Abstract
Understanding the regulated inter- and intra-cellular metal circulation is one of the challenges in the field of metal homeostasis. Inside organisms metal ions are bound to organic ligands to prevent their uncontrolled reactivity and to increase their solubility. Nicotianamine (NA) is one of the important ligands. This non-proteinogenic amino acid is synthesized by nicotianamine synthase (NAS). NA is involved in mobilization, uptake, transport, storage, and detoxification of metals. Much of the progress in understanding NA function has been achieved by studying mutants with altered nicotianamine levels. Mild and strong Arabidopsis mutants impaired in nicotianamine synthesis have been identified and characterized, namely nas4x-1 and nas4x-2. Arabidopsis thaliana has four NAS genes. In this review, we summarize the structure and evolution of the NAS genes in the Arabidopsis genome. We summarize previous results and present novel evidence that the four NAS genes have partially overlapping functions when plants are exposed to Fe deficiency and nickel supply. We compare the phenotypes of nas4x-1 and nas4x-2 and summarize the functions of NAS genes and NA as deduced from the studies of mutant phenotypes.
Keywords: nicotianamine, metal binding, chelation, gene family, multiple mutant
Introduction
Iron (Fe) and copper (Cu) are essential elements for all living organisms because of their unique property of being able to catalyze oxidation/reduction reactions. Conversely, an excess of Fe, especially Fe2+, is detrimental since it catalyzes the production of reactive oxygen species (ROS) in the Fenton reaction (Fenton, 1894; Hell and Stephan, 2003). For this reason, free metal ions are not likely to exist in large amounts in cells. Indeed, Fe and other metals are mainly present in stable complexes with organic ligands or inorganic phosphates (Haydon and Cobbett, 2007).
Nicotianamine (NA) is one of the most investigated metal chelator molecules in plants. NA is a non-proteinogenic amino acid and it results from the enzymatic condensation of three S-adenosyl methionine molecules (SAM) catalyzed by nicotianamine synthase (NAS; Herbik et al., 1999; Ling et al., 1999; Takahashi et al., 1999). NA is able to form stable complexes with Mn2+, Fe2+, Co2+, Zn2+, Ni2+, and Cu2+ (Benes et al., 1983; Anderegg and Ripperger, 1989). Moreover, NA has a high capacity to chelate Fe3+ (von Wiren et al., 1999; Weber et al., 2006; Rellan-Alvarez et al., 2008). For all the metals considered, the stability of the NA–metal complexes had its maximum at pH values above 6.5, suggesting that NA is more likely a symplastic chelator of metals and therefore would bind metals predominately within cells and the phloem (von Wiren et al., 1999). Cu2+ is an exception among the essential metals, since the Cu2+–NA complex is very stable in mild acidic conditions, which is a strong argument in favor of the possible occurrence of Cu2+–NA complexes in the apoplastic environment as prevailing in the xylem sap (von Wiren et al., 1999). Nicotianamine can be transported to the various organs and tissues via oligopeptide transporters, such as yellowstripe1-like (YSL) proteins (Curie et al., 2001, 2009). Rice ENA1 and ENA2 transporters were just recently described to mediate NA export from cells (Nozoye et al., 2011).
Studies of solanaceous and graminaceous plants as well as of hyperaccumulators showed that NA functions in long-distance transport of Cu (Pich and Scholz, 1996), short-distance and intracellular transport of Fe (Becker et al., 1995; Curie and Briat, 2003), plant reproduction (Takahashi et al., 2006), detoxification of heavy metals like Ni (Douchkov et al., 2005; Kim et al., 2005; Pianelli et al., 2005; Mari et al., 2006; Ouerdane et al., 2006; van de Mortel et al., 2006; Callahan et al., 2007) and Zn (Becher et al., 2004; Weber et al., 2004; Talke et al., 2006; van de Mortel et al., 2006), and in grasses as a precursor in the biosynthesis of phytosiderophores (Mori and Nishizawa, 1987). Several studies suggested that NA could be involved in iron mobilization and accumulation in plant roots and seeds (Douchkov et al., 2001, 2005; Cheng et al., 2007; Lee et al., 2009). NA is beneficial for increased bioavailability of Fe in foods (Maurer et al., 2010; Zheng et al., 2010).
Taken together, NA is a key compound of metal homeostasis in plants contributing to mobilization, uptake, transport, storage, and detoxification of metals. Since NA was found to be an important biofortification factor for essential nutrients like Fe and Zn in edible portions of crop plants (Zheng et al., 2010), further knowledge about the functions of NA and the characterization of the essential genes for its production are of high relevance.
NAS Gene Family of A. thaliana
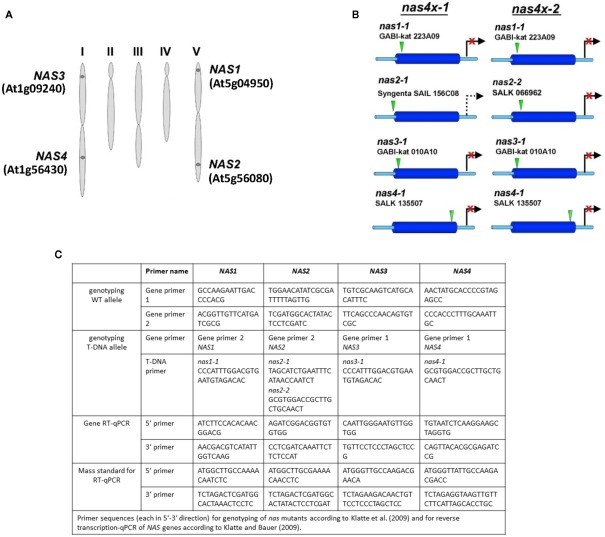
The Arabidopsis system provides all tools that allow combining genetic studies with physiological analyzes and global gene expression experiments. This species was therefore utilized to investigate NA function. While not all plant species have multiple NAS genes, the Arabidopsis genome harbors a NAS gene family comprising four members (Bauer et al., 2004). NAS1 and NAS2 are located on chromosome V, while NAS3 and NAS4 are located on chromosome I (Figure 1A). Multiple alignment (CLUSTALW) showed a close relation between NAS genes located on the same chromosome with more than 80% identity while alignment of genes belonging to separate chromosomes showed an identity of about 70% (Bauer et al., 2004). Gene mapping between Solanum esculentum and A. thaliana suggested that the four Arabidopsis genes, as well as the single tomato NAS gene originated from a common ancestor NAS gene. This finding is in agreement with a first genome duplication event in the evolution of Arabidopsis, followed later by two independent duplication events (Bauer et al., 2004).
Figure 1.
Genomic organization of NAS genes and tools for analyzing NAS gene function. (A) Scheme representing the five chromosomes of Arabidopsis, indicating the location of the four NAS genes and their gene identity numbers (Bauer et al., 2004). (B) Scheme representing the location of T-DNA insertions, indicating the allele names. It is shown which alleles are combined in the quadruple mutants nas4x-1 and nas4x-2 (Klatte et al., 2009). (C) Table listing the primers used for genotyping as described in (Klatte et al., 2009) and the primers used for gene expression analysis by reverse transcription-qPCR according to (Klatte and Bauer, 2009).
T-DNA insertion lines of all four NAS genes were identified and crossed to each other (Figure 1B; primer sequences for genotyping are found in Figure 1C). Under regular growth conditions single, double, and triple mutants did not show any obvious phenotypes suggesting functional redundancy (Klatte et al., 2009). Single mutants had similar NA contents as wild type. Triple nas mutants had NA levels that were reduced to 30–40% of wild type levels (Klatte et al., 2009). Since NA can be transported short and long-distance in plants, severe metal homeostasis phenotypes are not expected in the presence of a functional NAS gene. Interestingly, however, upon exposure to modestly toxic Ni supply, nas4-1 had a more chlorotic phenotype than nas3-1, while nas1-1 and nas2-1 had mild phenotypes like the wild type (Klatte et al., 2009). With increasing number of NAS knockout alleles, the NA contents decreased in the mutants while the severity of the leaf chlorosis was enhanced (Klatte et al., 2009). This suggests that NA contents correlate with Ni tolerance. Here, we show the seedling growth responses of single and multiple mutants in response to Fe deficiency. We found that all single nas mutants tested had a stronger leaf chlorosis than wild type plants upon Fe deficiency (Figure 2A). nas4-1 Mutants had the strongest leaf chlorosis among the tested single mutants (Figure 2A). It can therefore be concluded that the NAS gene functions are partially non-overlapping. Perhaps the location of NA production is important. Alternatively, the NAS isoforms might have different enzyme activities, perhaps under specific conditions like Fe deficiency and Ni supply.
Figure 2.
Analysis of multiple nas mutants. (A) Percentage of leaf chlorosis phenotypes of multiple nas mutants, combining the alleles nas1-1, nas2-1, nas3-1, and nas4-1, germinated for 2 weeks on Hoagland agar medium devoid of Fe; the medium is described in (Jakoby et al., 2004). The colors indicate the percentage of plants with light green leaves (weak leaf chlorosis, black), light green intercostal areas (intermediate degree of leaf chlorosis, light gray), yellow intercostal areas (strong leaf chlorosis, dark gray). The numbers on the right side indicate the number of seedlings examined; * indicates a strong phenotype. (B) Table summarizing the gene expression results from (Bauer et al., 2004; Klatte et al., 2009) and Schuler et al. (in preparation).
Partial non-redundancy is further confirmed by the fact that the NAS genes are differentially regulated in plants (Bauer et al., 2004; Klatte et al., 2009; summarized in Figure 2B; primers for gene expression analysis in Figure 1C). NAS1, NAS2, and NAS4 were found expressed in roots, where they were induced by Ni supply. NAS2 was also up-regulated by Fe deficiency. NAS1 and NAS4 were expressed in leaves, and NAS4 could be induced by Fe deficiency and Ni in leaves. NAS2 was not expressed in leaves. On the other hand, NAS3 was expressed in leaves where it was repressed by Fe deficiency but strongly induced by Ni supply. In flowers, NAS3 was expressed in sepals and petals while NAS1, NAS2, and NAS4 were not expressed (Schuler et al., in preparation).
Taken together, NAS genes evolved as a gene family in Arabidopsis where they acquired overlapping and specific functions in metal homeostasis as well as differential gene regulation in response to metals. NAS3 seems important for leaf and flower nicotianamine production upon Fe supply as well as Ni tolerance, while NAS4 was more important for Fe deficiency in leaves and perhaps in roots. NAS2 might be especially relevant for Fe deficiency responses in roots.
Physiological Analysis of Quadruple NAS Mutants
Mutant analysis showed that all four NAS genes are functional, so that quadruple mutant analysis was needed to study NA function. The leaf chlorosis phenotypes of quadruple nas mutants were more severe than those of single mutants. Quadruple nas1-1 nas2-1 nas3-1 nas4-1 mutants (termed nas4x-1) were found to have a stronger reduction of NA levels than all triple mutant combinations analyzed, namely to approximately 15% in vegetative leaves and 30% in seeds compared to wild type (Klatte et al., 2009). While nas4x-1 plants had a residual NA level in leaves at the vegetative stage, this was not the case in the reproductive stage in leaves. Full loss of function nas1-1 nas2-2 nas3-1 nas4-1 mutants (termed nas4x-2) did not contain any NA (Klatte et al., 2009). The morphological phenotypes of nas4x-1 and nas4x-2 were compared (summarized in Table 1).
Table 1.
Comparison of nas4x-1 and nas4x-2.
| nas4x-1 | nas4x-2* | Reference | |
|---|---|---|---|
| Genotype | nas1-1 nas2-1 nas3-1 nas4-1 | nas1-1 nas2-2 nas3-1 nas4-1 | Klatte et al. (2009) |
| Full-length NAS transcripts | NAS2 | none | Klatte et al. (2009) |
| NA content, versus WT | 10% in leaves, vegetative stage; 0% in leaves, reproductive stage; 40% in seeds | 0% in all tested leaves; no seeds produced | Klatte et al. (2009) |
| Interveinal leaf chlorosis | Starts during reproductive phase | Strong during vegetative and reproductive phase; more severe in young leaves than in older leaves | Klatte et al. (2009) |
| Numerous small rosette leaves | Yes | Yes | Klatte et al. (2009) |
| Flowers | Normal | Sterile | Klatte et al. (2009) |
| Flowering time and senescence | Normal | Delayed | Klatte et al. (2009) |
| Global gene expression | Categories affected: metal homeostasis, biotic stress responses, leaf photosystem and root carbohydrate metabolism | n.a. | Schuler et al. (2011) |
*A more detailed investigation of nas4x-2 phenotypes will be presented in Schuler et al. (in preparation); n.a. = not analyzed.
nas4x-1 plants appeared nearly normal during the vegetative stage, unless they were grown under Fe deficiency or Ni supply. However, nas4x-1 plants showed an interveinal leaf chlorosis upon transition to the reproductive growth stage, and Fe contents were increased in leaves at this stage compared to wild type (Klatte et al., 2009). Mobilization of Fe by nas4x-1 roots was up-regulated at this stage which accounts for the increased Fe contents (Klatte et al., 2009). Presumably, intercostal leaf areas with mesophyll cells did not acquire Fe in sufficient amounts and may have emitted a long-distance Fe deficiency signal that stimulated root Fe uptake. nas4x-1 plants are still fertile, yet flowers and seeds were found to contain less Fe than in the wild type (Klatte et al., 2009). nas4x-1 mutant plants are valuable models to study NA function in late phases of plant development. These plants also served to perform a transcriptome analysis of roots and leaves upon Fe supply and Fe deficiency (Schuler et al., 2011). A comparison with the wild type transcriptomes confirmed that nas4x-1 was affected in metal homeostasis since a high number of genes of this category was hit by differential expression. Besides this category, the mutant was also affected in biotic stress responses, leaf photosystem organization, and root carbohydrate metabolism (Schuler et al., 2011). Significantly more genes of these four biological categories were affected by differential expression between mutant and wild type compared to all genes analyzed in the microarray study. A change of expression of genes from these categories can be explained as an adaptation response to altered Fe levels.
Nas4x-2, on the other hand, is a severely affected mutant. Leaf chlorosis started during the vegetative phase (Klatte et al., 2009). Closer inspection of this mutant showed that NA was involved in the long-distance transport of Fe to young leaves presumably using the phloem, while older leaves received Fe from citrate-mediated transport in the xylem (Schuler et al., in preparation). The leaf chlorosis was due to Fe accumulation in the vascular system suggesting that NA is involved in lateral transport of Fe from vascular tissues to mesophyll (Schuler et al., in preparation). Furthermore, nas4x-2 mutants were affected in pollination (Schuler et al., in preparation).
Concluding Remarks
Arabidopsis served as a model for the study of nicotianamine function in plants. The split of a NAS locus to four NAS genes in Arabidopsis resulted in a partial non-redundant specialization of NAS gene functions. These are conferred at least partly by differential gene expression of the NAS genes in response to developmental cues, tissue specificity, and metals. It has not been investigated yet whether the enzyme activities of the NAS isoforms are differentially regulated by metal supply. Using the mild and severe nas4x-1 and nas4x-2 mutants novel nicotianamine functions were uncovered, such as seed Fe loading, long-distance Fe transport to leaves, short-distance transport from vascular tissues to mesophyll, and in pollination.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the DFG grants (Ba 1610/4–1, 4–4, 6–1) within the framework of the Arabidopsis Functional Genomics Network (AFGN).
Abbreviations
NA, nicotianamine; NAS, nicotianamine synthase.
References
- Anderegg G., Ripperger H. (1989). Correlation between metal complex formation and biological activity of nicotianamine analogues. J. Chem. Soc. Chem. Commun. 647–650 10.1039/c39890000647 [DOI] [Google Scholar]
- Bauer P., Thiel T., Klatte M., Bereczky Z., Brumbarova T., Hell R., Grosse I. (2004). Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol. 136, 4169–4183 10.1104/pp.104.047233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M., Talke I. N., Krall L., Krämer U. (2004). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37, 251–268 10.1046/j.1365-313X.2003.01959.x [DOI] [PubMed] [Google Scholar]
- Becker R., Fritz E., Manteuffel R. (1995). Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva. Plant Physiol. 108, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes I., Schreiber K., Rippberger H., Kircheiss A. (1983). Metal complex formation of nicotianamine, a posible phytosiderophore. Experientia 39, 261–262 10.1007/BF01955293 [DOI] [Google Scholar]
- Callahan D. L., Kolev S. D., O’Hair R. A. J., Salt D. E., Baker A. J. M. (2007). Relationships of nicotianamine and other amino acids with nickel, zinc and iron in Thlaspi hyperaccumulators. New Phytol. 176, 836–848 10.1111/j.1469-8137.2007.02216.x [DOI] [PubMed] [Google Scholar]
- Cheng L. J., Wang F., Shou H. X., Huang F. L., Zheng L. Q., He F., Li J. H., Zhao F. J., Ueno D., Ma J. F., Wu P. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 145, 1647–1657 10.1104/pp.107.107912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Briat J.-F. (2003). Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54, 183–206 10.1146/annurev.arplant.54.031902.135018 [DOI] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., Misson J., Schikora A., Czernic P., Mari S. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11 10.1093/aob/mcn207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- Douchkov D., Gryczka C., Stephan U. W., Hell R., Baumlein H. (2005). Ectopic expression of nicotianamine synthase genes results in improved iron accumulation and increased nickel tolerance in transgenic tobacco. Plant Cell Environ. 28, 365–374 10.1111/j.1365-3040.2005.01273.x [DOI] [Google Scholar]
- Douchkov D., Hell R., Stephan U. W., Baumlein H. (2001). Increased iron efficiency in transgenic plants due to ectopic expression of nicotianamine synthase. Plant Nutr. 92, 54–55 10.1007/0-306-47624-X_25 [DOI] [Google Scholar]
- Fenton H. J. H. (1894). Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 65, 899–910 10.1039/ct8946500899 [DOI] [Google Scholar]
- Haydon M. J., Cobbett C. S. (2007). Transporters of ligands for essential metal ions in plants. New Phytol. 174, 499–506 10.1111/j.1469-8137.2007.02051.x [DOI] [PubMed] [Google Scholar]
- Hell R., Stephan U. W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216, 541–551 [DOI] [PubMed] [Google Scholar]
- Herbik A., Koch G., Mock H. P., Dushkov D., Czihal A., Thielmann J., Stephan U. W., Baumlein H. (1999). Isolation, characterization and cDNA cloning of nicotianamine synthase from barley – a key enzyme for iron homeostasis in plants. Eur. J. Biochem. 265, 231–239 10.1046/j.1432-1327.1999.00717.x [DOI] [PubMed] [Google Scholar]
- Jakoby M., Wang H. Y., Reidt W., Weisshaar B., Bauer P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534 10.1016/j.febslet.2004.10.062 [DOI] [PubMed] [Google Scholar]
- Kim S., Takahashi M., Higuchi K., Tsunoda K., Nakanishi H., Yoshimura E., Mori S., Nishizawa N. K. (2005). Increased nicotianamine biosynthesis confers enhanced tolerance of high levels of metals, in particular nickel, to plants. Plant Cell Physiol. 46, 1809–1818 10.1093/pcp/pci141 [DOI] [PubMed] [Google Scholar]
- Klatte M., Schuler M., Wirtz M., Fink-Straube C., Hell R., Bauer P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150, 257–271 10.1104/pp.109.136374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M., Bauer P. (2009). Accurate real-time reverse transcription quantitative PCR. Methods Mol. Biol. 479, 61–77 [DOI] [PubMed] [Google Scholar]
- Lee S., Jeon U. S., Lee S. J., Kim Y.-K., Persson D. P., Husted S., Schjorring J. K., Kakei Y., Masuda H., Nishizawa N. K., An G. (2009). Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. U.S.A. 106, 22014–22019 10.1073/pnas.0809284106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H., Koch Q., Baumlein G. H., Ganal M. W. (1999). Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc. Natl. Acad. Sci. U.S.A. 96, 7098–7103 10.1073/pnas.96.14.7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari S., Gendre D., Pianelli K., Ouerdane L., Lobinski R., Briat J. F., Lebrun M., Czernic P. (2006). Root-to-shoot long distance circulation of nicotianamine and nicotianamine-nickel chelates in the metal hyperaccumulator Thlaspi caerulescens. J. Exp. Bot. 57, 4111–4122 10.1093/jxb/erl184 [DOI] [PubMed] [Google Scholar]
- Maurer F., Daum N., Schaefer U. F., Lehr C. M., Bauer P. (2010). Plant genetic factors for iron homeostasis affect iron bioavailability in Caco-2 cells. Food Res. Int. 43, 1661–1665 10.1016/j.foodres.2010.05.008 [DOI] [Google Scholar]
- Mori S., Nishizawa N. K. (1987). Methionine as a dominant precursor of phytosiderophores in Gramineae plants. Plant Cell Physiol. 28, 1081–1092 [Google Scholar]
- Nozoye T., Nagasaka S., Kobayashi T., Takahashi M., Sato Y., Sato Y., Uozumi N., Nakanishi H., Nishizawa N. K. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454 10.1074/jbc.M110.180026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouerdane L., Mari S., Czernic P., Lebrun M., Lobinski R. (2006). Speciation of non-covalent nickel species in plant tissue extracts by electrospray Q-TOFMS/MS after their isolation by 2D size exclusion-hydrophilic interaction LC (SEC-HILIC) monitored by ICP-MS. J. Anal. At. Spectrom 21, 676. 10.1039/b602689c [DOI] [Google Scholar]
- Pianelli K., Mari S., Marquès L., Lebrun M., Czernic P. (2005). Nicotianamine over-accumulation confers resistance to nickel in Arabidopsis thaliana. Transgenic Res. 14, 739–748 10.1007/s11248-005-7159-3 [DOI] [PubMed] [Google Scholar]
- Pich A., Scholz G. (1996). Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum Mill.): nicotianamine-stimulated copper transport in the xylem. J. Exp. Bot. 47, 41–47 10.1093/jxb/47.1.41 [DOI] [Google Scholar]
- Rellan-Alvarez R., AbadiÂa J., Alvarez-Fernandez A. (2008). Formation of metal-nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange. A study by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 22, 1553–1562 10.1002/rcm.3523 [DOI] [PubMed] [Google Scholar]
- Schuler M., Keller A., Backes C., Phillipar K., Lenhof H. P., Bauer P. (2011). Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol. 11, 87. 10.1186/1471-2229-11-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Inoue H., Ishimaru Y., Nakanishi H., Mori S., Nishizawa N. (2006). The role of nicotianamine and mugineic acid in metal transport at reproductive stage. Plant Cell Physiol. 47, S230–S230 10.1093/pcp/pcl017 [DOI] [Google Scholar]
- Takahashi M., Yamaguchi H., Nakanishi H., Shioiri T., Nishizawa N. K., Mori S. (1999). Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (strategy II) in graminaceous plants. Plant Physiol. 121, 947–956 10.1104/pp.121.3.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke I. N., Hanikenne M., Kramer U. (2006). Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyper accumulator Arabidopsis halleri. Plant Physiol. 142, 148–167 10.1104/pp.105.076232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel J. E., Villanueva L. A., Schat H., Kwekkeboom J., Coughlan S., Moerland P. D., van Themaat E. V. L., Koornneef M., Aarts M. G. M. (2006). Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142, 1127–1147 10.1104/pp.106.082073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N., Klair S., Bansal S., Briat J. F., Khodr H., Shioiri T., Leigh R. A., Hider R. C. (1999). Nicotianamine chelates both Fe-III and Fe-II. Implications for metal transport in plants. Plant Physiol. 119, 1107–1114 10.1104/pp.119.3.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., von Wiren N., Hayen H. (2006). Analysis of iron (II)/iron(III) phytosiderophore complexes by nano-electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom 20, 973–980 10.1002/rcm.2402 [DOI] [PubMed] [Google Scholar]
- Weber M., Harada E., Vess C., Roepenack-Lahaye E. V., Clemens S. (2004). Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyper accumulating factors. Plant J. 37, 269–281 10.1046/j.1365-313X.2003.01960.x [DOI] [PubMed] [Google Scholar]
- Zheng L., Cheng Z., Ai C., Jiang X., Bei X., Zheng Y., Glahn R. P., Welch R. M., Miller D. D., Lei X. G., Shou H. (2010). Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS ONE 5, e10190. 10.1371/journal.pone.0012555 [DOI] [PMC free article] [PubMed] [Google Scholar]