyte infection and to nail down certain phenotypes clearly to the colonization by one defined (or a defined combination of) strain(s) of endophytes.
At the in silico level, there is a large pool of potential information in all the genomic sequencing projects that have become realizable due to affordable next-generation sequencing techniques. In the first evaluation step of any genome sequencing project that is realized with an eukaryotic organism, genes of putatively microbial origin are considered “contaminations” and therefore are routinely removed from the raw datasets. However, the microorganisms that carry these genes are likely to represent an integrative part of the functioning entire organism, rather than contaminations. Any bacterium or fungus that is regularly infecting (or otherwise associated with) a certain host affects the phenotype of its host in a predictable manner and is likely to co-evolve with its host. Discarding these sequences from further analysis prevents us from a more complete understanding of the true subject of our scientific effort: the entire plant as it exists in nature.
The taxonomy and evolution of endophytic microbes
As most of the above-mentioned methods produce sequence data for the microbial strains that form the community within a given host plant, the information obtained can be used to place the discovered taxa within the broader phylogenetic context: a crucial step if one aims at understanding the evolution of endophytes. Are mutualistic or commensalistic microorganisms former pathogens that have lost their virulence to some degree but maintained the capacity to invade the tissue of living plants? Or are mutualists and antagonists derived from different phylogenetic clades? Considering the general conditionality in all these interactions and the existence of mutualistic and pathogenic taxa within the same genera (see above, and the multiple fungal taxa that form ectomycorrhiza: A. E. Bennett, pers. comm.), a discovery of major clades that are characterized by a well defined life history appears to be unlikely. As mentioned, many endophytes are believed to have evolved from former insect parasites and the Clavicipitaceae comprise, besides mutualistic plant endophytes, also entomopathogens, plant pathogens, and saprophytes (Rodriguez et al., 2009). Because the sexual phase of Neotyphodium, Epichloë, has a parasitic nature (Muller and Krauss, 2005), it appears tempting to speculate that endophytes are former pathogens that have successfully completed the common tendency in host–parasite coevolution: decreasing their virulence until a balanced, asymptomatic interaction has been reached. Intriguingly, one single mutation was sufficient to convert a pathogenic strain of Colletotrichum magna into a non-pathogenic endophyte that had the capacity to protect its host from infection by fungal pathogens (Freeman and Rodriguez, 1993). Similarly, the endophyte Guignardia mangiferae (Botryosphaeriaceae) and the citrus pathogen G. citricarpa differ only in a limited number of enzymes and the endophyte appeared phylogenetically derived from the pathogen (Romao et al., 2011). Thus, current evidence leads to the conclusion that endophytes commonly represent former pathogens that have reduced their virulence to a degree that allows asymptomatic life within their host.
However, the patterns at the lowest taxonomic levels might look completely different. Specific strains of what is currently considered as one species might be well specialized to usually cause either a pathogenic or an asymptomatic infection or to preferably infect different hosts (Ormond et al., 2010). Direct comparisons of related strains with contrasting strategies such as those presented by (Romao et al., 2011) would greatly help in the understanding of factors that plant-infecting microorganisms employ to successfully infect their host and to elicit, suppress, or avoid major host resistance responses.
Population ecology
We have stated above that a completely endophyte-free plant is unlikely to be able to survive under natural conditions (Figure 1). Unfortunately, as to our very best knowledge, no empirical data exist to support this statement. Studies on class I endophytes routinely produce “endophyte-free” plants and investigate their behavior under field conditions. However, the controls are only checked for the absence of the (vertically transmitted) clavicipitaceous fungus in the beginning of the experiment, but not for other, horizontally acquired endophytes. It would be highly interesting to produce complete endophyte-free plants and study their phenotype and ecological success under controlled conditions as well as in the field. These studies should be accompanied with the above-mentioned cultivation-independent screening techniques, in order to monitor for the occurrence of horizontally acquired endophytes in the originally endophyte-free plants. Two aspects appear to be of particularly crucial importance if we aim at understanding the role of endophytes in any ecosystem: (1) the population densities reached by endophytes and (2) the temporal order at which the symbioses of a plant with various microbial partners are usually established.
That microbial population density matters is nicely illustrated by a recent report on the effects of different inoculum densities used to infect Datura stramonium with various Glomus species: the positive effects on seed set of the infection followed an optimum curve with increasing infection intensity, whereas plant tolerance to defoliation was linearly and negatively correlated with infection intensity (Garrido et al., 2010a). Such patterns are likely to be common, because any endophyte represents a cost to its host plant (see above). Therefore, too high densities can be expected to cause negative effects under most conditions.
We follow Gange and Ayres (1999) and use an easy graphical model (Figure 3) to illustrate the importance of the quantity of any given endophyte for the net effect of the infection. Because every cell of living endophyte has its own metabolic demands, the direct costs of endophyte infection should be linearly and positively correlated to the amount of endophyte that must me nourished by a host plant. By contrast, the effects of many, if not all, endophytes are likely to follow a saturation curve: increased nitrogen supply from Rhizobia or increased supply of phosphorous from mycorrhizal fungi benefits plants only until all needs are saturated and other factors become limiting. Likewise, any resistance factor will reach an optimum level of concentration above which an increase in the concentration of the compound does not further enhance the level of phenotypic resistance. As the net effect of the endophyte can be calculated by its benefits minus its costs (see above), we predict that the net positive effects of endophyte infection in relation to their quantity follow an optimum curve, as it has been indeed been found empirically by Garrido et al. (2010a).
Data obtained in the above-mentioned screening processes can also be directly used to answer critical questions: how common are endophytes in natural populations? how specific are they? what determines the endophytic flora, particularly for the horizontally transmitted species: host or ecosystem? That is, are two plants more likely to share common endophytes when they are (a) taxonomically related or (b) growing in the same habitat? Both host species and season affected the distribution of genetically defined strains of B., bassiana (Ormond et al., 2010) and such patterns are likely to be more pertinent at higher taxonomic levels. Data of this type represent an essential basis for the formulation of further, more detailed research questions aimed at improving our knowledge on the ecological role of endophytes in natural ecosystems.
Another important question concerns the temporal order in which plants become infected with the different types of endophytes, and the temporal relations among these infections and the numerous further ecological interactions of a plant (Van Dam and Heil, 2011). Erb et al. (2011) have nicely demonstrated that the sequence of arrival matters for the plant-mediated interactions among two herbivores. Similar patterns were found for the mutualistic outcome of the infection of plants with certain Colletotrichum strains (Redman et al., 2001). In fact, the sequence of arrival becomes crucial when we consider endophyte-mediated changes in the resistance phenotype of a plant. Most beneficial effects of endophytes must be considered preventive rather than curative, but can we expect the infection by endophytes always to precede the attack of a plant by its enemies? Vertically transmitted endophytes infect a plant throughout its entire life cycle: infection with clavicipitaceous endophytes, thus, should normally precede any other ecological interaction of the host plant. It appears also to be likely that plant roots acquire nodulating bacteria and mycorrhizal fungi at early stages of root development, soon after germination. The situation is, however, much less clear for the leaf-colonizing type II endophytes. In one study, the distance from a putative source tree influenced endophyte colonization, suggesting a highly dynamic infection process (Arnold and Herre, 2003). Type II endophytes might therefore have to infect a plant after its encounters with pathogens or herbivores, with as yet unknown consequences for all partners involved. Given the conditionality of all these interactions, the temporal order at which a plant normally establishes the interactions with its various horizontally transmitted microbial symbionts certainly requires future investigation. Again, the above-mentioned molecular methods could be applied to plants under natural conditions to characterize the endophytic flora in different organs, in plants of different developmental stages and in con- and heterospecific plants that grow at different sites.
Effectors as determinants of the specificity and net effects of endophytes
As mentioned above (see Balanced Antagonism: The Molecular Perspective), endophytes require virulence factors to overcome the resistance of their host (Zamioudis and Pieterse, 2011). The balance between host resistance and endophyte virulence is likely to be a central determinator of endophyte density, with all its consequences on host development and fitness (Eaton et al., 2011). Effectors play a crucial role in the microbial invasion process and are likely to be central determinants of the host spectrum of pathogens (Schulze-Lefert and Panstruga, 2011). Similarly, endophytes employ effectors to suppress plant immunity and colonize the host plant (Zamioudis and Pieterse, 2011). As suggested for the investigation of generalist vs. specialist pathogens (Barrett and Heil, 2012), comparing the genomes and phenotypes of taxonomically related pathogens and mutualistic endophytes will allow to directly investigate the genetic differences that are contributing to these different microbial life styles. Next-generation high-throughput sequencing should also be applied to mRNA, because the detailed balance between the infection by the endophyte and its control by the host is maintained mainly via inducible responses, that is, at the transcriptional level (Eaton et al., 2011). Considering the arguments presented above, this type of experiments will benefit strongly from a detailed experimental control at the qualitative and quantitative level over the endophyte population of the host in combination with comparisons under different environmental conditions and of different host ontogenetic stages.
Concluding Remarks
Plant-infecting microorganisms are ubiquitous and affect many vitally important plant traits, particularly those that control the interactions of plants with their abiotic and biotic environment. Undoubtedly, some microorganisms will usually have negative effects on the fitness of their hosts and thus should be termed pathogens, whereas other microorganisms that colonize the tissue of plants usually have positive effects and then represent mutualistic endophytes. However, all plant–microbe interactions can shift along a mutualism–antagonism continuum. To understand their effects on plant ecology, plant fitness, and ultimately plant evolution, we therefore need to know the net effects that the typical microbial flora of a plant has under the average environmental conditions and understand the molecular mechanisms that cause the observed phenotypic effects. Most importantly, a microbe-free plant is not what we normally see in nature. It is likely that all plants that grow under natural conditions are, or have been in their past, colonized by several microorganisms, which exert highly conditional, but vitally important effects on their host. Considering, and ideally controlling for, the presence of these microorganisms will benefit future research into our study object of interest: the “normal” plant that is growing in – and interacting with – its natural environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to Alejandro de León for preparing Figures 1 and 2 and to Priscila Chaverri and Romina O. Gazis for sharing their unpublished data set on the co-occurrence of endophytes in rubber tree samples. Helen E. Roy, Alison E. Bennett, and our referees, Juriaan Ton and Toby Kiers provided us with many helpful comments on an earlier version of this manuscript. Financial support by CONACyT de México is gratefully acknowledged.
Glossary
Benefits, direct – all direct contributions to the growth, development, or reproduction of the host organism
Benefits, ecological – all effects on ecological interactions of the host with abiotic or abiotic factors that exert a beneficial net effect on host fitness (for example, an enhancement of host resistance to herbivores benefits the host only in the presence of herbivores)
Costs, direct – have direct negative effects on the fitness of the organism. Direct costs can result from the allocation of limited resources to the trait, autotoxic effects of the trait or its direct, genetic coupling to another trait with negative net effects (pleiotropic costs)
Costs, ecological – all negative effects of the expression of a given trait or interaction that result only from an negative interaction among the trait in question and other, ecologically relevant interactions (for example, the attraction of root herbivores to N-fixing nodules represents an ecological cost of nitrogen fixation)
Effector – molecule secreted by a plant enemy to manipulate host resistance
Endophyte – a microorganism that lives at least during a part of its life cycle inside living plant tissue without causing visible disease symptoms
Endophyte, Type I – the ‘singlbaseclassical’ fungal endophytes: vertically transmitted asexual fungi (usually genus Neotyphodium, Clavicipitaceae) in grasses that cause no visible disease symptoms under most growing conditions and that usually exert some kind of positive effect on their host
Endophyte, Type II – horizontally transmitted bacterial or fungal endophytes
Induction – the augmentation of gene expression by exogenous factors
Pathogen – an endophytic microorganism that causes phenotypically visible disease symptoms, thereby negatively affecting plant fitness
Phenotype, extended – sensu (Dawkins, 1999) the sum of ALL effects of a gene, including those elicited in other organisms. In the context of the present work, those effects of the plant phenotype that are caused by the interaction of the genome of the endophytic microorganism with the plant genome and its environment
Priming – the preparation of a gene by an exogenous factor for faster and stronger expression
Resistance mechanism, direct – in the context of the present article all negative effects on the enemies of the host plant that are caused by direct antagonisms between endophyte and plant enemy (most commonly mediated by the production of compounds that deter or harm the enemy or by direct endophyte-enemy competition)
Resistance mechanism, indirect – in the context of the present article all negative effects on the enemies of the host plant that are caused by stimulation (induction or priming) of host resistance genes or that are mediated via interactions with further organisms (such as, for example, vectors)
References
- Albrectsen B. R., Bjorken L., Varad A., Hagner A., Wedin M., Karlsson J., Jansson S. (2010). Endophytic fungi in European aspen (Populus tremula) leaves-diversity, detection, and a suggested correlation with herbivory resistance. Fungal Divers. 41, 17–28 10.1007/s13225-009-0011-y [DOI] [Google Scholar]
- Alejo-Iturvide F., Marquez-Lucio M. A., Morales-Ramirez I., Vazquez-Garciduenas M. S., Olalde-Portugal V. (2008). Mycorrhizal protection of chili plants challenged by Phytophthora capsici. Eur. J. Plant Pathol. 120, 13–20 10.1007/s10658-007-9188-7 [DOI] [Google Scholar]
- Álvarez-Loayza P., White J. F. J., Torres M. S., Balslev H., Kristiansen T., Svenning J.-C., Gil N. (2011). Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS ONE 6, e16386. 10.1371/journal.pone.0016386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A. E., Herre E. A. (2003). Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae). Mycologia 95, 388–398 10.2307/3761880 [DOI] [PubMed] [Google Scholar]
- Arnold A. E., Maynard Z., Gilbert G. S., Coley P. D., Kursar T. A. (2000). Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 3, 267–274 10.1046/j.1461-0248.2000.00159.x [DOI] [Google Scholar]
- Arnold A. E., Mejia L. C., Kyllo D., Rojas E. I., Maynard Z., Robbins N., Herre E. A. (2003). Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. U.S.A. 100, 15649–15654 10.1073/pnas.2533483100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazani O., Benderoth M., Groten K., Kuhlemeier C., Baldwin I. T. (2005). Piriformospora indica and Sebacina vermifera increase growth performance at the expense of herbivore resistance in Nicotiana attenuata. Oecologia 146, 234–243 [DOI] [PubMed] [Google Scholar]
- Barrett L., Heil M. (2012). Unifying concepts and mechanisms in the specificity of plant-enemy interactions. Trends Plant Sci. (in press). [DOI] [PubMed] [Google Scholar]
- Becklin K. M., Gamez G., Uelk B., Raguso R. A., Galen C. (2011). Soils fungal effects on floral signals, rewards, and aboveground interactions in an alpine pollination web. Am. J. Bot. 98, 1299–1308 10.3732/ajb.1000450 [DOI] [PubMed] [Google Scholar]
- Bennett A. E., Bever J. D. (2007). Mycorrhizal species differentially alter plant growth and response to herbivory. Ecology 88, 210–218 10.1890/0012-9658(2007)88[210:MSDAPG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bent S. J., Forney L. J. (2008). The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2, 689–695 10.1038/ismej.2008.44 [DOI] [PubMed] [Google Scholar]
- Bianciotto V., Lumini E., Bonfante P., Vandamme P. (2003). ‘Candidatus Glomeribacter gigasporarum’ gen. nov., sp nov., an endosymbiont of arbuscular mycorrhizal fungi. Int. J. Syst. Evol. Microbiol. 53, 121–124 10.1099/ijs.0.02382-0 [DOI] [PubMed] [Google Scholar]
- Binladen J., Gilbert M. T. P., Bollback J. P., Panitz F., Bendixen C., Nielsen R., Willerslev E. (2007). The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2, e197. 10.1371/journal.pone.0000197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P. (2010). “Plant–fungal interactions in Mycorrhizas,” in Encyclopedia of Life Sciences (John Wiley & Sons, Ltd; ) [Google Scholar]
- Bonfante P., Genre A. (2008). Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends Plant Sci. 13, 492–498 10.1016/j.tplants.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Borowicz V. A. (2001). Do arbuscular mycorrhizal fungi alter plant-pathogen relations? Ecology 82, 3057–3068 10.2307/2679834 [DOI] [Google Scholar]
- Brevin N. (2010). “Root nodules (legume–rhizobium symbiosis),” in Encyclopaedia of Life Sciences, pub2 10.1002/9780470015902.a0003720 [DOI] [Google Scholar]
- Brundrett M. C. (2002). Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304 10.1046/j.1469-8137.2002.00397.x [DOI] [PubMed] [Google Scholar]
- Cheplick G. P., Clay K., Marks S. (1989). Interactions between infection by endophytic fungi and nutrient limitation in the grasses Lolium perenne and Festuca arundinacea. New Phytol. 111, 89–97 10.1111/j.1469-8137.1989.tb04222.x [DOI] [Google Scholar]
- Clay K. (1990). Fungal endophytes of grasses. Annu. Rev. Ecol. Syst. 21, 275–297 10.1146/annurev.es.21.110190.001423 [DOI] [Google Scholar]
- Currie A. F., Murray P. J., Gange A. C. (2011). Is a specialist root-feeding insect affected by arbuscular mycorrhizal fungi? Appl. Soil Ecol. 47, 77–83 10.1016/j.apsoil.2010.12.002 [DOI] [Google Scholar]
- Dawkins R. (1999). The Extended Phenotype. Oxford: Oxford University Press [Google Scholar]
- De Deyn G. B., Biere A., Van Der Putten W. H., Wagenaar R., Klironomos J. N. (2009). Chemical defense, mycorrhizal colonization and growth responses in Plantago lanceolata L. Oecologia 160, 433–442 10.1007/s00442-009-1312-2 [DOI] [PubMed] [Google Scholar]
- de Román M., Fernández I., Wyatt T., Sahrawy M., Heil M., Pozo M.-J. (2011). Elicitation of foliar resistance mechanisms transiently impairs root association with arbuscular mycorrhizal fungi. J. Ecol. 99, 36–45 10.1111/j.1365-2745.2010.01752.x [DOI] [Google Scholar]
- Dean J. M., Mescher M. C., De Moraes C. M. (2009). Plant-rhizobia mutualism influences aphid abundance on soybean. Plant Soil 323, 187–196 10.1007/s11104-009-9924-1 [DOI] [Google Scholar]
- Denison R. F., Kiers E. T. (2004). Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol. Lett. 237, 187–193 10.1111/j.1574-6968.2004.tb09695.x [DOI] [PubMed] [Google Scholar]
- Eaton C. J., Cox M. P., Scott B. (2011). What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci. 180, 190–195 10.1016/j.plantsci.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Elsen A., Gervacio D., Swennen R., De Waele D. (2008). AMF-induced biocontrol against plant parasitic nematodes in Musa sp.: a systemic effect. Mycorrhiza 18, 251–256 10.1007/s00572-008-0173-6 [DOI] [PubMed] [Google Scholar]
- Erb M., Robert C. A. M., Hibbart B. E., Turlings T. C. J. (2011). Sequence of arrival determines plant-mediated interactions between herbivores. J. Ecol. 99, 7–15 10.1111/j.1365-2745.2010.01757.x [DOI] [Google Scholar]
- Ercolin F., Reinhardt D. (2011). Successful joint ventures of plants: arbuscular mycorrhiza and beyond. Trends Plant Sci. 16, 356–362 10.1016/j.tplants.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Faeth S. H. (2002). Are endophytic fungi defensive plant mutualists? Oikos 98, 25–36 10.1034/j.1600-0706.2002.980103.x [DOI] [Google Scholar]
- Faeth S. H., Fagan W. F. (2002). Fungal endophytes: common host plant symbionts but uncommon mutualists. Integr. Comp. Biol. 42, 360–368 10.1093/icb/42.2.360 [DOI] [PubMed] [Google Scholar]
- Faeth S. H., Shochat E. (2010). Inherited microbial symbionts increase herbivore abundances and alter arthropod diversity on a native grass. Ecology 91, 1329–1343 10.1890/08-2123.1 [DOI] [PubMed] [Google Scholar]
- Faeth S. H., Sullivan T. J. (2003). Mutualistic asexual endophytes in a native grass are usually parasitic. Am. Nat. 161, 310–325 10.1086/345937 [DOI] [PubMed] [Google Scholar]
- Ferreira A., Quecine M. C., Lacava P. T., Oda S., Azevedo J. L., Araujo W. L. (2008). Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 287, 8–14 10.1111/j.1574-6968.2008.01258.x [DOI] [PubMed] [Google Scholar]
- Fisher B. L. (1992). Facultative ant association benefits a neotropical orchid. J. Trop. Ecol. 8, 109–114 10.1017/S0266467400006179 [DOI] [Google Scholar]
- Fisher P. J., Petrini O., Petrini L. E., Sutton B. C. (1994). Fungal endophytes from the leaves and twigs of Quercus ilex L. from England, Majorca and Switzerland. New Phytol. 127, 133–137 10.1111/j.1469-8137.1994.tb04267.x [DOI] [PubMed] [Google Scholar]
- Fornoni J. (2011). Ecological and evolutionary implications of plant tolerance to herbivory. Funct. Ecol. 25, 399–407 10.1111/j.1365-2435.2010.01805.x [DOI] [Google Scholar]
- Francis R., Read D. J. (1995). Mutualism and antagonism in the mycorrhizal symbiosis, with special reference to impacts on plant community structure. Can. J. Bot. 73, 1301–1309 10.1139/b95-391 [DOI] [Google Scholar]
- Freeman S., Rodriguez R. J. (1993). Genetic conversion of a fungal pathogen to a nonpathogenic, endophytic mutualist. Science 260, 75–78 10.1126/science.260.5104.75 [DOI] [PubMed] [Google Scholar]
- Fritz M., Jakobsen I., Lyngkjaer M. F., Thordal-Christensen H., Pons-Kuhnemann J. (2006). Arbuscular mycorrhiza reduces susceptibility of tomato to Alternaria solani. Mycorrhiza 16, 413–419 10.1007/s00572-006-0051-z [DOI] [PubMed] [Google Scholar]
- Frohlich J., Hyde K. D., Petrini O. (2000). Endophytic fungi associated with palms. Mycol. Res. 104, 1202–1212 10.1017/S095375620000263X [DOI] [Google Scholar]
- Gange A. C., Ayres R. L. (1999). On the relation between arbuscular mycorrhizal colonization and plant ‘benefit.’ Oikos 87, 615–621 10.2307/3546829 [DOI] [Google Scholar]
- Gao F. K., Dai C. C., Liu X. Z. (2010). Mechanisms of fungal endophytes in plant protection against pathogens. Afr. J. Microbiol. Res. 4, 1346–1351 [Google Scholar]
- Garcia-Garrido J. M., Ocampo J. A. (2002). Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J. Exp. Bot. 53, 1377–1386 10.1093/jexbot/53.373.1377 [DOI] [PubMed] [Google Scholar]
- Garrido E., Bennett A. E., Fornoni J., Strauss S. Y. (2010a). The dark side of the mycorrhiza. Plant Signal. Behav. 5, 1019–1021 10.4161/psb.5.8.12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido E., Bennett A. E., Fornoni J., Strauss S. Y. (2010b). Variation in arbuscular mycorrhizal fungi colonization modifies the expression of tolerance to above-ground defoliation. J. Ecol. 98, 43–49 10.1111/j.1365-2745.2009.01586.x [DOI] [Google Scholar]
- Gazis R., Chaverri P. (2010). Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 3, 240–254 10.1016/j.funeco.2009.12.001 [DOI] [Google Scholar]
- Gehring C., Bennett A. (2009). Mycorrhizal fungal-plant-insect interactions: the importance of a community approach. Environ. Entomol. 38, 93–102 10.1603/022.038.0111 [DOI] [PubMed] [Google Scholar]
- Ghimire S. R., Charlton N. D., Bell J. D., Krishnamurthy Y. L., Craven K. D. (2011). Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatum L.) growing in the native tallgrass prairie of northern Oklahoma. Fungal Divers. 47, 19–27 10.1007/s13225-010-0085-6 [DOI] [Google Scholar]
- Gilbert S. F., Mcdonald E., Boyle N., Buttino N., Gyi L., Mai M., Prakash N., Robinson J. (2010). Symbiosis as a source of selectable epigenetic variation: taking the heat for the big guy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 671–678 10.1098/rstb.2009.0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurulingappa P., Sword G. A., Murdoch G., Mcgee P. A. (2010). Colonization of crop plants by fungal entomopathogens and their effects on two insect pests when in planta. Biol. Control 55, 34–41 10.1016/j.biocontrol.2010.06.011 [DOI] [Google Scholar]
- Hartley S. E., Gange A. C. (2009). Impacts of plant symbiotic fungi on insect herbivores: mutualism in a multitrophic context. Annu. Rev. Entomol. 54, 323–342 10.1146/annurev.ento.54.110807.090614 [DOI] [PubMed] [Google Scholar]
- Heil M. (2002). Ecological costs of induced resistance. Curr. Opin. Plant Biol. 5, 345–350 10.1016/S1369-5266(02)00267-4 [DOI] [PubMed] [Google Scholar]
- Heil M. (2008). Indirect defence via tritrophic interactions. New Phytol. 178, 41–61 10.1111/j.1469-8137.2007.02330.x [DOI] [PubMed] [Google Scholar]
- Heil M., Baldwin I. T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67 10.1016/S1360-1385(01)02186-0 [DOI] [PubMed] [Google Scholar]
- Herms D. A., Mattson W. J. (1992). The dilemma of plants: to grow or to defend. Q. Rev. Biol. 67, 283–335 10.1086/417659 [DOI] [Google Scholar]
- Higgins K. L., Coley P. D., Kursar T. A., Arnold A. E. (2011). Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia 103, 247–260 10.3852/09-158 [DOI] [PubMed] [Google Scholar]
- Hoeksema J. D., Chaudhary V. B., Gehring C. A., Johnson N. C., Karst J., Koide R. T., Pringle A., Zabinski C., Bever J. D., Moore J. C., Wilson G. W. T., Klironomos J. N., Umbanhowar J. (2010). A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol. Lett. 13, 394–407 10.1111/j.1461-0248.2009.01430.x [DOI] [PubMed] [Google Scholar]
- Hoffman M. T., Arnold A. E. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76, 4063–4075 10.1128/AEM.01404-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys C. P., Franks P. J., Rees M., Bidartondo M. I., Leake J. R., Beerling D. J. (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 1. 10.1038/ncomms1105 [DOI] [PubMed] [Google Scholar]
- Huys G., Vanhoutte T., Vandamme P. (2008). Application of sequence-dependent electrophoresis fingerprinting in exploring biodiversity and population dynamics of human intestinal microbiota: what can be revealed? Interdiscip. Perspect. Infect. Dis. article ID 597603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber L. R., Vidal S. (2010). Fungal endophyte negative effects on herbivory are enhanced on intact plants and maintained in a subsequent generation. Ecol. Entomol. 35, 25–36 10.1111/j.1365-2311.2009.01152.x [DOI] [Google Scholar]
- Jani A. J., Faeth S. H., Gardner D. (2010). Asexual endophytes and associated alkaloids alter arthropod community structure and increase herbivore abundances on a native grass. Ecol. Lett. 13, 106–117 10.1111/j.1461-0248.2009.01401.x [DOI] [PubMed] [Google Scholar]
- Johnson N. C., Graham J. H., Smith F. A. (1997). Functioning of mycorrhiza associations along the mutualism-parasitism continuum. New Phytol. 135, 575–585 10.1046/j.1469-8137.1997.00729.x [DOI] [Google Scholar]
- Johnson S. N., Gregory P. J., Greenham J. R., Zhang X., Murray P. J. (2005). Attractive properties of an isoflavonoid found in white clover root nodules on the clover root weevil. J. Chem. Ecol. 31, 2223–2229 10.1007/s10886-005-6355-1 [DOI] [PubMed] [Google Scholar]
- Johnston-Monje D., Raizada M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6, e20396. 10.1371/journal.pone.0020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama N., Zhang Z. Q., Ohgushi T. (2011). Community-wide effects of below-ground rhizobia on above-ground arthropods. Ecol. Entomol. 36, 43–51 10.1111/j.1365-2311.2010.01242.x [DOI] [Google Scholar]
- Kempel A., Schmidt A. K., Brandl R., Schadler M. (2010). Support from the underground: induced plant resistance depends on arbuscular mycorrhizal fungi. Funct. Ecol. 24, 293–300 10.1111/j.1365-2435.2009.01647.x [DOI] [Google Scholar]
- Kiers E. T., Denison R. F. (2008). Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu. Rev. Ecol. Evol. Syst. 39, 215–236 10.1146/annurev.ecolsys.39.110707.173423 [DOI] [Google Scholar]
- Koltai H., Kapulnik Y. (eds.). (2010). Arbuscular Mycorrhizas: Physiology and Function, 2nd Edn Berlin: Springer [Google Scholar]
- Koricheva J., Gange A. G., Jones T. (2009). Effects of mycorrhizal fungi on insect herbivores: a meta-analysis. Ecology 90, 2088–2097 10.1890/08-1555.1 [DOI] [PubMed] [Google Scholar]
- Koricheva J., Larsson S., Haukioja E., Keinänen M. (1998). Regulation of woody plant secondary metabolism by resource availability: hypothesis testing by means of meta-analysis. Oikos 83, 212–226 10.2307/3546833 [DOI] [Google Scholar]
- Kula A. A. R., Hartnett D. C., Wilson G. W. T. (2005). Effects of mycorrhizal symbiosis on tallgrass prairie plant-herbivore interactions. Ecol. Lett. 8, 61–69 10.1111/j.1461-0248.2004.00690.x [DOI] [Google Scholar]
- Lehtonen P., Helander M., Saikkonen K. (2005). Are endophyte-mediated effects on herbivores conditional on soil nutrients? Oecologia 142, 38–45 10.1007/s00442-004-1701-5 [DOI] [PubMed] [Google Scholar]
- Lehtonen P., Helander M., Siddiqui S. A., Lehto K., Saikkonen K. (2006). Endopyhtic fungus decreases plant virus infections in meadow rygerass (Lolium pratense). Biol. Lett. 2, 620–623 10.1098/rsbl.2006.0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Y., Maldonado-Mendoza I., Lopez-Meyer M., Cheung F., Town C. D., Harrison M. J. (2007). Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 50, 529–544 10.1111/j.1365-313X.2007.03069.x [DOI] [PubMed] [Google Scholar]
- Markmann K., Parniske M. (2009). Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci. 14, 77–86 10.1016/j.tplants.2008.11.009 [DOI] [PubMed] [Google Scholar]
- Márquez L. M., Redman R. S., Rodriguez R. J., Roossinck M. J. (2007). A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515 10.1126/science.1136237 [DOI] [PubMed] [Google Scholar]
- Mithöfer A. (2002). Suppression of plant defence in rhizobia-legume symbiosis. Trends Plant Sci. 10, 440–444 10.1016/S1360-1385(02)02336-1 [DOI] [PubMed] [Google Scholar]
- Moreira V. S. S., del Claro K. (2005). The outcomes of an ant-treehopper association on Solanum lycocarpum St. Hill: increased membracid fecundity and reduced damage by chewing herbivores. Neotrop. Entomol. 34, 881–887 10.1590/S1519-566X2005000600017 [DOI] [Google Scholar]
- Muller C. B., Krauss J. (2005). Symbiosis between grasses and asexual fungal endophytes. Curr. Opin. Plant Biol. 8, 450–456 10.1016/j.pbi.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Naumann M., Schuessler A., Bonfante P. (2010). The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J. 4, 862–871 10.1038/ismej.2010.21 [DOI] [PubMed] [Google Scholar]
- Nocker A., Burr M., Camper A. K. (2007). Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54, 276–289 10.1007/s00248-007-9236-z [DOI] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Downie J. M. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546 10.1146/annurev.arplant.59.032607.092839 [DOI] [PubMed] [Google Scholar]
- Ormond E. L., Thomas A. P. M., Pugh P. J. A., Pell J. K., Roy H. E. (2010). A fungal pathogen in time and space: the population dynamics of Beauveria bassiana in a conifer forest. FEMS Microbiol. Ecol. 74, 146–154 10.1111/j.1574-6941.2010.00939.x [DOI] [PubMed] [Google Scholar]
- Ownley B. H., Gwinn K. D., Vega F. E. (2010). Endophytic fungal entomopathogens with activity against plant pathogens: ecology and evolution. BioControl 55, 113–128 10.1007/s10526-009-9241-x [DOI] [Google Scholar]
- Partida-Martinez L. P., Hertweck C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 10.1038/nature03997 [DOI] [PubMed] [Google Scholar]
- Partida-Martinez L. P., Monajembashi S., Greulich K.-O., Hertweck C. (2007). Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17, 773–777 10.1016/j.cub.2007.06.054 [DOI] [PubMed] [Google Scholar]
- Paszkowski U. (2006). Mutualism and parasitism: the yin and yang of plant symbioses. Curr. Opin. Plant Biol. 9, 364–370 10.1016/j.pbi.2006.05.008 [DOI] [PubMed] [Google Scholar]
- Pavlo A., Leonid O., Iryna Z., Natalia K., Maria P. A. (2011). Endophytic bacteria enhancing growth and disease resistance of potato (Solanum tuberosum L.). Biol. Control 56, 43–49 10.1016/j.biocontrol.2010.09.014 [DOI] [Google Scholar]
- Pieterse C. M. J., Dicke M. (2007). Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 12, 564–569 10.1016/j.tplants.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Van Pelt J. A., Van Wees S. C. M., Ton J., Léon-Kloosterziel K., Keurentjes J. J. B., Verhagen B. W. M., Knoester M., Van Der Sluis I., Bakker P. A. H. M., Van Loon L. C. (2001). Rhizobacteria-mediated induced systemic resistance: triggering, signalling and expression. Eur. J. Plant Pathol. 107, 51–61 10.1023/A:1008747926678 [DOI] [Google Scholar]
- Pineda A., Zheng S.-J., Van Loon J. J. A., Pieterse C. M. J., Dicke M. (2010). Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15, 507–514 10.1016/j.tplants.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Pozo M. J., Azcón-Aguilar C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398 10.1016/j.pbi.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Redecker D., Kodner R., Graham L. E. (2000). Glomalean fungi from the Ordovician. Science 289, 1920–1921 10.1126/science.289.5486.1920 [DOI] [PubMed] [Google Scholar]
- Redman R. S., Dunigan D. D., Rodriguez R. J. (2001). Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 151, 705–716 10.1046/j.0028-646x.2001.00210.x [DOI] [PubMed] [Google Scholar]
- Reeder J., Knight R. (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7, 668–669 10.1038/nmeth0910-668b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy W., Taylor T. N., Hass H., Kerp H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U.S.A. 91, 11841–11843 10.1073/pnas.91.25.11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. J., White J. F., Arnold A. E., Redman R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330 10.1111/j.1469-8137.2009.02773.x [DOI] [PubMed] [Google Scholar]
- Romao A. S., Sposito M. B., Andreote F. D., Azevedo J. L., Araujo W. L. (2011). Enzymatic differences between the endophyte Guignardia mangiferae (Botryosphaeriaceae) and the citrus pathogen G. citricarpa. Genet. Mol. Res. 10, 243–252 [DOI] [PubMed] [Google Scholar]
- Romero G. Q., Koricheva J. (2011). Contrasting cascade effects of carnivores on plant fitness: a meta-analysis. J. Anim. Ecol. 80, 696–704 10.1111/j.1365-2656.2011.01808.x [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Roy H. E., Brodie E. L., Chandler D., Goettel M. S., Pell J. K., Wajnberg E., Vega F. E. (2010). Deep space and hidden depths: understanding the evolution and ecology of fungal entomopathogens. BioControl 55, 1–6 10.1007/s10526-009-9244-7 [DOI] [Google Scholar]
- Rudgers J. A., Afkhami M. E., Rúa M. A., Davitt A. J., Hammer S., Huguet V. M. (2009). A fungus among us: broad patterns of endophyte distribution in the grasses. Ecology 80, 1531–1539 10.1890/08-0116.1 [DOI] [PubMed] [Google Scholar]
- Saari S., Helander M., Lehtonen P., Wallius E., Saikkonen K. (2010). Fungal endophytes reduce regrowth and affect competitiveness of meadow fescue in early succession of pastures. Grass Forage Sci. 65, 287–295 [Google Scholar]
- Saikkonen K. (2007). Forest structure and fungal endophytes. Fungal Biol. Rev. 21, 67–74 10.1016/j.fbr.2007.05.001 [DOI] [Google Scholar]
- Saikkonen K., Lehtonen P., Helander M., Koricheva J., Faeth S. H. (2006). Model systems in ecology: dissecting the endophyte-grasss literature. Trends Plant Sci. 11, 428–433 10.1016/j.tplants.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Saikkonen K., Saari S., Helander M. (2010). Defensive mutualism between plants and endophytic fungi? Fungal Divers. 41, 101–113 10.1007/s13225-010-0023-7 [DOI] [Google Scholar]
- Schardl C. L., Leuchtmann A., Spiering M. J. (2004). Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 55, 315–340 10.1146/annurev.arplant.55.031903.141735 [DOI] [PubMed] [Google Scholar]
- Schoettner S., Hoffmann F., Wild C., Rapp H. T., Boetius A., Ramette A. (2009). Inter- and intra-habitat bacterial diversity associated with cold-water corals. ISME J. 3, 756–759 10.1038/ismej.2009.15 [DOI] [PubMed] [Google Scholar]
- Schulz B., Boyle C. (2005). The endophytic continuum. Mycol. Res. 109, 661–686 10.1017/S095375620500273X [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P., Panstruga R. (2011). A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16, 117–125 10.1016/j.tplants.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Shi J., Liu A., Li X., Feng S., Chen W. (2011). Inhibitory mechanisms induced by the endophytic bacterium MGY2 in controlling anthracnose of papaya. Biol. Control 56, 2–8 10.1016/j.biocontrol.2010.09.012 [DOI] [Google Scholar]
- Singh B. K., Millard P., Whiteley A. S., Murrell J. C. (2004). Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol. 12, 386–393 10.1016/j.tim.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Smith S. E., Read D. J. (2008). Mycorrhizal Symbiosis, 3rd Edn London: Academic Press [Google Scholar]
- Song Y. Y., Zeng R. S., Xu J. F., Li J., Shen X., Yihdego W. G. (2010). Interplant communication of tomato plants through underground common mycorrhizal networks. PLoS ONE 5, e13324. 10.1371/journal.pone.0013324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrsky J. D., Eubanks M. D. (2007). Ecological consequences of interactions between ants and honeydew-producing insects. Proc. Biol. Sci. 274, 151–164 10.1098/rspb.2006.3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam N. M., Heil M. (2011). Multitrophic interactions below and above ground: en route to the next level. J. Ecol. 99, 77–88 10.1111/j.1365-2745.2010.01761.x [DOI] [Google Scholar]
- Van Moelken T. (2009). Pros and Cons of Virus Infection in Plant – An Ecological Perspective. Ph.D. thesis, Radboud University, Nijmegen [Google Scholar]
- van Wees S. C. M. (1999). Rhizobacteria-Mediated Induced Systemic Resistance in Arabidopsis: Signal Transduction and Expression Ph.D. thesis, University of Utrecht, Utrecht. [Google Scholar]
- Vannette R. L., Hunter M. D. (2011). Plant defense theory re-examined: non-linear expectations based on the cost and benefits of resource mutualisms. J. Ecol. 99, 66–76 10.1111/j.1365-2745.2010.01755.x [DOI] [Google Scholar]
- Vega F. E., Goettel M. S., Blackwell M., Chandler D., Jackson M. A., Keller S., Koike M., Maniania N. K., Rangel D. E. N., Roy H. E. (2009). Fungal entomopathogens: new insights on their ecology. Fungal Ecol. 2, 149–159 10.1016/j.funeco.2009.05.001 [DOI] [Google Scholar]
- Verhagen B. W. M., Glazebrook J., Zhu T., Chang H.-S., Van Loon L. C., Pieterse C. M. J. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant Microbe Interact. 17, 895–908 10.1094/MPMI.2004.17.8.895 [DOI] [PubMed] [Google Scholar]
- Vesterlund S. R., Helander M., Faeth S. H., Hyvonen T., Saikkonen K. (2011). Environmental conditions and host plant origin override endophyte effects on invertebrate communities. Fungal Divers. 47, 109–118 10.1007/s13225-011-0089-x [DOI] [Google Scholar]
- Weyens N., Van Der Lelie D., Taghavi S., Newman L., Vangronsveld J. (2009). Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 27, 591–598 10.1016/j.tibtech.2009.07.006 [DOI] [PubMed] [Google Scholar]
- White J. F., Sullivan R., Balady G., Gianfagna T., Yue Q., Meyer W., Cabral D. (2001). A fungal endosymbiont of the grass Bromus setifolius: distribution in some Andean populations, identification, and examination of beneficial properties. Symbiosis 31, 241–257 [Google Scholar]
- Wilson D. (1995). Endophyte – the evolution of a term, and clarification of its use and definition. Oikos 73, 274–276 10.2307/3545919 [DOI] [Google Scholar]
- Yang J.-W., Yi H.-S., Kim H., Lee B., Lee S., Ghim S.-Y., Ryu C.-M. (2011). Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J. Ecol. 99, 46–56 10.1111/j.1365-2745.2010.01756.x [DOI] [Google Scholar]
- Yuan Z. L., Zhang C. L., Lin F. C. (2010). Role of diverse non-systemic fungal endophytes in plant performance and response to stress: progress and approaches. J. Plant Growth Regul. 29, 116–126 10.1007/s00344-009-9112-9 [DOI] [Google Scholar]
- Zamioudis C., Pieterse C. M. J. (2011). Modulation of host immunity by geneficial microbes. Mol. Plant Microbe Interact. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]