Abstract
Correct interpretation of the coding capacity of RNA polymerase II transcribed eukaryotic genes is determined by the recognition and removal of intronic sequences of pre-mRNAs by the spliceosome. Our current knowledge on dynamic assembly and subunit interactions of the spliceosome mostly derived from the characterization of yeast, Drosophila, and human spliceosomal complexes formed on model pre-mRNA templates in cell extracts. In addition to sequential structural rearrangements catalyzed by ATP-dependent DExH/D-box RNA helicases, catalytic activation of the spliceosome is critically dependent on its association with the NineTeen Complex (NTC) named after its core E3 ubiquitin ligase subunit PRP19. NTC, isolated recently from Arabidopsis, occurs in a complex with the essential RNA helicase and GTPase subunits of the U5 small nuclear RNA particle that are required for both transesterification reactions of splicing. A compilation of mass spectrometry data available on the composition of NTC and spliceosome complexes purified from different organisms indicates that about half of their conserved homologs are encoded by duplicated genes in Arabidopsis. Thus, while mutations of single genes encoding essential spliceosome and NTC components lead to cell death in other organisms, differential regulation of some of their functionally redundant Arabidopsis homologs permits the isolation of partial loss of function mutations. Non-lethal pleiotropic defects of these mutations provide a unique means for studying the roles of NTC in co-transcriptional assembly of the spliceosome and its crosstalk with DNA repair and cell death signaling pathways.
Keywords: spliceosome, NineTeen complex, co-transcriptional splicing, DNA repair, cell death signaling, Arabidopsis
Introduction
Intron sequences of RNA polymerase II (RNAPII) transcribed nuclear pre-mRNAs in eukaryotes are removed by the spliceosome to produce mature mRNAs. The spliceosome is composed of U-rich U1, U2, U4, U5, and U6 small nuclear snRNAs, which form snRNP particles with specific sets of proteins. Spliceosomal snRNPs undergo in cell extracts timely ordered assembly on model pre-mRNA templates and recruit numerous stage specific auxiliary regulatory factors, the complexity of which increases in metazoan’s evolution (Wahl et al., 2009). Except for plants, where thus far no suitable in vitro splicing assay is available, past studies of in vitro spliceosome assembly have generated a wealth of mass spectrometry, RNA cross-linking, and crystallographic data on basic functions, binding specificities, and interactions of core spliceosomal proteins. Other studies have independently identified the functions of numerous auxiliary factors that co-purify with different snRNPs. The emerging regulatory interactions reveal a huge complexity and gene/intron specific variation of in vivo spliceosome interactions with the RNAPII transcription initiation, capping, elongation, and polyadenylation/cleavage complexes, as well as subunits of the exon junction, pre-mRNA splicing and retention (RES), mRNA export (THO/TREX), exosome, microRNA processing, nuclear pore, nonsense-mediated decay (NMD), and chromatin remodeling/modifying complexes. In fact, accumulating data on transcript specific splicing kinetics and alternative splicing demonstrate that these interactions govern co-transcriptional assembly, selectivity, and progressivity of the spliceosome, which are also remarkably dependent on changes in epigenetic modifications of histones during different phases of transcription (Oesterreich et al., 2011). Co-transcriptional coupling and regulation of the spliceosome activity is further indicated by the fact that introns are removed from the majority of pre-mRNAs before completion of transcription in yeast and mammalian cells. In general, splicing of introns close to the 5′-ends of transcripts is completed earlier compared to the removal of 3′ introns. Due to different sequence features of introns, their splicing does not however follow always the order of their transcription. Some “difficult” introns might remain completely or partially unspliced upon transcription termination. Partially spliced pre-mRNAs accumulate together with hyperphosphorylated RNAPII, spliceosome components, and auxiliary factors in nuclear speckles, especially in differentiated cells types, where their processing occurs post-transcriptionally (Han et al., 2011). Thus, while spliceosome assembly appears to be regulated co-transcriptionally, splicing per se does not necessarily depend on active transcription.
Spliceosomal snRNPs
As in other metazoans, the majority of introns are recognized by the U2-type spliceosome in Arabidopsis. Regulatory features and targets of Arabidopsis U12-specific minor spliceosome have been reviewed recently (Simpson and Brown, 2008). Here we provide a compilation of proteins identified by mass spectrometry in 27 purified yeast, Drosophila, and human U2-type spliceosome complexes and annotation of corresponding Arabidopsis homologs (Table S1 in Supplementary Material). Yeast spliceosome complexes are composed of 50–60 core snRNP subunits and about hundred additional splicing-related factors, most of which are conserved in metazoans (Fabrizio et al., 2009). In comparison, the total number of proteins identified in purified Drosophila and human spliceosomal complexes is about 260 and 400, respectively (Rappsilber et al., 2002; Herold et al., 2009; Will and Lührmann, 2011). Based on comparative sequence analysis, Wang and Brendel (2004) predicted 395 genes encoding splicing-related proteins in Arabidopsis, including all putative RNA-binding factors identified earlier by Lorkovic and Barta (2002). Our compilation, based on comparative analysis of mass spectrometry data using the TAIR10 version of reannotated genome sequence, indicates a conservation of about 430 spliceosomal factors in Arabidopsis (Table S1 in Supplementary Material). However, in some cases the relationships between yeast, Arabidopsis, and other metazoan homologs are suggested only by conservation of certain functionally important domains. Thus, verification of composition of Arabidopsis core snRNPs and auxiliary factors of various spliceosomal complexes awaits further analysis of their subunits by mass spectrometry.
In yeast and metazoans, the U1, 2, 4, and 5 snRNAs associate through their U-rich PuAU4–6GPu motives to a heptamer of Sm proteins, while U6 is bound to a similar Sm-like LSm complex (Tharun, 2009; Weber et al., 2010). In Arabidopsis all seven Sm proteins and LSm1, 3, and 6 are encoded by duplicated genes (Figure 1). In addition to their U6 snRNP-specific roles, the LSm proteins target aberrant transcripts for decapping and 5′ to 3′ decay. Mutation of Arabidopsis LSm4 (emb1644) causes embryonic lethality, while the lsm5/sad1 point mutation results in a partial loss of function conferring supersensitivity to abscisic acid (ABA) and drought (Xiong et al., 2001).
Figure 1.
Spliceosomal U snRNP components conserved in Arabidopsis. Subunits encoded by two or more genes are highlighted in bold and characterized gene mutations are indicated in red.
Subunits of snRNPs show weaker sequence conservation but similar redundancy in Arabidopsis. From the U1 snRNP-associated proteins, several are encoded by at least two genes, while Arabidopsis lacks Prp42 and Snu56 homologs (Figure 1). Similarly, the U2AF35, U2AF65, and polypyrimidin tract-binding (PTB) proteins from the splice-site selecting U2 components, as well as numerous U2 snRNP subunits and U2-related factors are encoded by duplicated genes in Arabidopsis. Spliceosome association of other factors that share only domain homology with the U2 snRNP components is nonetheless uncertain and should be confirmed by further proteomics analyses. Genetic studies indicate that inactivation of PRP39a component of U1 snRNP confers late flowering by upregulating the transcription of FLC flowering time repressor (Wang et al., 2007). In contrast, the elf9 mutation of Arabidopsis ortholog of tat-SF1 U2 snRNP component upregulates transcription of a defective splice-isoform of suppressor of overexpression of CO1 (SOC1) causing early flowering (Song et al., 2009). On the other hand, the U2 SPF45 subunit is implicated in DNA recombination and repair both in Arabidopsis and Drosophila, where it is an interacting partner of RAD201, a member of the RecA/Rad51 family (Chaouki and Salz, 2006).
In the case of U5 snRNP, there are multiple Arabidopsis homologs of Prp8 and Brr2 RNA helicases and Snu114 GTPase that play important roles in both activation and dissociation of the spliceosome (Wahl et al., 2009). Nonetheless, analysis of the Arabidopsis NineTeen Complex (NTC) complex (Monaghan et al., 2009) and genetic data suggest that BRR2 is represented probably by only a single locus, while PRP8 and SNU114 are encoded by two differentially regulated genes. The prp8a/sus2 mutation results in a cell division defect of embryonic suspensor, while the emb1507 embryo lethal mutation is located in the BRR2 gene. Different genetic screens led to the identification of maternal effect of embryo arrest 5 (mee5), clotho (clo1), and gametophytic factor 1 (gfa1) mutations of SNU114, which is required for specification of egg cell fate and floral organ number and identity (Liu et al., 2009; Yagi et al., 2009). Similarly, the atropos (ato) mutation of U2 snRNP SF3a60/PRP9 gene compromises egg and central cell fate, and SNU114 is necessary for tissue specific expression of Lachesis (LIS) that encodes the U4/U6 snRNP subunit PRP4 (Moll et al., 2008). A common consequence of all these mutations is the abortion of gametophyte. A potential link to cell death regulation is suggested by high level induction of Yellow-Leaf-Specific Gene 8 (U5-15/YLS8) during late leaf senescence (Yoshida et al., 2001). Expression of the U5 snRNA subunit gene U5-102/PRP6 is cold stimulated and its stabilized 1 (sta1)/emb2770 mutation results in defective splicing and stabilization of the cold-induced COR15A and other unstable transcripts leading to a range of pleiotropic developmental and stress response defects (Lee et al., 2006). Like the ELF9/tat-SF1 subunit of U2 snRNP, PRP6 might function in interaction with the exosome and exon junction complex (EJC) implicated in the recruitment of NMD regulators (Gehring et al., 2005).
The U4/U6 subunits PRP3 and Tri-20 are represented by two Arabidopsis homologs, while U4/U6-15.5/SNU13 is likely encoded by three genes as the Tri65 subunit of U4/U6.U5 tri-snRNP. Mutation of U4/U6 subunits Prp24 and Prp31 lead to embryo lethality (emb140 and emb1220), while inactivation of the RS-domain of Tri120/SNU66 by the meristem defective (mdf) mutation stimulates ectopic meristem formation in vegetative tissues but inhibits root and shoot meristem activities. The mdf mutation reduces the transcript levels of PIN2 and PIN4 auxin influx carriers, as well as those of Plethora, Scarecrow, and Shortroot in the root and Wuschel (WUS) in the shoot meristem (Casson et al., 2009), likely due to splicing defects and destabilization of these transcripts. The mdf mutation is allelic with defectively organized tributaries 2 (dot2) that causes altered vein differentiation pattern in juvenile leaves consistently with defects of auxin transport (Petricka et al., 2008).
Spliceosome Catalytic Cycle
The U1 and U2 snRNPs recognize the 5′and 3′ splice-sites (5′SS and 3′SS) and conserved branch sites (BS: AG) of introns. BS is followed by a polypyrimidine tract (PPT) upstream of the 3′SS in metazoans. BS and PPT are poorly conserved in plants (Brown and Simpson, 1998; Simpson et al., 2002), although orthologs of BS-interacting U2AF and PTB proteins are present in Arabidopsis. In vitro assembly studies indicate that the U1 snRNP binds first through U1-C and U1-70K to the 5′SS. Subsequently, PPT and 3′SS are bound respectively by the U2 auxiliary factors U2AF65 and U2AF35 that interact with SF1 at the BP, as well as with a range of SR, hnRNP, Transformer (Tra), and EJC proteins that recognize exonic and intronic splicing enhancer and silencer sequences, contributing to the definition of exons’ positions (for review see Will and Lührmann, 2011). The SR-related Arabidopsis SR45a/Tra-2b factor interacts, for example, with U1-70K and U2F35a to assist splice-site selection, as well as with PRP38 during spliceosome activation (Tanabe et al., 2009). From the 19 Arabidopsis SR-proteins classified into seven subfamilies (Barta et al., 2010), yeast and human orthologs of SR1, SC35, RS33, and RSZ33, as well as the SR-related proteins Tra-1A, B1/2, and SRm160 were identified in purified spliceosomal complexes (Table S1 in Supplementary Material). Transcription and alternative splicing of SR-protein genes is regulated by a multitude of stress and hormonal stimuli, and their known mutations result in pleiotropic regulatory defects (Reddy, 2007). The activity and stability of SR-proteins is regulated by phosphorylation including the Lammer/CLK, SRPK1, and SRPK2 kinases families, as well as by several PRMT arginine methylases that also recognize other classes (e.g., Sm, Lsm, hnRNP, etc.) of spliceosomal proteins (Fluhr, 2008). Arabidopsis PRMT5 was recently demonstrated to methylate several Sm and LSm factors (Deng et al., 2010). The prmt5 mutation results in defective splicing of FLK/hnRNP-E pre-mRNA and late flowering by increasing the FLC transcript level, as well as alters 5′SS recognition leading to aberrant processing of pre-mRNAs encoding components of the circadian clock (Sanchez et al., 2010).
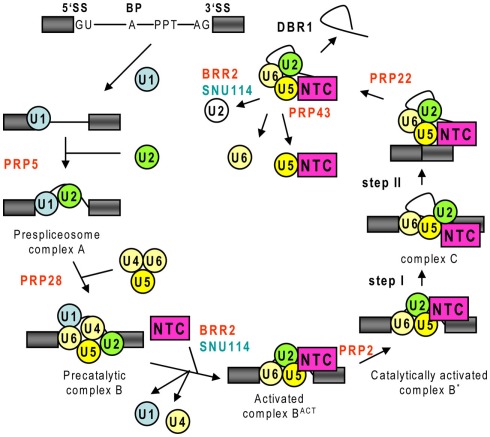
Interaction of U1 with the U2AF-recruited U2 snRNP is stabilized by the ATP-dependent DExH/D-box RNA helicase Prp5. Displacement of SF1 by the SF3b14a subunit of PPT-binding U2-SF3a/b complex is stimulated by Prp5 leading to the formation of prespliceosome complex A (Behzadnia et al., 2007; Figure 2). Prp5 also facilitates annealing the U2 snRNA with BS, which bulges out an A residue of the intron for the first transesterification reaction. Subsequent recruitment of the U4/U6.U5 tri-snRNP results in the assembly of U1/2/4/5/6 penta-snRNP in the precatalytic complex B (Deckert et al., 2006). Penta-snRNP can be purified from yeast but it is inactive and requires additional factors, in particular the NTC, to form an activated BACT complex (Stevens et al., 2002).
Figure 2.
Schematic presentation of spliceosomal assembly and catalytic cycle. The scheme is drawn according to Wahl et al. (2009) indicating the assembly phase specific regulatory roles of key ATP-dependent DExH/D-box RNA helicases (in red) and the spliceosome-activating NTC complex. 5′ and 3′ splice site (5′SS and 3′SS), branch point (BP), and polypyrimidine tract (PPT). Exons are indicated by gray boxes, while thin black lines show intron and intron lariat.
During complex B to BACT transition, interaction of U1 and U1-C with the 5′SS is interrupted by the Prp28/U5-100 helicase, which is activated by the U4/U6.U5 tri-snRNP-associated kinase SRPK2. Subsequently, unwinding the based-paired U4/U6 snRNAs by the Brr2/U5-200 helicase facilitates U6 snRNA interactions with the 5′SS and U2 snRNA. This stimulates the release of U1 and U4 snRNPs, U6 specific Lsm, and Prp24 proteins, as well as the formation of an intramolecular stem-loop (ISL) in U6, which represents the metal-binding catalytic center of the spliceosome. Brr2 is controlled by the interacting Snu114 GTPase and Prp8 U5 subunit. Removal of U2-associated SF3a/b proteins by the Prp2 helicase exposes BS in the remodeled catalytically active complex B*. During step I of splicing, the 2′-OH of BS adenosine residue attacks and forms a covalent bond with the U6 ISL-cleaved 5′SS yielding the 5′-exon and lariat intron-3′exon intermediates.
Catalytic activation of the spliceosome critically depends on its association with the NTC before or during unwinding of the U4 and U6 snRNAs. The NTC regulates the interactions of U5 and U6 with the pre-mRNA before and after step I, as well as formation of the spliceosome’s catalytic center (Chan et al., 2003; Chan and Cheng, 2005). Nucleophilic attack of 3′-OH of 5′ exon at the 3′SS in step II requires further structural rearrangements by multiple factors leading to the formation of complex C. Prp8, anchoring the 5′exon and lariat intron-3′ exon intermediates (Grainger and Beggs, 2005), the Prp16 ATPase, and the NTC subunit Isy1 are involved in monitoring completion of step I and displacing U6 from the 5′SS to liberate it for the catalytic step II. The Prp18 helicase and loop 1 of U5 snRNA juxtapose for ligation of the 5′ exon and 3′SS bound by the interacting NTC subunits Slu7 and Prp22 helicase (Smith et al., 2008). Next, Prp22 deposited downstream of the exon–exon junction disrupts the interaction of Prp8 and U5 with exon sequences, releasing the spliced mRNA from complex C. Dissociation of U2, U5, and U6 is catalyzed by the Prp43 helicase, which is encoded by three candidates genes in Arabidopsis, while Brr2 and Snu114 are thought to unwind and separate the U2 and U6 components of the post-splicing complex (Valadkhan and Jaladat, 2010).
NTC Subunit Composition
NTC is a regulatory non-snRNP complex that is essential for catalytic activation of the spliceosome (Hogg et al., 2010). When isolated under stringent conditions from in vitro assembly reactions, yeast NTC contains eight core subunits (Prp19, Cef1/Ccd5, Snt309/Spf27, Syf1, Syf2, Syf3/Clf1, Isy1, and Ntc20 listed in Figure 3; Fabrizio et al., 2009). In contrast, tandem affinity purification of NTC by its tagged Cef1/Cdc5 subunit from budding and fission yeast cell extracts indicates a conservation of at least 26 NTC-associated proteins (Ohi et al., 2002). Recent mass spectrometry analysis of NTC complexes purified by the help of TAP-tagged Prp19, Prp17, and Cwc2 subunits also shows that the eight core subunits are associated in vivo with several Sm proteins, components of the U2 and U5 snRNPs, and at least 30 non-snRNP proteins (Ren et al., 2011). Many of the latter NTC-associated factors were previously found to co-purify with spliceosome A, B, B*, or C, as well as with the RES and THO/TREX complexes. The human core NTC assembled on RNA templates in vitro and purified at high stringency shares only PRP19, CDC5L, and SPF27 with the yeast NTC. The human NTC core carries PRL1/PRLG1 and AD002/HSPC148 orthologs of NTC-associated yeast proteins Prp46 and Cwc15, respectively, and two human specific subunits CTNNBL1/NAP and HSP73 (Grote et al., 2010; Figure 3).
Figure 3.
Conserved components of NTC core and associated subunits detected in purified yeast, human, and Arabidopsis complexes. Proposed NTC core subunits are highlighted in bold, brackets indicate factors that are not present in purified NTC complexes but encoded by corresponding homologous genes in the yeast, human, and Arabidopsis genomes.
Detailed interaction studies between the NTC subunits demonstrate that the PRP19 WD-40-protein forms a homotetrameric platform, in which its U-box domain interacts with SPF27 and the C-terminus of CDC5. Due to heterogeneity of N-terminal sequences of Prp46/PRL1 orthologs, the N-terminus of yeast Prp46 interacts only with Cef1/Cdc5, while N-terminus of human PRL1 binds to PRP19, which might explain why yeast Prp46/PRL1 dissociates from the NTC core under stringent conditions (Ohi and Gould, 2002; Grote et al., 2010). Arabidopsis PRP19, CDC5, and PRL1 show analogous interactions as their human orthologs (Palma et al., 2007). Comparison of mass spectrometry data (Table S1 in Supplementary Material) indicates that nearly all components of larger NTC complexes purified by TAP-tagging from yeast are present in different yeast, Drosophila, and human spliceosome complexes isolated either by immunoprecipitation or in vitro assembly on model pre-mRNA templates (Ajuh et al., 2000; Makarov et al., 2002; Stevens et al., 2002; Zhou et al., 2002; Makarova et al., 2004; Deckert et al., 2006; Behzadnia et al., 2007; Bessonov et al., 2008, 2010; Fabrizio et al., 2009; Grillari et al., 2009; Herold et al., 2009). Remarkably, several components of the NTC core and some associated factors were also detected in the human prespliceosome complex A (Behzadnia et al., 2007). Nonetheless, semi-quantitative mass spectrometry and western blotting studies indicate that the representation of NTC components increases from complex A/B to C dramatically (Makarova et al., 2004; Agafonov et al., 2011).
Arabidopsis NTC purified recently by its tagged SPF27/MOS4 subunit (Monaghan et al., 2009) contains six subunit orthologs of the yeast NTC core [CDC5/Cef1 (MAC1), Prp19a/b (MAC3A/B), SPF27 (MOS4), SYF1 (MAC9), SYF3/CRN1c (MAC10), and ISY1 (MAC8) listed in Figure 3]. In addition, PRL1 (MAC2) is present in the Arabidopsis complex as in the human NTC core. Intriguingly, Arabidopsis NTC carries two PRL19 paralogs encoded by similarly regulated duplicated genes in Arabidopsis. This suggests that they might form a heterotetramer in analogy to yeast and human NTCs. Compared to yeast, the Arabidopsis complex lacks Syf2, although it has a plant homolog, and also Ntc20, which is a unique component of yeast NTC. Although encoded in the Arabidopsis genome, homologs of human AD002/HSPC148, CTNNBL1/NAP, and HSP73 NTC subunits are also missing from the complex. By contrast, several proteins found in association with the NTC core in yeast and humans, including PRP45/SKIP (MAC6), ECM2-1a/b (MAC5A/B), and PRP17/CDC40 (MAC12), and the complex BACT and C specific factors PRP2a (MAC15), and Aquarius (AQR/MAC7) are parts of the Arabidopsis NTC. More remarkably, U5-220/PRP8a (MAC16), U5-200-2a/BRR2 (MAC14), U5-116-1a/SNU114 (MAC11), and the PRP8-binding WD-40 factor U5-40 (MAC17), which are essential for spliceosome activation, co-purify with NTC from Arabidopsis. Composition of Arabidopsis NTC thus resembles to that of the human 35S U5 snRNA complex characterized earlier by Makarov et al. (2002). While the human 35S U5 snRNP and affinity purified yeast NTCs contain a full set of Sm proteins and other NTC-associated factors that are likely part of functional NTC in vivo, their corresponding homologs are absent from the Arabidopsis NTC.
NTC Subunit Functions
Precursor RNA processing 19 (Prp19) was first identified in yeast and found in stable association with the spliceosome after dissociation of U1 and U4 snRNPs. The yeast prp19 mutation blocks step I of splicing and results in a temperature-dependent growth defect, likely due to defective splicing of α-tubulin and Bub1 pre-mRNAs (Tarn et al., 1993; Song et al., 2011). Inactivation of mice Prp19/SNEV leads to early embryonic lethality in the blastocyst stage (Fortschegger et al., 2007). Recently, a prp19a prp19b (mac3a mac4b, modifier of SNC1) double T-DNA insertion mutation was reported to suppress innate immunity conferred by a dominant mutation of suppressor of NPR1-1 constitutive 1 (SNC1) without affecting normal plant development (Monaghan et al., 2009). However, other combinations of prp19a and prp19b mutations cannot be isolated in homozygous form, which suggests that PRP19 is also an essential gene in Arabidopsis.
Prp19 carries an N-terminal U-box/RING-finger domain, which confers E3 ubiquitin ligase activity and mediates the formation of ubiquitin K63 chain on the U4 snRNP subunit Ppr3 by enhancing its interaction with Prp8 in the U4/U6.U5 tri-snRNP. Ubiquitinated Prp3 is recognized by the U4/U6 snRNP subunit Prp24/SART3 and deubiquitinated by Usp4 facilitating Brr2/Snu114-mediated dissociation of U4 components before spliceosome activation (Song et al., 2011). While K63 ubiquitin chain formation does not target proteins for proteasomal degradation, intriguingly both yeast and human Prp19 interact in vivo with the β7 catalytic subunit of 20S proteasome suggesting that PRP19 could mediate degradation of some NTC-associated factors (Löscher et al., 2005; Sihn et al., 2007). The central coiled-coil domain of Prp19 is required for its tetramerization, which is essential for NTC and spliceosome assembly (Ohi et al., 2005; Grote et al., 2010). Prp19 tetramerization creates a central stalk of U-boxes and exposes the periferial WD-40 domains for interaction with yeast Cwc2 (i.e., a binding partner of ubiquitin conjugase Ubc3) and Prp17 (Ren et al., 2011). Mapping of NTC subunit interactions shows that the central U-box domain is required for binding Snt309/SPF27, as well as the C-terminus of Cef1/CDC5 and N-terminus of human PRL1 (Chen et al., 2002; Ohi and Gould, 2002; Grote et al., 2010). The presence of PRP19, CDC5, PRL1, and SPF27 in the Arabidopsis NTC suggests that these molecular interactions are likely conserved. Yeast prp19 and snt309 mutants accumulate free U4 snRNA, while U6 snRNA is depleted in the snt309 background. Suppression of growth defect of snt309 mutant by overexpression of U6 snRNA indicates that NTC also controls recycling of spliceosomal snRNPs (Chen et al., 2006).
Human Snt309 is named Breast Cancer Amplified Sequence 2 (BCAS2) and involved as transcriptional co-activator of the estrogen receptor in the regulation of p53-dependent induction of apoptosis (Kuo et al., 2009). Mutation of Arabidopsis Snt309/BCAS2 homolog MOS4 suppresses constitutive activation of innate immunity pathway in the snc1 mutant similarly to mutations of other plant NTC subunits (Palma et al., 2007).
Snt309/BCAS2 is required for stable interaction of Prp19 with CDC5, a member of the Myb transcription factor family. CDC5 shows specific DNA-binding in Arabidopsis, while its mutations leads to accumulation of retained introns in partially spliced pre-mRNAs and G2 cell cycle arrest in yeast (Hirayama and Shinozaki, 1996; Burns et al., 1999). Cell cycle arrest of the yeast cdc5 mutant is suppressed by removal of the intron from the α-tubulin (Tub1) gene (Burns et al., 2002). Cell cycle dependent phosphorylation of CDC5 appears to be an important modulator of NTC-mediated activation of the spliceosome (Gräub et al., 2008). Human CDC5 interacts with NIPP1 (nuclear inhibitor of protein phosphatase 1 absent from Arabidopsis), which recognizes the SF3b155 subunit of U2-SF3a/b complex that becomes hyperphosphorylated during the first splicing reaction. Interaction of CDC5 with NIPP1 probably targets a PP1 phosphatase to the SF1a/b complex leading to its destabilization, which is required for the second step of splicing (Tanuma et al., 2008). Furthermore, human CDC5 directly interacts with hLodestar/HuF2, which is an SF2-like ATP-dependent helicase subunit of SWI/SNF chromatin remodeling complex implicated in transcription termination (Leonard et al., 2003).
C-terminus of CDC5 binds the C-terminal WD-40 domain of Prp46/PRL1 in yeast and human NTCs (Ajuh et al., 2001; Ohi and Gould, 2002). PRL1 encoded by the Pleiotropic Regulatory Locus 1 was first identified in Arabidopsis by a T-DNA insertion mutation causing highly pleiotropic defects characterized by altered regulation of root and leaf development, flowering time, sugar, cold, ethylene, cytokinin, and auxin responses (Németh et al., 1998). Arabidopsis has a PRL1 paralog, PRL2, which is transcribed at low levels in vegetative tissues but shows much higher expression compared to PRL1 during embryogenesis and seed development. Low PRL2 expression in the prl1 mutant thus appears to be sufficient for maintaining plant viability but leads to highly pleiotropic defects. Nuclear import of PRL1 is aided by its interaction with a specific member of the α-importin family, ATHKAP2/IMPA-3 (Németh et al., 1998). Inactivation of IMPA-3 in the Arabidopsis mos6 mutant suppresses constitutive activation of innate immunity by the snc1 mutation similarly to the mos4 (snt309), cdc5 (mac1), and prl1 (mac2) mutations (Palma et al., 2005, 2007). In comparison, human CDC5 is imported into the nucleus by the CTNNBL1 (catenin-β-like 1) armadillo repeat protein, which remains in CDC5L-bound form in the human core NTC and mediates interaction with the PRP31 subunit of U4/U6 snRNP (Grote et al., 2010). Temperature sensitive mutations of fission yeast Cwf1/CDC5 and Prp5/PRL1 show strong negative genetic interactions (i.e., synthetic lethality at permissive temperature) suggesting overlapping functions (McDonald et al., 1999). Removal of the C-terminus of CDC5 results in dissociation of PRL1 from the yeast NTC (Ohi and Gould, 2002) but it is yet unclear whether this leads to overall destabilization of the complex. Virus-induced gene silencing of Arabidopsis CDC5 is reported to activate early senescence causing accelerated cell death independently of salicylic acid (SA) signaling (Lin et al., 2007a). According to Palma et al. (2007), however, the cdc5, prl1, and mos4 mutations stimulate SA production but show npr1-independent activation of the PR genes. RNAi-silencing of CDC5 was also suggested to result in a G2/M cell cycle defect by affecting Shoot Meristemless (STM) and WUS transcription and meristem development. Furthermore, the GABI_278B09 cdc5 T-DNA insertion mutation was found to cause embryo lethality by Lin et al. (2007b). The latter phenotypic trait is however probably caused by an unrelated mutation, since Palma et al. (2007) found that removal of C-terminal PRL1-binding domain from the CDC5 coding sequence by the same T-DNA insertion mutation results in viable plants showing a partial loss of function phenotype similar to that of the prl1 mutant.
Arabidopsis PRL1 interacts with and inhibits in vitro the activity of Snf1-related protein kinase AKIN10 (Bhalerao et al., 1999), a functional homolog of yeast Snf1 that represents a regulatory partner of GCN5 histone acetylase in the yeast RNAPII SAGA co-activator complex (Lo et al., 2001; Liu et al., 2010). Whereas association of SnRK1 with SAGA in plants awaits further confirmation, SnRK1 AKIN10 was found to interact in vivo with the common SKP1 subunits of SCF (Skp1–cullin1–F-box) E3 ubiquitin ligases by targeting them to the α4/PAD1 subunit of 20S proteasome catalytic cylinder (Farrás et al., 2001). Arabidopsis α4/PAD1 is an ortholog of human proteasome subunit α7, which is the closest neighbor of β7 subunit targeted by Prp19 (Löscher et al., 2005). Recently, PRL1 was found to function as substrate receptor subunit of a Cul4–DDB1 ubiquitin ligase, and the prl1 mutation was reported to stabilize SnRK1 AKIN10. It is remarkable that in addition to PRL1, WD-40 repeats of spliceosome-associated THOC6 and FY subunits of the THO/TREX mRNA export, and polyadenylation/cleavage complexes also contain DDB1-binding DWD motives are detected in association with Cul4 and DDB1 in Arabidopsis (Lee et al., 2008). Knockout of mouse PRLG1/PRL1 results in early embryonic lethality as inactivation of mouse PRP19/Pso4 (Löscher et al., 2005; Kleinridders et al., 2009), whereas no homozygous prl1, prl2 and cdc5 null mutants can be obtained in Arabidopsis. These results are consistent with the observations of Ohi et al. (2002) indicating that Cef1/Cdc5, Prp46/PRL1, and Prp19 are essential genes in yeast. In contrast, the yeast snt309 and Arabidopsis mos4 mutations do no cause lethality (Palma et al., 2007). Ecm2/Slt11 is also not essential in yeast but its mutation causes cold sensitivity (i.e., as the prl1 mutation in Arabidopsis; Németh et al., 1998) and impaired splicing (Xu and Friesen, 2001). However, a combination of mutations of corresponding Arabidopsis homologs ECM2A and B (MAC5A/B) is lethal (Monaghan et al., 2010). Ecm2, as the homologous human RBM22 zinc-finger RNA-binding protein is controlled by sumoylation and involved in U2/U6 helix II formation required for spliceosome activation. Ecm2/RBM22 interacts with the step II splicing factors Slu7, Prp16, and Prp17 and its inactivation in zebrafish leads to early embryo lethality as in Arabidopsis (He et al., 2009).
From the remaining conserved NTC components, Syf1/Ntc90/XAB2/MAC9 is essential in yeast and interacts with most NTC subunits, except Prp19, and SPF27/Ntc25. Syf1 is required for recruitment of the coiled-coil factor Yju2/Cwc16/CCDC130, which is transiently associated with NTC before step I of splicing together with Cwc22, and facilitates the first catalytic reaction after Prp2-mediated remodeling of the spliceosome (Chang et al., 2009). Cwc22 is essential for Prp2-mediated release of U2-SF3a/b complex before step I of splicing (Yeh et al., 2011). Prp45/SKIP/MAC6 is essential in budding and fission yeast, shows genetic interactions with step II splicing factors and is required for recruitment of Prp22 helicase mediating spliceosome dissociation (Gahura et al., 2009). In addition, Prp45 interacts in fission yeast with the splice-site selecting U2AF35 factor suggesting an early role in spliceosome assembly (Ambrozková et al., 2001). Transcription of Arabidopsis SKIP/Prp45 is stimulated by ABA, salt, and osmotic stress and its overexpression confers salt and osmotic stress tolerance (Lim et al., 2010).
The Syf3/CRN1/MAC10 tetratricopeptide repeat (TRP) protein is one of the earliest acting NTC proteins, which is essential for loading the U4/U6.U5 tri-snRNP during complex A to B transition. In Drosophila, mutation of CRN1/Syf3 (crooked neck) causes early embryonic lethality (Chung et al., 2002). Isy1/MAC8 interacts with the Prp16 ATPase required for proper recognition of 3′SS and implicated in remodeling of U6. Isy1 also interacts with Cwc21/SRm300, as well a with the trimethylguanosine synthase capping enzyme during early stage of spliceosome assembly (Villa and Guthrie, 2005; Hausmann et al., 2008; Khanna et al., 2009). The PRL1-binding non-essential second step factor Prp17/CDC40/MAC17 is a WD-40-protein, which functions together with Prp16, Prp8, and the SR protein kinase Sky1 in the recognition of 3′SS. Prp17 is essential for splicing of introns longer than 200 nucleotides and shows co-immunoprecipitation with lariat intron–exon intermediates (Dagher and Fu, 2001; Sapra et al., 2008). Finally, from the conserved NTC-associated proteins Aquarius/AQR/MAC7 is found to bind intron sequences in the vicinity of BP and it is identified by the Arabidopsis by the embryo defective 2765 mutation (Hirose et al., 2006). The key step II RNA helicase Prp2 is represented by three potential homologs in Arabidopsis, from which the eps3 mutation of PRP2/MAC5 confers enhanced gene silencing, probably due to destabilization of aberrantly spliced pre-mRNAs and subsequent generation of derived siRNAs (Herr et al., 2006). The maternal embryo effect 29 (mee29) mutation in its paralog Prp2b results in deficient gametophyte/embryo development (Pagnussat et al., 2005).
Taken together, the available genetic data indicate that mutations of essential Arabidopsis NTC components result in lethality. In those cases, where one of the duplicated genes is preferentially expressed during embryo and seed development whereas the other shows higher expression in vegetative tissues, the corresponding single gene mutations yield embryo lethality and pleiotropic effects in seedlings, respectively. Although not all NTC mutants were characterized so far for their pleiotropic defects in such a detail as prl1, all studied NTC deficiencies appear to function as mos suppressors of snc1-induced innate immunity, which is coupled to the induction of cell death in pathogen infected tissues. Recently, the prl1 mutation was also demonstrated to suppress the induction of cell death by singlet oxygen generated through the accumulation of free protochlorophyllide in the fluorescent in blue light (flu) mutant (Baruah et al., 2009). Furthermore, the Arabidopsis mos2 mutation, which has not been connected so far to NTC, is located in a gene encoding a spliceosome complex C specific factor Spp2/GPKOW, which is a binding partner of yeast Prp2 and human protein kinase Cβ2 that shows interaction with PRL1 in human cells (Roy et al., 1995; Németh et al., 1998; Zhang et al., 2005a; Aksaas et al., 2011). Another candidate MOS factor is the complex C specific protein RUVBL1, a chromatin remodeling ATPase inhibitor of apoptosis (Taniue et al., 2011). The Arabidopsis RUVBL1 homolog, TIP49A/RIN1, is an interacting partner of RPM1 receptor required for disease resistance against Pseudomonas syringae pv. maculicola (Holt et al., 2002). It is also apparent that some pathogen produced effector molecules could directly modulate the activity of the spliceosome. Thus, a natural antitumor compound isolated from Pseudomonas functions as spliceostatin A that specifically targets the U2 snRNP SF3b complex and thereby inhibits splicing and nuclear retention of unspliced pre-mRNAs (Kaida et al., 2007).
NTC Provides a Link between Splicing and Regulation of DNA Repair, Recombination, and Cell Death
In addition to controlling spliceosome assembly and activation, NTC is emerging to play a central role in the regulation of DNA damage responses, which is consistent with the effects of NTC subunit mutations observed in Arabidopsis. In yeast, Prp19 was identified as Pso4, mutation of which causes hypersensitivity to the DNA cross-linking agent psoralen, as well as to a broad range of other DNA damaging agents (see for review Legerski, 2009). Expression of human PRP19/Pso4/SNEV is induced by DNA damage and its down-regulation by RNAi results in the accumulation of double-stranded DNA breaks and apoptosis (Mahajan and Mitchell, 2003). Using a DNA interstand cross-link repair assay, Pso4 was purified together with the NTC subunits CDC5, PRL1, and SPF27, from which CDC5 directly interacts with the Werner syndrome DNA helicase (WRN) in complex with the replication protein A (RPA, Zhang et al., 2005b). RPA is essential for stabilization of single-stranded gap structure arising from dual incision of damaged DNA strand, while WRN assists DNA repair replication.
DNA damage causing an arrest of transcription elongation and DNA replication leads to activation of the DNA repair checkpoint, which prevents cells entering or leaving the S-phase. Mutations inactivating the regulators of this checkpoint lead to persistent activation of p53 tumor suppressor stimulating cell death and apoptosis in mammals. Activation of transcription-independent global genome and transcription-coupled nucleotide excision repair (GG-NER and TC-NER) pathways is mediated by different sensory but common executing components (Lagerwerf et al., 2011). In GG-NER, various DNA lesions are recognized by the UV-DDB (Cul4–DDB1–DDB2) and XPC–Rad23B complexes. In TC-NER, the hyperphosphorylated RNAPII stalled by the DNA damage is recognized the Cockayne Syndrome CSB chromatin remodeling ATPase, which subsequently recruits the Cul4–DDB1–CSA E3 ubiquitin ligase and GCN5/p300 histone acetylase complexes. Remarkably, DDB2 and CSA/ERCC8 are WD-40 proteins that in analogy to PRL1 are recruited to DDB1. This suggests that PRL1 in the Pso4 complex might perform a similar substrate receptor function although its target is unknown so far. Cul4–DDB1–DDB2 ubiquitinates histones H3 and H4 contributing to opening the damaged chromatin site. On the other hand, the CSA WD-40 protein is a substrate receptor of CSB, which is being released and degraded before subsequent loading of the TFIIH RNAPII transcription initiation/DNA repair complex to the damaged site. In the case of TC-NER, loading of TFIIH requires the recognition of CSA/CSB by the XAB2/Syf1 NTC subunit, as well as interaction of the XAB2 complex with the transcription elongation factor TFIIS and high mobility group chromatin factor HMGN1 (Fousteri et al., 2006).
The human XAB2 complex contains Aquarius, PRP19, CCDC16/ZNF830 (a component of spliceosome complex B, Table S1 in Supplementary Material), ISY1 and the NTC-associated PPIE peptidyl-prolyl cis–trans isomerase (Kuraoka et al., 2008). Differences between subunit composition of the Pso4 and XAB2 complexes is probably explained by the observation that in response to DNA damage PRP19 undergoes self-ubiquitination, which stimulates the release of CDC5 and PRL1 (Lu and Legerski, 2007). Whether this event leads to switching the role of NTC between splicing and DNA repair remains however to be determined.
Mutations inactivating essential components of the DNA repair pathways lead to prolonged presence of the ATR checkpoint kinase at the damaged DNA sites. Chronic activation of ATR, as well as inhibition of transcription elongation, causes a dramatic increase in phosphorylated forms of p53 and histone H2AX, and ubiquitination of H2A (Lagerwerf et al., 2011). Although not detected in either Pso4 or XAB2 complex, the NTC subunit Syf2 is found in association with the MCM3 and PCNA DNA replication factors and is required for activation of ATR (Chu et al., 2006). The yeast NTC subunit Cfl1/CRN1 is similarly found in a complex with the DNA replication factor Orc2 and its mutation results in delayed entry into the S-phase, indicating a direct role in DNA replication (Zhu et al., 2002). Human CDC5 interacts with and phosphorylated by ATR, and is required for the activation of components of downstream S-phase checkpoint pathway blocking cell cycle progression (Zhang et al., 2009). Knockouts of mouse and zebrafish PRLG1/PRL1 result in nuclear to cytoplasmic translocation of CDC5, stimulation of p53 and histone H2AX phosphorylation, and induction of apoptosis (Kleinridders et al., 2009). This indicates that, despite their lack in the XAB2 complex, both PRL1 and CDC5 are essential for activation of the DNA repair checkpoint and their absence results in stimulation of default cell death pathway in mammals.
The human NTC subunit BCAS2/SPF27 (MOS4) directly interacts with and inhibits the activity of p53. As the prl1 mutation, inactivation of BCAS2 stimulates nuclear retention and phosphorylation of p53 leading to apoptosis, while in p53 mutant cells permits normal G2/M cell cycle arrest (Kuo et al., 2009). Another NTC subunit, SKIP/Prp45 is specifically required for proper splicing of the cell cycle arrest factor p21Cip1. Thus, inactivation of SKIP leads to deregulation of the cell cycle and activation of p53 even in the absence of DNA damage (Chen et al., 2011). Remarkably, PRP19/SNEV/PSO4 overexpression confers tolerance to reactive oxygen species and DNA damaging agents prolonging the life span of human cells (Voglauer et al., 2006). Whether this overexpression effect is due to accumulation of free PRP19, which is not incorporated into NTC, remains a question to answer. In any case, in Arabidopsis that lacks a functional p53 homolog the effects of NTC mutations are just opposite compared to yeast and mammals, as instead of stimulating apoptosis they function as suppressors of induced cell death. It is thus important to clarify how these mutations affect the DNA repair pathways and how do they modulate splicing of downstream effectors of signaling pathways that are activated by the snc1 and flu mutations of pathogen and oxidative stress response pathways.
Role of NTC in Co-Transcriptional Spliceosome Assembly
While splicing defects caused by the Arabidopsis NTC mutations are poorly studied so far, it was observed early on that the Arabidopsis prl1 mutation results in both up and down-regulation of transcription of many stress regulated genes, which is also observable in nuclear run-on transcription assays (Németh et al., 1998). The conclusion that NTC plays a direct role in the regulation of RNAPII transcription and simultaneous co-transcriptional spliceosome assembly is supported now by an overwhelming amount of data (see for reviews e.g., Perales and Bentley, 2009; Muñoz et al., 2010; Oesterreich et al., 2011).
From the core NTC subunits, BCAS2/Snt309, and SKIP/Prp45 are directly recruited to promoters of RNAPII transcribed genes. BCAS2/Snt309 is a transcription co-activator of human estrogen receptor, while SKIP/Prp45 is a common co-activator of several nuclear receptors and a binding partner of multiple histone deacetylases and nuclear co-repressors. SKIP plays a pivotal role in the regulation of transcription in response to Notch and Wnt/β-catenin signaling, and acts as an inhibitor of NADH+-dependent SIRT deacetylase by controlling senescence and aging in conjunction with the retinoic acid receptor (Zhou and Hayward, 2001; Kang et al., 2010; Wang et al., 2010).
Other NTC components are recruited to RNAPII by their contacts with initiation and elongation factors, and spliceosome components. The carboxy-terminal Y1S2P3T4S5P6S7 heptapeptide repeat domain of RNAPII largest subunit (RNAPII CTD) serves as platform for binding and assembly of these regulatory factors (Buratowski, 2009). During transcription initiation, the RNAPII CTD undergoes S5/S7 phosphorylation by the TFIIH-associated Kin28/CDK7 kinases (Akhtar et al., 2009). Phosphorylation of the S5 CTD-residue mediates the recruitment of the capping enzyme, which interacts with the NTC component Isy1 (Hausmann et al., 2008). Subsequent formation of the cap-binding complex CBC and interaction of the U1 snRNP subunit Prp40 with the phosphorylated CTD mediates RNAPII loading of the U1 snRNP through the CBC-binding of U1 subunit Luc7. At the same time, PRP40 recruits the NTC subunit Clf1/Syf3 (Fortes et al., 1999). Arabidopsis PRP40 homologs show similar recognition of RNAPII CTD (Kang et al., 2009) and mutations of the CBC subunits result in the accumulation of retained introns in pre-mRNAs (Laubinger et al., 2008). As U1 snRNP interaction can also be observed with the RNAPII on intronless pre-mRNAs, the U1 snRNP-5′SS commitment complex is only stabilized when the branch site is bound by the U2AF complex. Interaction of the NTC subunit Syf3/Clf1 with U2AF65 and recognition of BP-bound factor SF1 by Prp40 forms a platform for further loading of the U2 snRNP (Chung et al., 2011). In addition, U2AF65 and its interacting SR protein partner SC35 directly bind to RNAPII CTD, and thus play a role in co-transcriptional assembly of the spliceosome (Spiluttini et al., 2010).
Recently, U2AF65 was reported to co-purify with the NTC components PRP19, CDC5, PRL1, and SPF27 and shown to facilitate RNAPII CTD-dependent NTC-mediated activation of splicing (David et al., 2011). The U2 subunit SF3b130 and Cul4–DDB1 are associated with GCN5/p300 histone acetylases of yeast and human SAGA/STAGA RNAPII co-activator complexes. Similarly to activation of NTC-modulated DNA repair, histone acetylation is essential for stabilization of the U2 snRNP in the prespliceosome (Martinez et al., 2001; Gunderson and Johnson, 2009).
Phosphoserine-5 mark of the RNAPII CTD also provides a signal for recruitment of SET1-type histone methyltransferases (Buratowski, 2009). H3K4-trimethyl histone marks deposited by Set1 are recognized by the human SAGA-associated chromodomain protein CHD1, which specifically interacts with the SF3a subcomplex of U2 snRNP (Sims et al., 2007). The U2 snRNP SF3b complex recruits the NTC-associated factor Bud31 and the Ist3 subunit of mRNA retention RES complex, which remain then associated with the spliceosome (Wang et al., 2005). The U2-associated human factor tat-SF1 together with the cap-binding complex plays a role in recruitment of the CDK9/P-TEFb kinase, which stimulates transcription elongation by phosphorylating the S2 residues of RNAPII CTD.
P-TEFb directly interacts with human SKIP and Menin H3K4 methyltransferase (Brès et al., 2009), as well as with exonic splicing enhancer- and silencer-binding SR-proteins, such as SF2/ASF and SRp20. In complex with the HP1 adaptor protein, SRp20 is involved in the recognition of histone H3K9 trimethylation and regulation of alternative splicing (for review see Lenasi and Barboric, 2010). In addition, P-TEFb-mediated phosphorylation of RNAPII CTD S2 residue results in the recruitment of Set2-type histone methyltransferases that deposit H3K36-trimethyl histone marks preferentially on exon sequences. The H3K36me3 chromatin mark is recognized by the adaptor protein MRG15, which in complex with the PPT-binding protein PTB acts as important regulator of alternative splicing (see for review Luco et al., 2011). Interaction of NTC with RNAPII also critically affects transcriptional elongation by recruiting components of the THO/TREX complex required for the formation and nuclear export of messenger ribonucleoproteins (mRNPs, Chanarat et al., 2011).
Conclusion and Perspectives
In this review, we took a glimpse at molecular mechanisms underlying the emerging central regulatory functions of NTC in coordination of spliceosome assembly/activation with transcription, DNA repair/replication, and stress responses related to aging and cell death. In particular, we wished to highlight recent advances in understanding the regulatory functions of Arabidopsis NTC and spliceosome components embedded in a frame of current knowledge derived from similar yeast, Drosophila, and human studies. In the absence of corresponding Arabidopsis data on NTC connections to the regulation of mitotic and meiotic recombination, chromosome segregation, chromatin remodeling, temperature sensing, Polycomb and small RNA-mediated gene silencing, nuclear mRNA export, nonsense mediated decay, and several other essential processes, these subjects remained to be covered by a next timely overview. Compilation of available data on yeast, Drosophila, and human NTC and spliceosome-associated proteins and their conserved plant homologs in the Supplement intends to assist further research to fill this gap.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00009/abstract
Compilation of mass spectrometry data on protein composition of purified yeast, Drosophila, human, and Arabidopsis spliceosome and NTC complexes. Phosphorylated human proteins reported by Agafonov et al. (2011) are highlighted in red, Arabidopsis factors with indentified regulatory functions are indicated in bold. Core subunits of yeast, human, and Arabidopsis NTC complexes are shaded in gray. References describing the purification of various spliceosomal and NTC complexes are given at the end of the Table corresponding to numbers in bracket indicated in the heading. Brackets within the table mark unique fission yeast factors, which have either Drosophila or human or Arabidopsis homologs. For each factor, the corresponding NCBI gene accession number is depicted and in all cases frequently used alternative designations are listed.
Acknowledgments
We apologize for those many colleagues whose work we could not cite due to space limitation. Our studies related to this review were funded by the DFG grant KO 1438/12-1 within the Arabidopsis Functional Genomics Network (AFGN).
References
- Agafonov D. E., Deckert J., Wolf E., Odenwälder P., Bessonov S., Will C. L., Urlaub H., Lührmann R. (2011). Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol. Cell. Biol. 31, 2667–2682 10.1128/MCB.05266-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh P., Kuster B., Panov K., Zomerdijk J. C., Mann M., Lamond A. I. (2000). Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19, 6569–6581 10.1093/emboj/19.23.6569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh P., Sleeman J., Chusainow J., Lamond A. I. (2001). A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 276, 42370–42381 10.1074/jbc.M105453200 [DOI] [PubMed] [Google Scholar]
- Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009). TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34, 387–393 10.1016/j.molcel.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksaas A. K., Larsen A. C., Rogne M., Rosendal K., Kvissel A. K., Skålhegg B. S. (2011). G-patch domain and KOW motifs-containing protein, GPKOW; a nuclear RNA-binding protein regulated by protein kinase A. J. Mol. Signal. 6, 10. 10.1186/1750-2187-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrozková M., Puta F., Fuková I., Skruzný M., Brábek J., Folk P. (2001). The fission yeast ortholog of the coregulator SKIP interacts with the small subunit of U2AF. Biochem. Biophys. Res. Commun. 284, 1148–1154 10.1006/bbrc.2001.5108 [DOI] [PubMed] [Google Scholar]
- Barta A., Kalyna M., Reddy A. S. (2010). Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22, 2926–2929 10.1105/tpc.110.078352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah A., Simková K., Hincha D. K., Apel K., Laloi C. (2009). Modulation of O-mediated retrograde signaling by the PLEIOTROPIC RESPONSE LOCUS 1 (PRL1) protein, a central integrator of stress and energy signaling. Plant J. 60, 22–32 10.1111/j.1365-313X.2009.03935.x [DOI] [PubMed] [Google Scholar]
- Behzadnia N., Golas M. M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C. L., Urlaub H., Stark H., Lührmann R. (2007). Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26, 1737–1748 10.1038/sj.emboj.7601631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Krasauskas A., Golas M. M., Sander B., Will C. L., Urlaub H., Stark H., Lührmann R. (2010). Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16, 2384–2403 10.1261/rna.2456210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008). Isolation of an active step I spliceosome and composition of its RNP core. Nature 452, 846–850 10.1038/nature06842 [DOI] [PubMed] [Google Scholar]
- Bhalerao R. P., Salchert K., Bakó L., Ökrész L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. U.S.A. 96, 5322–5327 10.1073/pnas.96.9.5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V., Yoshida T., Pickle L., Jones K. A. (2009). SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell 36, 75–87 10.1016/j.molcel.2009.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Simpson C. G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 77–95 10.1146/annurev.arplant.49.1.77 [DOI] [PubMed] [Google Scholar]
- Buratowski S. (2009). Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. G., Ohi R., Krainer A. R., Gould K. L. (1999). Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 96, 13789–13794 10.1073/pnas.96.25.14499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C. G., Ohi R., Mehta S., O’Toole E. T., Winey M., Clark T. A., Sugnet C. W., Ares M., Jr., Gould K. L. (2002). Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 801–815 10.1128/MCB.22.3.801-815.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S. A., Topping J. F., Lindsey K. (2009). MERISTEM-DEFECTIVE, an RS domain protein, is required for the correct meristem patterning and function in Arabidopsis. Plant J. 57, 857–869 10.1111/j.1365-313X.2008.03738.x [DOI] [PubMed] [Google Scholar]
- Chan S. P., Cheng S. C. (2005). The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280, 31190–31199 10.1074/jbc.M502767200 [DOI] [PubMed] [Google Scholar]
- Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. (2003). The Prp19p-associated complex in spliceosome activation. Science 302, 279–282 10.1126/science.1086602 [DOI] [PubMed] [Google Scholar]
- Chanarat S., Seizl M., Sträber K. (2011). The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 25, 1147–1158 10.1101/gad.623411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. J., Chen H. C., Cheng S. C. (2009). Ntc90 is required for recruiting first step factor Yju2 but not for spliceosome activation. RNA 15, 1729–1739 10.1261/rna.1625309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouki A. S., Salz H. K. (2006). Drosophila SPF45: a bifunctional protein with roles in both splicing and DNA repair. PLoS Genet. 2, e178. 10.1371/journal.pgen.0020178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Kao D. I., Chan S. P., Kao T. C., Lin J. Y., Cheng S. C. (2006). Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA 12, 765–774 10.1261/rna.2292106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Yu W. C., Tsao T. Y., Wang L. Y., Chen H. R., Lin J. Y., Tsai W. Y., Cheng S. C. (2002). Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30, 1029–1037 10.1093/nar/30.2.592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang L., Jones K. A. (2011). SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 25, 701–716 10.1101/gad.2002611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P. C., Yang Y. C., Lu Y. T., Chen H. T., Yu L. C., Chang M. S. (2006). Silencing of p29 affects DNA damage responses with UV irradiation. Cancer Res. 66, 8484–8491 10.1158/0008-5472.CAN-05-3229 [DOI] [PubMed] [Google Scholar]
- Chung S., McLean M. R., Rymond B. C. (2011). Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5, 1042–1054 10.1017/S1355838299990635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Zhou Z., Huddleston K. A., Harrison D. A., Reed R., Coleman T. A., Rymond B. C. (2002). Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim. Biophys. Acta 1576, 287–297 [DOI] [PubMed] [Google Scholar]
- Dagher S. F., Fu X. D. (2001). Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA 7, 1284–1297 10.1017/S1355838201016077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. J., Boyne A. R., Millhouse S. R., Manley J. L. (2011). The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 25, 972–983 10.1101/gad.2038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543 10.1128/MCB.00582-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 107, 19114–19119 10.1073/pnas.1007883107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Lührmann R. (2009). The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36, 593–608 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- Farrás R., Ferrando A., Jásik J., Kleinow T., Ökrész L., Tiburcio A., Salchert K., del Pozo C., Schell J., Koncz C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756 10.1093/emboj/20.11.2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr R. (2008). Regulation of splicing by protein phosphorylation. Curr. Top. Microbiol. Immunol. 326, 119–138 10.1007/978-3-540-76776-3_7 [DOI] [PubMed] [Google Scholar]
- Fortes P., Bilbao-Cortés D., Fornerod M., Rigaut G., Raymond W., Séraphin B., Mattaj I. W. (1999). Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 13, 2425–2438 10.1101/gad.13.18.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortschegger K., Wagner B., Voglauer R., Katinger H., Sibilia M., Grillari J. (2007). Early embryonic lethality of mice lacking the essential protein SNEV. Mol. Cell. Biol. 27, 3123–3130 10.1128/MCB.01188-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M., Vermeulen W., van Zeeland A. A., Mullenders L. H. (2006). Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23, 471–482 10.1016/j.molcel.2006.06.029 [DOI] [PubMed] [Google Scholar]
- Gahura O., Abrhámová K., Skruzný M., Valentová A., Munzarová V., Folk P., Puta F. (2009). Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J. Cell. Biochem. 106, 139–151 10.1002/jcb.21989 [DOI] [PubMed] [Google Scholar]
- Gehring N. H., Kunz J. B., Neu-Yilik G., Breit S., Viegas M. H., Hentze M. W., Kulozik A. E. (2005). Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65–75 10.1016/j.molcel.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Grainger R. J., Beggs J. D. (2005). Prp8 protein: at the heart of the spliceosome. RNA 11, 533–557 10.1261/rna.2220705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräub R., Lancero H., Pedersen A., Chu M., Padmanabhan K., Xu X. Q., Spitz P., Chalkley R., Burlingame A. L., Stokoe D., Bernstein H. S. (2008). Cell cycle-dependent phosphorylation of human CDC5 regulates RNA processing. Cell Cycle 7, 1795–1803 10.4161/cc.7.12.6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J., Löscher M., Denegri M., Lee K., Fortschegger K., Eisenhaber F., Ajuh P., Lamond A. I., Katinger H., Grillari-Voglauer R. (2009). Blom7alpha is a novel heterogeneous nuclear ribonucleoprotein K homology domain protein involved in pre-mRNA splicing that interacts with SNEVPrp19-Pso4. J. Biol. Chem. 284, 29193–29204 10.1074/jbc.M109.036632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M., Wolf E., Will C. L., Lemm I., Agafonov D. E., Schomburg A., Fischle W., Urlaub H., Lührmann R. (2010). Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell. Biol. 30, 2105–2119 10.1128/MCB.01505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson F. Q., Johnson T. L. (2009). Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 5, e1000682. 10.1371/journal.pgen.1000682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Xiong J., Wang D., Fu X. D. (2011). Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 21, 336–343 10.1016/j.tcb.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S., Zheng S., Costanzo M., Brost R. L., Garcin D., Boone C., Shuman S., Schwer B. (2008). Genetic and biochemical analysis of yeast and human cap trimethylguanosine synthase: functional overlap of 2,2,7-trimethylguanosine caps, small nuclear ribonucleoprotein components, pre-mRNA splicing factors, and RNA decay pathways. J. Biol. Chem. 283, 31706–31718 10.1074/jbc.M806127200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wang C. T., Gou L. T. (2009). RNA-binding motif protein RBM22 is required for normal development of zebrafish embryos. Genet. Mol. Res. 8, 1466–1473 10.4238/vol8-4gmr683 [DOI] [PubMed] [Google Scholar]
- Herold N., Will C. L., Wolf E., Kastner B., Urlaub H., Lührmann R. (2009). Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol. Cell. Biol. 29, 281–301 10.1128/MCB.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Molnàr A., Jones A., Baulcombe D. C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 14994–15001 10.1073/pnas.0606536103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (1996). A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 13371–13376 10.1073/pnas.93.23.13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Ideue T., Nagai M., Hagiwara M., Shu M. D., Steitz J. A. (2006) A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol. Cell 23, 673–684 10.1016/j.molcel.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Hogg R., McGrail J. C., O’Keefe R. T. (2010). The function of the nineteen complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem. Soc. Trans. 38, 1110–1115 10.1042/BST0381110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt B. F., III, Boyes D. C., Ellerström M., Siefers N., Wiig A., Kauffman S., Grant M. R., Dangl J. L. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817 10.1016/S1534-5807(02)00174-0 [DOI] [PubMed] [Google Scholar]
- Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H., Tani T., Horinouchi S., Yoshida M. (2007). Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 10.1038/nchembio.2007.18 [DOI] [PubMed] [Google Scholar]
- Kang C. H., Feng Y., Vikram M., Jeong I. S., Lee J. R., Bahk J. D., Yun D. J., Lee S. Y., Koiwa H. (2009). Arabidopsis thaliana PRP40s are RNA polymerase II C-terminal domain-associating proteins. Arch. Biochem. Biophys. 484, 30–38 10.1016/j.abb.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Kang M. R., Lee S. W., Um E., Kang H. T., Hwang E. S., Kim E. J., Um S. J. (2010). Reciprocal roles of SIRT1 and SKIP in the regulation of RAR activity: implication in the retinoic acid-induced neuronal differentiation of P19 cells. Nucleic Acids Res. 38, 822–831 10.1093/nar/gkp805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M., Van Bakel H., Tang X., Calarco J. A., Babak T., Guo G., Emili A., Greenblatt J. F., Hughes T. R., Krogan N. J., Blencowe B. J. (2009). A systematic characterization of Cwc21, the yeast ortholog of the human spliceosomal protein SRm300. RNA 15, 2174–2185 10.1261/rna.1790509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinridders A., Pogoda H. M., Irlenbusch S., Smyth N., Koncz C., Hammerschmidt M., Brüning J. C. (2009). PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol. Cell. Biol. 29, 3173–3185 10.1128/MCB.01807-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P. C., Tsao Y. P., Chang H. W., Chen P. H., Huang C. W., Lin S. T., Weng Y. T., Tsai T. C., Shieh S. Y., Chen S. L. (2009). Breast cancer amplified sequence 2, a novel negative regulator of the p53 tumor suppressor. Cancer Res. 69, 8877–8885 10.1158/0008-5472.CAN-09-2023 [DOI] [PubMed] [Google Scholar]
- Kuraoka I., Ito S., Wada T., Hayashida M., Lee L., Saijo M., Nakatsu Y., Matsumoto M., Matsunaga T., Handa H., Qin J., Nakatani Y., Tanaka K. (2008). Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 283, 940–950 10.1074/jbc.M706647200 [DOI] [PubMed] [Google Scholar]
- Lagerwerf S., Vrouwe M. G., Overmeer R. M., Fousteri M. I., Mullenders L. H. (2011). DNA damage response and transcription. DNA Repair (Amst.) 10, 743–750 10.1016/j.dnarep.2011.04.024 [DOI] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J. U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 8795–8800 10.1073/pnas.0802493105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Kapoor A., Zhu J., Zhu J. K. (2006). Stabilized1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18, 1736–1749 10.1105/tpc.106.043042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Terzaghi W., Gusmaroli G., Charron J. B., Yoon H. J., Chen H., He Y. J., Xiong Y., Deng X. W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20, 152–167 10.1105/tpc.108.060541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J. (2009). The Pso4 complex splices into the DNA damage response. Cell Cycle 8, 3448–3449 10.4161/cc.8.21.9760 [DOI] [PubMed] [Google Scholar]
- Lenasi T., Barboric M. (2010). P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol. 7, 145–150 10.4161/rna.7.2.11057 [DOI] [PubMed] [Google Scholar]
- Leonard D., Ajuh P., Lamond A. I., Legerski R. J. (2003). hLodestar/HuF2 interacts with CDC5L and is involved in pre-mRNA splicing. Biochem. Biophys. Res. Commun. 308, 793–801 10.1016/S0006-291X(03)01486-4 [DOI] [PubMed] [Google Scholar]
- Lim G. H., Zhang X., Chung M. S., Lee D. J., Woo Y. M., Cheong H. S., Kim C. S. (2010). A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 185, 103–113 10.1111/j.1469-8137.2009.03032.x [DOI] [PubMed] [Google Scholar]
- Lin Z., Yin K., Wang X., Liu M., Chen Z., Gu H., Qu L. J. (2007a). Virus induced gene silencing of AtCDC5 results in accelerated cell death in Arabidopsis leaves. Plant Physiol. Biochem. 45, 87–94 10.1016/j.plaphy.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Lin Z., Yin K., Zhu D., Chen Z., Gu H., Qu L. J. (2007b). AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 17, 815–828 10.1038/cr.2007.71 [DOI] [PubMed] [Google Scholar]
- Liu M., Yuan L., Liu N. Y., Shi D. Q., Liu J., Yang W. C. (2009). GAMETOPHYTIC FACTOR 1, involved in pre-mRNA splicing, is essential for megagametogenesis and embryogenesis in Arabidopsis. J. Integr. Plant Biol. 51, 261–271 10.1111/j.1744-7909.2008.00783.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Xu X., Kuo M. H. (2010). Snf1p regulates Gcn5p transcriptional activity by antagonizing Spt3p. Genetics 184, 91–105 10.1534/genetics.110.114017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Duggan L., Emre N. C., Belotserkovskya R., Lane W. S., Shiekhattar R., Berger S. L. (2001). Snf1-a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293, 1142–1146 10.1126/science.1062322 [DOI] [PubMed] [Google Scholar]
- Lorkovic Z. J., Barta A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635 10.1093/nar/30.3.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher M., Fortschegger K., Ritter G., Wostry M., Voglauer R., Schmid J. A., Watters S., Rivett A. J., Ajuh P., Lamond A. I., Katinger H., Grillari J. (2005). Interaction of U-box E3 ligase SNEV with PSMB4, the beta7 subunit of the 20 S proteasome. Biochem. J. 388, 593–603 10.1042/BJ20041517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Legerski R. J. (2007). The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem. Biophys. Res. Commun. 354, 968–974 10.1016/j.bbrc.2007.01.097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Allo M., Schor I. E., Kornblihtt A. R., Misteli T. (2011). Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan K. N., Mitchell B. S. (2003). Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. U.S.A. 100, 10746–10751 10.1073/pnas.1631060100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002). Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298, 2205–2208 10.1126/science.1077783 [DOI] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Urlaub H., Will C. L., Gentzel M., Wilm M., Lührmann R. (2004). A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23, 2381–2391 10.1038/sj.emboj.7600241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E., Palhan V. B., Tjernberg A., Lymar E. S., Gamper A. M., Kundu T. K., Chait B. T., Roeder R. G. (2001). Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21, 6782–6795 10.1128/MCB.21.20.6782-6795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald W. H., Ohi R., Smelkova N., Frendewey D., Gould K. L. (1999). Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19, 5352–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll C., von Lyncker L., Zimmermann S., Kägi C., Baumann N., Twell D., Grossniklaus U., Gross-Hardt R. (2008). CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 56, 913–921 10.1111/j.1365-313X.2008.03650.x [DOI] [PubMed] [Google Scholar]
- Monaghan J., Xu F., Gao M., Zhao Q., Palma K., Long C., Chen S., Zhang Y., Li X. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5, e1000526. 10.1371/journal.ppat.1000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Xu F., Xu S., Zhang Y., Li X. (2010). Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant Physiol. 154, 1783–1793 10.1104/pp.110.158931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M. J., de la Mata M., Kornblihtt A. R. (2010). The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 35, 497–504 10.1016/j.tibs.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Németh K., Salchert K., Putnoky P., Bhalerao R., Koncz-Kálmán Z., Stankovic-Stangeland B., Bakó L., Mathur J., Ökrész L., Stabel S., Geigenberger P., Stitt M., Rédei G. P., Schell J., Koncz C. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12, 3059–3073 10.1101/gad.12.19.3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterreich F. C., Bieberstein N., Neugebauer K. M. (2011). Pause locally, splice globally. Trends Cell Biol. 21, 328–335 10.1016/j.tcb.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Ohi M. D., Gould K. L. (2002). Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8, 798–815 10.1017/S1355838202025050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M. D., Link A. J., Ren L., Jennings J. L., McDonald W. H., Gould K. L. (2002). Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22, 2011–2024 10.1128/MCB.22.7.2011-2024.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Ren L., Hirsch J. P., Chazin W. J., Walz T., Gould K. L. (2005). Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 25, 451–460 10.1128/MCB.25.1.451-460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G. C., Yu H. J., Ngo Q. A., Rajani S., Mayalagu S., Johnson C. S., Capron A., Xie L. F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614 10.1242/dev.01595 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhang Y., Li X. (2005). An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15, 1129–1135 10.1016/j.cub.2005.05.022 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhao Q., Cheng Y. T., Bi D., Monaghan J., Cheng W., Zhang Y., Li X. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493 10.1101/gad.1559607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R., Bentley D. (2009). “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36, 178–191 10.1016/j.molcel.2009.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J. J., Clay N. K., Nelson T. M. (2008). Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56, 251–263 10.1111/j.1365-313X.2008.03595.x [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245 10.1101/gr.473902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58, 267–294 10.1146/annurev.arplant.58.032806.103754 [DOI] [PubMed] [Google Scholar]
- Ren L., McLean J. R., Hazbun T. R., Fields S., Van der Kooi C., Ohi M. D., Gould K. L. (2011). Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS ONE 6, e16719. 10.1371/journal.pone.0016719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J., Kim K., Maddock J. R., Anthony J. G., Woolford J. L., Jr. (1995). The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1, 375–390 [PMC free article] [PubMed] [Google Scholar]
- Sanchez S. E., Petrillo E., Beckwith E. J., Zhang X., Rugnone M. L., Hernando C. E., Cuevas J. C., Godoy Herz M. A., Depetris-Chauvin A., Simpson C. G., Brown J. W., Cerdán P. D., Borevitz J. O., Mas P., Ceriani M. F., Kornblihtt A. R., Yanovsky M. J. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 10.1038/nature09470 [DOI] [PubMed] [Google Scholar]
- Sapra A. K., Khandelia P., Vijayraghavan U. (2008). The splicing factor Prp17 interacts with the U2, U5 and U6 snRNPs and associates with the spliceosome pre- and post-catalysis. Biochem. J. 416, 365–374 10.1042/BJ20081195 [DOI] [PubMed] [Google Scholar]
- Sihn C. R., Cho S. Y., Lee J. H., Lee T. R., Kim S. H. (2007). Mouse homologue of yeast Prp19 interacts with mouse SUG1, the regulatory subunit of 26S proteasome. Biochem. Biophys. Res. Commun. 356, 175–180 10.1016/j.bbrc.2007.02.134 [DOI] [PubMed] [Google Scholar]
- Simpson C. G., Brown J. W. (2008). U12-dependent intron splicing in plants. Curr. Top. Microbiol. Immunol. 326, 61–82 10.1007/978-3-540-76776-3_4 [DOI] [PubMed] [Google Scholar]
- Simpson C. G., Thow G., Clark G. P., Jennings S. N., Watters J. A., Brown J. W. (2002). Mutational analysis of a plant branch point and polypyrimidine tract required for constitutive splicing of a mini-exon. RNA 8, 47–56 10.1017/S1355838202015546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R. J., Millhouse S., Chen C. F., Lewis B. A., Erdjument-Bromage H., Tempst P., Manley J. L., Reinberg D. (2007). Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription post initiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676 10.1016/j.molcel.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. J., Query C. C., Konarska M. M. (2008). “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell 30, 657–666 10.1016/j.molcel.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., Rape M. (2011). The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447 10.1101/gad.1925010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. R., Song J. D., Cho J. N., Amasino R. M., Noh B., Noh Y. S. (2009). The RNA binding protein ELF9 directly reduces suppressor of overexpression of CO1 transcript levels in Arabidopsis, possibly via nonsense-mediated mRNA decay. Plant Cell 21, 1195–1211 10.1105/tpc.108.064774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiluttini B., Gu B., Belagal P., Smirnova A. S., Nguyen V. T., Hébert C., Schmidt U., Bertrand E., Darzacq X., Bensaude O. (2010). Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J. Cell. Sci. 123, 2085–2093 10.1242/jcs.061358 [DOI] [PubMed] [Google Scholar]
- Stevens S. W., Ryan D. E., Ge H. Y., Moore R. E., Young M. K., Lee T. D., Abelson J. (2002). Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9, 31–44 10.1016/S1097-2765(02)00436-7 [DOI] [PubMed] [Google Scholar]
- Tanabe N., Kimura A., Yoshimura K., Shigeoka S. (2009). Plant-specific SR-related protein atSR45a interacts with spliceosomal proteins in plant nucleus. Plant Mol. Biol. 70, 241–252 10.1007/s11103-009-9469-y [DOI] [PubMed] [Google Scholar]
- Taniue K., Oda T., Hayashi T., Okuno M., Akiyama T. (2011). A member of the ETS family, EHF, and the ATPase RUVBL1 inhibit p53-mediated apoptosis. EMBO Rep. 12, 682–689 10.1038/embor.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanuma N., Kim S. E., Beullens M., Tsubaki Y., Mitsuhashi S., Nomura M., Kawamura T., Isono K., Koseki H., Sato M., Bollen M., Kikuchi K., Shima H. (2008). Nuclear inhibitor of protein phosphatase-1 (NIPP1) directs protein phosphatase-1 (PP1) to dephosphorylate the U2 small nuclear ribonucleoprotein particle (snRNP) component, spliceosome-associated protein 155 (Sap155). J. Biol. Chem. 283, 35805–35814 10.1074/jbc.M805468200 [DOI] [PubMed] [Google Scholar]
- Tarn W. Y., Lee K. R., Cheng S. C. (1993). Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 90, 10821–10825 10.1073/pnas.90.22.10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S. (2009). Roles of eukaryotic Lsm proteins in the regulation of mRNA function. Int. Rev. Cell Mol. Biol. 272, 149–189 10.1016/S1937-6448(08)01604-3 [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Jaladat Y. (2010). The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics 10, 4128–4141 10.1002/pmic.201000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Guthrie C. (2005). The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 19, 1894–1904 10.1101/gad.1336305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglauer R., Chang M. W., Dampier B., Wieser M., Baumann K., Sterovsky T., Schreiber M., Katinger H., Grillari J. (2006). SNEV overexpression extends the life span of human endothelial cells. Exp. Cell Res. 312, 746–759 10.1016/j.yexcr.2005.11.025 [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Will C. L., Lührmann R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wang B. B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5, R102. 10.1186/gb-2004-5-2-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Tian Q., Hou Z., Mucha M., Aukerman M., Olsen O. A. (2007). The Arabidopsis thaliana ATPRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 26, 1357–1366 10.1007/s00299-006-0276-5 [DOI] [PubMed] [Google Scholar]
- Wang Q., He J., Lynn B., Rymond B. C. (2005). Interactions of the yeast SF3b splicing factor. Mol. Cell. Biol. 25, 10745–10754 10.1128/MCB.25.11.4455-4465.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fu Y., Gao L., Zhu G., Liang J., Gao C., Huang B., Fenger U., Niehrs C., Chen Y. G., Wu W. (2010). Xenopus skip modulates Wnt/beta-catenin signaling and functions in neural crest induction. J. Biol. Chem. 285, 10890–10901 10.1074/jbc.M109.072421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., Trowitzsch S., Kastner B., Lührmann R., Wahl M. C. (2010). Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29, 4172–4184 10.1038/emboj.2010.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C. L., Lührmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Gong Z., Rock C. D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J. K. (2001). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1, 771–781 10.1016/S1534-5807(01)00087-9 [DOI] [PubMed] [Google Scholar]
- Xu D., Friesen J. D. (2001). Splicing factor slt11p and its involvement in formation of U2/U6 helix II in activation of the yeast spliceosome. Mol. Cell. Biol. 21, 1011–1023 10.1128/MCB.21.9.2981-2990.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi N., Takeda S., Matsumoto N., Okada K. (2009). VAJ/GFA1/CLO is involved in the directional control of floral organ growth. Plant Cell Physiol. 50, 515–527 10.1093/pcp/pcp011 [DOI] [PubMed] [Google Scholar]
- Yeh T. C., Liu H. L., Chung C. S., Wu N. Y., Liu Y. C., Cheng S. C. (2011). Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol. Cell. Biol. 31, 43–53 10.1128/MCB.00801-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ito M., Nishida I., Watanabe A. (2001). Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana. Plant Cell Physiol. 42, 170–178 10.1093/pcp/pce021 [DOI] [PubMed] [Google Scholar]
- Zhang N., Kaur R., Akhter S., Legerski R. J. (2009). Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 10, 1029–1035 10.1038/embor.2009.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y. T., Bi D., Palma K., Li X. (2005a). MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr. Biol. 15, 1936–1942 10.1016/j.cub.2005.09.038 [DOI] [PubMed] [Google Scholar]
- Zhang N., Kaur R., Lu X., Shen X., Li L., Legerski R. J. (2005b). The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 280, 40559–40567 10.1074/jbc.M500397200 [DOI] [PubMed] [Google Scholar]
- Zhou S., Hayward S. D. (2001). Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol. 21, 6222–6232 10.1128/MCB.21.17.5970-5978.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Licklider L. J., Gygi S. P., Reed R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185 10.1038/nature01031 [DOI] [PubMed] [Google Scholar]
- Zhu W., Rainville I. R., Ding M., Bolus M., Heintz N. H., Pederson D. S. (2002). Evidence that the pre-mRNA splicing factor Clf1p plays a role in DNA replication in Saccharomyces cerevisiae. Genetics 160, 1319–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Compilation of mass spectrometry data on protein composition of purified yeast, Drosophila, human, and Arabidopsis spliceosome and NTC complexes. Phosphorylated human proteins reported by Agafonov et al. (2011) are highlighted in red, Arabidopsis factors with indentified regulatory functions are indicated in bold. Core subunits of yeast, human, and Arabidopsis NTC complexes are shaded in gray. References describing the purification of various spliceosomal and NTC complexes are given at the end of the Table corresponding to numbers in bracket indicated in the heading. Brackets within the table mark unique fission yeast factors, which have either Drosophila or human or Arabidopsis homologs. For each factor, the corresponding NCBI gene accession number is depicted and in all cases frequently used alternative designations are listed.