Abstract
Alternative precursor mRNA splicing is a widespread phenomenon in multicellular eukaryotes and represents a major means for functional expansion of the transcriptome. While several recent studies have revealed an important link between splicing regulation and fundamental biological processes in plants, many important aspects, such as the underlying splicing regulatory mechanisms, are so far not well understood. Splicing decisions are in general based on a splicing code that is determined by the dynamic interplay of splicing–controlling factors and cis-regulatory elements. Several members of the group of heterogeneous nuclear ribonucleoprotein (hnRNP) proteins are well known regulators of splicing in animals and the comparatively few reports on some of their plant homologs revealed similar functions. This also applies to polypyrimidine tract-binding proteins, a thoroughly investigated class of hnRNP proteins with splicing regulatory functions in both animals and plants. Further examples from plants are auto- and cross-regulatory splicing circuits of glycine-rich RNA binding proteins and splicing enhancement by oligouridylate binding proteins. Besides their role in defining splice site choice, hnRNP proteins are also involved in multiple other steps of nucleic acid metabolism, highlighting the functional versatility of this group of proteins in higher eukaryotes.
Keywords: alternative splicing, splicing regulation, polypyrimidine tract-binding protein, PTB, heterogeneous nuclear ribonucleoprotein, hnRNP
Introduction
The majority of plant genes contain intronic regions, which need to be removed with a high degree of precision and efficiency in the process of precursor messenger RNA (pre-mRNA) splicing. In the recent years, the tremendous increase in plant transcript data has revealed that a significant proportion of all genes generates transcript variants due to alternative splicing (AS) of pre-mRNAs (Wang and Brendel, 2006; Filichkin et al., 2010; Zhang et al., 2010). This enormous expansion of the transcriptome has major implications for both proteome diversity and gene regulation via altered mRNA features, such as the mRNA turnover rate, and essential functions have been ascribed to splicing variants in the context of manifold biological processes in plants (Reddy, 2007).
Numerous AS events have been demonstrated to be regulated in a spatial–temporal manner or by exogenous stimuli, requiring complex control mechanisms for correct splice site choice. While regulation of AS has been intensively studied in animals (Chen and Manley, 2009), only few reports are available that describe cis-regulatory sequences and trans-acting factors controlling AS in plants (Reddy, 2007; Barbazuk et al., 2008; Wachter, 2010). Two major classes of universal splicing regulatory factors are constituted by the serine/arginine-rich (SR) proteins and members of the heterogeneous nuclear ribonucleoprotein (hnRNP) protein family, which have been reported to act antagonistically in several AS events. For example, in case of the Dscam exon 6 cluster, the specific interplay between the hnRNP protein HRP36 and an SR protein is required for correct inclusion of a single cassette exon (Olson et al., 2007). Whereas earlier studies had indicated that hnRNP and SR proteins typically act as repressors and activators of splicing, respectively, it is now becoming evident that the mode of action of splicing regulatory factors often is context-dependent. On one hand, the effect of splicing factors can vary dependent on their binding position within the pre-mRNA. On the other hand, the splicing outcome is often established by the interaction of multiple components of the splicing machinery and additional regulatory factors.
The original definition of hnRNP proteins was based on their experimental identification as major protein constituents of high molecular weight RNA complexes. Subsequently, further proteins with similar characteristics or homology to a previously described hnRNP protein were added to this list, which nowadays comprises a diverse group of proteins that do not share one certain structural or functional feature (Martinez-Contreras et al., 2007; Han et al., 2010). However, given their ability to associate with RNA and single-stranded (ss) DNA, all hnRNP proteins contain RNA recognition motifs (RRMs) or other functionally equivalent domains, including KH domains, quasi-RRMs, and additional types of atypical RRMs (Han et al., 2010). Due to the presence of those domains, most hnRNP proteins can bind to a broad spectrum of ss nucleic acid, however, more stringent experimental conditions allowed the definition of specific binding sequences (Martinez-Contreras et al., 2007; Han et al., 2010). Furthermore, numerous cellular parameters such as the physico-chemical environment, local protein concentration, and interaction with additional factors can critically alter the binding characteristics of hnRNP proteins, thereby providing additional layers of their regulatory potential. Interestingly, a recent study by Mackereth et al. (2011) has revealed that the U2 auxiliary splicing factor (U2AF) 65 undergoes multi-domain conformational selection, dependent on the strength of the bound polypyrimidine tract. According to their model, with increasing strength of the polypyrimidine tract, a larger fraction of U2AF65 proteins is captured in their open conformation, which represents the domain arrangement that efficiently triggers spliceosomal assembly. A similar mechanism might also be found for some hnRNP proteins, given their nature as multi-domain proteins and their ability to bind diverse sequence motifs. Besides nucleic acid binding domains, most hnRNP proteins contain additional domains linked to their multifaceted functions in nucleic acid metabolism, including chromatin remodeling, pre-mRNA splicing, control of mRNA stability and modifications, and mRNA transport (Martinez-Contreras et al., 2007; Han et al., 2010). This complexity is further increased by the occurrence of both AS of many hnRNP pre-mRNAs and different types of posttranslational protein modifications with important implications for hnRNP functions (Martinez-Contreras et al., 2007; Han et al., 2010).
Most of the currently available information on hnRNP proteins is based on studies in the mammalian system. However, homology-based searches for related proteins from plants (Wang and Brendel, 2004) and first reports on the functional characterization of some of them have highlighted the presence and important roles of hnRNP proteins in plants. In this review, we will discuss the potential of hnRNP proteins in plant splicing regulation. First, a brief insight into the most prevalent splicing regulatory mechanisms of hnRNP proteins will be given. Most of these studies have been performed in human or animal systems, however, it seems likely that similar mechanisms are also used by plant hnRNP proteins. Second, the current state of the art on the functions and biological implications of polypyrimidine tract-binding (PTBs) proteins, an example of extensively studied hnRNP proteins with splicing regulatory functions in both animals and plants, will be reviewed. Third, an overview of the experimental reports on other plant hnRNP proteins, with a focus on their role in the regulation of splicing, will be presented. In summary, this review will highlight the recent progress in studying the role of hnRNP proteins in plant splicing regulation. Further, by comparison with the mammalian system, it will be pointed out that hnRNP proteins might play an even more widespread role in controlling splicing decisions in plants, than reflected by the currently available data.
Mechanisms of hnRNP Protein-Mediated Splicing Regulation
Several types of hnRNP proteins are well known regulators of AS and some of them have also been reported to play a role in constitutive splicing. The depletion of hnRNP F (Gamberi et al., 1997) or hnRNP Q (Mourelatos et al., 2001), for instance, resulted in diminished in vitro splicing activity. Furthermore, using mass spectrometry, hnRNP proteins were identified as components of purified ribosomes in mammals (Neubauer et al., 1998). A role of hnRNP proteins in generic splicing is also supported by the finding that introduction of intronic binding sites for hnRNPs A/B and F/H triggered splicing in vitro and in vivo, and that computational prediction revealed an overrepresentation of the corresponding binding sites at both ends of human introns (Martinez-Contreras et al., 2006). Based on these discoveries, it was suggested that some hnRNP proteins might play a more general role in the recognition of splice sites that could be brought together by the interaction of hnRNP proteins bound to flanking intronic regions.
The regulatory outcome of splicing factor binding can be highly position-dependent, as was revealed by several recent studies. Transcriptome-wide maps of RNA binding sites for the neuron-specific splicing factor Nova, for instance, revealed that binding around the alternative 5′ or constitutive 3′ splice site typically induces exon inclusion, whereas exon skipping is often associated with binding sites around the constitutive 5′ splice site or the alternative exon (Ule et al., 2006; Licatalosi et al., 2008). Binding position-dependent splicing regulatory functions have also been described for FOX proteins (Zhang et al., 2008; Venables et al., 2009; Yeo et al., 2009), Mbnl1 (Du et al., 2010), and PTB (Xue et al., 2009; Llorian et al., 2010). Furthermore, in the case of mutually exclusive splicing, where only one of two or more alternative exons is included, hnRNP proteins can induce skipping of one exon and stimulate inclusion of another one. This has been reported for the human pyruvate kinase pre-mRNA, the mutually exclusive splicing of which is regulated by hnRNPs A1 and A2, and PTB (David et al., 2010). It is also more frequently observed that two or more types of hnRNP proteins act in a combinatorial manner, further expanding the repertoire of hnRNP protein-mediated modes of action and functions.
Precursor messenger RNA splicing is confined to the nuclear compartment, in which some hnRNP proteins are highly abundant. However, many hnRNP proteins display nucleo-cytoplasmic shuttling that has been demonstrated to be regulated by posttranslational modifications. Shuttling between those compartments might represent a means to limit their splicing regulatory activity in the nucleus. Furthermore, transport of hnRNPs out of the nucleus might be a prerequisite for the cytosolic functions, ascribed to some hnRNP proteins. For example, mammalian PTB was shown to be specifically phosphorylated by the 3′,5′-cAMP-dependent protein kinase, which is required for export of the protein into the cytosol (Xie et al., 2003). In addition to protein phosphorylation, the methylation of arginine residues (Chang et al., 2011) has also been described as a type of posttranslational modification with implications for the subcellular distribution of hnRNP proteins. It can be concluded that the splicing regulatory activity of hnRNP proteins can be subject to tight control, and, furthermore, that hnRNP proteins typically act together with other splicing factors to alter splicing decisions. Numerous models for mechanisms of AS control by hnRNP proteins have been suggested and, based on recent findings, it seems likely that they are not mutually exclusive, but rather reflect the versatility of splicing regulation. In the following paragraphs, the most commonly proposed modes of action will be briefly discussed.
hnRNP-mediated displacement or recruitment of splicing factors
The most straightforward mode of action is based on a competition between hnRNP proteins and other splicing factors for binding to cis-elements having a splicing regulatory function (Figure 1A). Negative regulation can be achieved by occlusion of 5′ or 3′ splice sites, branch point and polypyrimidine tract, or binding sites of activating SR proteins. For example, hnRNP L has been shown to suppress the recognition of either 5′ or 3′ splice sites by binding to adjacent, intronic CA-rich motifs (Heiner et al., 2010). In contrast, hnRNP L binding to an intronic CA repeat within human endothelial nitric oxide synthase pre-mRNA stimulates splicing from a nearby 5′ splice site (Hui et al., 2003). This might be explained by an hnRNP protein-mediated recruitment of the U1 small nuclear ribonucleoprotein (snRNP) complex (Figure 1B), as shown for hnRNP H-regulated splicing of HIV-1 (Caputi and Zahler, 2002). Alternatively, a positive effect might also be caused by displacement of a splicing repressor, as it has been reported for PTB that can antagonize the splicing suppressing function of SRp30c in the hnRNP A1 pre-mRNA (Paradis et al., 2007). Thus, depending on the binding position, hnRNP proteins can displace or recruit spliceosomal core components and other regulatory factors, thereby altering the splicing outcome.
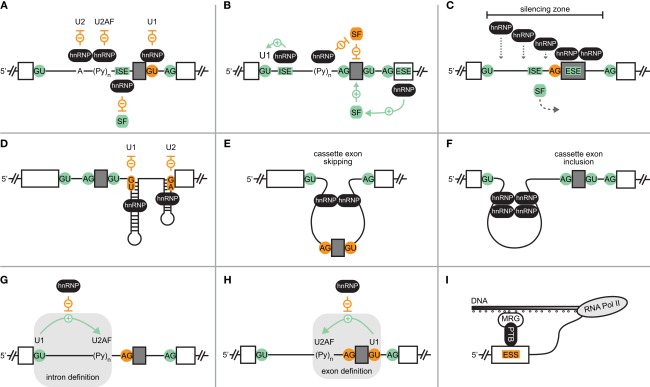
Figure 1.
Mechanisms of hnRNP-mediated splicing regulation. (A) Competition of hnRNP proteins with components of the spliceosome (U1, U2, U2AF) or a positive splicing factor (SF) results in skipping of a cassette exon (gray box). Exons and introns are depicted as boxes and lines, respectively. Negative interactions and disabled splice sites are shown in orange, positive interactions and activating factors or elements are in green. ISE, intronic splicing enhancer. Symbols and color code apply to all panels. (B) Activation of cassette exon inclusion by hnRNP-mediated recruitment of spliceosomal components or positive factors, or by displacement of a negative factor. ESE, exonic splicing enhancer. (C) Cooperative binding of hnRNP proteins establishes a zone of silencing, inducing cassette exon skipping. (D) Occlusion of splice sites by RNA structures stabilized by hnRNP proteins. (E) hnRNP-mediated looping of the cassette exon prevents its inclusion, whereas looping of a region upstream of the cassette exon can bring splice sites together and induce exon inclusion (F). (G) Interference of an hnRNP protein with intron definition interactions between U1 and downstream bound U2AF, or inhibition of exon definition (H). (I) Histone modification (small circles below DNA)-dependent recruitment of the hnRNP protein PTB via MRG to an exonic splicing silencer (ESS).
Often, splicing control by hnRNP proteins not only involves a single binding site within the pre-mRNA, but rather several motifs, for which cooperative binding behavior can occur (Figure 1C). For example, upon hnRNP A1 binding to a high-affinity site within HIV-1 pre-mRNA, cooperative binding to additional sites was observed (Okunola and Krainer, 2009). The cooperative spreading of hnRNP A1 preferentially takes place in a 3′- to 5′-direction, resulting in unwinding of RNA structures and removal of bound proteins, thereby establishing a zone of silencing.
Modulation of pre-mRNA structures by hnRNP proteins
Several recent studies argue for a more widespread role of pre-mRNA structures in the regulation of splicing (Warf and Berglund, 2010). Eukaryotic riboswitches represent one example, in which alternate RNA structures play an active role in both signal sensing and splicing regulation via controlling splice site accessibility (Wachter, 2010). In most other reported cases of structure-dependent splicing, however, alternate RNA folds are enforced by the binding of proteins, including members of the group of hnRNP proteins. Looping of RNA regions can lead either to activation or suppression of a certain splicing event. Accordingly, negative control often is mediated by occlusion of cis-elements that need to be recognized to enable a certain splicing outcome (Figure 1D). In contrast, RNA structures can also disable binding of splicing suppressors, or expose splicing enhancers, resulting in splicing activation. Finally, formation of RNA folds can alter critical distances, thereby influencing the recognition of splice sites by the spliceosomal machinery (Figures 1E,F). Interestingly, RNA looping might be stabilized by a single hnRNP protein containing several RRMs, as suggested for PTB, the RRMs 3 and 4 of which were shown to interact with each other on their dorsal faces, resulting in an antiparallel orientation of bound RNA (Oberstrass et al., 2005; Vitali et al., 2006). Structure-dependent AS control was also proposed for hnRNP C, which can trigger cassette exon skipping or inclusion in a binding position-dependent manner (Konig et al., 2010).
Interference of hnRNPs with interactions between spliceosomal components
Besides competition with or recruitment of singular factors, hnRNPs can also interfere with protein interactions that are required for the formation of a functional spliceosomal complex. Different steps in spliceosome assembly can be affected, as nicely illustrated by PTB-regulated AS events. On one hand, PTB can suppress intron definition (Figure 1G) by preventing the 5′ splice site-dependent assembly of U2AF into the spliceosomal E complex (Sharma et al., 2005). On the other hand, PTB can also inhibit exon definition (Figure 1H), as in the example of Fas exon 6 (Izquierdo et al., 2005). The inclusion of this cassette exon is stimulated by an interaction of U1 snRNP at the 5′ splice site and U2AF at the 3′ splice site, downstream and upstream, respectively. Furthermore, splicing can also be controlled in the phase of transition from exon definition to an intron-defined spliceosome, as was proposed based on proteomic analyses of spliceosomal complexes formed in the presence of PTB (Sharma et al., 2008).
Coupling of splicing with other steps in gene expression
Numerous studies have revealed that splicing can occur co-transcriptionally, and, more recently, examples for functional coupling of splicing and transcription have been described (Oesterreich et al., 2011). Importantly, the rate of transcriptional elongation can influence the availability of competing splice sites and other cis-regulatory elements. For example, RNA polymerase II pausing can delay transcription of competing splice sites, thereby triggering inclusion of a cassette exon with weak splice sites. Several RNA binding proteins, including PTB, were found to be associated with the transcriptional complex, with striking differences in their distribution along different regions of the genes (Swinburne et al., 2006). Furthermore, a correlation between the association of PTB and RNA polymerase II was revealed, and it was hypothesized that transcriptional complexes containing certain RNA binding proteins might display altered elongation rates, having important implications for AS decisions (Swinburne et al., 2006). Similarly, hnRNP proteins might also interfere with other steps in mRNA processing, such as 3′ end processing, and thereby regulate AS.
Furthermore, a recent study also described a direct link between epigenetic modifications and PTB-dependent splicing regulation in human (Luco et al., 2010). Luco et al. (2010) found a correlation between certain histone modifications and the AS outcome for various genes, which was attributed to the histone mark-specific recruitment of PTB via the chromatin-binding protein MRG15 that serves as adaptor (Figure 1I). Based on these findings, it is tempting to speculate that further links between epigenetic modifications and AS might exist.
Splicing Regulatory Functions of Polypyrimidine Tract-Binding Proteins
Polypyrimidine tract-binding proteins represent one of the best studied families of hnRNPs that fulfill diverse functions in mRNA metabolism (Spellman and Smith, 2006; Sawicka et al., 2008), including regulation of pre-mRNA splicing and polyadenylation, translation of viral RNAs from internal ribosomal entry sites (IRES), and mRNA transport. Numerous reports have highlighted the critical role of PTBs in controlling AS decisions, thereby providing important insights into both mechanistic aspects and functional implications of regulated AS events.
In line with the findings for other splicing regulatory factors, the mode of action of PTB is highly dependent on the binding position within the pre-mRNA. Best studied is splicing inhibition by PTB, resulting in skipping of cassette exons (Spellman and Smith, 2006). For some other AS events, however, a stimulatory effect of PTB on exon inclusion was revealed, as for example in the case of the pre-mRNA of calcitonin and calcitonin gene-related peptide (Lou et al., 1999). To address the binding position-dependent splicing action of PTB, Xue et al. (2009) applied cross-linking immunoprecipitation coupled with high-throughput sequencing (Clip-seq) to identify PTB binding sites within the transcriptome. Based on their findings, it was suggested that PTB binding close to alternative sites generally induces exon skipping, whereas binding close to the constitutive sites typically is linked to inclusion of an alternative exon. This model is in line with the classical view of PTB as a general splicing repressor, where weakening of constitutive splice sites allows inclusion of a cassette exon. However, a different model of position-dependent AS regulation by PTB was put forward by Llorian et al. (2010), who employed high-resolution splice-sensitive microarrays to study AS upon PTB knockdown. A majority of the discovered PTB-dependent AS events were PTB-repressed cassette exons, which typically had PTB binding sites upstream of or within the respective exon. In contrast, PTB-stimulated exons possessed polypyrimidine motifs downstream of the cassette exon, indicating a direct activation of exon inclusion by PTB rather than acting by weakening of neighboring constitutive sites. The discrepancies between these two studies might be explained, at least partially, by the different methods that were applied, as well as by the distinct sets of analyzed AS events. Accordingly, analysis of the Clip-seq data from the study by Xue et al. (2009) with respect to the PTB regulation targets identified by Llorian et al. (2010) further supported the model suggested in the latter work. Moreover, previous studies had highlighted that PTB can regulate splicing via different mechanisms, and, similarly, also the position-dependent splicing outcome might vary between AS events.
Most of the work on the mechanistic aspects of splicing regulation by PTB has focused on the repression of exon inclusion (Spellman and Smith, 2006), which still appears to be the dominant type of PTB-controlled AS. Several of the aforementioned mechanisms of AS control have also been proposed to underlie PTB-mediated splicing control, the studies of which have made major contributions to our current understanding of regulated splicing. Again, it should be highlighted that these models are not mutually exclusive, but rather might be implemented in the context of different AS events. PTB displays a high-affinity for pyrimidine-rich sequences, and, therefore, can compete with U2AF65 for binding to the polypyrimidine tract (Singh et al., 1995; Sauliere et al., 2006), which is typically present in the region between the intronic branch point and the 3′ splice site. However, given that most pre-mRNAs with PTB-regulated exons contain several PTB motifs, many of which are not located within the polypyrimidine tract (Wagner and Garcia-Blanco, 2001; Amir-Ahmady et al., 2005), additional mechanisms of PTB-mediated AS must exist. Proposed models include propagation of PTB protein binding between high-affinity sites on the pre-mRNA, and RNA looping, both of which can establish a zone of silencing, where cis-regulatory elements are masked, leading to skipping of the respective exon (Wagner and Garcia-Blanco, 2001; Spellman and Smith, 2006). Due to its RRM domain organization (Oberstrass et al., 2005; Vitali et al., 2006), PTB might cause RNA looping by binding to two distinct sites within the same pre-mRNA molecule. However, artificial tethering of PTB domains to a pre-mRNA via the phage MS2 RNA-protein interaction system preserved PTB-mediated splicing control, indicating that, at least in some instances, PTB action can be uncoupled from the mode of RNA binding (Robinson and Smith, 2006). Besides direct occlusion of cis-elements, PTB was demonstrated to disturb the interaction of protein factors at different steps of spliceosome assembly, thereby suppressing splicing (see above). While several models exist that describe splicing inhibition by PTB, only few studies have addressed the mode of action of this protein as a positive splicing regulator, as in the case of the alternative exon 7B of the hnRNP A1 pre-mRNA (Paradis et al., 2007). Here, the stimulatory effect was found to originate from PTB counteracting the splicing inhibitory activity of the SRp30c protein. Mechanistic studies of further PTB-stimulated exons might reveal if this anti-repressor activity of PTB is more widespread in AS control.
Polypyrimidine tract-binding protein contributes to the regulation of numerous AS events in mammals, as revealed by studies applying splice-sensitive microarrays to compare transcriptome-wide splicing patterns between control and PTB knockdown samples (Boutz et al., 2007; Xing et al., 2008; Llorian et al., 2010). Upon down-regulation of PTB, Llorian et al. (2010), for instance, identified 196 repressed and 67 activated exons, a major fraction of which showed neuronal and striated muscle specificity. Further important insights into biological functions of PTB-controlled AS programs in mammals were gained by analysis of tissue-specific expression of PTB and its neuronal homolog nPTB. In neuronal cells, PTB is down-regulated by a tissue-specific microRNA (Makeyev et al., 2007), leading to a switch from PTB to nPTB, coinciding with an altered splicing program in developing neurons (Boutz et al., 2007; Makeyev et al., 2007). Direct comparison of RNA binding affinities and in vitro splicing regulation of PTB and nPTB (Markovtsov et al., 2000), as well as knockdown of individual or in parallel both PTB isoforms followed by transcriptome-wide splicing studies revealed both specific and redundant functions of those proteins (Boutz et al., 2007; Spellman et al., 2007). Interestingly, PTB and nPTB levels are subject to feedback control via auto-regulation, whereas PTB also negatively regulates nPTB expression (Wollerton et al., 2004; Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007). These auto-and cross-regulatory circuits involve formation of splicing variants that are targeted by nonsense-mediated decay (NMD). In addition, evidence for splicing-independent repression of PTB protein generation has been provided, however, the underlying mechanism remained ill-defined. Moreover, cross-regulation of mammalian PTBs is not limited to PTB and nPTB, but also includes the less-studied, hematopoietic cell-specific PTB homolog ROD1, the splicing of which is altered upon simultaneous knockdown of PTB and nPTB (Spellman et al., 2007).
By far most of the work on PTBs was performed in mammalian systems, however, homologs of those proteins are also present and have been studied in other eukaryotic clades. For example, in Drosophila, the PTB gene was shown to encode a germline-specific mRNA isoform that is required for male fertility by contributing to spermatid individualization (Robida et al., 2010). Recently, PTBs have also been described in plants (Ham et al., 2009; Wang and Okamoto, 2009) and, in the case of three PTB homologs from Arabidopsis, evidence for their splicing regulatory functions has been provided (Stauffer et al., 2010). Ham et al. (2009) identified a pumpkin PTB as a constituent of a phloem-mobile ribonucleoprotein complex, the assembly of which depends on PTB phosphorylation (Li et al., 2011). A study of the two closely related Arabidopsis PTB homologs, AtPTB1 and AtPTB2, suggested a role of those proteins in pollen germination, as mutants displayed diminished germination efficiency (Wang and Okamoto, 2009). However, the molecular basis of this phenotype and the mode of action of AtPTB1 and 2 remained unsolved.
The work by Stauffer et al. (2010) delivered the first insights into the splicing regulatory potential of all three Arabidopsis PTB homologs, the expression of which is controlled by auto- and cross-regulation, akin to the mammalian PTBs. Analysis of the splicing patterns for all three genes revealed the existence of two major splicing products, of which one encodes the full-length protein, whereas the other contains a premature termination codon (PTC) and is targeted by NMD (Figure 2). Splicing to the NMD target transcript can be triggered in an auto-regulatory feedback loop, where an elevated level of one PTB alters splicing of its corresponding pre-mRNA. Additionally, cross-regulation between the two close homologs AtPTB1 and 2, but not the distantly related AtPTB3, was observed. These findings revealed not only the splicing control functions of AtPTBs, but also that regulation of PTBs via auto- and cross-regulatory feedback loops is present in both mammals and plants. Future work will reveal if plant PTBs, as their mammalian counterparts, also contribute to complex splicing programs, and, if so, what the biological implications of those AS events are. AtPTB1 and 2 are close homologs, with more than 80% of their amino acid residues conserved, and, in both cases, retention of a cassette exon with a PTC results in formation of the NMD-targeted splicing variant (Stauffer et al., 2010). In contrast, AtPTB3 is more distantly related, displaying a comparable degree of protein homology of ∼50% to the other two Arabidopsis and the human PTB homologs. Furthermore, as for human PTB, feedback regulation of AtPTB3 gives rise to skipping of a cassette exon, resulting in a downstream PTC and NMD targeting. Thus, AS of plant PTB pre-mRNAs can serve as a model to study both positive and negative effects of PTBs on exon inclusion. Interestingly, in vivo splicing reporter assays indicated that AtPTB2- and AtPTB3-mediated exon inclusion and skipping, respectively, can be counteracted by elevated levels of U2AF65 (Stauffer et al., 2010). These findings are in line with competition between PTBs und U2AF65 in splice site choice, but further work is needed to elucidate the molecular basis of those findings.
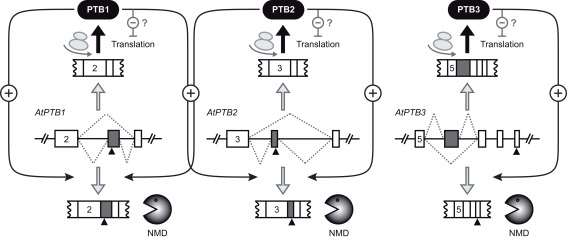
Figure 2.
Auto- and cross-regulation of the ArabidopsisPTB homologs. Exon-intron organizations of the Arabidopsis genes AtPTB1, AtPTB2, and AtPTB3 are shown for the regions giving rise to alternative splicing (dotted lines). Exons and introns are depicted as boxes and lines, respectively. Gray boxes refer to cassette exons, which are either skipped or included. Numbers of first displayed exons are given for all three genes. Black triangles indicate premature termination codons, rendering the respective splicing variants degradation via nonsense-mediated decay (NMD, bottom). The splicing variants shown on top of each gene model are translated into PTB proteins, which can interfere with translation of PTB mRNAs (for simplicity, only the effect on their own mRNAs is shown). PTB proteins also trigger splicing of their own pre-mRNAs to the NMD target variants (auto-regulation). Additionally, cross-regulation between PTB1 and PTB2 occurs. The model is based on the work from Stauffer et al. (2010).
Auto- and cross-regulation of Arabidopsis PTB homologs does not only affect AS, but also acts downstream of it. Co-expression of PTBs with reporter constructs containing the 5′ region of mature PTB mRNAs resulted in reduced accumulation of the translation products, whereas processing and levels of the corresponding transcripts appeared unchanged (Stauffer et al., 2010). Interestingly, Arabidopsis PTBs fused to fluorescent proteins were found to localize to the nucleus, cytosol, and processing bodies (P-bodies). Thus, as an intriguing possibility, PTBs might interfere with expression of their own and also other mRNAs by retracting them from the polysomal pool, followed by storage in P-bodies. Splicing-independent functions of PTBs have also been described in other species, including repression of translation in Drosophila (Besse et al., 2009) and human (Boutz et al., 2007), as well as the widespread positive role in translation from IRES (Auweter and Allain, 2008; Sawicka et al., 2008). Further work will be required to understand those diverse regulatory functions of PTBs in more detail.
Splicing Regulation and Further Functions of Other Plant hnRNP Proteins
Besides PTB homologs, several other types of hnRNP proteins from different plant species have been investigated. These studies provided important insights into the mode of action and biological functions of this versatile group of proteins, and, given that many other hnRNP homologs from plants still are at best partially characterized, an even wider scope of hnRNP-dependent functions in plants can be anticipated. As reported for their animal counterparts, hnRNP proteins from plants can regulate splicing, but also affect other nucleic acid metabolic processes, a brief summary of which will be provided in the following sections.
Regulation of alternative splicing by AtGRP7 and AtGRP8
AtGRP7 and AtGRP8 are glycine-rich RNA binding proteins (GRPs) from Arabidopsis that regulate AS of their corresponding pre-mRNAs in auto- and cross-regulatory circuits (Staiger et al., 2003; Schoning et al., 2008) in a similar manner as described in the previous section for PTBs. Constitutive overexpression of AtGRP7 promotes usage of a cryptic splice site within an intron of the pre-mRNA derived from the endogenous AtGRP7 locus, giving rise to a splicing variant with a decreased half-life (Staiger et al., 2003). The authors further demonstrated that AtGRP7 overexpression affects the AS of the AtGRP8 pre-mRNA, which encodes an RNA binding protein closely related to AtGRP7 and exhibits a cryptic splice site conserved with that of AtGRP7. Similarly to the AtGRP7 regulatory circuit, AtGRP8 can regulate splicing of its own and the AtGRP7 pre-mRNA by triggering the use of an alternative splice site (Schoning et al., 2008). Both AS variants derived from AtGRP7 and AtGRP8 harbor PTCs, targeting them for rapid degradation via NMD, as confirmed by their accumulation in NMD-impaired mutants and upon cycloheximide treatment (Staiger et al., 2003; Schoning et al., 2007). By promoting unproductive splicing coupled to a decay of transcripts via the NMD pathway, the interlocked regulatory circuits might be a means to integrate diverse stimuli, thereby fine tuning the expression of their components, and even to influence common downstream targets. Moreover, Schoning et al. (2007) could substantiate that the binding of AtGRP7 to its target transcripts is necessary for the described negative regulation. By introducing a single arginine to glutamine point mutation into the AtGRP7 RRM, the in vitro RNA binding affinity of the protein was reduced, disrupting the AtGRP7-mediated auto-regulation as well as the regulation of downstream targets in vivo (Schoning et al., 2007).
AtGRP7 has been demonstrated to be regulated by the circadian clock and contributes to the control of flowering time. Both, an atgrp7 T-DNA insertion mutant, and independent RNA interference lines with reduced levels of AtGRP7 and the closely related AtGRP8, showed a late flowering phenotype, whereas AtGRP7 overexpressing plants were reported to flower early (Streitner et al., 2008). Changes in flowering time were found to be mediated by altered transcript levels of the key regulator FLOWERING LOCUS C (FLC), displaying elevated, and diminished FLC transcript levels, respectively, in AtGRP7 loss-of-function and overexpression mutants (Streitner et al., 2008). Interestingly, disturbing AtGRP7 levels did not interfere with the photoperiodic response and the effects on flowering time were overridden by vernalization, both of which are features of the autonomous pathway.
To gain further insights into the functions of AtGRP7, global transcript profiles of an overexpression mutant in comparison to wild type plants were analyzed (Streitner et al., 2010). Thereby, around 300 transcripts showing altered levels upon AtGRP7 overexpression were found, of which one third is under control of the circadian clock. Additionally, transcripts responsive to abiotic and biotic stimuli were identified as putative targets of AtGRP7, as well as components involved in ribosome function and RNA metabolism, consistent with its role in post-transcriptional regulation. These findings suggest a complex set of AtGRP7 regulation targets, the number of which might further increase, if splicing-sensitive transcriptome studies would be applied. An involvement of AtGRP7 in diverse biological processes is also supported by the altered performance of misexpression lines under biotic and abiotic stress conditions. AtGRP7 plays a role in plant immunity, as knockout of this hnRNP resulted in increased susceptibility to the pathogen Pseudomonas syringae (Fu et al., 2007). Fu et al. (2007) found that suppression of plant immunity by P. syringae involves a mono-ADP-ribosyltransferase as an effector protein, which modifies RNA binding proteins including AtGRP7. Recently, Jeong et al. (2011) identified a conserved arginine residue within the RRM of AtGRP7 as the site of ADP-ribosylation, resulting in diminished RNA binding affinity in vitro. Interestingly, earlier studies had revealed that mutation of this residue also disrupts the activities of AtGRP7 in vivo (Schoning et al., 2007), highlighting its critical role in protein functioning. In addition to its role under biotic stress, AtGRP7 also has been reported to play a role in seed germination, seedling growth, and under various abiotic stress conditions, including high salt and freezing (Kim et al., 2008b).
Arginine methylation affects AS in plants
Arginine methylation occurs not only in histones, but also has been described for many other protein classes, including transcription factors and certain types of hnRNP proteins (Lee and Stallcup, 2009). In Arabidopsis, mutation of the arginine methyltransferase 5 (AtPRMT5) was found to cause pleiotropic phenotypes, including late flowering, growth retardation, and a reduced vernalization sensitivity (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008). Deng et al. (2010) demonstrated that AtPRMT5 not only methylates histones but also has a variety of non-histone substrates, including RNA binding or processing factors, like the hnRNPs AtGRP7 and AtGRP8, as well as several U snRNP core proteins. Using a high-throughput sequencing approach, hundreds of genes involved in multiple biological processes, displaying splicing changes upon mutation of AtPRMT5 were identified (Deng et al., 2010). Based on these results, the authors could attribute the late flowering phenotype of atprmt5 mutants to AS of transcripts encoding for RNA processing factors involved in flowering time regulation (Figure 3). In particular, the described AS event resulted in decreased levels of the transcript variant encoding the functional form of the hnRNP E homolog FLK, an autonomous pathway protein known to promote flowering by repressing the expression of FLC (Lim et al., 2004; Mockler et al., 2004). Therefore, in addition to regulating transcription through histone modifications, AtPRMT5 may contribute directly or indirectly to the regulation of pre-mRNA splicing through modifications of non-histone proteins.
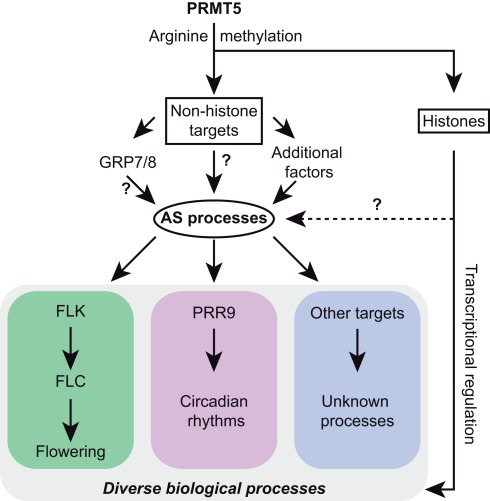
Figure 3.
Arginine methylation and alternative splicing are linked in Arabidopsis. The arginine methyltransferase PRMT5 modifies both histone and non-histone targets, including the splicing factors GRP7 and GRP8 (Deng et al., 2010). Changes in alternative splicing (AS) affect diverse biological processes, such as FLK/FLC-mediated flowering control (Deng et al., 2010) and regulation of circadian rhythms via PRR9 (Sanchez et al., 2010). A connection between histone modifications and AS has been demonstrated in mammals (Luco et al., 2010), but so far not in plants.
In another study of the same arginine methyltransferase, Sanchez et al. (2010) showed that AtPRMT5 also provides a link between AS and the circadian clock (Figure 3). In line with the results obtained by Deng et al. (2010), Sanchez et al. (2010) uncovered numerous splicing changes in the atprmt5 mutant, including AS of the core-clock gene PSEUDO RESPONSE REGULATOR 9 (PRR9). The altered splicing of PRR9 might at least partially explain the impairment of several circadian rhythms in the atprmt5 mutant. Interestingly, the expression of AtPRMT5 underlies circadian oscillations, which are reflected in expression and splicing patterns of a subset of genes. The authors further found an enrichment of alternative 5′ splice sites among the splicing events altered in the atprmt5 mutant, indicating a role of this factor in 5′ splice site recognition. Thus, analyses of splicing patterns in atprmt5 mutants provided an intriguing link between this protein and AS control in plants. However, the molecular mechanisms of this post-transcriptional regulation as well as further physiological processes that AtPRMT5 might be involved in, remain to be uncovered.
UBP1 and UBP1-associated proteins
The oligouridylatebinding protein 1 (UBP1) from Nicotiana plumbaginifolia has been characterized as a nuclear RNA binding protein that associates with polyA-RNA in vivo and can be cross-linked to U-rich intron and UTR sequences in vitro (Lambermon et al., 2000). UBP1 was found to enhance the splicing efficiency of otherwise inefficiently processed introns, as well as to increase the accumulation of reporter mRNAs that are intronless or bear suboptimal introns. Only the UBP1-mediated accumulation of reporter mRNAs was shown to be promoter dependent, suggesting independent activities of UBP1 at more than one level of plant nuclear pre-mRNA maturation. UBP1 homologs are also present in Arabidopsis, however, so far no splicing functions of these proteins have been reported. In a follow-up study by Lambermon et al. (2002), two UBP1-associated proteins, UBA1a and UBA2a, were identified that also can stabilize mRNAs in the nucleus. However, in contrast to UBP1, these hnRNP proteins show no effect on pre-mRNA splicing. Expression studies of three UBA2 genes from Arabidopsis revealed AS of their corresponding pre-mRNAs, which is regulated in response to wounding and alters the 3′ UTRs of the respective transcripts (Bove et al., 2008).
Further investigations of hnRNP proteins from N. plumbaginifolia resulted in the identification of the two structurally related proteins RBP45 and RBP47, which display, similar to UBP1, specificity for oligouridylates (Lorkovic et al., 2000). RBP45 and RBP47 are also localized in the nucleus and associate with polyA-RNA. However, no stimulation of splicing or accumulation of mRNAs could be observed upon overexpression of RBP45 and RBP47 in a protoplast system (Lorkovic et al., 2000), suggesting a participation of these proteins in a different step of pre-mRNA maturation. Alternatively, the absence of splicing effects might also be explained by saturating levels of the respective factors already without their overexpression in this particular experimental system, and, thus, does not generally exclude a role of those proteins in splicing. In line with this hypothesis, both UBP1 and RBP45 were able to enhance intron recognition upon overexpression in an artificial mini-exon system (Simpson et al., 2004). Furthermore, in an approach to determine proteins affecting plant U12-dependent intron splicing, no effect of RBP45 or UBP1 on splicing efficiency for this rare intron type was observed (Lewandowska et al., 2004).
Plant hnRNP proteins with unknown or splicing-unrelated modes of function
Biological roles have been ascribed to several other plant hnRNP proteins, e.g., by analyzing biological consequences upon altering their expression. The mode of action for many of these factors, however, remains largely unknown. For example, the AAPK-interacting protein 1 (AKIP1) of Vicia faba, a protein with sequence homology to hnRNP A/B, was revealed to be a substrate of the abscisic acid-activated protein kinase (AAPK; Li et al., 2002). This kinase is localized in guard cells, where it regulates plasma membrane ion channels in response to the stress hormone abscisic acid (ABA). Both AKIP1 and AAPK were shown to be nuclear localized, but upon in vivo treatment with ABA, AKIP1 increasingly localized to speckle-like structures within the nucleus. Additionally, upon ABA-mediated phosphorylation of AKIP1 by AAPK, AKIP1 displayed an increased affinity for its interaction target, the mRNA of dehydrin, a protein reported to be involved in cell protection under stress conditions. As mentioned before, posttranslational modifications have been described for several other hnRNP proteins and, thus, might play a more widespread role in defining their RNA target specificity.
The closest homolog of V. faba AKIP1 is the previously described protein UBA2a from Arabidopsis, showing a similar nuclear reorganization to speckles in response to ABA (Riera et al., 2006). However, in contrast to AKIP1, UBA2a is, if at all, only weakly phosphorylated by OPEN STOMATA 1 (OST1), the Arabidopsis ortholog of AAPK. Overexpression of the three Arabidopsis UBA2a genes was demonstrated to be lethal, while expression under control of an inducible system triggered leaf senescence and hypersensitive-like cell death (Kim et al., 2008a). Further work will be required to reveal if these phenotypes are linked to altered metabolism of distinct target transcripts.
A possible link between polycomb regulation, which is based on polycomb complex-mediated control of gene expression via modulation of epigenetic patterns (Schwartz and Pirrotta, 2007), and RNA processing was indicated by investigation of LIF2, a putative RNA binding protein of the hnRNP family. LIF2 interacts with the chromo domain protein Like Heterochromatin Protein 1 (LHP1; Latrasse et al., 2011), which is a subunit of a polycomb repressive complex (PRC) in Arabidopsis. LHP1 recognizes histone H3 lysine 27 trimethylation, an epigenetic silencing signal, deposited by the PRC2 complex. Its interaction partner LIF2 was described to be able to either antagonize or act with LHP1, suggesting that it may modulate LHP1-activity at specific loci or in response to environmental changes, in order to control cell fate determination.
Besides their function in RNA metabolism, hnRNP proteins can also bind to DNA and affect diverse processes, such as replication, DNA repair, and transcription. One example from plants is the G-strand specific single-stranded telomere binding protein GTBP1 from Nicotiana tabacum, which negatively regulates telomere length (Lee and Kim, 2010). Using an RNA interference approach to downregulate expression of GTBP1, Lee and Kim (2010) found severe developmental abnormalities in the mutant plants, as well as signs of genome instability, including longer telomeres, formation of extrachromosomal telomeric circles and abnormal anaphase bridges. Subsequently, they could reveal a function of GTBP1 in inhibiting telomeric strand invasion, the initial step in interchromosomal homologous recombination, thus indicating that GTBP1 is a negative regulator of telomere length, being essential for both structure and function of tobacco telomeres.
Interestingly, hnRNP protein homologs are also found in plant organelles. Chloroplastidic ribonucleoproteins (cpRNPs) have been described to play a role in chloroplast RNA processing steps (Tillich et al., 2010), such as 3′ end processing (Schuster and Gruissem, 1991) and RNA editing (Hirose and Sugiura, 2001), as well as in regulating transcript stability (Nakamura et al., 2001). For example, knockout of the cpRNP C31A gave rise to alterations in both editing and steady state levels of distinct sets of chloroplast mRNAs (Tillich et al., 2009). cpRNPs possess twin RRMs and are subject to light-dependent phosphorylation (Tillich et al., 2010), which has been demonstrated to reduce the RNA in vitro binding affinity in case of CP28 (Lisitsky and Schuster, 1995). Furthermore, analysis of mutants suggests that cpRNPs have both specific and combinatorial functions, as it is also observed for many hnRNPs.
Conclusion
Global analyses of transcript data have revealed the widespread occurrence of AS in plants, bearing an enormous potential for a functional expansion of the transcriptome. Although at this point it remains unclear which fraction of all AS events plays a role in plant gene expression, a steadily growing list of studies provides compelling evidence for important implications of AS in numerous fundamental biological processes. Thus, one of the most intriguing questions in current AS research in plants is how AS can be regulated and how it can be coordinated with other processes to trigger specific splicing programs in response to internal and external cues. Important lessons can be learned from splicing research performed in animal systems, however, previous studies also have highlighted numerous distinct features of splicing in plants and animals. Recent progress in the characterization of SR and hnRNP proteins from plants represents an important step toward deciphering the plant splicing code. It can be anticipated that the combinatorial action of those splicing factors play a pivotal role in determining the complex AS patterns observed in plants, and thereby critically contributes to the regulation of gene expression in the context of diverse intrinsic processes as well as in response to external signals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Gabriele Drechsel for critical reading of the manuscript. Splicing research in the Wachter laboratory is supported by an Emmy Noether fellowship and research grant WA 2167/4-1 of the German Research Foundation.
References
- Amir-Ahmady B., Boutz P. L., Markovtsov V., Phillips M. L., Black D. L. (2005). Exon repression by polypyrimidine tract binding protein. RNA 11, 699–716 10.1261/rna.2250405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter S. D., Allain F. H. (2008). Structure-function relationships of the polypyrimidine tract binding protein. Cell. Mol. Life Sci. 65, 516–527 10.1007/s00018-007-7378-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W. B., Fu Y., Mcginnis K. M. (2008). Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 18, 1381–1392 10.1101/gr.053678.106 [DOI] [PubMed] [Google Scholar]
- Besse F., Lopez De Quinto S., Marchand V., Trucco A., Ephrussi A. (2009). Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 23, 195–207 10.1101/gad.505709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz P. L., Stoilov P., Li Q., Lin C. H., Chawla G., Ostrow K., Shiue L., Ares M., Jr., Black D. L. (2007). A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 21, 1636–1652 10.1101/gad.1558107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J., Kim C. Y., Gibson C. A., Assmann S. M. (2008). Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 67, 71–88 10.1007/s11103-008-9302-z [DOI] [PubMed] [Google Scholar]
- Caputi M., Zahler A. M. (2002). SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21, 845–855 10.1093/emboj/21.4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. I., Hsu S. C., Chau G. Y., Huang C. Y., Sung J. S., Hua W. K., Lin W. J. (2011). Identification of the methylation preference region in heterogeneous nuclear ribonucleoprotein K by protein arginine methyltransferase 1 and its implication in regulating nuclear/cytoplasmic distribution. Biochem. Biophys. Res. Commun. 404, 865–869 10.1016/j.bbrc.2010.12.076 [DOI] [PubMed] [Google Scholar]
- Chen M., Manley J. L. (2009). Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10, 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. J., Chen M., Assanah M., Canoll P., Manley J. L. (2010). HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 107, 19114–19119 10.1073/pnas.1007883107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Cline M. S., Osborne R. J., Tuttle D. L., Clark T. A., Donohue J. P., Hall M. P., Shiue L., Swanson M. S., Thornton C. A., Ares M., Jr. (2010). Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17, 187–193 10.1038/nsmb.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Priest H. D., Givan S. A., Shen R., Bryant D. W., Fox S. E., Wong W. K., Mockler T. C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58 10.1101/gr.093302.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Guo M., Jeong B. R., Tian F., Elthon T. E., Cerny R. L., Staiger D., Alfano J. R. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447, 284–288 10.1038/nature05737 [DOI] [PubMed] [Google Scholar]
- Gamberi C., Izaurralde E., Beisel C., Mattaj I. W. (1997). Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol. 17, 2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham B. K., Brandom J. L., Xoconostle-Cazares B., Ringgold V., Lough T. J., Lucas W. J. (2009). A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215 10.1105/tpc.108.061317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. P., Tang Y. H., Smith R. (2010). Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392 10.1042/BJ20100396 [DOI] [PubMed] [Google Scholar]
- Heiner M., Hui J., Schreiner S., Hung L. H., Bindereif A. (2010). HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 7, 56–64 10.4161/rna.7.1.10402 [DOI] [PubMed] [Google Scholar]
- Hirose T., Sugiura M. (2001). Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J. 20, 1144–1152 10.1093/emboj/20.5.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J., Stangl K., Lane W. S., Bindereif A. (2003). HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10, 33–37 10.1038/nsb875 [DOI] [PubMed] [Google Scholar]
- Izquierdo J. M., Majos N., Bonnal S., Martinez C., Castelo R., Guigo R., Bilbao D., Valcarcel J. (2005). Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 19, 475–484 10.1016/j.molcel.2005.06.015 [DOI] [PubMed] [Google Scholar]
- Jeong B. R., Lin Y., Joe A., Guo M., Korneli C., Yang H., Wang P., Yu M., Cerny R. L., Staiger D., Alfano J. R., Xu Y. (2011). Structure function analysis of an ADP-ribosyltransferase type III effector and its RNA-binding target in plant immunity. J. Biol. Chem. 286, 43272–43281 10.1074/jbc.M111.290122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. Y., Bove J., Assmann S. M. (2008a). Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol. 180, 57–70 10.1111/j.1469-8137.2008.02557.x [DOI] [PubMed] [Google Scholar]
- Kim J. S., Jung H. J., Lee H. J., Kim K. A., Goh C. H., Woo Y., Oh S. H., Han Y. S., Kang H. (2008b). Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J. 55, 455–466 10.1111/j.1365-313X.2008.03518.x [DOI] [PubMed] [Google Scholar]
- Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D. J., Luscombe N. M., Ule J. (2010). iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambermon M. H., Fu Y., Wieczorek Kirk D. A., Dupasquier M., Filipowicz W., Lorkovic Z. J. (2002). UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell. Biol. 22, 4346–4357 10.1128/MCB.22.12.4346-4357.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambermon M. H., Simpson G. G., Wieczorek Kirk D. A., Hemmings-Mieszczak M., Klahre U., Filipowicz W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19, 1638–1649 10.1093/emboj/19.7.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D., Germann S., Houba-Herin N., Dubois E., Bui-Prodhomme D., Hourcade D., Juul-Jensen T., Le Roux C., Majira A., Simoncello N., Granier F., Taconnat L., Renou J. P., Gaudin V. (2011). Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS ONE 6, e16592. 10.1371/journal.pone.0016592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Stallcup M. R. (2009). Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol. Endocrinol. 23, 425–433 10.1210/me.2007-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. W., Kim W. T. (2010). Tobacco GTBP1, a homolog of human heterogeneous nuclear ribonucleoprotein, protects telomeres from aberrant homologous recombination. Plant Cell 22, 2781–2795 10.1105/tpc.110.077958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska D., Simpson C. G., Clark G. P., Jennings N. S., Barciszewska-Pacak M., Lin C. F., Makalowski W., Brown J. W., Jarmolowski A. (2004). Determinants of plant U12-dependent intron splicing efficiency. Plant Cell 16, 1340–1352 10.1105/tpc.020743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Kinoshita T., Pandey S., Ng C. K., Gygi S. P., Shimazaki K., Assmann S. M. (2002). Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418, 793–797 10.1038/nature00865 [DOI] [PubMed] [Google Scholar]
- Li P., Ham B. K., Lucas W. J. (2011). CmRBP50 protein phosphorylation is essential for assembly of a stable phloem-mobile high-affinity ribonucleoprotein complex. J. Biol. Chem. 286, 23142–23149 10.1074/jbc.M111.229781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D. D., Mele A., Fak J. J., Ule J., Kayikci M., Chi S. W., Clark T. A., Schweitzer A. C., Blume J. E., Wang X., Darnell J. C., Darnell R. B. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 10.1038/nature07488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M. H., Kim J., Kim Y. S., Chung K. S., Seo Y. H., Lee I., Hong C. B., Kim H. J., Park C. M. (2004). A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16, 731–740 10.1105/tpc.019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I., Schuster G. (1995). Phosphorylation of a chloroplast RNA-binding protein changes its affinity to RNA. Nucleic Acids Res. 23, 2506–2511 10.1093/nar/23.13.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M., Schwartz S., Clark T. A., Hollander D., Tan L. Y., Spellman R., Gordon A., Schweitzer A. C., De La Grange P., Ast G., Smith C. W. (2010). Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 17, 1114–1123 10.1038/nsmb.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z. J., Wieczorek Kirk D. A., Klahre U., Hemmings-Mieszczak M., Filipowicz W. (2000). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6, 1610–1624 10.1017/S1355838200001163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H., Helfman D. M., Gagel R. F., Berget S. M. (1999). Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19, 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., Misteli T. (2010). Regulation of alternative splicing by histone modifications. Science 327, 996–1000 10.1126/science.1184208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth C. D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., Rybin V., Valcarcel J., Sattler M. (2011). Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475, 408–411 10.1038/nature10171 [DOI] [PubMed] [Google Scholar]
- Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 10.1016/j.molcel.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V., Nikolic J. M., Goldman J. A., Turck C. W., Chou M. Y., Black D. L. (2000). Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20, 7463–7479 10.1128/MCB.20.20.7463-7479.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R., Cloutier P., Shkreta L., Fisette J. F., Revil T., Chabot B. (2007). hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 623, 123–147 10.1007/978-0-387-77374-2_8 [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R., Fisette J. F., Nasim F. U., Madden R., Cordeau M., Chabot B. (2006). Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 4, e21. 10.1371/journal.pbio.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T. C., Yu X., Shalitin D., Parikh D., Michael T. P., Liou J., Huang J., Smith Z., Alonso J. M., Ecker J. R., Chory J., Lin C. (2004). Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12759–12764 10.1073/pnas.0404552101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z., Abel L., Yong J., Kataoka N., Dreyfuss G. (2001). SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20, 5443–5452 10.1093/emboj/20.19.5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Ohta M., Sugiura M., Sugita M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276, 147–152 10.1074/jbc.M108594200 [DOI] [PubMed] [Google Scholar]
- Neubauer G., King A., Rappsilber J., Calvio C., Watson M., Ajuh P., Sleeman J., Lamond A., Mann M. (1998). Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20, 46–50 10.1038/1700 [DOI] [PubMed] [Google Scholar]
- Oberstrass F. C., Auweter S. D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D. L., Allain F. H. (2005). Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309, 2054–2057 10.1126/science.1114066 [DOI] [PubMed] [Google Scholar]
- Oesterreich F. C., Bieberstein N., Neugebauer K. M. (2011). Pause locally, splice globally. Trends Cell Biol. 21, 328–335 10.1016/j.tcb.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Okunola H. L., Krainer A. R. (2009). Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol. Cell. Biol. 29, 5620–5631 10.1128/MCB.01678-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S., Blanchette M., Park J., Savva Y., Yeo G. W., Yeakley J. M., Rio D. C., Graveley B. R. (2007). A regulator of Dscam mutually exclusive splicing fidelity. Nat. Struct. Mol. Biol. 14, 1134–1140 10.1038/nsmb1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis C., Cloutier P., Shkreta L., Toutant J., Klarskov K., Chabot B. (2007). hnRNP I/PTB can antagonize the splicing repressor activity of SRp30c. RNA 13, 1287–1300 10.1261/rna.403607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Niu L., Lu F., Liu C., Zhai J., Kong X., Cao X. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144, 1913–1923 10.1104/pp.107.099531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58, 267–294 10.1146/annurev.arplant.58.032806.103754 [DOI] [PubMed] [Google Scholar]
- Riera M., Redko Y., Leung J. (2006). Arabidopsis RNA-binding protein UBA2a relocalizes into nuclear speckles in response to abscisic acid. FEBS Lett. 580, 4160–4165 10.1016/j.febslet.2006.06.064 [DOI] [PubMed] [Google Scholar]
- Robida M., Sridharan V., Morgan S., Rao T., Singh R. (2010). Drosophila polypyrimidine tract-binding protein is necessary for spermatid individualization. Proc. Natl. Acad. Sci. U.S.A. 107, 12570–12575 10.1073/pnas.1007935107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson F., Smith C. W. (2006). A splicing repressor domain in polypyrimidine tract-binding protein. J. Biol. Chem. 281, 800–806 10.1074/jbc.M510578200 [DOI] [PubMed] [Google Scholar]
- Sanchez S. E., Petrillo E., Beckwith E. J., Zhang X., Rugnone M. L., Hernando C. E., Cuevas J. C., Godoy Herz M. A., Depetris-Chauvin A., Simpson C. G., Brown J. W., Cerdan P. D., Borevitz J. O., Mas P., Ceriani M. F., Kornblihtt A. R., Yanovsky M. J. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 10.1038/nature09470 [DOI] [PubMed] [Google Scholar]
- Sauliere J., Sureau A., Expert-Bezancon A., Marie J. (2006). The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 26, 8755–8769 10.1128/MCB.00893-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka K., Bushell M., Spriggs K. A., Willis A. E. (2008). Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 36, 641–647 10.1042/BST0360641 [DOI] [PubMed] [Google Scholar]
- Schmitz R. J., Sung S., Amasino R. M. (2008). Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 411–416 10.1073/pnas.0710423104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning J. C., Streitner C., Meyer I. M., Gao Y., Staiger D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36, 6977–6987 10.1093/nar/gkn847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning J. C., Streitner C., Page D. R., Hennig S., Uchida K., Wolf E., Furuya M., Staiger D. (2007). Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52, 1119–1130 10.1111/j.1365-313X.2007.03302.x [DOI] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. (1991). Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 10, 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V. (2007). Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8, 9–22 10.1038/nrm2080 [DOI] [PubMed] [Google Scholar]
- Sharma S., Falick A. M., Black D. L. (2005). Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell 19, 485–496 10.1016/j.molcel.2005.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kohlstaedt L. A., Damianov A., Rio D. C., Black D. L. (2008). Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 15, 183–191 10.1038/nsmb.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C. G., Jennings S. N., Clark G. P., Thow G., Brown J. W. (2004). Dual functionality of a plant U-rich intronic sequence element. Plant J. 37, 82–91 10.1046/j.1365-313X.2003.01941.x [DOI] [PubMed] [Google Scholar]
- Singh R., Valcarcel J., Green M. R. (1995). Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268, 1173–1176 10.1126/science.7761834 [DOI] [PubMed] [Google Scholar]
- Spellman R., Llorian M., Smith C. W. (2007). Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 27, 420–434 10.1016/j.molcel.2007.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Smith C. W. (2006). Novel modes of splicing repression by PTB. Trends Biochem. Sci. 31, 73–76 10.1016/j.tibs.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Staiger D., Zecca L., Wieczorek Kirk D. A., Apel K., Eckstein L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361–371 10.1046/j.1365-313X.2003.01629.x [DOI] [PubMed] [Google Scholar]
- Stauffer E., Westermann A., Wagner G., Wachter A. (2010). Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 64, 243–255 10.1111/j.1365-313X.2010.04321.x [DOI] [PubMed] [Google Scholar]
- Streitner C., Danisman S., Wehrle F., Schoning J. C., Alfano J. R., Staiger D. (2008). The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J. 56, 239–250 10.1111/j.1365-313X.2008.03591.x [DOI] [PubMed] [Google Scholar]
- Streitner C., Hennig L., Korneli C., Staiger D. (2010). Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 10, 221. 10.1186/1471-2229-10-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne I. A., Meyer C. A., Liu X. S., Silver P. A., Brodsky A. S. (2006). Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription. Genome Res. 16, 912–921 10.1101/gr.5211806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Beick S., Schmitz-Linneweber C. (2010). Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biol. 7, 172–178 10.4161/rna.7.2.11090 [DOI] [PubMed] [Google Scholar]
- Tillich M., Hardel S. L., Kupsch C., Armbruster U., Delannoy E., Gualberto J. M., Lehwark P., Leister D., Small I. D., Schmitz-Linneweber C. (2009). Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 6002–6007 10.1073/pnas.0808529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Stefani G., Mele A., Ruggiu M., Wang X., Taneri B., Gaasterland T., Blencowe B. J., Darnell R. B. (2006). An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586 10.1038/nature05304 [DOI] [PubMed] [Google Scholar]
- Venables J. P., Klinck R., Koh C., Gervais-Bird J., Bramard A., Inkel L., Durand M., Couture S., Froehlich U., Lapointe E., Lucier J. F., Thibault P., Rancourt C., Tremblay K., Prinos P., Chabot B., Elela S. A. (2009). Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16, 670–676 10.1038/nsmb.1608 [DOI] [PubMed] [Google Scholar]
- Vitali F., Henning A., Oberstrass F. C., Hargous Y., Auweter S. D., Erat M., Allain F. H. (2006). Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 25, 150–162 10.1038/sj.emboj.7600911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A. (2010). Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 7, 67–76 10.4161/rna.7.1.10489 [DOI] [PubMed] [Google Scholar]
- Wagner E. J., Garcia-Blanco M. A. (2001). Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21, 3281–3288 10.1128/MCB.21.10.3281-3288.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. B., Brendel V. (2004). The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 5, R102. 10.1186/gb-2004-5-2-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. B., Brendel V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 103, 7175–7180 10.1073/pnas.0602073103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Okamoto T. (2009). Involvement of polypyrimidine tract-binding protein (PTB)-related proteins in pollen germination in Arabidopsis. Plant Cell Physiol. 50, 179–190 10.1093/pcp/pcn207 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26, 1934–1941 10.1038/sj.emboj.7601701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf M. B., Berglund J. A. (2010). Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 35, 169–178 10.1016/j.tibs.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., Smith C. W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 10.1016/S1097-2765(03)00502-1 [DOI] [PubMed] [Google Scholar]
- Xie J., Lee J. A., Kress T. L., Mowry K. L., Black D. L. (2003). Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U.S.A. 100, 8776–8781 10.1073/pnas.0438010100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Stoilov P., Kapur K., Han A., Jiang H., Shen S., Black D. L., Wong W. H. (2008). MADS: a new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA 14, 1470–1479 10.1261/rna.1070208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhou Y., Wu T., Zhu T., Ji X., Kwon Y. S., Zhang C., Yeo G., Black D. L., Sun H., Fu X. D., Zhang Y. (2009). Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36, 996–1006 10.1016/j.molcel.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G. W., Coufal N. G., Liang T. Y., Peng G. E., Fu X. D., Gage F. H. (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137 10.1038/nsmb.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang Z., Castle J., Sun S., Johnson J., Krainer A. R., Zhang M. Q. (2008). Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22, 2550–2563 10.1101/gad.1703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Guo G., Hu X., Zhang Y., Li Q., Li R., Zhuang R., Lu Z., He Z., Fang X., Chen L., Tian W., Tao Y., Kristiansen K., Zhang X., Li S., Yang H., Wang J. (2010). Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 20, 646–654 10.1101/gr.107334.110 [DOI] [PMC free article] [PubMed] [Google Scholar]