Abstract
Diacylglycerol kinase (DGK) is a pivotal enzyme that phosphorylates diacylglycerol (DAG) to form phosphatidic acid (PA). The production of PA from phospholipase D (PLD) and the coupled phospholipase C/DGK route is an important signaling process in animal and plant cells. In this study, we report a genomic analysis of eight putative rice DGKs encoded by a gene family (OsDGKs) grouped into three clusters. To further investigate the functions of the OsDGKs, a double-stranded RNA (dsRNA)-induced RNA silencing method was established. Introduction of in vitro-synthesized dsRNAs corresponding to a unique or conserved region of OsDGKs into rice protoplasts abolished or diminished the expression of individual or multiple OsDGK genes. Suppressing the expression of OsDGKs resulted in a distinct depletion of the transcripts of the defense gene OsNPR1 and the salt-responsive gene OsCIPK15. Our primary results suggest that OsDGKs are involved in the signaling of stress responses.
Keywords: diacylglycerol kinase, double-stranded RNA, rice, RNA interference
Introduction
Phospholipase C (PLC) catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] to produce diacylglycerol (DAG), which is phosphorylated to phosphatidic acid (PA) by diacylglycerol kinases (DGKs; Arisz et al., 2009). In animal cells, DAG and PA are important signaling molecules. DGK is thought to act as a conversional switch with two functional implications: the termination of DAG signaling and the initiation of PA signaling (Frere and Paolo, 2009). However, no direct DAG target (such as PKC in animal cells) has been found in plants, and the role for DAG as a plant signaling molecule has yet to been confirmed (Munnik and Testerink, 2009). In contrast, PA generated from both the PLD and PLC/DGK pathways is emerging as a stress signal molecule in plants (Munnik and Testerink, 2009; Hong et al., 2010; Zhang et al., 2010). PLD-derived PA has been found to regulate a series of developmental and environmental responses via its downstream targets (Li et al., 2009; Zhang et al., 2010). Relatively little genetic evidence for PA from PLC-coupled DGK regulating signaling in plant cells has been found (Munnik and Testerink, 2009).
Diacylglycerol kinase activity has been reported in several plant species, including tobacco, wheat, tomato, and Arabidopsis (Kamada and Muto, 1991; Lundberg and Sommarin, 1992; Wissing and Wagner, 1992; Katagiri et al., 1996; Snedden and Blumwald, 2000). Recently, multiple DGK-encoding genes have been isolated from plants (Snedden and Blumwald, 2000; Gómez-Merino et al., 2005; Chen et al., 2007). In Arabidopsis, AtDGK2 and AtDGK7 have been biochemically characterized (Gómez-Merino et al., 2004, 2005). A hydrophobic segment at the N-termini of AtDGK1 and AtDGK2 is necessary to target the resulting proteins to endoplasmic reticulum (ER) membranes (Vaultier et al., 2008).
In rice, pharmacological evidence indicates that PLC/DGK-mediated signaling is required for a benzothiadiazole-induced oxidative burst and hypersensitive cell death, and the transcription of one OsDGK is induced during this process (Chen et al., 2007). No genomic analysis of the rice DGK family has been reported. In this study, we report that OsDGKs are grouped into three clusters (I, II, and III) based on gene architecture, evolutionary relationships, and sequence identity. The transcription of OsDGKs was characterized using RT-PCR and real-time PCR, and their expression levels following treatment with xylanase or salt were analyzed. We established a transient double-stranded RNA (dsRNA)-induced RNA silencing assay that was used for rapid analysis of OsDGK functions in stress responses.
Materials and Methods
Suspension cell cultures and treatments
Rice (Oryza sativa L. Nipponbare ssp. japonica) suspension cells were initiated from embryogenic calli induced from mature rice scutella. The cells were grown in 100-ml conical flasks containing Murashige and Skoog liquid medium supplemented with 3% sucrose and 2 mg/l 2,4-D, and incubated on a rotary shaker (100–120 rpm) at 26 ± 2°C in darkness. The suspension cells were subcultured every 7 day.
Fresh suspension cells were collected by centrifugation after subculture for 4 to 5 day, and then treated with 200 μg/ml xylanase (Trichoderma viride; Fluka Bio-Chemika).
Rice seedling growth and protoplast isolation
Rice seedlings were grown in a growth room at 27°C for 2–3 weeks. Seedlings of 5–8′′ high were used for protoplast isolation according to the method of Sheen (2001) with modifications. Stems, including the sheaths of young seedlings, were cut into ∼0.5–1.0 mm segments using a razor blade, and immediately placed in a Petri dish containing K3 medium (see Table A1 in Appendix) supplemented with 1.5% cellulase R-10 (Yakult Honsha, Japan) and 0.3% macerozyme R-10 (Yakult Honsha). The segments were vacuum-infiltrated for 1 h at 20 mmHg and digested in darkness with gentle shaking (∼40 rpm) at room temperature for about 4 h. Following the incubation, the enzyme solution was gently removed using a glass pipette, and the same volume of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, 5 mM glucose, pH 5.6) was added to the Petri dish for further shaking (∼80 rpm) for 1 h to release the protoplasts. Both the enzyme solution and the W5 solution were filtered through a 35- to 75-μm nylon mesh. The protoplasts were collected by centrifugation at 150 × g for 4 min at room temperature. The pelleted protoplasts were washed twice with W5 solution and counted under a microscope using a hemocytometer.
Stress treatments
Protoplasts (1 × 105) were incubated in a six-well dish with each well containing 2 ml of WI culture medium (500 mM mannitol, 4 mM MES, 20 mM KCl, pH 5.6) supplemented with 150 μg/ml xylanase. For salt treatment, protoplasts (1 × 105) were incubated in modified WI culture medium containing 50 mM NaCl (400 mM mannitol, 4 mM MES, 20 mM KCl, 50 mM NaCl, pH 5.6). WI culture medium was used as a control. The protoplasts were incubated at 28°C in darkness for the indicated times, followed by harvesting with centrifugation at 200 × g for 5 min.
RNA isolation, RT-PCR, and real-time PCR
Total RNAs were isolated from suspension cells or protoplasts using Trizol reagent according to the manufacturer’s protocol (Takara, Japan). Reverse transcription was performed using Prime Script™ RT Reagent Kit (Takara). RT-PCR conditions were as follows: denaturing at 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. The primers used for RT-PCR analyses are described in Table A2 in Appendix. The OsGAPDH gene was amplified as an internal control.
Real-time PCR conditions were as follows: denaturing at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 58°C for 10 s, and 72°C for 10 s, and a final extension at 72°C for 5 min. The primers used for real-time PCR analyses are described in Table A3 in Appendix. The expression level of the OsActin gene detected with actin-specific primers was used to standardize the RNA sample for each real-time PCR. The real-time PCR was performed according to the manufacturer’s protocol (SYBR Premix Ex Taq™; Takara) in an ABI PRISM 7500 real-time PCR system.
In vitro synthesis of dsRNA
In vitro synthesis of dsRNA was carried out according to the method of Zhai et al. (2009) with minor modifications. DNA templates were synthesized by PCR from rice cDNA and engineered to contain the minimal T7 RNA polymerase promoter sequence (TAATACGACTCACTATAGGGAGG) at both the 5′ and 3′ ends. The primers used to amplify DNA from the targeted genes are listed in Table A4 in Appendix. The PCR conditions were as follows: denaturing at 94°C for 5 min, followed by 32 cycles of 94°C for 30 s, 62°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. dsRNAs were synthesized in vitro using the RiboMAX™ Large Scale RNA Production Systems T7 Kit (Promega) according to the manufacturer’s recommendations. DNA templates were removed using RNase-free DNase (Promega). dsRNA was purified using the RNeasy kit (Qiagen). The dsRNA was dissolved in DEPC-treated H2O, and its yield was measured using a UV spectrometer. Typical yields of RNA from 1 μg of DNA template were in the 80- to 100-μg range. The dsRNA was separated on a 1% agarose gel to check its integrity and size.
To prepare the fluorescent dsRNAs, a 40-bp of dsRNA directed against OsPLDα1 was synthesized, and labeled with FAM fluorescence at the 5′ end of sense strand. The sense and antisense strands of dsRNAs were: 5′-AGGCGCCACCAAGGUGUAUUCUACCA UUGAUCUGGAGAAA (sense); 5′-UUUCUCCAGAUCAAUGGUAGAAUACACCUU GGUGGCGCCU (antisense). The transfection of the dsRNA was done according to the protocol by the manufacturer (GenePharma, China). The fluorescence was visualized under a confocal microscope (TCS SP2, Leica, Germany).
Transfection of protoplasts with dsRNAs
Protoplasts (1 × 106 ml−1) in W5 solution were incubated on ice for 30 min. The protoplasts were pelleted and resuspended at 1 × 106 ml−1 in MMg solution (0.6 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.6). dsRNA (5–10 μg) was added to 100 μl of protoplasts (1 × 105 in MMg solution) to which an equal volume of PEG solution [40% (v/v) PEG4000, 0.4 M mannitol, 0.1 M CaCl2] was gradually added, and the mixture was incubated at room temperature in darkness for 15 min. The transfected protoplasts were collected by centrifugation for 2 min at 150 × g after diluting the transfection mixture with 600 μl of W5 solution. The protoplasts were resuspended in 1 ml of W5 solution and incubated in a six-well culture plate at 28°C in darkness for the indicated times.
RT-PCR analysis of gene expression in RNAi protoplasts
At the end of the transfection, the protoplasts were collected by centrifugation for 2 min at 150 × g. Total RNAs were isolated from protoplasts with the Trizol reagent according to the manufacturer’s protocol (Takara). Reverse transcription was performed using the Prime Script™ RT reagent Kit (Takara). The primers used for RT-PCR analyses are described in Table A2 in Appendix. The gene-silencing effect of the dsRNA was visualized in a 1% agarose gel, comparing the relative expression of RNAi-targeted genes to that of OsGAPDH. All photographs were taken in the Bio-Rad UV-Gel documentation system using Quantity-One analysis software.
Genomic search and sequence analysis
To identify DGK gene homologs in rice, BLAST searches were performed using the reported sequences of OsDGK (Os04g54200; Zhang et al., 2008) and AtDGK (Gómez-Merino et al., 2004) at the TIGR1 and NCBI2 web sites. Sequences of rice and Arabidopsis DGK (Gómez-Merino et al., 2004) proteins were aligned using CLUSTAL X (ver. 1.83), and a phylogenetic tree was constructed with the neighbor-joining method using the MEGA program3.
Domain analysis
Searches for conserved domains in the OsDGK proteins were carried out using SMART4 and PFAM5
Results
The DGK family in rice
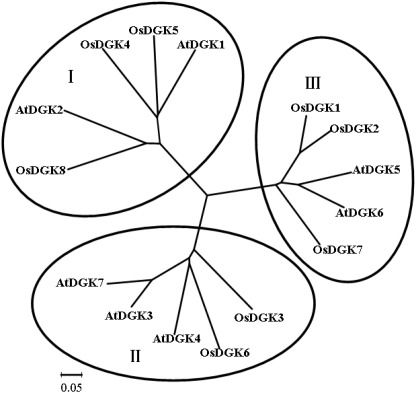
To identify members of the DGK family in rice (O. sativa), sequence information from Arabidopsis and rice was used to perform searches of relevant DNA databases and protein domains. Text searches using the keyword “diacylglycerol kinases” were also performed. Eight putative genes were identified in rice using these approaches. The obtained sequences were further analyzed for their potential to encode a DGK using the programs PFAM and SMART. To study the evolutionary relationships between different DGK members, the MEGA program was used to analyze the phylogenetic relationships of DGKs from rice and Arabidopsis (Figure 1). The results indicate that OsDGKs fall into three phylogenetic clusters, as also described for the AtDGKs (Gómez-Merino et al., 2004). Cluster I comprises OsDGK(4, 5, 8), and its closest homologs are AtDGK(1, 2); cluster II contains OsDGK(3, 6), and AtDGK(1, 4, 7) are within the same group; finally, the isoforms OsDGK(1, 2, 7) fall into cluster III, along with AtDGK(5, 6).
Figure 1.
Phylogenetic analysis of the DGK family in rice and Arabidopsis. Accession numbers for the DGKs are as follows: AtDGK1 (At5g07920), AtDGK2 (At5g63770), AtDGK3 (At2g18730), AtDGK4 (At5g57690), AtDGK5 (At2g20900), AtDGK6 (At4g28130), and AtDGK7 (At4g30340); OsDGK1 (Os04g54200), OsDGK2 (Os08g08110), OsDGK3 (Os02g54650), OsDGK4 (Os12g38780), OsDGK5 (Os03g31180), OsDGK6 (Os08g15090), OsDGK7 (Os01g57420), and OsDGK8 (Os12g12260). At, Arabidopsis thaliana; Os, Oryza sativa. The phylogenetic analysis was performed with the neighbor-joining method using the MEGA program.
Protein domains in OSDGKs
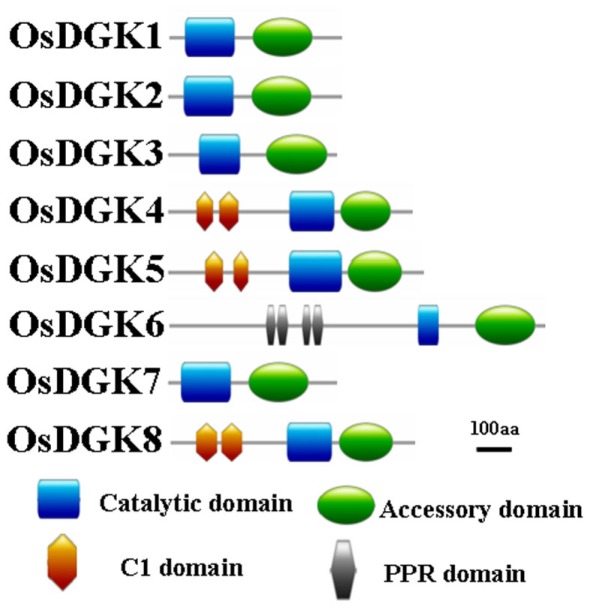
Domain analyses using the SMART and PFAM databases revealed that the rice DGK family contains a catalytic domain, an accessory domain, a C1 domain, and a PPR domain (Figure 2). In eukaryotic DGKs, the kinase domain contains a conserved catalytic domain with a presumed ATP binding site, and an accessory domain to make contact with the catalytic domain (Gómez-Merino et al., 2004; Arisz et al., 2009). The C1 domain contains a tandem Cys-rich sequence, which was first identified in PKC as binding to DAG and phorbol esters (PE; Azzi et al., 1992). This domain was suggested to exist in Arabidopsis DGKs (AtDGK1 and AtDGK2; Gómez-Merino et al., 2004; Arisz et al., 2009). The PPR (pentatricopeptide repeat) domain has been proposed to mediate macromolecular interactions. More than 400 genes encoding PPR proteins have been reported to exist in Arabidopsis, and most are predicted to reside in either mitochondria or chloroplasts (Small and Peeters, 2000; Lurin et al., 2004).
Figure 2.
Schematic presentation of the structure of OsDGK proteins. Conserved domains were identified by SMART and are highlighted in different colors.
All of the OsDGKs were found to contain a catalytic domain and an accessory domain. In addition, OsDGK(4, 5, 8) harbor two C1 domains, whereas OsDGK6 has four putative PPR domains (Figure 2). To our knowledge, among the reported plant DGKs, only OsDGK6 contains a PPR domain. It should be noted that OsDGK6 could be a unique (or putative) OsDGK, according to protein structure analysis (Figure 2), but it was nevertheless grouped into the same cluster (II) as OsDGK3 in a bioinformatics analysis (Figure 1). A similarity analysis revealed the highest similarity between OsDGK6 and OsDGK3 (Table A5 in Appendix). In addition, we isolated all OsDGK cDNAs except for that of OsDGK6. RT-PCR analysis did not detect the OsDGK6 transcript under normal or stress conditions (Figure 3); therefore, the existence and function in rice of this OsDGK have yet to be determined.
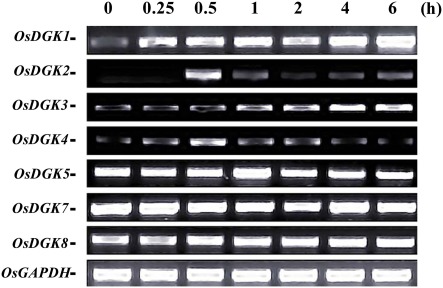
Figure 3.
RT-PCR analysis of OsDGK transcripts in rice cells treated with xylanase. Suspension cells were treated with 200 μg/ml xylanase for the indicated times, and OsDGK transcripts were monitored by RT-PCR. Approximately 0.1 μg of total RNA was used in each PCR. OsDGK6 was undetectable in cells with or without xylanase treatment. The OsGAPDH gene was used as an internal control.
Xylanase-induced expression of OsDGK in cells
The expression of OsDGK genes in suspension cells was analyzed using RT-PCR. Under control conditions, the transcription of six of the eight OsDGKs (all except for OsDGK2 and OsDGK6) was confirmed. The activation of PLC and DGK pathway has been reported in tomato cells treated with the fungal elicitor xylanase (Laxalt et al., 2007). We therefore explored whether expression of OsDGK(s) are activated by the xylanase. Treatment of cells with 200 μg/ml xylanase led to increases in the transcription of OsDGK(1, 2, 3, 4). Although the basal transcript levels of OsDGK(5, 7, 8) were high, their transcription was not affected by xylanase treatment. The transcription of OsDGK6 was undetectable with or without xylanase (Figure 3).
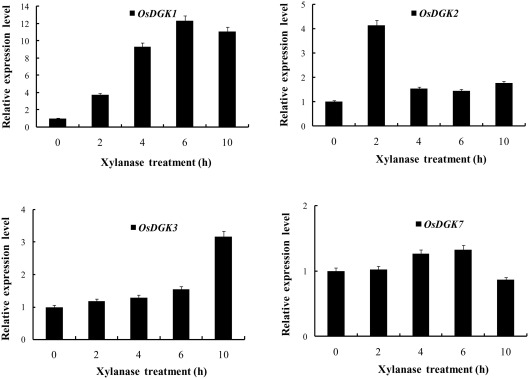
We selected OsDGK(1, 2, 3), which were induced by xylanase, and OsDGK7, which was not induced by xylanase, for further testing with real-time PCR. As shown in Figure 4, the transcription of both OsDGK1 and OsDGK3 gradually increased in xylanase-treated protoplasts. The transcription of OsDGK2 exhibited a sharp peak at 2 h after the addition of 150 μg/ml xylanase. A peak in the OsDGK2 transcript was also found in the RT-PCR analysis, but it appeared earlier, probably due to the higher concentration of xylanase (150 μg/ml there) used. The transcription of OsDGK7 was not affected by xylanase under the tested conditions.
Figure 4.
Real-time PCR analysis of the expression of OsDGK(1, 2, 3, 7). Rice protoplasts were treated with 150 μg/ml xylanase. The transcripts of four OsDGK genes were monitored with real-time PCR. The data shown are averages of three replicates, each with three PCR samples from the same cDNA archive.
Transient suppression of OsDGK expression by introduction of the corresponding dsRNA
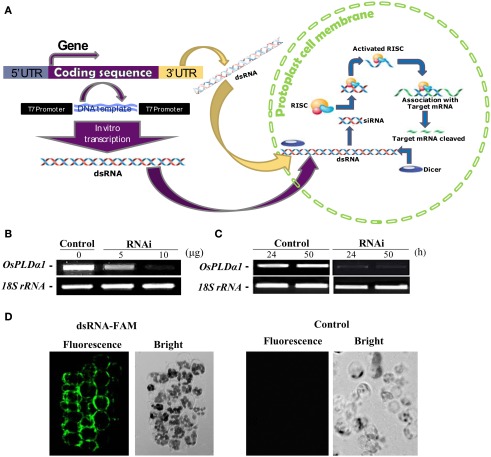
To study the functions of OsDGKs in rice cells, we established methods for dsRNA-induced RNA silencing according to published procedures (Endo et al., 2008; Zhai et al., 2009). Transfection with in vitro-synthesized dsRNAs against target genes into cells resulted in the depletion of their transcripts due to the combined actions of the DICER enzyme and the RNA-induced silencing complex (RISC) enzyme (Figure 5A).
Figure 5.
Double-stranded RNA-induced RNA silencing in rice protoplasts. (A) Simple illustration of dsRNA-induced RNA silencing. When a long dsRNA is introduced into plant cells, the endogenous DICER enzyme cuts long dsRNA into short siRNAs. The RNA-induced silencing complex (RISC) enzyme attaches to these siRNAs, forming the siRNA–RISC complex (Plasterk, 2002). The target mRNA is attached to the siRNA–RISC complex and cleaved into small pieces. This process inhibits the expression of target genes. (B) Dose dependence of the transient RNAi silencing of OsPLDα1 in rice protoplasts. Protoplasts were transfected with 0, 5, or 10 μg of dsRNA against OsPLDα1 (RNAi) or with sterile water (Control), followed by incubation in darkness for 24 h. The silencing effect of OsPLDα1 was monitored using RT-PCR. (C) Time dependence of transient RNAi silencing of OsPLDα1 in rice protoplasts. Protoplasts were transfected with 10 μg of a dsRNA against OsPLDα1 (RNAi) or with sterile water (Control). RNAi and control protoplast samples were collected at 24 and 50 h, respectively, and then analyzed with RT-PCR for OsPLDα1 transcripts. 18S rRNA was amplified as an internal control. (D) A 40-bp of synthesized dsRNA labeled with FAM fluorescence at the 5′ end of sense strand against OsPLDα1 was transfected into rice protoplasts. The fluorescence was visualized under a confocal microscope after 8 h incubation in darkness. Control, protoplasts were transfected with water.
We first selected OsPLDα1 to test because the expression of its mRNA was characterized in our previous work (Shen et al., 2011). A dsRNA against a 486-bp sequence corresponding to the OsPLDα1 coding sequence was synthesized in vitro. Rice protoplasts were transfected with 5 or 10 μg of dsRNA (RNAi) or with sterile water (control, as a mock transfection) and incubated for 24 h in darkness. The silencing effects of dsRNA were dose-dependent (Figure 5B). This inhibition of transcription lasted for at least 50 h (Figure 5C).
To prove the in vitro-synthesized dsRNA was transported into protoplasts, we synthesized a 40-bp dsRNA labeled with FAM fluorescence at the 5′ end of sense strand against OsPLDα1, and transfected into rice protoplasts. The transfected protoplasts were visualized under a fluorescent microscope after 8 h incubation in darkness. The fluorescence was found in protoplasts. As a control, no fluorescence was found in the protoplasts transfected with sterile water (Figure 5D). This together with the RT-PCR results suggest that in vitro-synthesized dsRNA had been transported into protoplasts and suppressed gene transcripts.
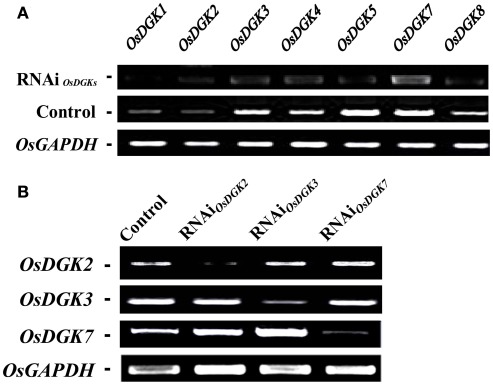
We then tried to silence expression of the OsDGK family. Two in vitro-synthesized dsRNAs against conserved regions of OsDGK(1, 2, 3, 7) and OsDGK(4, 5, 8) were simultaneously transfected into protoplasts. Mock-transfected protoplasts (control) and two dsRNA-transfected protoplasts (RNAi) were collected for RT-PCR analysis after incubation in darkness for 24 h. The abundance of the OsDGK transcripts after RT-PCR analysis was normalized to the internal standard gene OsGAPDH. As shown in Figure 6A, the transcription of seven OsDGKs was successfully repressed simultaneously by two dsRNAs as compared the mock-transfected control.
Figure 6.
Multi-gene and specific gene silencing of OsDGKs in protoplasts using transient RNAi. (A) Multi-gene silencing of OsDGKs in rice protoplasts using transient RNAi. RT-PCR analysis of OsDGKs was performed after transient RNAi by simultaneously introducing two in vitro-synthesized dsRNAs against conserved regions of OsDGK(1, 2, 3, 7) and OsDGK(4, 5, 8). Mock-transfected (Control) and two dsRNA-transfected protoplasts (RNAi) were incubated for 24 h, followed by protoplast collection for RNA isolation. OsGAPDH was amplified as an internal control. (B) Gene-specific silencing of OsDGKs in rice protoplasts with transient RNAi. RT-PCR analysis of OsDGK2, OsDGK3, and OsDGK7 was performed after transient RNAi using in vitro-synthesized dsRNAs against the 3′-UTR regions of each. dsRNA-transfected (RNAi) and mock-transfected (Control) protoplasts were incubated for 24 h, followed by protoplast collection for RT-PCR analysis. OsGAPDH was amplified as an internal control.
We next carried out gene-specific interference of the expression of OsDGKs. RT-PCR analysis was undertaken for OsDGK2, OsDGK3, and OsDGK7 after transient RNAi using in vitro-synthesized dsRNAs corresponding to their 3′-untranslated regions (3′-UTRs). The expression of OsDGK2 was repressed significantly by introducing in vitro-synthesized dsRNA directed against its 3′-UTR, whereas the expression of OsDGK3 and OsDGK7 was not affected (Figure 6B). Similarly, gene-specific interference was found for OsDGK3 versus OsDGK2 and OsDGK7, and OsDGK7 versus OSDGK2 and OsDGK3. Taken together, these results suggest that the introduction of dsRNAs is effective for targeted gene silencing in rice protoplasts.
Effect of the transient silencing of OsDGK on the transcription of genes related to stress tolerance
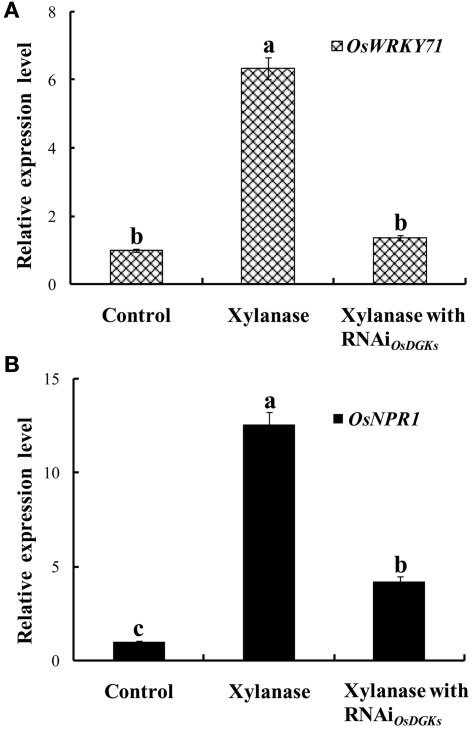
Using the transient RNA interference assay, we next explored the functions of the OsDGKs in stress responses. The WRKY transcriptional factors have been reported to be involved in various stress responses; in particular, overexpression of OsWRKY71 enhances resistance to virulent bacterial pathogens (Liu et al., 2007). Treatment of protoplasts with xylanase (150 μg/ml) induced an increase in the expression of OsWRKY71 and the pathogen-related gene OsNPR1 (Liu et al., 2005). Transient silencing of OsDGKs triggered by the introduction of two dsRNAs corresponding to the conserved regions of OsDGK(1, 2, 3, 7) and OsDGK(4, 5, 8; Figure 6A) prevented the increases in expression of OsWRKY71 and OsNPR1 induced by xylanase (Figures 7A,B).
Figure 7.
Effects of silencing of OsDGKs on OsWRKY71 and OsNPR1 transcription induced by xylanase in rice protoplasts. After transient RNAi using two in vitro-synthesized dsRNAs against conserved regions of OsDGK(1, 2, 3, 7) and OsDGK(4, 5, 8) for 24 h, the transfected protoplasts were treated with 150 μg/ml xylanase for 6 h. The protoplasts were collected for real-time PCR analysis to detect transcripts of OsWRKY71 (A) and OsNPR1 (B). Values followed by different letters differ significantly (P < 0.01).
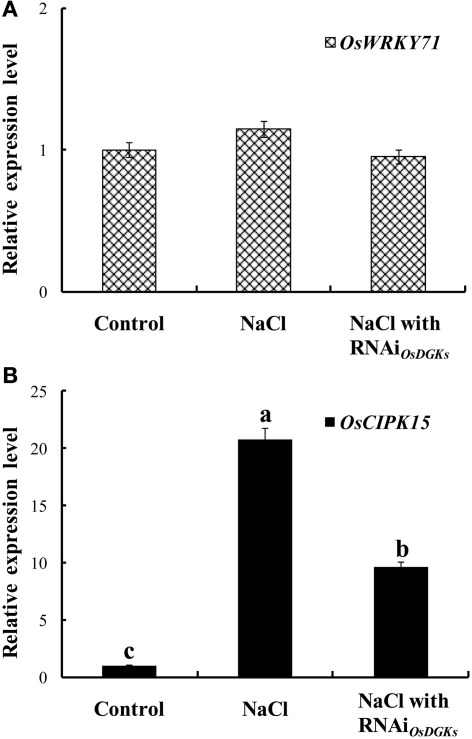
We next asked whether OsDGKs regulate abiotic stress responses. The expression of OsWRKY7 was not affected by salt stress; transient silencing of OsDGKs had no effect on OsWRKY7 expression (Figure 8A). However, under salt stress, the expression of OsCIPK15 (CIPK, for calcineurin B-like protein interaction protein kinase) was induced, and the overexpression of OsCIPK15 improved the salt tolerance of rice seedlings (Xiang et al., 2007). An increase in the transcription of OsCIPK15 was also observed in rice protoplasts exposed to NaCl solution. This NaCl-induced increase in OsCIPK15 was repressed in cells in which OsDGKs were transiently silenced (Figure 8B). These results suggest that OsDGKs regulate abiotic and biotic stresses through different signaling pathways.
Figure 8.
Effects of silencing of OsDGKs on transcription of OsWRKY71 and OsCIPK15 induced by NaCl in rice cells. Following transient RNAi using two in vitro-synthesized dsRNAs against conserved regions of OsDGK(1, 2, 3, 7) and OsDGK(4, 5, 8) for 24 h, the transfected protoplasts were treated with 50 mM NaCl for 6 h. The protoplasts were collected for real-time PCR analysis to detect transcripts of OsWRKY71 (A) and OsCIPK15 (B). Values followed by different letters differ significantly (P < 0.01).
Discussion
Diacylglycerol kinase phosphorylates DAG to generate PA, serving as a DAG consumer as well as a PA generator. Therefore, DGK is thought to regulate the balance between these two lipid messengers by catalyzing their interconversion (Frere and Paolo, 2009). The mammalian DGKs are a large enzyme family with 10 isozymes, which are subdivided into five groups according to their structural features (Sakane et al., 2007). In the Arabidopsis genome, seven DGK isoforms that form three clusters have been identified (Gómez-Merino et al., 2004). The rice DGKs also group into three clusters (Figure 1). Both Arabidopsis and rice DGKs contain catalytic and accessory domains (Gómez-Merino et al., 2004; Figure 2). OsDGK(4, 5, 8) and AtDGK(1, 2) belong to cluster I, which possesses two cysteine-rich domains conferring C1 domains. These domains are absent from other Arabidopsis and rice DGKs (Figure 2; Vaultier et al., 2008). The C1 domain can bind to proteins to modulate the interaction with lipids and proteins (Colon-Gonzalez and Kazanietz, 2006). AtDGK7 still displays kinase activity in the absence of the C1 domain (Gómez-Merino et al., 2005), suggesting that this domain is not necessary for its phosphorylation activity (Snedden and Blumwald, 2000). By fusion with fluorescent proteins, the hydrophobic segment in the amino-terminal region upstream of the C1 domain in AtDGK1 and AtDGK2 has been proven to be sufficient and necessary to sequester proteins to ER membranes (Vaultier et al., 2008). However, whether the final localization of full-length DGKs is identical with this segment is unclear. Much less is known about plant DGKs than animal DGKs.
Transient RNA interference caused the decreased expression of unique and multiple OsDGK genes in rice protoplasts (Figure 6). This approach has also been successfully used in Arabidopsis and Zinnia (Endo et al., 2008; Zhai et al., 2009). Introduction of in vitro-synthesized dsRNAs corresponding to the cellulose synthesis gene CesA into Zinnia cells repressed the expression of Zinnia CesA homologs. The repression phenocopies Arabidopsis cellulose synthase mutants that have defects in secondary cell wall synthesis and increased abnormal tracheary elements (Endo et al., 2008). In our work, repression of multiple OsDGK genes impaired stress-induced gene expression (Figures 7 and 8). The combination of transient RNA interference and DGK mutants will help to fully elucidate the functions of DGKs in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Science Foundation of China (31171461, 30625027), the Ministry of Science and Technology of China (2012CB11420), the Fundamental Research Funds for the Central Universities (KYT201001), the Education Department of Jiangsu (200910, PAPD), and from the Youth Sci-Tech Innovation Fund, NJAU (KJ09019).
Appendix
Table A1.
K3 medium used for rice protoplast system.
| K3 medium | 10 × B5 Macro, 100 × B5 micro (I), 1000 × B5 micro (II), 100 × B5 vitamins, 200 × MES (0.1 g/ml), 500 × myo-inositol (0.05 g/ml), 100 × NH3NO3 (25 mg/ml), 100 × CaCl2 (75 mg/ml), 100 × xylose (25 mg/ml), 0.4 M d-mannitol. pH is adjusted to 5.6 by 1 M KOH. |
| 10 × B5 Macro (1 l) | KNO3 25 g, (NH4)2SO4 1.34 g, CaCl2·2H2O 1.5 g, MgSO4·7H2O 2.5 g, NaH2PO4· H2O 1.5 g. |
| 100 × B5 Micro(I) (1 l) | MnSO4·H2O 0.78 g, ZnSO4·7H2O, 0.2 g, H3BO3 0.3 g, KI 0.075 g. |
| 1000 × B5 Micro(II) (1 l) | NaMoO4·2H2O 0.250 g, CuSO4·5H2O 25 mg, CoCl2·6H2O 25 mg. |
| 100 × B5 Vitamins (1 l) | Vitamin B1 (thiamine-HCl) 1 g, vitamin B6 (pyridoxine-HCl) 0.1 g, nicotinic acid 0.1 g. |
K3 medium was prepared according to the method by Chen et al. (2006) with minor modification.
Table A2.
List of oligos used for RT-PCR analysis.
| Genes | Directions | Oligos (5′–3′) |
|---|---|---|
| OsDGK1 | Forward | CTGGCACCAGGAAAGTACAAGATAGAGAC |
| Reverse | TCGTGGTTGCTACAGCACATCGG | |
| OsDGK2 | Forward | AGACTTATTGAGGTTGTTGGATTCCGTGAT |
| Reverse | CAGGGATCTTGAATGTATCTGCGGC | |
| OsDGK3 | Forward | GTTCTGAATGGGAGCAAGTTACAATGC |
| Reverse | AGAGAAGGGTAAGGAACTTTGTTTATCTCG | |
| OsDGK4 | Forward | ATCGCTCTGAGGAGGATTCTTTCTGC |
| Reverse | CTTCAATATCAGATGGCGGGTCAATAG | |
| OsDGK5 | Forward | TGAGATTCCAGAGGATTCAGAAGGTGTT |
| Reverse | CCTCTTCTGAGATGCAGTGATTAGGTGAC | |
| OsDGK6 | Forward | CCATTCGGATAGTCAAGAACCTC |
| Reverse | CCAACTATGCGGACTTAACCAG | |
| OsDGK7 | Forward | GGGGAAGAGAAATCCTGGAACAGATG |
| Reverse | ATTGGATGACATAGGGATGCACAGAAC | |
| OsDGK8 | Forward | TCTGTCTGTGAAAGAAGTTGCCCAAG |
| Reverse | TCTTGCCGTTAATGAAAACAAGCAGT | |
| OsPLDα1 | Forward | GGTAACCGTGAGGTGAAGCA |
| Reverse | GCATTCCCAGGTGCTCGTAC | |
| OsGAPDH | Forward | ACCACAAACTGCCTTGCTCC |
| Reverse | ATGCTCGACCTGCTGTCACC | |
| 18S rRNA | Forward | CCTATCAACTTTCGATGGTAGGATA |
| Reverse | CGTTAAGGGATTTAGATTGTACTCATT |
Table A3.
List of oligos used for real-time PCR analysis.
| Genes | Directions | Oligo (5′–3′) |
|---|---|---|
| OsDGK1 | Forward | GGCTGCTTGGTGTAGTTAGTG |
| Reverse | TCTTGGTCAGTGGTTGGGTT | |
| OsDGK2 | Forward | TCGTCTGTCTCAACCTGCCTAG |
| Reverse | CACGGAATCCAACAACCTCAAT | |
| OsDGK3 | Forward | GAATGGGAGCAAGTTACAATG |
| Reverse | ATCGGAATGAGCTTCGACAA | |
| OsDGK7 | Forward | TACAGTCAGTAAGACAAGCGAAAG |
| Reverse | CAGCGAATGAGGCAAATCCA | |
| OsWRKY71 | Forward | CGCCGACCCATCCGACCTCA |
| Reverse | TCTTGACAGGGCAGGCGGGA | |
| OsNPR1 | Forward | CCCGCGATGTTCGAACGTGC |
| Reverse | CGACGAGAGCCCCGACCTGT | |
| OsCIPK15 | Forward | GTTACCACTTCCTATCATATCATC |
| Reverse | CTAAACATCAACTCTCCAAATAC | |
| OsActin | Forward | AGGAAGGCTGGAAGAGGACC |
| Reverse | CGGGAAATTGTGAGGGACAT |
Table A4.
List of oligos used in amplifying DNA templates for in vitro dsRNA synthesis.
| Targeted Gene | Directions | Oligos (5′–3′) | Size of dsRNA (bp) |
|---|---|---|---|
| OsPLDα1 | Forward | GCTTAATACGACTCACTATAGGGAGGTTGACGATGAGTACATCATCATCGG | 486 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGCTATGAGGTGAGGATGGGGGGCATG | ||
| OsDGKs(1,2,3,7) | Forward | GCTTAATACGACTCACTATAGGGAGGTGTCGGCTTTCGCGATGCCT | 208 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGGGCTGCTTCCATGGCTCCCC | ||
| OsDGKs(4,5,8) | Forward | GCTTAATACGACTCACTATAGGGAGGCGCGCGCAGAGGTTAGCTCA | 229 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGGGAGGCCGCATGGCCGATAG | ||
| OsDGK2 (3′-UTR) | Forward | GCTTAATACGACTCACTATAGGGAGGAATTGGTATCTTTTCTAGGTTGCAT | 260 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGTCTGCTGAACAATAAACAAGAAATC | ||
| OsDGK3 (3′-UTR) | Forward | GCTTAATACGACTCACTATAGGGAGGAATCAGGGTCGTATTCTAGATCGTT | 303 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGCATAACAGGAGGGGAAATCTGAGTA | ||
| OsDGK7 (3′-UTR) | Forward | GCTTAATACGACTCACTATAGGGAGGTGACGAGGTTTTGTACGTATGGCTG | 342 |
| Reverse | GCTTAATACGACTCACTATAGGGAGGCGTGGAGGTATATTCTGCGGGTAGT |
Table A5.
Amino acid sequence identities among OsDGKs.
| OsDGK1 | OsDGK2 | OsDGK3 | OsDGK4 | OsDGK5 | OsDGK6 | OsDGK7 | OsDGK8 | |
|---|---|---|---|---|---|---|---|---|
| OsDGK1 | 81 | 32 | 27 | 25 | 29 | 63 | 25 | |
| OsDGK2 | 31 | 25 | 25 | 29 | 62 | 24 | ||
| OsDGK3 | 24 | 25 | 54 | 33 | 26 | |||
| OsDGK4 | 59 | 17 | 25 | 50 | ||||
| OsDGK5 | 16 | 23 | 52 | |||||
| OsDGK6 | 30 | 17 | ||||||
| OsDGK7 | 24 | |||||||
| OsDGK8 |
References
- Chen S., Tao L., Zeng L., Vega-Sanchez M. E., Umemura K., Wang G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427 10.1111/j.1364-3703.2006.00349.x [DOI] [PubMed] [Google Scholar]
Footnotes
References
- Arisz S. A., Testerink C., Munnik T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophy. Acta 179, 869–875 [DOI] [PubMed] [Google Scholar]
- Azzi A., Boscoboinik D., Hensey C. (1992). The protein kinase C family. Eur. J. Biochem. 208, 547–557 10.1111/j.1432-1033.1992.tb17219.x [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang W. D., Song F. M., Zheng Z. (2007). Phospholipase C/diacylglycerol kinase-mediated signalling is required for benzothiadiazole-induced oxidative burst and hypersensitive cell death in rice suspension-cultured cells. Protoplasma 230, 13–21 10.1007/s00709-006-0195-x [DOI] [PubMed] [Google Scholar]
- Colon-Gonzalez F., Kazanietz M. G. (2006). C1 domains exposed: from diacylglycerol binding to protein–protein interactions. Biochim. Biophys. Acta 1761, 827–837 [DOI] [PubMed] [Google Scholar]
- Endo S., Pesquet E., Tashiro G., Kuriyama H., Goffner D., Fukuda H., Demura T. (2008). Transient transformation and RNA silencing in Zinnia tracheary element differentiating cell cultures. Plant J. 53, 864–875 10.1111/j.1365-313X.2007.03377.x [DOI] [PubMed] [Google Scholar]
- Frere S. G., Paolo G. D. (2009). A lipid kinase controls the maintenance of dendritic spines. EMBO J. 28, 999–1000 10.1038/emboj.2009.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Merino F. C., Arana-Ceballos F. A., Trejo-Tëllez L. I., Skirycz A., Brearley C. A., Dörmann P., Mueller-Roeber B. (2005). Arabidopsis AtDGK7, the smallest member of plant diacylglycerol kinases (DGKs), displays unique biochemical features and saturates at low substrate concentration. J. Biol. Chem. 280, 34888–34899 10.1074/jbc.M506859200 [DOI] [PubMed] [Google Scholar]
- Gómez-Merino F. C., Brearley C. A., Ornatowska M., Abdel-Haliem M. E., Zanor M. I., Mueller-Roeber B. (2004). AtDGK2, a novel diacylglycerol kinase from Arabidopsis thaliana, phosphorylates 1-stearoyl-2-arachidonoyl-sn-glycerol and 1,2-dioleoyl-sn-glycerol and exhibits cold-inducible gene expression. J. Biol. Chem. 279, 8230–8241 10.1074/jbc.M312187200 [DOI] [PubMed] [Google Scholar]
- Hong Y., Zhang W., Wang X. (2010). Phospholipase D and phosphatidic acid signaling in plant response to drought and salinity. Plant Cell Environ. 33, 627–635 10.1111/j.1365-3040.2009.02087.x [DOI] [PubMed] [Google Scholar]
- Kamada Y., Muto S. (1991). Ca2+ regulation of phosphatidylinositol turnover in the plasma membrane of tobacco suspension culture cells. Biochim. Biophys. Acta 1093, 72–79 10.1016/0167-4889(91)90140-S [DOI] [PubMed] [Google Scholar]
- Katagiri T., Mizoguchi T., Shinozaki K. (1996). Molecular cloning of a cDNA encoding diacylglycerol kinase (DGK) in Arabidopsis thaliana. Plant Mol. Biol. 30, 647–653 10.1007/BF00049339 [DOI] [PubMed] [Google Scholar]
- Laxalt A. M., Raho N., Have A. T., Lamattina L. (2007). Nitric oxide is critical for inducing phosphatidic acid accumulation in xylanase-elicited tomato cells. J. Biol. Chem. 282, 21160–21168 10.1074/jbc.M701212200 [DOI] [PubMed] [Google Scholar]
- Li M., Hong Y., Wang X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791, 927–935 [DOI] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979 10.1016/j.jplph.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Liu X. Q., Bai X. Q., Qian Q., Wang X. J., Chen M. S., Chu C. C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603 10.1038/sj.cr.7290259 [DOI] [PubMed] [Google Scholar]
- Lundberg G. A., Sommarin M. (1992). Diacylglycerol kinase in plasma membranes from wheat. Biochim. Biophys. Acta 1123, 177–183 [DOI] [PubMed] [Google Scholar]
- Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Ret L. M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 10.1105/tpc.104.022236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Testerink C. (2009). Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. 50, S260–S265 10.1194/jlr.R800098-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H. A. (2002). RNA silencing: the genome’s immune system. Science 296, 1263–1265 10.1126/science.1072148 [DOI] [PubMed] [Google Scholar]
- Sakane F., Imai S., Kai M., Yasuda S., Kanoh H. (2007). Diacylglycerol kinases: why so many of them? Biochim. Biophy. Acta 1771, 793–806 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475 10.1104/pp.010820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Wang R., Jing W., Zhang W. (2011). Rice phospholipase Dα is involved in salt tolerance by the mediation of H+-ATPase activity and transcription. J. Int. Plant Biol. 53, 289–299 10.1111/j.1744-7909.2010.01021.x [DOI] [PubMed] [Google Scholar]
- Small I. D., Peeters N. (2000). The PPR motif-a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 10.1016/S0968-0004(99)01520-0 [DOI] [PubMed] [Google Scholar]
- Snedden W. A., Blumwald E. (2000). Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J. 24, 317–326 10.1046/j.1365-313x.2000.00877.x [DOI] [PubMed] [Google Scholar]
- Vaultier M. N., Cantrel C., Guerbette F., Boutte Y., Vergnolle C., Cicek C., Bolte S., Zachowski A., Ruelland E. (2008). The hydrophobic segment of Arabidopsis thaliana cluster I diacylglycerol kinases is sufficient to target the proteins to cell membranes. FEBS Lett. 582, 1743–1748 10.1016/j.febslet.2008.04.042 [DOI] [PubMed] [Google Scholar]
- Wissing J. B., Wagner K. G. (1992). Diacylglycerol kinase from suspension cultured plant cells: characterization and subcellular localization. Plant Physiol. 98, 1148–1153 10.1104/pp.98.3.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Huang Y., Xiong L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428 10.1104/pp.107.101295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Sooksa-Nguan T., Vatamaniuk O. K. (2009). Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol. 149, 642–652 10.1104/pp.108.130260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chen J., Zhang H., Song F. M. (2008). Overexpression of a rice diacylglycerol kinase gene OsBIDK1 enhances disease resistance in transgenic tobacco. Mol. Cells 26, 258–264 [PubMed] [Google Scholar]
- Zhang W., Wan X., Hong Y., Li W., Wang X. (2010). “Plant phospholipase.” in Lipid Signaling in Plants, Plant Cell Monographs, Vol. 16, ed. Munnik D. T. (Heidelberg: Springer-Verlag; ), 39–62 [Google Scholar]