Abstract
Plant viruses use several strategies to transport their nucleic acid genomes throughout the plants. Regardless of the movement mechanism, a universal major block to uninterrupted viral trafficking is the induction of antiviral silencing that degrades viral RNA. To counteract this defense, viruses encode suppressors that block certain steps in the RNA silencing pathway, and consequently these proteins allow viral spread to proceed. There is a constant battle between plants and viruses and sometimes viruses will succeed and invade the plants and in other cases the RNA silencing mechanism will override the virus. A key role in the silencing versus suppression conflict between plants and viruses is played by one or more members of the Argonaute protein (AGO) family encoded by plants. Here we review the mechanisms and effects of antiviral silencing with an emphasis on the contribution of AGOs, especially the recently discovered role of AGO2.
Keywords: plant, virus, Argonaute, RNA silencing, suppressor, movement
Plant Virus Infection Steps
There are several steps during the course of a virus infection that form crucial plant–viral interaction determinants that need to be compatible to ensure the establishment of a successful systemic invasion. Briefly, first the virus needs an entryway into the plant tissue, either via natural openings, through mechanical damage, or by biological vectors such as insects. Upon entry, a virus disassembly process is necessary for release of the genome to initiate expression of key proteins for replication. Some viral replicase proteins induce membrane invaginations, e.g., P33 from Tomato bushy stunt virus (TBSV; McCartney et al., 2005), that stimulate the formation of organelle-like RNA replication factories (Den Boon and Ahlquist, 2010). Subsequently, other virus proteins need to be expressed, for instance for viral cell-to-cell movement that in some instances may involve transport of aforementioned replication complexes (Kawakami et al., 2004; Guenoune-Gelbart et al., 2008). This movement originates from its intracellular location toward and through plasmodesmata for intercellular movement to neighboring cells, followed by systemic spread through the whole plant via the vascular system (Oparka and Santa-Cruz, 2000; Lucas and Lee, 2004; Ueki and Citovsky, 2007; Harries and Ding, 2011; Niehl and Heinlein, 2011).
At any step above, existing barriers or innate and induced plant immune responses can interfere with virus accumulation. A classic molecular interaction is illustrated by the elicitation of a gene-for-gene type hypersensitive response (HR) by specific virus proteins (e.g., replicase, coat protein, or movement protein), leading to the formation of local lesions that arrest further spread, in essence by walling off the virus from the surrounding healthy tissue. Another conserved strategic defense response, which is the focus of the present review, is based on RNA silencing that targets viral RNA (of both DNA and RNA viruses) for destruction, thereby eliminating further virus spread and permitting plants to recover.
RNA Silencing and Virus Movement
Systemic spread of most plant viruses is predicated on traversing different cell types: epidermal, mesophyll, bundle sheath, vascular parenchyma, companion cells, and sieve elements, to culminate in phloem-mediated viral movement to new shoots. To facilitate this trafficking, viruses encode specialized movement proteins (MP) that assist and protect viral nucleic acids while traveling through the cells. MPs of different viruses are quite variable but most possess functionally equivalent properties (Scholthof, 2005) such as: nucleic acid binding activity, ability to interact with plasmodesmata and change their size exclusion limit, and in some cases they exhibit pleiotropic roles in the infection process. Many viral proteins that in the past were characterized as proteins involved in systemic invasion, such as TBSV P19, Potato virus X (PVX) p25, Tobacco mosaic virus (TMV) 126-kDa protein, and the potyviral HC-Pro are now known to be suppressors of gene silencing (Ueki and Citovsky, 2007; Taliansky et al., 2008). Their identification as movement-related proteins prior to the advent of RNA silencing is currently understood to be related to the outcome that silencing against viruses is responsible for degradation of viral RNA. This inevitably causes the abolishment of viral transport throughout plants, and it follows that suppressors counteracting the degradation will let systemic spread take full effect (Scholthof, 2005). Suppressors of RNA silencing are also known to interfere with a “mobile” signal used by plants (Melnyk et al., 2011), possibly to prevent the alerting of distant plant organs of the viral infection (Ratcliff et al., 1997; Schwach et al., 2005). In summary, if a virus fails to express an effective suppressor, the plant can mount an attack using the antiviral RNA silencing process to eliminate virus accumulation and spread.
The Antiviral RNA Silencing Pathway
RNA interference (RNAi) or RNA silencing is a highly conserved molecular mechanism known to regulate gene expression and combat invasive nucleic acids across species, for instance in mammals (Liu et al., 2004), Caenorhabditis elegans (Fire et al., 1998), plants (Baulcombe, 2004; Voinnet, 2005; Li and Ding, 2006), fungi (Romano and Macino, 1992), and insects (Hammond et al., 2001). As currently understood, RNA silencing is triggered by the presence of double-stranded RNA (dsRNA) structures in the cell, for instance those that are transcribed as progenitor microRNAs (miRNAs), or that accumulate in the cell via viral infection or upon artificial introduction (Filipowicz, 2005). These dsRNAs are cleaved into smaller segments by a Dicer protein to yield short duplex RNAs such as miRNAs or short-interfering RNAs (siRNAs), and one of the strands is used by the cell to program an RNA-induced silencing complex (RISC) containing at least one Argonaute protein (AGO). This programmed RISC then targets single-stranded RNAs complementary to that incorporated siRNA for cleavage or translational repression, resulting in post-transcriptional gene silencing of specific genes (Alvarado and Scholthof, 2009).
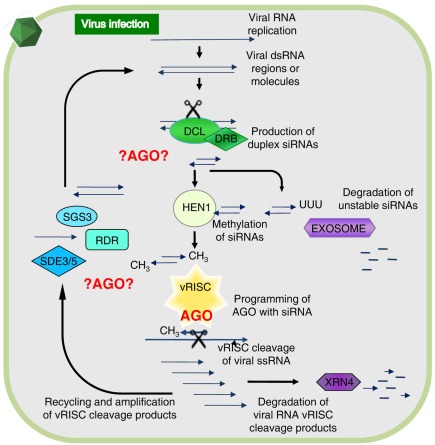
The individual components of the general RNA silencing pathway in plants (Figure 1) were mostly identified through genetic studies, but many of the potential biochemical interactions that could occur between these components remain to be elucidated. However, it is known that Dicer-like proteins (DCLs) are the first to interact with viral RNA. Specifically, DCL2 and DCL4 are necessary for antiviral silencing of Tobacco rattle virus (TRV; Deleris et al., 2006), as well as participating in other RNA silencing functions such as generation of trans-acting siRNAs and transitivity (Gasciolli et al., 2005; Mlotshwa et al., 2008). Double-stranded RNA binding proteins (DRBs) interact with DCLs to assist these with dsRNA recognition and cleavage, for instance DRB4 was shown to contribute to antiviral silencing (Curtin et al., 2008). The siRNAs are stabilized by methylation of their 3′-end by HEN1 methyl-transferase, preventing siRNAs from uridylation and degradation (Li et al., 2005; Yu et al., 2005), hence, ensuring maintenance of the siRNA signal and programming of AGOs. The primary siRNAs move cell-to-cell in short distance signaling (10–15 cells) with the aid of silencing movement deficient proteins (SMD1,2,3; Ding and Voinnet, 2007). Secondary siRNAs are generated by an RNA-dependent RNA (RDR) polymerase-dependent amplification cycle (Figure 1) and these are presumably used for long distance signaling (Dunoyer et al., 2010; Molnar et al., 2010). RDR activity results in the creation of long dsRNA targets that increase the levels of siRNAs processed by DCL proteins. Interestingly, three out of six RDRs in plants contain a C-terminal catalytic DLDGD peptide domain conserved in many eukaryotes (Wassenegger and Krczal, 2006). RDR6 was shown to have antiviral activities in Arabidopsis and Nicotiana benthamiana (Qu et al., 2005, 2008; Schwach et al., 2005) and requires SDE3, a protein with helicase activity, and SDE5 (Ding and Voinnet, 2007) for this amplification process (Vaistij et al., 2002). SGS3 binds and stabilizes RNA templates to initiate RDR6-mediated dsRNA synthesis, and these two proteins co-localize and interact in specific cytoplasmic granules referred to as SGS3/RDR6-bodies (Kumakura et al., 2009).
Figure 1.
The antiviral RNA silencing model as presently understood. During viral replication or transcription of DNA or RNA viruses, highly structured RNA or double-stranded (ds) RNA is formed providing a substrate for cleavage by Dicer-like proteins (DCL) associated with dsRNA binding protein (DRB). This generates duplex short-interfering RNAs (siRNAs) and upon methylation by HUA enhancer 1 (HEN1) these associate with an antiviral RNA-induced silencing complex (vRISC), or in absence of methylation these will be polyuridylated and degraded. One siRNA strand remains bound to an Argonaute protein (AGO) in vRISC and this operates as a search-and-strike module to cleave single-stranded (ss) RNA complementary to the siRNA. The resulting RNA fragments are either degraded via XRN4 or re-amplified by concerted action of SGS3, SDE3, SDE5, and an RNA-dependent RNA polymerase (RDR). The vRISC (star-shaped) associated AGO is indicated in red, whereas other possible AGO-mediated interactions alluded to in the text are denoted by ?AGO? Figure modified from Alvarado and Scholthof (2009).
There are a number of other RNA silencing-associated proteins not depicted in Figure 1 with accessory functions that can feasibly contribute to the silencing pathway. For instance, during the secondary amplification cycle a specific mRNA export complex (THO/TREX) might perform a function in transporting long RNA molecules to be used as templates for secondary siRNA production (Yelina et al., 2010). Furthermore, AGO-associated proteins such as cyclophilin 40 (Smith et al., 2009), and the heat shock protein HSP90 (Iki et al., 2010), assist with AGO1 conformational changes. Other examples are proteins that bind directly to AGOs via a conserved GW-rich domain “Ago hook” and their different functional interactions may play roles in distinct gene silencing pathways (Till and Ladurner, 2007). For instance, the NRPD1b subunit of the plant-specific RNA PolIVb/PolV (El-Shami et al., 2007) and another protein (SPT5-like) associate with AGO4 in Arabidopsis via a GW-rich repeat domain (Bies-Etheve et al., 2009).
Even though the common assumption is that an AGO forms a core constituent of the antiviral RISC (vRISC; Figure 1), it remains to be established which AGO(s) are recruited for vRISC activity. Additionally, considering the abovementioned alternative AGO-mediated interactions it is possible that different AGOs are engaged in the assembly of alternative antiviral silencing complexes to participate at different steps in the pathway as shown in red in Figure 1. Toward gaining a comprehensive understanding, investigations are ongoing to precisely elucidate which, and how, AGOs function in antiviral RNA silencing.
First There was AGO1
According to our present understanding, AGO1 is the primary Argonaute protein in the plant miRNA silencing pathway (Baumberger and Baulcombe, 2005). The miRNA process is employed by plants to regulate genes involved in numerous biological functions such as development, hormone regulation, biotic, and abiotic stress responses (Voinnet, 2009). Several lines of evidence also implicate AGO1 in antiviral responses; for instance, its transcript levels are induced upon virus inoculation (Varallyay et al., 2010) and some suppressors of gene silencing (e.g., Tombusvirus P19) apparently decrease AGO1 levels in infected tissue by enhancing expression of miR168 which regulates AGO1 through a feedback mechanism (Varallyay et al., 2010). Other suppressors mediate the targeted degradation of AGO1 protein (Polerovirus P0; Csorba et al., 2010) or inhibit its RNA cleavage activity, as shown for Cucumber mosaic virus (CMV) 2b (Zhang et al., 2006), and Turnip crinkle virus (TCV) P38 (Azevedo et al., 2010). Moreover, an ago1 hypomorphic Arabidopsis mutant was found to be more susceptible to virus infection than wild-type plants (Morel et al., 2002). All these findings point to the importance of AGO1 in plant silencing pathways involved in endogenous gene regulation as well as in host defenses. Nevertheless, a major question remains: is AGO1 the only AGO protein involved in antiviral defense mechanisms in plants?
Plants encode at least 10 AGOs that may well play specific parts in the silencing cascade to regulate endogenous gene expression, and that could potentially also be recruited for antiviral defense. Indirect support for this notion is derived from studies showing that Arabidopsis AGO2 and AGO5 bind CMV-derived viral siRNAs (Takeda et al., 2008), and are localized to the nucleus and cytoplasm. These AGOs exhibit a 5′-end nucleotide preference in relation to the type of associated siRNA. For example, AGO2 prefers a 5′-adenine nucleotide while AGO5 favors a 5′-cytosine (Takeda et al., 2008). Genetic studies also implicate other AGOs in addition to AGO1 in antiviral defense responses. For example, experiments involving Arabidopsis ago1 and ago7 mutants infected with suppressor-defective TCV point to complementary roles for AGO1 and AGO7 in viral defense responses (Qu et al., 2008; Azevedo et al., 2010). Another example is represented by AGO4, which was connected to virus defense, but solely in the context of the induction of a specific resistance gene-mediated response (Bhattacharjee et al., 2009). Moreover, the Polerovirus P0 suppressor not only targets AGO1 for degradation, it also affects AGO2, -4, -5, -6, and -9 (Baumberger et al., 2007). Collectively, these discoveries suggest that potentially multiple AGOs might be enlisted in the effort to target viral invaders for destruction.
Then Came AGO2
In support of the above notion that AGOs in addition to AGO1 may be a factor in the battle against viruses, four recent studies linked AGO2 in Arabidopsis and N. benthamiana with antiviral defense responses. In the first study, Arabidopsis AGO mutant knockouts (KOs) for each of the 10 AGOs, were challenged with TCV and CMV (Harvey et al., 2011). These viruses encode suppressors of gene silencing, P38 for TCV and 2b for CMV, which among other activities directly interfere with AGO1 function, resulting in the condition that regardless of the KO line, upon infection AGO1 activity was compromised by these viruses. Out of all the mutants tested within this experimental framework only the ago2 KO line showed enhanced susceptibility to infections with either virus. It appears that this novel antiviral role for Arabidopsis AGO2 is normally hidden in the presence of active AGO1, because the latter regulates the expression of AGO2 via the production of miR403 (Allen et al., 2005). But once AGO1 has vanished as a consequence of the activity of P38 or 2b, AGO2 is expressed at higher levels and then surfaces as an antiviral defense protein.
At the same time a similar role was discovered for Arabidopsis AGO2 in antiviral defense, but in this particular case it was found that AGO1 and AGO2 operate in a non-redundant and cooperative manner against a weak form of CMV not expressing the 2b suppressor (CMVΔ2b; Wang et al., 2011). Individual Arabidopsis lines bearing T-DNA insertional KOs to eliminate expression of specific AGO-encoding genes were infected with the CMVΔ2b variant. The outcome was that only ago1 and ago2 KO lines were susceptible to CMVΔ2b, while the double KO displayed even higher susceptibility. Moreover, an increase in CMVΔ2b viral siRNAs was observed in the single and double KO mutant lines of ago1 and ago2 when compared to wild-type plants, indicating that AGO1 and AGO2 are conceivably not required for the production of primary and secondary siRNAs in CMVΔ2b–Arabidopsis interactions, but they are involved in downstream antiviral activities. Pull down assays were also performed with FLAG-tagged AGO1 and AGO2 in order to analyze viral siRNA sorting preferences for these two proteins. The results showed that siRNAs with 5′-uracil nucleotides were preferred by AGO1 whereas 5′-adenine nucleotides were favored by AGO2, corroborating previous findings on AGO-dependent preferential sorting properties.
A third study associating Arabidopsis AGO2 with antiviral silencing identified a PVX “strain” capable of infecting Arabidopsis while this species is normally a non-host for this virus (Jaubert et al., 2011). However, it was determined that the “strain” actually comprised a mix of two separate viruses, PVX, and Pepper ring spot virus (PepRSV), a Tobravirus. In this scenario PepRSV acted by repressing viral RNA silencing, thereby creating a molecular milieu now conducive for PVX infection. Key to understanding the nature of the non-host behavior was the observation that PVX alone was competent for infecting an Arabidopsis triple dicer mutant (dcl2, dcl3, and dcl4) to cause severe disease symptoms along with systemic necrosis pointing to silencing as the main impediment for PVX to infect wild-type Arabidopsis plants (Jaubert et al., 2011). Subsequently it was investigated whether AGOs were involved in this non-host interaction, by challenging Arabidopsis ago mutant plants with PVX and observing that only ago2 plants allowed accumulation of PVX in systemic tissues. Sap from these PVX-infected Arabidopsis plants induced an HR in Rx2 N. benthamiana plants that are resistant to PVX, providing evidence that the original tissue contained virus. However, sap from upper leaves of wild-type Arabidopsis or from other ago mutant plants inoculated with PVX, failed to cause a local lesion response in the Rx2 N. benthamiana plants, indicating the absence of virus in that source. Thus, it was concluded that Arabidopsis AGO2 plays a major role in the antiviral defense mechanism against PVX.
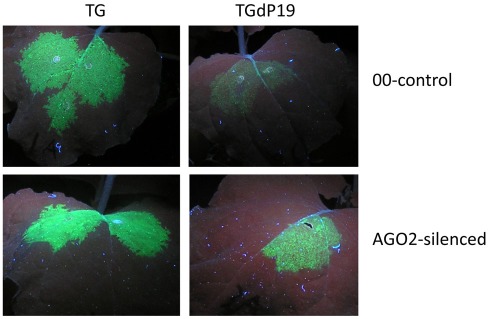
The final study identifying AGO2 as an important component of the antiviral silencing defense mechanism was not derived from work with Arabidopsis, but is instead based on studies with N. benthamiana infected with TBSV mutants (Scholthof et al., 2011). The purpose of the investigation was to identify which N. benthamiana AGOs are important for silencing TBSV RNA and that could perhaps be a component of vRISC (Omarov et al., 2007). Toward this, infectious TBSV clones were used that either expressed the P19 suppressor (TBSV) or that were defective for its translation (TdP19). Additional versions of the TBSV constructs expressed green fluorescence protein (TG and TGdP19) to visualize the effects of silencing on virus spread. In a normal wild-type TBSV infection in N. benthamiana, the plants succumb to a lethal necrosis about a week after inoculation, but plants infected with dP19 variants display much weaker symptoms and in fact eventually recover from the initial infections through the action of antiviral RNA silencing (Scholthof, 2006). It was reasoned that in plants inactivated for a crucial AGO, the dP19 mutants would not be targeted by silencing and yield a wild-type-like virus infection. To obtain AGO “KO” plants in N. benthamiana, TRV-based virus-induced gene silencing was used to down-regulate expression of AGO1, AGO2, or AGO4, and after the onset of AGO-silencing, those plants were subsequently challenged with TBSV, TdP19, TG, or TGdP19. Non-silenced control plants, or plants silenced for AGO1 or AGO4, recovered from virus infections with TdP19 as a result of anti-TBSV silencing. Similarly, in such plants TGdP19 initially expressed GFP but at reduced intensity compared to TG infections (Figure 2), and a few days later GFP expression by TGdP19 had entirely disappeared due to antiviral RNA silencing. In contrast, AGO2-inactivated N. benthamiana plants supported wild-type-like infections by the TdP19 virus and moreover, these plants exhibited no difference when infected with TG or TGdP19 because GFP intensity and duration was the same for both (Figure 2). In short, inactivation of AGO2 in N. benthamiana compromised the ability of the plants to mount an effective antiviral silencing defense against TBSV.
Figure 2.
AGO2-mediated silencing of Tomato bushy stunt virus RNA in Nicotiana benthamiana. Plants were agroinfiltrated to express Tobacco rattle virus (TRV) as a virus-induced gene silencing vector containing either no insert (00-control) or an AGO2 fragment (AGO2-silenced). At ~40 days post-infiltration, upper leaves of these plants were infiltrated to express TBSV–GFP (TG) or TBSV–GFP devoid of P19 translation (TGdP19); images of leaves were captured 5 days later under UV illumination. Evidently when AGO2 expression is silenced, GFP expression from TGdP19 (only infiltrated in one half of the leaf) is maintained indicating the absence of effective silencing when compared to that in the 00-control. Details can be found in Scholthof et al. (2011), that also describes the eventual complete abolishment of GFP expression in TGdP19-infected 00-control plants at later time points, while expression in AGO2-silenced plants is maintained stably.
Prospects
It is evident that plants evolved RNA silencing not only to regulate endogenous gene expression but also as an important strategy for defense against invasions by foreign nucleic acids. An increasing body of evidence points to vital antiviral roles not only for AGO1 but most recently also for AGO2. Probably additional or maybe even all AGOs have overlapping and/or epistatic functions relating to antiviral defense. The type of viral infection (e.g., localization or formation of specific intracellular structures) and the nature of the suppressors, could possibly also determine which AGOs are recruited most effectively by the plant for an antiviral defense function under the prevailing conditions. Moreover, host-dependent interactions might also influence what type of AGO is engaged against any particular virus, and to either target the viral RNA for destruction or perhaps translational repression. In the end it is possible that the plant will assemble several complexes (Figure 1) composed of one or more AGOs that under the existing circumstances are the most effective in compromising the accumulation and spread of viruses in the plants in their constant battle with the negating effect of suppressors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Alvarado V., Scholthof H. B. (2009). Plant responses against invasive nucleic acids: RNA silencing and its suppression by plant viral pathogens. Semin. Cell Dev. Biol. 20, 1032–1040 10.1016/j.semcdb.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., Braun L., Bergdoll M., Mohamed A. H., Lagrange T., Voinnet O. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24, 904–915 10.1101/gad.1908710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431, 356–363 10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 11928–11933 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D. C. (2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614 10.1016/j.cub.2007.08.039 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Zamora A., Azhar M. T., Sacco M. A., Lambert L. H., Moffett P. (2009). Virus resistance induced by NB–LRR proteins involves Argonaute4-dependent translational control. Plant J. 58, 940–951 10.1111/j.1365-313X.2009.03832.x [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T. (2009). RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 10, 649–654 10.1038/embor.2009.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Lózsa R., Hutvágner G., Burgyán J. (2010). Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472 10.1111/j.1365-313X.2010.04163.x [DOI] [PubMed] [Google Scholar]
- Curtin S. J., Watson J. M., Smith N. A., Eamens A. L., Blanchard C. L., Waterhouse P. M. (2008). The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 582, 2753–2760 10.1016/j.febslet.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Deleris A., Gallego-Bartolome J., Bao J., Kasschau K. D., Carrington J. C., Voinnet O. (2006). Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science 313, 68–71 10.1126/science.1128214 [DOI] [PubMed] [Google Scholar]
- Den Boon J. A., Ahlquist P. (2010). Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64, 241. 10.1146/annurev.micro.112408.134012 [DOI] [PubMed] [Google Scholar]
- Ding S.-W., Voinnet O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426 10.1016/j.cell.2007.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J. C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328, 912–916 10.1126/science.1185880 [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.-A., Jacobsen S. E., Cooke R., Lagrange T. (2007). Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21, 2539–2544 10.1101/gad.451207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. (2005). RNAi: the nuts and bolts of the RISC machine. Cell 122, 17–20 10.1016/j.cell.2005.06.023 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Gasciolli V., Mallory A. C., Bartel D. P., Vaucheret H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500 10.1016/j.cub.2005.07.024 [DOI] [PubMed] [Google Scholar]
- Guenoune-Gelbart D., Elbaum M., Sagi G., Levy A., Epel B. L. (2008). Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol. Plant Microbe Interact. 21, 335–345 [DOI] [PubMed] [Google Scholar]
- Hammond S. M., Boettcher G. S., Caudy A. A., Kobayashi T. R., Hannon G. J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150 10.1126/science.1064023 [DOI] [PubMed] [Google Scholar]
- Harries P., Ding B. (2011). Cellular factors in plant virus movement: at the leading edge of macromolecular trafficking in plants. Virology 411, 237–243 10.1016/j.virol.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Harvey J. J. W., Lewsey M. G., Patel K., Westwood J., Heimsta S., Carr J. P., Baulcombe D. C. (2011). An antiviral defense role of AGO2 in plants. PLoS ONE 6, e14639. 10.1371/journal.pone.0014639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M. C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. (2010). In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell 39, 282–291 10.1016/j.molcel.2010.05.014 [DOI] [PubMed] [Google Scholar]
- Jaubert M., Bhattacharjee S., Mello A. F. S., Perry K. L., Moffett P. (2011). ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 156, 1556–1564 10.1104/pp.111.178012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami S., Watanabe Y., Beachy R. N. (2004). Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. U.S.A. 101, 6291–6296 10.1073/pnas.0401221101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R., Watanabe Y. (2009). SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 583, 1261–1266 10.1016/j.febslet.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Li F., Ding S. W. (2006). Virus counter defense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60, 503–531 10.1146/annurev.micro.60.080805.142205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507 10.1016/j.cub.2005.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J.-J., Hammond S. M., Joshua-Tor L., Hannon G. J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Lucas W. J., Lee J.-Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5, 712–726 10.1038/nrm1470 [DOI] [PubMed] [Google Scholar]
- McCartney A. W., Greenwood J. S., Fabian M. R., White K. A., Mullen R. T. (2005). Localization of the Tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17, 3513–3531 10.1105/tpc.105.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk C. W., Molnar A., Baulcombe D. C. (2011). Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563 10.1038/emboj.2011.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S., Pruss G. J., Peragine A., Endres M. W., Li J., Chen X., Poethig R. S., Bowman L. H., Vance V. (2008). DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 3, e1755. 10.1371/journal.pone.0001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A., Melnyk C. W., Bassett A., Hardcastle T. J., Dunn R., Baulcombe D. C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 10.1126/science.1187959 [DOI] [PubMed] [Google Scholar]
- Morel J. B., Godon C., Mourrain P., Béclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 629–639 10.1105/tpc.010358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A., Heinlein M. (2011). Cellular pathways for viral transport through plasmodesmata. Protoplasma 248, 75–99 10.1007/s00709-010-0246-1 [DOI] [PubMed] [Google Scholar]
- Omarov R. T., Ciomperlik J. J., Scholthof H. B. (2007). RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc. Natl. Acad. Sci. U.S.A. 104, 1714–1719 10.1073/pnas.0608117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka K. J., Santa-Cruz S. (2000). The great escape: phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 323–347 10.1146/annurev.arplant.51.1.323 [DOI] [PubMed] [Google Scholar]
- Qu F., Ye X., Hou G., Sato S., Clemente T. E., Morris T. J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217 10.1128/JVI.79.24.15209-15217.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F., Ye X., Morris T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737 10.1073/pnas.0801436105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Harrison B. D., Baulcombe D. C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560 10.1126/science.276.5318.1558 [DOI] [PubMed] [Google Scholar]
- Romano N., Macino G. (1992). Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353 10.1111/j.1365-2958.1992.tb02202.x [DOI] [PubMed] [Google Scholar]
- Scholthof H. B. (2005). Plant virus transport: motions of functional equivalence. Trends Plant Sci. 10, 376–382 10.1016/j.tplants.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Scholthof H. B. (2006). The Tombusvirus-encoded P19: from irrelevance to elegance. Nat. Rev. Microbiol. 4, 405–411 10.1038/nrmicro1395 [DOI] [PubMed] [Google Scholar]
- Scholthof H. B., Alvarado V. Y., Vega-Arreguin J. C., Ciomperlik J., Odokonyero D., Brosseau C., Jaubert M., Zamora A., Moffett P. (2011). Identification of an ARGONAUTE for antiviral RNA silencing in Nicotiana benthamiana. Plant Physiol. 156, 1548–1555 10.1104/pp.111.178764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F., Vaistij F. E., Jones L., Baulcombe D. C. (2005). An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852 10.1104/pp.105.063537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Willmann M. R., Wu G., Berardini T. Z., Möller B., Weijers D., Poethig R. S. (2009). Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5424–5429 10.1073/pnas.0908706106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 49, 493–500 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- Taliansky M., Torrance L., Kalinina N. O. (2008). “Role of plant virus movement proteins,” in Plant Virology Protocols, eds Foster G. D., Johansen I. E., Hong Y., Nagy P. D.(New York: Humana Press; ), 33–54 [DOI] [PubMed] [Google Scholar]
- Till S., Ladurner A. G. (2007). RNA Pol IV plays catch with Argonaute 4. Cell 131, 643–645 10.1016/j.cell.2007.10.044 [DOI] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2007). “Spread throughout the plant: systemic transport of viruses,” in Viral Transport in Plants, eds Waigmann E., Heinlein M. (New York: Springer; ), 85–118 [Google Scholar]
- Vaistij F. E., Jones L., Baulcombe D. C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867 10.1105/tpc.010480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay E., Valoczi A., Agyi A., Burgyan J., Havelda Z. (2010). Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519 10.1038/emboj.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220 10.1038/nrg1555 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang X.-B., Jovel J., Udomporn P., Wang Y., Wu Q., Li W.-X., Gasciolli V., Vaucheret H., Ding S.-W. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative Argonautes in Arabidopsis thaliana. Plant Cell 23, 1625–1638 10.1105/tpc.111.083618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Krczal G. (2006). Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 11, 142–151 10.1016/j.tplants.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Yelina N. E., Smith L. M., Jones A. M. E., Patel K., Kelly K. A., Baulcombe D. C. (2010). Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13948–13953 10.1073/pnas.0911341107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R. W., Steward R., Chen X. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 10.1126/science.1107130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Y. R., Pei Y., Lin S. S., Tuschl T., Patel D. J., Chua N. H. (2006). Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268 10.1101/gad.1426906 [DOI] [PMC free article] [PubMed] [Google Scholar]