Abstract
The genetic set up and the enzymes that define the O-glycosylation sites and transfer the activated sugars to cell wall glycoprotein Extensins (EXTs) have remained unknown for a long time. We are now beginning to see the emerging components of the molecular machinery that assembles these complex O-glycoproteins on the plant cell wall. Genes conferring the posttranslational modifications, i.e., proline hydroxylation and subsequent O-glycosylation, of the EXTs have been recently identified. In this review we summarize the enzymes that define the O-glycosylation sites on the O-glycoproteins, i.e., the prolyl 4-hydroxylases (P4Hs), the glycosyltransferases that transfer arabinose units (named arabinosyltransferases, AraTs), and the one responsible for transferring a single galactose (galactosyltransferase, GalT) on the protein EXT backbones. We discuss the effects of posttranslational modifications on the structure and function of extensins in plant cell walls.
Keywords: plant cell wall, O-glycosylation, proline hydroxylation, O-glycoproteins, extensins, cell expansion
Introduction
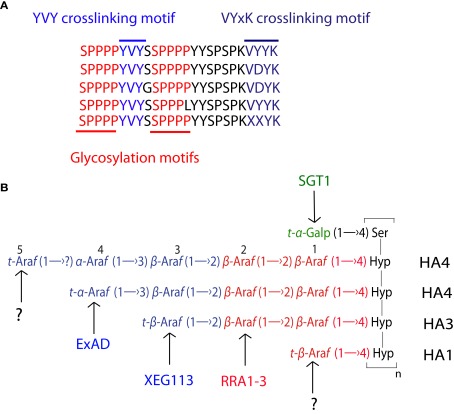
The major components of primary plant cell walls include a complex composite of networks of cellulose microfibrils, branched xyloglucans, and a diverse pectin matrix (Somerville et al., 2004; Cosgrove, 2005). In addition, a structural role has been clearly assigned to the hydroxyproline-rich O-glycoprotein extensins (EXTs) in building and maintaining the growing cell wall (Lamport and Northcote, 1960; Hall and Cannon, 2002; Held et al., 2004; Cannon et al., 2008; Lamport et al., 2011; Velasquez et al., 2011a,b). EXTs are plant cell wall O-glycoproteins that belong to the hydroxyproline-rich glycoproteins (HRGPs) superfamily with a modular highly repetitive sequence that includes the O-glycosylated Ser-(Hyp)3–4 repeats and some of the crosslinking motifs Val-Tyr-Lys, Val-Tyr-Lys-Tyr-Lys, Tyr-Tyr-Tyr-Lys, or Tyr-X-Tyr, all of a hydrophobic nature (Figure 1A; Fry, 1982; Brady et al., 1998; Held et al., 2004; Cannon et al., 2008). EXTs can also occur in the context of natural, chimeric glycosylated polypeptides, e.g., EXT-arabinogalactan proteins (Lind et al., 1994), leucine-rich repeat (LRR) EXT proteins (Rubinstein et al., 1995), or lectin-EXTs (Kieliszewski and Lamport, 1994). Here in this review, we will highlight the major breakthroughs of the posttranslational modifications of EXTs and how these modifications affect the function of EXTs within the primary plant cell wall.
Figure 1.
(A) Predominant repetitive modular sequences in certain most common EXTs with isodityrosine (YVY) and other crosslinking sites (VYxK) as well as the Ser-(Hyp)4, which is O-glycosylated. (B) O-glycans present in the Ser-(Hyp)4 motif. In addition, the glycosyltransferases that transfer specific sugars on the EXT backbone are indicated in the figure.
Although the protein sequences that define the O-glycosylation site on HRGPs are partially known (Kieliszewski and Lamport, 1994; Shpak et al., 1999, 2001), the glycosyltransferases that transfer the activated sugars to the protein backbone (arabinose and galactose) have remained a mystery for decades. Now we are beginning to perceive the emerging picture of the molecular machinery that assembles sugars into these complex O-glycoproteins which comprises an important part of the cell wall. Most of the genes involved in the posttranslational modification of EXTs that includes proline hydroxylation and subsequent O-glycosylation are now being identified (Egelund et al., 2007; Cannon et al., 2008; Gille et al., 2009; Velasquez et al., 2011a), increasing our understanding of plant cell wall EXTs synthesis and their roles in cell expansion during growth.
While several aspects of EXTs have been reviewed recently (Kieliszewski et al., 2010; Showalter et al., 2010; Lamport et al., 2011), the present review focuses specifically on the enzymes that define the O-glycosylation sites, namely the prolyl 4-hydroxylases (P4Hs) and the glycosyltransferases that transfer arabinosyl units (named arabinosyltransferases, AraTs) or a single galactosyl residue (galactosyltransferase, GalT) to the protein EXT backbones. Finally, the structure and functions of the O-glycoprotein EXTs are discussed in the context of cell wall expansion during growth.
Steps to Build Mature, Functional EXTs
Structural O-glycoproteins EXTs that are secreted into plant cell walls are first shaped by several posttranslational modifications that include signal peptide processing in the ER, hydroxylation of peptidyl-proline residues into hydroxyproline (Hyp) in the ER-Golgi apparatus (Lamport and Northcote, 1960; Lamport, 1967; Yuasa et al., 2005; Velasquez et al., 2011a), O-glycosylation on Hyp and Ser by the action of several glycosyltranferases, mostly in the Golgi apparatus, although early steps could be taking place in the ER (Figure 1B; Shimma et al., 2011; Velasquez et al., 2011a), and finally, covalent intra- and inter-molecular crosslinking at the plant cell wall level (Fry, 1982; Held et al., 2004). All these posttranslational modifications of EXTs are crucial for the function of EXTs in the plant cell wall during cell development and growth (Hall and Cannon, 2002; Cannon et al., 2008; Velasquez et al., 2011a,b).
Two P4H families, the collagen P4Hs (C-P4Hs) and the hypoxia-inducible transcription factor (HIF)-associated P4Hs, are responsible for the hydroxylation of collagens and HIF, respectively (Myllyharju, 2003, 2008). The vertebrate C-P4Hs are α2β2 tetramers, in which the α-subunits possess the enzymatic activity and the β-subunits are identical to the enzyme disulfide isomerase. The tetrameric assembly is required for stability and full activity. In contrast, plant P4Hs and also HIF-P4Hs are monomers (Hieta and Myllyharju, 2002; Myllyharju, 2003; Koski et al., 2007, 2009). Plant P4Hs are membrane-bound enzymes that catalyze the hydroxylation of peptidyl-proline to yield hydroxyproline by introducing an oxygen atom between the hydrogen and carbon atoms of the peptidyl-proline adding merely 16 atomic mass units (Lamport, 1963). From four possible isomers of hydroxyproline, the most common isomer in nature is (2S,4R)-4-hydroxyproline, herein after referred to as Hyp (trans-4-hydroxyproline) in reference to the trans-relationship between the –COOH and –OH substituents on the pyrrolidine ring (Hieta and Myllyharju, 2002; Myllyharju, 2003). In addition, the cis-hydroxyproline (2S,4S)-4-hydroxyproline is reported to be present in the toxic cyclic peptides from Amanita mushrooms (e.g., alpha-amanitin and phalloidin; Alexopoulos et al., 1996). The P4Hs belong to a family of 2-oxoglutarate-dependent dioxygenases that require 2-oxoglutarate and O2 as co-substrates, Fe2+ as a cofactor and ascorbate to prevent the rapid inactivation by self-oxidation (Hieta and Myllyharju, 2002; Koski et al., 2007, 2009). From a mechanistic point of view, α-ketoglutarate is oxidatively decarboxylated to produce succinate and CO2. The conversion of Pro to Hyp in the secretory pathway provides reactive hydroxyl groups for further modification such as O-glycosylation. In addition, the product of this reaction, poly-(4-hydroxyproline), has an even greater tendency than polyproline to adopt a helical polyproline II-type conformation (Lamport, 1977, 1980; Van Holst and Varner, 1984; Valentin et al., 2010).
In vitro and in vivo characterization of plant P4Hs has been carried out in Arabidopsis thaliana (Hieta and Myllyharju, 2002; Tiainen et al., 2005; Vlad et al., 2007; Asif et al., 2009; Velasquez et al., 2011a), Nicotiana tabacum (Yuasa et al., 2005), Dianthus caryophyllus (Vlad et al., 2010), and the green alga Chlamydomonas reinhardtii (Keskiaho et al., 2007). Only the activity of AtP4H1 and AtP4H2, out of the 13 P4Hs encoded in the Arabidopsis genome, have been fully characterized in vitro, and only CrP4H1 has been crystallized, without and with the (Ser-Pro)5 peptide as a substrate (Koski et al., 2007, 2009). Both AtP4Hs hydroxylate proline-rich peptides of both extreme types: arabinogalactan proteins (AGPs) and EXTs. However, AtP4H5 seems to be more specific as yeast two-hybrid assays and molecular modeling strongly suggest its preference for polyproline II peptide instead of the AGP-type substrate (Velasquez et al., 2011a). Moreover, three AtP4H T-DNA insertional mutants (Atp4h2,5,13) exhibited a clear short root hair phenotype and reduced root Hyp levels as well. Remarkably, overexpression of these P4Hs displayed extra long root hairs (Velasquez et al., 2011a,b) and much higher Hyp contents in roots when compared to WT Col-0 (Estevez et al., unpublished results). Interestingly, heterologous peptides that mimic collagen (X–Pro–Gly)n are also hydroxylated by some plant P4Hs, though inefficiently (Tanaka et al., 1981; Kaska et al., 1988).
Based on multiple pieces of evidence, it became clear that proline hydroxylation of HRGPs, and specifically EXTs, are essential for polarized cell expansion in root hairs. Therefore, it is tempting to imagine that homologous P4Hs and HRGPs will also be relevant in cell expansion in other rapidly expanding cell types such as pollen tubes or dark-grown hypocotyls. On the other hand, suppression of the gene encoding 1 out of 10 Chlamydomonas reinhardtii prolyl 4-hydroxylases (CrP4H1), which hydroxylates algal HRGP sequences, led to a defective cell wall (Keskiaho et al., 2007), suggesting a major unique structural role of certain HRGPs as targets of CrP4H1 in this unicellular green algae. In two other studies, a direct role of P4Hs in hypoxia stress and during different stages of plant growth and development is speculated, suggesting that P4Hs could be acting as putative oxygen sensors (Vlad et al., 2007; Asif et al., 2009). Nevertheless, further studies are needed to confirm this hypothesis.
The extent and type of O-glycosylation in HRGPs, and specifically in EXTs, can be predicted by the “Hyp contiguity” hypothesis, according to which O-glycosylation correlates with the primary sequence of the protein where location and context of Hyp residues are crucial (Kieliszewski and Lamport, 1994; Kieliszewski 2001; Shpak et al., 1999, 2001). Generally, wherever Hyp residues are contiguous in the amino acid sequence, arabinosylation is predominant in EXTs, whereas arabinogalactans are rather added to clustered, non-contiguous Hyp residues in AGPs. However, it is worth mentioning that isolated Hyp units were also arabinogalactosylated in some cases (Zhao et al., 2002). Another clear exception to this hypothesis is exemplified by the small CLE-like peptides (e.g., Tob/Tom-HypSys, PSY1, CLV3, and CLE2) where single proline units are modified with β-linked-L-arabinofuranosides with identical linkages/stereochemistry of the innermost three arabinose units found in the EXTs (Ryan and Pearce, 2003; Ito et al., 2006; Kondo et al., 2006; Ohyama et al., 2009; Matsuzaki et al., 2010), suggesting that similar P4Hs and glycosyltransferases participate in their posttranslational modifications. It is important to note that Hyp O-glycosylation has not been found in animal cells, which makes HRGPs unique in their structure and functions for plant cells.
EXT O-glycosylation in the Ser-(Hyp)3–4 repeats involves a series a glycosyltransferases that add a single galactose unit to serine and 4–5 arabinose units, in a linear fashion, to Hyp. The minimal set of activities necessary for synthesis of the entire glycomodule in EXTs (Figure 1B) may comprise: one Ser-α-D-galactosyltransferase acting exclusively on serine, one Hyp-β-L-arabinofuranosyltransferase, two β-(1 → 2)-L-arabinofuranosyltransferases, one α-(1 → 3)-L-arabinofuranosyltransferase and a last arabinosyltransferase adding the fifth arabinofuranose with a yet unknown regio-chemistry (Shimma et al., 2011; Velasquez et al., 2011a). Three glycosyltransferases involved in extensins maturation have been recently reported after characterizing EXT O-glycosylation in T-DNA mutant lines of the corresponding genes in Arabidopsis (Egelund et al., 2007; Gille et al., 2009; Shimma et al., 2011; Velasquez et al., 2011a). The arabinosyltransferase that would add the second β-(1 → 2)-arabinosyl unit in the Hyp-O-arabinosides is the proposed Reduced Residual Arabinose 3 (RRA3). The one likely to transfer the third β-(1 → 2)-arabinose was named as XyloEndoGlucanase (XEG113), both belonging to the GT77 Carbohydrate Active EnZymes (CAZY) family (Egelund et al., 2007; Gille et al., 2009; Velasquez et al., 2011a). Finally, the AraT that would transfer the fourth α-(1 → 3)-L-arabinose moiety was identified very recently as Extensin Arabinose Deficient transferase (ExAD) within the GT47 family (Petersen et al. unpublished results). In addition, the galactosyltransferase that adds galactose on serine in the Ser-(Hyp)3–4 repeats was recently reported as SGT1 (Shimma et al., 2011). To the present day, the arabinosyltransferase that adds the first and the last (fifth) arabinose remains unknown (Figure 1B). Remarkably, all the AraTs T-DNA mutants identified so far with deficient O-arabinosylation in EXT backbones displayed a clear root hair phenotype, suggesting that O-glycosylation on EXT is crucial on EXT function during cell expansion at least in this cell type and also in dark-grown hypocotyls for the mutant xeg113-2 (Gille et al., 2009; Velasquez et al., 2011a,b). The evolutionary conservation from unicellular green algae to vascular plants of the Ser-(Hyp)3–4 glycomodule implies a essential function of O-glycosylation in EXT shape and biological role.
Impact of Hydroxylation and O-glycosylation on EXT Function
It has been suggested that the O-glycans increase HRGP solubility, resistance to proteolytic degradation and thermal stability (Kieliszewski et al., 1989, 2010; Ferris et al., 2001; Shpak et al., 2001; Lamport et al., 2011). The structural and functional consequences of O-glycosylation in polyproline EXTs remained unclear although some evidence suggested that this posttranslational modification could provide conformational stability to these macromolecules (Lamport, 1980; Van Holst and Varner, 1984; Stafstrom and Staehelin, 1986). To understand the biological meaning of the lack of hydroxylation/O-glycosylation on EXTs, possible conformational changes induced by proline hydroxylation and subsequent O-arabinosylation on the minimal EXT repeat Ser(Pro)4 and Ser(Hyp)4 were tested by molecular dynamics modeling. In fact, O-glycosylation clearly appears to stabilize the helical conformation of the EXT minimal peptide whereas incomplete hydroxylation and O-glycosylation favors its flexible and disorganized conformation (Velasquez et al., 2011a) as it was previously suggested by circular dichroism on both short and repetitive EXT synthetic peptides and native EXTs (Van Holst and Varner, 1984; Shpak et al., 2001). In another recent study, it was found that O-glycosylation of Hyp residues causes a dramatic increase in the thermal stability of the polyproline type-II (PP-II) helix, which may be explained by a network of inter-glycan and glycan-peptide hydrogen bonds (Owens et al., 2010). Analysis of the β-O-glycosylated polyproline model peptides indicates that Hyp O-glycosylation leads to a significant increase in conformational stability (Owens et al., 2010; Velasquez et al., 2011a). Since contiguous O-glycosylation seems to bring about a rise in conformational stability, similar increases in the conformational stability of EXTs are likely to occur. Based on this, we theorize that EXTs with incomplete hydroxylation/O-glycosylation have an enormous negative impact on EXTs overall shape, affecting EXTs interactions with the surrounding environment, including those with other EXTs in the network or with other modifying enzymes such as EXT-specific peroxidases (PERext; Velasquez et al., 2011a). The latter belong to the type-III apoplastic peroxidases that form a large multigene family present in all land plants (Passardi et al., 2004).
Based on the in vitro activity of several peroxidases (Everdeen et al., 1988; Brownleader et al., 1995; Schnabelrauch et al., 1996; Magliano and Casal, 1998; Jackson et al., 2001; Price et al., 2003), it is proposed that the in vivo crosslinking of EXTs might involve multiple PERext, although their molecular roles and specific targets remain unknown. Some of these enzymes could catalyze the polymerization of EXT monomers via di-isodityrosine and pulcherosine, a tetramer and trimer of tyrosine, respectively (Fry, 1982; Brady et al., 1996, 1998) to form a covalent cross-linked network at the plant cell wall level (Cannon et al., 2008). For example, tomato PERext exhibited a strong activity onto diverse types of extensins (Schnabelrauch et al., 1996). Moreover, PERext activity might be modulated in vivo, as shown by a study that used a natural inhibitor of extensin crosslinking isolated from a cell wall compartment containing extensins and peroxidases (Brownleader and Dey, 1993; Brownleader et al., 2000). The high number of genes encoding type-III- peroxidases, in addition to the biochemical variety of their substrates, is still one of the main obstacles for the assigning of their specific functions in vivo. However, in Arabidopsis, by means of co-expression analysis of well known root hair cell wall-associated genes, a few putative EXT peroxidases such as PER13 and PER73 were identified, suggesting that at least some of them are responsible for the crosslinking of EXTs specifically in the root hair cell walls (Velasquez et al., 2011a).
Extensin Function
The known roles of extensins in cell wall assembly, cell shape and growth raise the question about the function for each individual extensin molecule (Hall and Cannon, 2002; Cannon et al., 2008; Velasquez et al., 2011a). Recently, it was reported that EXTs can form, at least in vitro, a tridimensional covalent network through Tyr linkages mediated by EXT peroxidases between individual EXT molecules and also via self-recognition and alignment of hydrophilic O-glycosylated Ser-(Hyp)3–4 repeats and hydrophobic peptide-crosslinking modules (Cannon et al., 2008). Thus, the ordered EXT monomers assembly in the plant cell wall would involve a zipper-like endwise association via crosslinking at the ends of the molecules (Kieliszewski et al., 2010; Lamport et al., 2011). It is also proposed that the oligomeric extensin interacts with pectins by a simple acid-base reaction forming a supramolecular ionic structure in the nascent cell wall, which would serve as a template for further cell wall deposition (Cannon et al., 2008; Kieliszewski et al., 2010). This hypothesis was recently confirmed in vitro by using a layer-by-layer method to construct a multilayer thin film used to investigate the interactions between extensin and pectin. Multilayer formation was successfully produced, thereby evidencing the occurrence of interactions between EXTs and pectin (Valentin et al., 2010). In addition, covalent EXT-pectin crosslinks were also suggested (Qi et al., 1995; Nuñez et al., 2009).
Although the Arabidopsis genome encodes up to 63 putative extensins of which 20 would be defined as classical extensins, where most of them share both motifs for O-glycosylation and Try-mediated crosslinks (Figure 1A), 12 are shorter extensins and 31 are extensin-like chimeras and hybrid-extensins that could contain other protein domains (Cannon et al., 2008; Showalter et al., 2010; Lamport et al., 2011), only one extensin mutant, root-, shoot-, hypocotyl-defective (rsh) ext3, has been shown to have a nearly lethal phenotype (Hall and Cannon, 2002; Cannon et al., 2008). However, only two other single ext mutants, such as the leucine-rich repeat extensins 1 and 2 (lrx1 and lrx2) with aberrant root hairs (Baumberger et al., 2001, 2003a,b; Ringli, 2010) and the proline-rich extensin-like receptor kinase perk13 displaying longer root hairs (Humphrey et al., 2007), have been reported, thus suggesting a low degree of EXT genetic redundancy. In support of this notion, a number of root hair-relevant extensins have been found taking advantage of network analysis of co-expression with other cell wall genes known to be important for root hair growth, like expansins EXP7 and EXP18 (Won et al., 2009), proline-rich proteins PRP1 and PRP3 (Fowler et al., 1999; Bernhardt and Tierney, 2000) and leucine-rich extensin protein LRX1 (Baumberger et al., 2003a,b). Consistently, the same EXTs were identified by functional genomics on root hair mutants (Baumberger et al., 2003a,b; Jones et al., 2006; Deal and Henikoff, 2010). Through these two approaches, it was possible to identify several EXTs that seem to be important for root hair growth. This was later confirmed when T-DNA homozygous mutant lines (ext6, ext7, ext10, etc.) of these EXTs turned out to exhibit drastically shorter root hairs (Velasquez et al., 2011a,b). All these EXT-related mutants highlight an essential role of each individual EXT type on cell expansion and growth, specifically in root hairs. This suggests that location and function of each individual EXT type within the network at the cell wall are relevant to the whole EXT network assembly.
Finally, from an evolutionary perspective, highly organized but non-covalent EXT-like cell wall networks are ancient, as they occur in several unicellular and multicellular green algae (Sumper and Hallmann, 1998; Ferris et al., 2001; Bollig et al., 2007; Lee et al., 2007; Sørensen et al., 2011). In agreement, type-III peroxidase genes seem to be absent in green algae genomes (Passardi et al., 2004). Hence, a covalently cross-linked EXT network seems to have been a secondary development in evolution that led to strong plant cell walls, enabling terrestrial colonization, and turgor-driven growth in land plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by PIP-CONICET 2010-071, Argentina and PICT FONCyT 2010-0658, Argentina. We thank N. D. Iusem for a critical reading of this review.
References
- Alexopoulos C. J., Mims C. W., Blackwell M. (1996). Introductory Mycology, 4th Edn New York: John Wiley & Sons, 687–688 [Google Scholar]
- Asif M. H., Trivedi P. K., Misra P., Nath P. (2009). Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Funct. Integr. Genomics 9, 525–535 10.1007/s10142-009-0118-y [DOI] [PubMed] [Google Scholar]
- Baumberger N., Doesseger B., Guyot R., Diet A., Parsons R. L., Clark M. A., Simmons M. P., Bedinger P., Goff S. A., Ringli C., Keller B. (2003a). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131, 1313–1326 10.1104/pp.102.014928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Steiner M., Ryser U., Keller B., Ringli C. (2003b). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35, 71–81 10.1046/j.1365-313X.2003.01784.x [DOI] [PubMed] [Google Scholar]
- Baumberger N., Ringli C., Keller B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15, 1128–1139 10.1101/gad.200201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C., Tierney M. L. (2000). Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 122, 705–714 10.1104/pp.122.3.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig K., Lamshoft M., Schweimer K., Marner F. J., Budzikiewicz H., Waffenschmidt S. (2007). Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii – conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 342, 2557–2566 10.1016/j.carres.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Brady J. D., Sadler I. H., Fry S. C. (1996). Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: a new potential cross-link. Biochem. J. 315(Pt 1), 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J. D., Sadler I. H., Fry S. C. (1998). Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: its role in cross-link formation. Phytochemistry 47, 349–353 10.1016/S0031-9422(97)00592-X [DOI] [PubMed] [Google Scholar]
- Brownleader M. D., Ahmed N., Trevan M., Chaplin M. F., Dey P. M. (1995). Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 109, 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownleader M. D., Dey P. M. (1993). Purification of extension from cell walls of tomato (hybrid of Lycopersicon esculentum and L. peruvianum) cells in suspension culture. Planta 191, 457–469 10.1007/BF00195747 [DOI] [PubMed] [Google Scholar]
- Brownleader M. D., Hopkins J., Mobasheri A., Dey P. M., Jackson P., Trevan M. (2000). Role of extensin peroxidase in tomato (Lycopersicon esculentum Mill.) seedling growth. Planta 210, 668–676 10.1007/s004250050058 [DOI] [PubMed] [Google Scholar]
- Cannon M. C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B. L., Chen L., Lamport D. T. A., Chen Y., Kieliszewski M. J. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. U.S.A. 105, 2226–2231 10.1073/pnas.0711980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 11, 850–861 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Deal R. B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 10.1016/j.devcel.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., Larsen Petersen B. (2007). Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64, 439–449 10.1007/s11103-007-9162-y [DOI] [PubMed] [Google Scholar]
- Everdeen D. S., Kiefer S., Willard J. J., Muldoon E. P., Dey P. M., Li X.-B., Lamport D. T. A. (1988). Enzymic cross-linkage of monomeric extensin precursors in vitro. Plant Physiol. 87, 616–621 10.1104/pp.87.3.616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Woessner J. P., Waffenschmidt S., Kilz S., Drees J., Goodenough U. W. (2001). Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40, 2978–2987 10.1021/bi0023605 [DOI] [PubMed] [Google Scholar]
- Fowler T. J., Bernhardt C., Tierney M. L. (1999). Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol. 121, 1081–1092 10.1104/pp.121.4.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S. C. (1982). Isodityrosine, a new cross-linking amino acid from plant cell wall glycoprotein. Biochem. J. 204, 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Hänsel U., Ziemann M., Pauly M. (2009). Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 14699–704 10.1073/pnas.0905434106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall Q., Cannon M. C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14, 1161–1172 10.1105/tpc.140720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M. A., Tan L., Kamyab A., Hare M., Shpak E., Kieliszewski M. J. (2004). Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J. Biol. Chem. 279, 55474–55482 10.1074/jbc.M408396200 [DOI] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002). Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 277, 23965–23971 10.1074/jbc.M201865200 [DOI] [PubMed] [Google Scholar]
- Humphrey T. V., Bonetta D. T., Goring D. R. (2007). Sentinels at the wall: cell wall receptors and sensors. New Phytol. 176, 7–21 10.1111/j.1469-8137.2007.02192.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845 10.1126/science.1128436 [DOI] [PubMed] [Google Scholar]
- Jackson P. A., Galinha C. I., Pereira C. S., Fortunato A., Soares N. C., Amâncio S. B., Pinto Ricardo C. P. (2001). Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kiloDalton peroxidase. Plant Physiol. 127, 1065–1076 10.1104/pp.010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A., Raymond M. J., Smirnoff N. (2006). Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45, 83–100 [DOI] [PubMed] [Google Scholar]
- Kaska D. D., Myllylä R., Günzler V., Gibor A., Kivirikko K. I. (1988). Prolyl 4-hydroxylase from Volvox carteri. A low-Mr enzyme antigenically related to the a subunit of the vertebrate enzyme. Biochem. J. 256, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K., Hieta R., Sormunen R., Myllyharju J. (2007). Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19, 256–269 10.1105/tpc.106.042739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M. J. (2001). The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57, 319–323 10.1016/S0031-9422(01)00029-2 [DOI] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T. A. (1994). Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172 [DOI] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T. A., Tan L., Cannon M. C. (2010). “Hydroxyproline-rich glycoproteins: form and function,” in Annual Plant Reviews: Plant Polysaccharides, Biosynthesis and Bioengineering, Vol. 41, ed. Ulvskov P. (Oxford: Wiley-Blackwell; ). 10.1002/9781444391015.ch13 [DOI] [Google Scholar]
- Kieliszewski M. J., Leykam J. F., Lamport D. T. A. (1989). Trypsin cleaves lysylproline in a hydroxyproline-rich glycoprotein from Zea mays. Pept. Res. 2, 246–248 [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagam Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 10.1126/science.1128439 [DOI] [PubMed] [Google Scholar]
- Koski M. K., Hieta R., Böllner C., Kivirikko K. I., Myllyharju J., Wierenga R. K. (2007). The active site of an algal prolyl 4-hydroxylase has a large structural plasticity. The active site of an algal prolyl 4-hydroxylase has a large structural plasticity. J. Biol. Chem. 282, 37112–37123 10.1074/jbc.M706554200 [DOI] [PubMed] [Google Scholar]
- Koski M. K., Hieta R., Hirsilä M., Rönkä A., Myllyharju J., Wierenga R. K. (2009). The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J. Biol. Chem. 284, 25290–25301 10.1074/jbc.M109.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T., Kieliszewski M. J., Chen Y., Cannon M. C. (2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156, 11–19 10.1104/pp.110.169011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T. A. (1963). Oxygen fixation into hydroxyproline of plant cell wall protein. J. Biol. Chem. 238, 1438–1440 [PubMed] [Google Scholar]
- Lamport D. T. A. (1967). Hydroxyproline-O-glycosidic linkage of the plant cell wall glycoprotein extensin. Nature 216, 1322–1324 10.1038/2161322a0 [DOI] [Google Scholar]
- Lamport D. T. A. (1977). “Structure, biosynthesis and significance of cell wall glycoproteins,” in Recent Advances in Phytochemistry, eds Loewus F. A., Runeckles V. C. (Plenum Publishing, New York: ), 79–115 [Google Scholar]
- Lamport D. T. A. (1980). Structure, and Function of Plant Glycoproteins in the Biochemistry of Plants, Vol. 3 New York: Academic Press, 501–540 [Google Scholar]
- Lamport D. T. A., Northcote D. H. (1960). Hydroxyproline in primary cell walls of higher plants. Nature 188, 665–666 10.1038/188665a0 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Waffenschmidt S., Small L., Goodenough U. (2007). Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol. 144, 1813–1826 10.1104/pp.107.100891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind J. L., Bacic A., Clarke A. E., Anderson M. A. (1994). A style-specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. Plant J. 6, 491–502 10.1046/j.1365-313X.1994.6040491.x [DOI] [PubMed] [Google Scholar]
- Magliano T. M. A., Casal J. J. (1998). In vitro cross-linking of extensin precursors by mustard extracellular isoforms of peroxidase that respond either to phytochrome or to wounding. J. Exp. Bot. 49, 1491–1499 10.1093/jexbot/49.326.1491 [DOI] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067 10.1126/science.1191132 [DOI] [PubMed] [Google Scholar]
- Myllyharju J. (2003). Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 22, 15–24 10.1016/S0945-053X(03)00006-4 [DOI] [PubMed] [Google Scholar]
- Myllyharju J. (2008). Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann. Med. 40, 402–417 10.1080/07853890801986594 [DOI] [PubMed] [Google Scholar]
- Nuñez A., Fishman M. L., Fortis L. L., Cooke P. H., Hotchkiss A. T., Jr. (2009). Identification of extensin protein associated with sugar beet pectin. J. Agric. Food Chem. 57, 10951–10958 10.1021/jf902162t [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580 10.1038/nchembio.182 [DOI] [PubMed] [Google Scholar]
- Owens N. W., Stetefeld J., Lattová E., Schweizer F. (2010). Contiguous O-galactosylation of 4(R)-hydroxy-l-proline residues forms very stable polyproline II helices. J. Am. Chem. Soc. 132, 5036–5042 10.1021/ja905724d [DOI] [PubMed] [Google Scholar]
- Passardi F., Longet D., Penel C., Dunand C. (2004). The plant peroxidase multigenic family in rice and its evolution in green plants. Phytochemistry 65, 1879–1893 10.1016/j.phytochem.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Price N. J., Pinheiro C., Soares C. M., Ashford D. A., Ricardo C. P., Jackson P. A. (2003). A biochemical and molecular characterization of LEP1, an extensin peroxidase from lupin. J. Biol. Chem. 278, 41389–41399 10.1074/jbc.M304519200 [DOI] [PubMed] [Google Scholar]
- Qi X., Behrens B. X., West P. R., Mort A. J. (1995). Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures: evidence for a covalent cross-link between extensin and pectin. Plant Physiol. 108, 1691–1701 10.1104/pp.108.4.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. (2010). The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J. 63, 662–669 [DOI] [PubMed] [Google Scholar]
- Rubinstein A. L., Broadwater A. H., Lowrey K. B., Bedinger P. A. (1995). PEX1, a pollen-specific gene with an extensin-like domain. Proc. Natl. Acad. Sci. U.S.A. 92, 3086–3090 10.1073/pnas.92.8.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C. A., Pearce G. (2003). Systemic signaling in tomato plants for defense against herbivores: isolation and characterization of three novel defense-signaling glycopeptides hormones coded in a single precursor gene. J. Biol. Chem. 278, 30044–30050 10.1074/jbc.M303552200 [DOI] [PubMed] [Google Scholar]
- Schnabelrauch L. S., Kieliszewski M., Upham B. L., Alizedeh H., Lamport D. T. (1996). Isolation of pl 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 4, 477–489 [DOI] [PubMed] [Google Scholar]
- Shimma Y., Saito F., Suyama A., Oka T., Yoko-o T., Matsuoka K., Jigami Y. (2011). “Identification of novel peptidyl serine O-galactosyltransferase gene family in plants,” in XVIII International Botanical Congress, Melbourne, July 23–30, IBC2011 Abstract 116. [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001). Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278 10.1074/jbc.M011323200 [DOI] [PubMed] [Google Scholar]
- Shpak E., Leykam J. F., Kieliszewski M. J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. U.S.A. 96, 14736–14741 10.1073/pnas.96.26.14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 10.1126/science.1102765 [DOI] [PubMed] [Google Scholar]
- Sørensen I., Pettolino F. A., Bacic A., Ralph J., Lu F. C., O’Neill M. A., Fei Z. Z., Rose J. K. C., Domozych D. S., Willats W. G. T. (2011). The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 68, 201–211 10.1111/j.1365-313X.2011.04686.x [DOI] [PubMed] [Google Scholar]
- Stafstrom J. P., Staehelin L. A. (1986). The role of carbohydrate in maintaining extensin in an extended conformation. Plant Physiol. 81, 242–246 10.1104/pp.81.1.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper M., Hallmann A. (1998). Biochemistry of the extracellular matrix of Volvox. Int. Rev. Cytol. 180, 51–85 10.1016/S0074-7696(08)61770-2 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Sato K., Uchida T. (1981). Plant prolyl hydroxylase recognizes poly(l-proline) II helix. J. Biol. Chem. 256, 11397–11400 [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. (2005). Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280, 1142–1148 10.1074/jbc.M411109200 [DOI] [PubMed] [Google Scholar]
- Valentin R., Cerclier C., Geneix N., Aguié-Béhin V., Gaillard C., Ralet M. C., Cathala B. (2010). Elaboration of extensin-pectin thin film model of primary plant cell wall. Langmuir 26, 9891–9898 10.1021/la100265d [DOI] [PubMed] [Google Scholar]
- Van Holst G. J., Varner J. E. (1984). Reinforced polyproline II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol. 74, 247–251 10.1104/pp.74.2.247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S. M., Ricardi M. M., Gloazzo Dorosz J., Fernandez P. V., Nadra A. D., Pol-Fachin L., Egelund J., Gille S., Ciancia M., Verli H., Pauly M., Bacic A., Erik Olsen C., Ulvskov P., Larsen Petersen B., Somerville C., Iusem N. D., Estevez J. M. (2011a). O-glycosylated cell wall extensins are essential in root hair growth. Science 33, 1401–1403 [DOI] [PubMed] [Google Scholar]
- Velasquez S. M., Iusem N. D., Estevez J. M., Root hair sweet growth. (2011b). Plant Signal. Behav. 6, 1600–1602 10.4161/psb.6.10.17135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Spano T., Vlad D., Bou Daher F., Ouelhadj A., Kalaitzis P. (2007). Arabidopsis prolyl 4-hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiol. Plant 130, 471–483 10.1111/j.1399-3054.2007.00915.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Tiainen P., Owen C., Spano T., Bou Daher T., Oualid F., Senol N. O., Vlad D., Myllyharju J., Kalaitzis P. (2010). Characterization of two carnation petal prolyl 4 hydroxylases. Physiol. Plant 140, 199–207 10.1111/j.1399-3054.2010.01390.x [DOI] [PubMed] [Google Scholar]
- Won S. K., Lee Y. J., Yeon Lee H., Kyung Heo Y., Cho M., Hyung-Taeg C. (2009). Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 150, 1459–1473 10.1104/pp.109.140905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005). Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41, 81–94 10.1111/j.1365-313X.2004.02279.x [DOI] [PubMed] [Google Scholar]
- Zhao Z. D., Tan L., Showalter A. M., Lamport D. T. A., Kieliszewski M. J. (2002). Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J. 31, 431–444 10.1046/j.1365-313X.2002.01365.x [DOI] [PubMed] [Google Scholar]