Abstract
Plant defenses aimed at necrotrophic pathogens appear to be genetically complex. Despite the apparent lack of a specific recognition of such necrotrophs by products of major R genes, biochemical, molecular, and genetic studies, in particular using the model plant Arabidopsis, have uncovered numerous host components critical for the outcome of such interactions. Although the JA signaling pathway plays a central role in plant defense toward necrotrophs additional signaling pathways contribute to the plant response network. Transcriptional reprogramming is a vital part of the host defense machinery and several key regulators have recently been identified. Some of these transcription factors positively affect plant resistance whereas others play a role in enhancing host susceptibility toward these phytopathogens.
Keywords: Arabidopsis, necrotrophs, transcriptional regulation, WRKY transcription factors
Introduction
Plants have developed a highly sophisticated immune system that enables them to perceive potential invaders and to respond accordingly to ensure host survival. Depending on the modus by which pathogens are recognized, two branches of plant immunity are currently distinguished based mainly on studies with the model plant Arabidopsis thaliana (Dodds and Rathjen, 2010). Pattern triggered immunity (PTI) is initiated by recognition of molecular structures characteristic of microbes (designated microbe-associated molecular patterns; MAMPS) by means of plasma membrane localized pattern recognition receptors (PRRs; Boller and Felix, 2009). In the case of effector triggered immunity (ETI), products of major resistance (R) genes, usually intracellular receptors, recognize corresponding effector molecules delivered by the pathogen into the host cell (Block et al., 2008). Although the molecular connections are not well understood, PTI and ETI share numerous components and give rise to similar qualitative responses. In both cases massive transcriptional reprogramming is a key step to initiate host defenses (Eulgem, 2005). Indeed, studies of the complex network properties of plant immunity have illustrated that it is comprised of distinct signaling sectors that interact with each other in a complex fashion (Tsuda et al., 2009; Sato et al., 2010). Plant immunity is also regulated by several phytohormones, including salicylic acid (SA), jasmonic acid (JA), and ethylene (ET; Glazebrook, 2005). In general, SA signaling sectors are essential for resistance toward biotrophic and hemibiotrophic pathogens whereas the JA and ET sectors are important for immunity toward necrotrophs (Pieterse et al., 2009). In this article we report on what is currently known on critical plant responses upon challenge mainly with necrotrophic fungi. Our focus of attention is directed toward the identified regulatory factors that modulate host transcriptional outputs in such interactions.
Arabidopsis–Necrotroph Pathogen Interactions
Necrotrophic fungi, including Alternaria brassicicola, Botrytis cinerea, Fusarium oxysporum, and Sclerotinia sclerotiorum constitute the largest class of fungal plant pathogens and are responsible for severe crop losses worldwide. Whereas resistance toward biotrophic pathogens is predominantly mediated by the recognition of pathogen effectors by R gene-encoded intracellular receptors, no R gene-dependent resistance toward necrotrophic fungi has been identified. This may in part be a consequence of the different strategies used by these phytopathogens. Colonization by biotrophic pathogens requires maintenance of host cell integrity at least for a restricted period of time, while the lifestyle of necrotrophic pathogens is geared to quickly killing host cells. They do so by employing toxins, various lytic enzymes, and additional molecules to destroy and decompose plant tissue (van Kan, 2006; Łaźniewska et al., 2010).
In Arabidopsis, resistance to B. cinerea appears to be under complex genetic control (Rowe and Kliebenstein, 2008). Genetic and pharmacological studies have identified plant genes and compounds that influence the outcome of host–B. cinerea interactions. Mutations in Arabidopsis genes encoding enzymes involved in secondary cell wall formation and cutin biosynthesis, in a pectin methylesterase, and in a novel membrane localized protein all enhanced resistance toward this pathogen (Hernandez-Blanco et al., 2007; Tang et al., 2007; Mang et al., 2009; Raiola et al., 2011). In contrast, mutations in BIK1, encoding a receptor-like kinase that functions in ethylene signaling and PTI, and in several autophagy genes increase plant susceptibility toward B. cinerea (Veronese et al., 2006; Lai et al., 2011). Similarly, the importance of the JA- and ET-signaling pathways in plant defense toward B. cinerea has been inferred from numerous mutant studies, but conflicting reports and identification of additional cross-communicating pathways suggest that our understanding of the specific host programs required for defense toward this necrotroph remains fragmentary, and in many cases is also affected by the pathogen isolate (Ferrari et al., 2007; Llorente et al., 2008; Pieterse et al., 2009; Rowe et al., 2010; Łaźniewska et al., 2010; El Oirdi et al., 2011). Part of the contradictory results may in part be due to differences in the assays used (infections on detached leaves versus on intact plants), since detachment of leaves also triggers the leaf senescence program, thereby complicating interpretations (Liu et al., 2007).
One critical component appears to be camalexin, the major phytoalexin of Arabidopsis. Plants deficient in the cytochrome P450 monooxygenase CYP71B15 (PAD3) that catalyzes the final step in camalexin biosynthesis are highly susceptible to B. cinerea infection (Ferrari et al., 2007; Figure 1). Moreover, plants exposed to treatments that drastically increase camalexin levels (i.e., UV-C light) are more resistant to this necrotroph (Stefanato et al., 2009). However, recent studies show that whereas resistance to some B. cinerea isolates is dependent on the accumulation of camalexin via JA signaling, this is not the case for other isolates, indicating that other signaling pathways can be utilized and that additional defenses are required (Rowe et al., 2010). Moreover, several mutants including bik1 have been identified that show enhanced B. cinerea susceptibility despite having wildtype levels of camalexin or remain fully resistant despite low levels of this phytoalexin (Veronese et al., 2004, 2006; Denby et al., 2005; Staal et al., 2008; Walley et al., 2008; Berr et al., 2010).
Figure 1.
Leaves of 4-week-old wt, wrky33 and pad3 plants were infected with droplets containing B. cinerea spores of the isolate 2100 from the Spanish Type Culture Collection. After 3 days leaves were detached and pictures taken. wrky33 and pad3 exhibited a severe phenotype with fast growing lesions.
Transcriptional Regulators Influencing Host–Necrotroph Interactions
Induced plant defense components may be uncovered by identifying transcriptional modulators controlling the expression of downstream regulatory circuits. Indeed, isolation of a JA-insensitive mutant designated jin1 revealed that the underlying gene encodes a basic-helix-loop-helix leucine zipper transcription factor named MYC2 (Lorenzo et al., 2004). MYC2 was found to antagonistically regulate two distinct branches of the JA pathway. Loss-of-MYC2 function rendered plants more resistant to both biotrophic and necrotrophic pathogens (Lorenzo et al., 2004; Nickstadt et al., 2004). Several whole-genome transcriptional profiles of Arabidopsis leaves inoculated with B. cinerea have been performed (AbuQamar et al., 2006; Ferrari et al., 2007; Rowe et al., 2010). These studies revealed massive B. cinerea-induced transcriptional reprogramming in the host affecting up to 20% (>4700) of the genes represented on the arrays. Among these are numerous genes encoding transcription factors. AbuQamar et al. (2006) identified 30 putative DNA-binding protein genes that were induced upon B. cinerea infection. Subsequent analyses of 14 loss-of-function mutants of these genes revealed that ZFAR1, encoding a novel zinc-finger protein with an ankyrin repeat, is required for resistance against this pathogen. Transcriptional profiling also revealed several gene members of the AP2/ERF-type transcription factor family to be strongly induced upon pathogen challenge. Members of this family have been shown to modulate expression of JA- and ET-response genes (Gutterson and Reuber, 2004; Pieterse et al., 2009). Transgenic Arabidopsis lines over-expressing ERF1 were sufficient to confer resistance toward the necrotrophic fungi B. cinerea, F. oxysporum, and P. cucumerina (Berrocal-Lobo and Solano, 2002; Berrocal-Lobo and Molina, 2004). Similarly, over-expression of ERF59/ORA59 increased resistance toward B. cinerea whereas RNAi-ORA59 silenced lines were more susceptible (Pré et al., 2008). Both ERF1 and ERF59/ORA59 appear to be key integrators of the JA and ET-signaling pathways (Pieterse et al., 2009). Moreover, ORA59 was recently shown to be a key mediator that counteracts SA-mediated suppression of JA/ET-response genes (Leon-Reyes et al., 2010). The studies on ORA59 function nicely exemplify the interplay between SA, JA and ET-signaling, and highlight the importance of hormone concentrations and in particular of the kinetics of phytohormone biosynthesis and signaling in determining the final outcome of a particular plant–pathogen interaction.
Two MYB-type Arabidopsis transcription factors (BOS1/AtMY B108 and MYB46) have been identified that regulate distinct host transcriptional responses toward B. cinerea. MYB46 modulates secondary cell wall biosynthesis in the vasculature of the stem but also appears to play a role in disease susceptibility since myb46 mutants show increased disease resistance toward B. cinerea (Ramírez et al., 2011). Elevated resistance however was not directly correlated with any major alterations in cell wall polymer constituents. Transcriptomic analysis revealed that most of the differentially expressed genes in the myb46 mutant were down-regulated, with a significant number of genes predicted to function in cell wall metabolism and extracellular matrix remodeling and plant defenses (in particular numerous class III peroxidases, but also in extracellular plant defenses). A few of the peroxidase genes have been shown to enhance resistance to B. cinerea upon over-expression (Chassot et al., 2007).
In contrast, loss-of-BOS1/AtMYB108 function resulted in increased plant susceptibility to B. cinerea and A. brassicicola infection (Mengiste et al., 2003). Pathogen-induced activation of BOS1/AtMYB108 expression partly requires an intact JA signaling pathway. BOS1/AtMYB108 physically interacts with and is ubiquitinated by a RING E3 ligase designated BOI (Luo et al., 2010). This regulatory relationship between these two proteins appears to be important since RNAi-BOI silenced lines were equally susceptible to B. cinerea as were bos1 mutants.
Two NAC family proteins, ANAC019 and ANAC055, were demonstrated to function as activators of JA-induced defense genes downstream of MYC2 (Bu et al., 2008). An anac019 anac055 double mutant showed strong resistance toward B. cinerea infection similar to that observed for myc2 mutant plants, whereas ANAC019 and ANAC055 over-expressor lines showed increased susceptibility. In a second study, the NAC transcription factor ATAF1 (ANAC002) was also found to be a negative regulator of defense responses against necrotrophic fungi (Wang et al., 2009). B. cinerea growth was retarded in the ataf1-2 mutant, while over-expression of ATAF1 resulted in severe susceptibility to B. cinerea and to A. brassicicola. Where within the genetic network ATAF1 acts however remains to be determined.
Alterations in local chromatin structure underlying promoters can be a key component involved in controlling highly restricted expression of genes. Two such players, namely SPLAYED (SYD), a SWI/SNF class chromatin remodeling ATPase, and SET DOMAIN GROUP8 (SDG8), a histone methyltransferase were identified that affect distinct plant defense responses. SYD was found to be directly recruited to promoters of several JA/ET-response genes, and syd mutants were susceptible to B. cinerea but not to the biotrophic bacterium Pseudomonas syringae (Walley et al., 2008). Similarly, SDG8 was required to induce a subset of JA/ET-response genes and sdg8-1 mutant plants showed reduced resistance toward A. brassicicola and B. cinerea (Berr et al., 2010). Interestingly, syd and sdg8 mutants both have wildtype-like levels of camalexin.
WRKY Transcription Factors in Defense Toward Necrotrophs
Numerous members of the large zinc-finger-type WRKY transcription factor family have been identified to play key roles in regulating defense responses in various plant species to different pathogens (Pandey and Somssich, 2009). Arabidopsis WRKY70 was shown to be important for resistance against the necrotrophic bacterium Pectobacterium carotovorum (formerly Erwinia carotovora), and it was proposed to act as an integrator of SA and JA signaling (Li et al., 2004). This gene was also identified among the 30 putative DNA-binding protein genes that were induced upon B. cinerea infection, and wrky70 mutants exhibited enhanced susceptibility to B. cinerea, although interestingly, these plants remained fully resistant to the necrotroph A. brassicicola (AbuQamar et al., 2006).
In contrast, wrky33 mutants were found to be highly susceptible to both B. cinerea and A. brassicicola indicating that WRKY33 is a key positive regulator of defenses toward these necrotrophic fungi (Zheng et al., 2006; Figures 1 and 2). Recent studies are beginning to uncover the mode of action by which WRKY33 integrates host signaling to confer resistance upon pathogen challenge. WRKY33 has been shown to interact with MAP KINASE 4 (MPK4) and the MPK4 substrate MKS1 within the nucleus (Andreasson et al., 2005; Qiu et al., 2008). Upon challenge with P. syringae or the MAMP flg22, MPK4 phosphorylates MKS1, which results in the release of MKS1 and WRKY33 from the complex, and binding of WRKY33 to the PAD3 promoter. Activation of PAD3 results in the increased synthesis of camalexin. The importance of WRKY33 for camalexin biosynthesis was substantiated in later experiments employing B. cinerea (Mao et al., 2011). However, in this case activation of WRKY33 expression and WRKY33 phosphorylation were dependent on the MAP KINASES 3 (MPK3) and 6 (MPK6). Whether distinct MAP kinase pathways are employed dependent on MAMP treatment or on the type of pathogen used for infection, or whether differences in experimental design led to these conflicting results remains to be clarified. That the former may be the case is exemplified by the latest observations demonstrating that whereas MPK3 plays a major role in maintenance of basal resistance toward B. cinerea it is MPK6 that is the major player in MAMP-triggered resistance against the necrotrophic pathogen (Galletti et al., 2011). One should note however that the vast majority of such studies have been performed on Arabidopsis seedlings and it remains to be determined whether the same signaling pathways are utilized in mature plants.
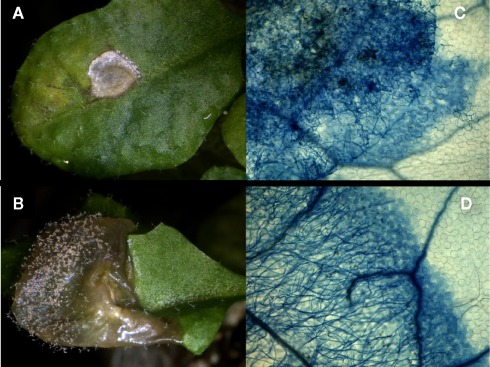
Figure 2.
Leaves of 4-week-old wt (A,C) and wrky33 (B,D) plants were infected with droplets containing B. cinerea spores. Macroscopic pictures (A,B) were taken 7 day past infection and show conidiophores growing only on wrky33. For the micrographs (C,D) leaves were stained with trypan blue 64 h past infection to visualize growing fungal mycelium and dying cells. While on wt the fungus died within the boundaries of the droplet, fast growing mycelium was observed on wrky33.
Interestingly, induced expression of WRKY33 itself appears to be regulated by WRKY factors including autoregulation by WRKY33 (Lippok et al., 2007; Mao et al., 2011). Recently two other WRKY factors, WRKY50 and WRKY51, have been identified that negatively influence the outcome of Arabidopsis–B. cinerea interactions (Gao et al., 2011). Mutations in SSI1, encoding a plastid-localized stearoyl-acyl-carrier protein desaturase, render plants susceptible to B. cinerea. WRKY50 and WRKY51 contribute to this susceptibility since ssi1 wrky50 wrky51 triple mutants were as resistant toward this pathogen as wild type plants. Genetic studies suggest that WRKY50/51 mediate SA-dependent repression of JA inducible defense responses but the mechanisms how this is achieved remain elusive.
Summary and Perspectives
The molecular basis for resistance toward necrotrophic pathogens is still mostly unknown despite recent advances that have uncovered distinct signaling pathways, enzymes, and key regulatory factors involved in this process. Since comprehensive transcriptional reprogramming is a major determinant in this process we need to unravel the interwoven regulatory circuits and define key regulatory nodes that ultimately influence proper host defense gene expression. In the case of WRKY33 we are currently using global expression microarrays and chromatin immunoprecipitation combined with next generation sequencing (ChIP-Seq) to identify downstream target genes. Our particular focus is on uncovering direct targets encoding additional transcriptional regulators that may define such regulatory sub-nodes. These studies should allow us to define the networks in which WRKY33 acts to maintain robust plant immunity toward B. cinerea infection.
Equally important however will be to uncover the numerous mechanisms employed by necrotrophic pathogens to overcome plant resistance. We still know every little about the evolutionary forces that shape plant–necrotrophic fungi interactions. As recently outlined by Rowe and Kliebenstein (2010) one reason for this lack of knowledge has to due with our previous failure to consider and use necrotrophic diversity in former studies. Thus future research needs to embrace the intraspecific variation that exists in these pathogens to fully grasp the biological complexity of plant–necrotroph interactions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the DFG Grant SO235/7-1 within the framework of the Arabidopsis Functional Genomics Network (AFGN).
References
- AbuQamar S., Chen X., Dhawan R., Bluhm B., Salmeron J., Lam S., Dietrich R. A., Mengiste T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48, 28–44 10.1111/j.1365-313X.2006.02849.x [DOI] [PubMed] [Google Scholar]
- Andreasson E., Jenkins T., Brodersen P., Thorgrimsen S., Petersen N. H. T., Zhu S., Qiu J.-L., Micheelsen P., Rocher A., Petersen M., Newman M.-A., Nielsen H. B., Hirt H., Somssich I., Mattsson O., Mundy J. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589 10.1038/sj.emboj.7600737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Mccallum E. J., Alioua A., Heintz D., Heitz T., Shen W.-H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414 10.1104/pp.110.161497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soil borne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17, 763–770 10.1094/MPMI.2004.17.7.763 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32 10.1046/j.1365-313x.2002.01191.x [DOI] [PubMed] [Google Scholar]
- Block A., Li G., Fu Z. Q., Alfano J. R. (2008). Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403 10.1016/j.pbi.2008.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- Bu Q., Jiang H., Li C.-B., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Li C. (2008). Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18, 756–767 10.1038/cr.2008.53 [DOI] [PubMed] [Google Scholar]
- Chassot C., Nawrath C., Metraux J.-P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49, 972–980 10.1111/j.1365-313X.2006.03017.x [DOI] [PubMed] [Google Scholar]
- Denby K. J., Jason L. J. M., Murray S. L., Last R. L. (2005). ups1, an Arabidopsis thaliana camalexin accumulation mutant defective in multiple defence signalling pathways. Plant J. 41, 673–684 10.1111/j.1365-313X.2005.02327.x [DOI] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 10.1038/nrm2939 [DOI] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T. A., Rigano L., El Hadrami A., Rodriguez M. C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23, 2405–2421 10.1105/tpc.111.083394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78 10.1016/j.tplants.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Galletti R., Denoux C., De Lorenzo G., Ausubel F. M., Dewdney J. (2007). Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 144, 367–379 10.1104/pp.107.095596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814 10.1104/pp.111.174003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.-M., Venugopal S., Navarre D., Kachroo A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476 10.1104/pp.110.166876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Gutterson N., Reuber L. T. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7, 465–471 10.1016/j.pbi.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Hernandez-Blanco C., Feng D. X., Hu J., Sanchez-Vallet A., Deslandes L., Llorente F., Berrocal-Lobo M., Keller H., Barlet X., Sanchez-Rodriguez C., Anderson L. K., Somerville S., Marco Y., Molina A. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903 10.1105/tpc.106.048058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968 10.1111/j.1365-313X.2011.04553.x [DOI] [PubMed] [Google Scholar]
- Łaźniewska J., Macioszek V., Lawrence C., Kononowicz A. (2010). Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens. Acta Physiol. Plant 32, 1–10 10.1007/s11738-009-0372-6 [DOI] [Google Scholar]
- Leon-Reyes A., Du Y., Koorneef A., Proietti S., Körbes A. P., Memelink J., Pieterse C. M. J., Ritsema T. (2010). Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 23, 187–197 10.1094/MPMI-23-2-0187 [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331 10.1105/tpc.017954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok B., Birkenbihl R. P., Rivory G., Brümmer J., Schmelzer E., Logemann E., Somssich I. E. (2007). Expression of AtWRKY33 encoding a pathogen-/PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20, 420–429 10.1094/MPMI-20-4-0420 [DOI] [PubMed] [Google Scholar]
- Liu G., Kennedy R., Greenshields D. L., Peng G., Forseille L., Selvaraj G., Wei Y. (2007). Detached and attached Arabidopsis leaf assays reveal distinctive defense responses against hemibiotrophic Colletotrichum spp. Mol. Plant Microbe Interact. 20, 1308–1313 10.1094/MPMI-20-10-1308 [DOI] [PubMed] [Google Scholar]
- Llorente F., Muskett P., Sanchez-Vallet A., Lopez G., Ramos B., Sanchez-Rodriguez C., Jorda L., Parker J., Molina A. (2008). Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 1, 496–509 10.1093/mp/ssn025 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J. M., Sanchez-Serrano J. J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Laluk K., Lai Z., Veronese P., Song F., Mengiste T. (2010). The Arabidopsis Botrytis susceptible1 interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 154, 1766–1782 10.1104/pp.110.163915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang H. G., Laluk K. A., Parsons E. P., Kosma D. K., Cooper B. R., Park H. C., Abuqamar S., Boccongelli C., Miyazaki S., Consiglio F., Chilosi G., Bohnert H. J., Bressan R. A., Mengiste T., Jenks M. A. (2009). The Arabidopsis RESURRECTION1 gene regulates a novel antagonistic interaction in plant defense to biotrophs and necrotrophs. Plant Physiol. 151, 290–305 10.1104/pp.109.142158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653 10.1105/tpc.111.084996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15, 2551–2565 10.1105/tpc.014167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickstadt A., Thomma B. P. H. J., Feussner I., Kangasjärvi J., Zeier J., Loeffler C., Scheel D., Berger S. (2004). The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol. Plant Pathol. 5, 425–434 10.1111/j.1364-3703.2004.00242.x [DOI] [PubMed] [Google Scholar]
- Pandey S. P., Somssich I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van Der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C. M. J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357 10.1104/pp.108.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.-L., Fiil B.-K., Petersen K., Nielsen H. B., Botanga C. J., Thorgrimsen S., Palma K., Suarez-Rodriguez M. C., Sandbech-Clausen S., Lichota J., Brodersen P., Grasser K. D., Mattsson O., Glazebrook J., Mundy J., Petersen M. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221 10.1038/emboj.2008.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiola A., Lionetti V., Elmaghraby I., Immerzeel P., Mellerowicz E. J., Salvi G., Cervone F., Bellincampi D. (2011). Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol. Plant Microbe Interact. 24, 432–440 10.1094/MPMI-07-10-0157 [DOI] [PubMed] [Google Scholar]
- Ramírez V., Agorio A., Coego A., García-Andrade J., Hernández M. J., Balaguer B., Ouwerkerk P. B. F., Zarra I., Vera P. (2011). MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 155, 1920–1935 10.1104/pp.110.171843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. C., Kliebenstein D. J. (2008). Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180, 2237–2250 10.1534/genetics.108.091439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. C., Kliebenstein D. J. (2010). All mold is not alike: the importance of intraspecific diversity in necrotrophic plant pathogens. PLoS Pathog. 6, e1000759. 10.1371/journal.ppat.1000759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. C., Walley J. W., Corwin J., Chan E. K. F., Dehesh K., Kliebenstein D. J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6, e1000861. 10.1371/journal.ppat.1000861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Tsuda K., Wang L., Coller J., Watanabe Y., Glazebrook J., Katagiri F. (2010). Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6, e1001011. 10.1371/journal.ppat.1001011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal J., Kaliff M., Dewaele E., Persson M., Dixelius C. (2008). RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 55, 188–200 10.1111/j.1365-313X.2008.03503.x [DOI] [PubMed] [Google Scholar]
- Stefanato F. L., Abou-Mansour E., Buchala A., Kretschmer M., Mosbach A., Hahn M., Bochet C. G., Métraux J.-P., Schoonbeek H.-J. (2009). The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 58, 499–510 10.1111/j.1365-313X.2009.03794.x [DOI] [PubMed] [Google Scholar]
- Tang D., Simonich M. T., Innes R. W. (2007). Mutations in LACS2, a long-chain Acyl-Coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol. 144, 1093–1103 10.1104/pp.106.094318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. 10.1371/journal.pgen.1000772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kan J. A. L. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253 10.1016/j.tplants.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Veronese P., Chen X., Bluhm B., Salmeron J., Dietrich R., Mengiste T. (2004). The BOS loci of Arabidopsis are required for resistance to Botrytis cinerea infection. Plant J. 40, 558–574 10.1111/j.1365-313X.2004.02232.x [DOI] [PubMed] [Google Scholar]
- Veronese P., Nakagami H., Bluhm B., Abuqamar S., Chen X., Salmeron J., Dietrich R. A., Hirt H., Mengiste T. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273 10.1105/tpc.105.035576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J. W., Rowe H. C., Xiao Y., Chehab E. W., Kliebenstein D. J., Wagner D., Dehesh K. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4, e1000237. 10.1371/journal.ppat.1000237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. E., Basnayake B. M. V. S., Zhang H., Li G., Li W., Virk N., Mengiste T., Song F. (2009). The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant Microbe Interact. 22, 1227–1238 10.1094/MPMI-22-5-0498 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S. A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 10.1111/j.1365-313X.2006.02901.x [DOI] [PubMed] [Google Scholar]