Abstract
Plants are sessile and cannot move to appropriate hiding places or feeding grounds to escape adverse conditions. As a consequence, they evolved mechanisms to detect changes in their environment, communicate these to different organs, and adjust development accordingly. These adaptations include two long-distance transport systems which are essential in plants: the xylem and the phloem. The phloem serves as a major trafficking pathway for assimilates, viruses, RNA, plant hormones, metabolites, and proteins with functions ranging from synthesis to metabolism to signaling. The study of signaling compounds within the phloem is essential for our understanding of plant communication of environmental cues. Determining the nature of signals and the mechanisms by which they are communicated through the phloem will lead to a more complete understanding of plant development and plant responses to stress. In our analysis of Arabidopsis phloem exudates, we had identified several lipid-binding proteins as well as fatty acids and lipids. The latter are not typically expected in the aqueous environment of sieve elements. Hence, lipid transport in the phloem has been given little attention until now. Long-distance transport of hydrophobic compounds in an aqueous system is not without precedence in biological systems: a variety of lipids is found in human blood and is often bound to proteins. Some lipid–protein complexes are transported to other tissues for storage, use, modification, or degradation; others serve as messengers and modulate transcription factor activity. By simple analogy it raises the possibility that lipids and the respective lipid-binding proteins in the phloem serve similar functions in plants and play an important role in stress and developmental signaling. Here, we introduce the lipid-binding proteins and the lipids we found in the phloem and discuss the possibility that they may play an important role in developmental and stress signaling.
Keywords: long-distance lipid signaling, phloem, lipid-binding proteins
Phloem Contents, Structure, and Function
Plants contain two long-distance transport systems: the xylem for water and nutrient transport, and the phloem. During the last 10–15 years, the view of the phloem function has changed from that of simple assimilate transport to a trafficking system for stress signals and developmental regulators (Citovsky and Zambryski, 2000; Wu et al., 2003; Ding et al., 2003; Haywood et al., 2005). It is now accepted that the phloem is a conduit system which is crucial for the transport of mineral nutrients, plant viruses, virus-induced silencing, defense against pathogen infection, and signaling of environmental conditions, and developmental changes (Lucas et al., 1995; Ryabov et al., 1999; Burton et al., 2000; Seo et al., 2001; Yoo et al., 2004; Suárez-López, 2005; Kehr, 2006; Lough and Lucas, 2006). It contains small molecules (Chen et al., 2001; Corbesier et al., 2003; Maeda et al., 2006), peptides and proteins (Fisher et al., 1992; Sakuth et al., 1993; Schobert et al., 1995; Kühn et al., 1997; Marentes and Grusak, 1998; Kehr et al., 1999; Xoconostle-Cazares et al., 1999; Haebel and Kehr, 2001; Hoffmann-Benning et al., 2002; Giavalisco et al., 2006; Lin et al., 2009), nucleic acids (Kühn et al., 1997; Ruiz-Medrano et al., 1999; Yoo et al., 2004; Haywood et al., 2005), and lipids (Madey et al., 2002; Guelette et al., 2007, 2012). Furthermore, phloem transport is likely a regulated process, dependent on interactions between several proteins or RNA and proteins (Xoconostle-Cazares et al., 1999; Aoki et al., 2002, 2005; Yoo et al., 2004; Lee et al., 2005; Kehr and Butz, 2008; Ham et al., 2009). Phloem movement occurs in the sieve elements, which have evolved to optimize longitudinal flow by removing any obstacles in the form of organelles and by increasing the porosity of the cell walls at the longitudinal ends (van Bel and Knoblauch, 2000). While sieve elements may still contain nuclei, vacuole, and organelles during their early development, those cell components disintegrate leaving the sieve elements with only the plasma membrane and a thin cytoplasm containing endoplasmic reticulum, phloem-specific plastids, and a few, often dilated, mitochondria (Shah and Jacob, 1969; Cronshaw, 1981; Behnke and Schulz, 1983; Behnke, 1991; van Bel and Knoblauch, 2000). As a result, it had long been assumed that most sieve-tube proteins are synthesized in the companion cells and transported into the sieve elements via plasmodesmata. However, recent findings of ribosome subunits in sieve-tube elements have reopened the debate on whether a functional protein synthesis machinery exists in the sieve elements (Raven, 1991; Hayashi et al., 2000; Lin et al., 2009).
Lipids in the Phloem
While the role of RNA, viruses, and many proteins in the phloem is extensively studied, lipophilic compounds were not expected in an aqueous environment like the phloem sap. Consequently, long-distance lipid transport or signaling has been given little attention until now. Yet, lipids ranging in complexity from simple lipids like Jasmonic acid (JA) to phytosterols, to more complex glycolipids have been described in the phloem of canola, Perilla, Arabidopsis, cabbage, tomato, and tobacco among others (Madey et al., 2002; Guelette et al., 2007, 2012; Behmer et al., 2011).
Lipids like oxylipins are synthesized in response to pathogen attack and act as messengers during systemic acquired resistance. One well studied example is the fatty acid derivative jasmonic acid (JA), an oxylipin, which has been shown to be transported in the phloem in response to pathogen infection (Schilmiller and Howe, 2005; Lough and Lucas, 2006; Thorpe et al., 2007; Truman et al., 2007). It may act via a non-autonomous (slow; phloem-mediated) and an autonomous (fast; possible electric or hydraulic signal; see Koo and Howe, 2009) pathway. Three enzymes which are essential for JA biosynthesis (allene oxide synthase, allene oxide cyclase, and lipoxygenase) are also present in sieve elements, indicating that its synthesis occurs, at least in part, within the sieve elements (Hause et al., 2003).
Recently, Behmer et al. (2011) suggested the presence of phytosteroids and cholesterol in the phloem sap. Thus far their role remains unknown.
Additional experiments have shown that canola phloem sap not only contains lipid droplets, but that the fatty acid composition of these lipids is distinct from those typically found in membranes (Madey et al., 2002) and includes short- and medium-chain as well as odd-carbon-number fatty acids. This suggests that lipids are not only present in the phloem but also that they are phloem-specific rather than membrane degradation products.
Our analysis of phloem exudates harvested from 6-week-old Arabidopsis thaliana (Guelette et al., 2012; see Materials and Methods in Appendix and 8-week-old Perilla crispa, Hoffmann-Benning et al., 2002) revealed the presence of lipids (Guelette et al., 2007, 2012), several of which were unusual due to their shortness or odd number of carbons (propanoic acid, nonanoic acid, dodecanoic acid, tetradecanoic acid, pentadecanoic acid). Saturated, even-numbered fatty acids make up 10–40% of the total fatty acids in most naturally occurring lipids. Fatty acyl groups of less than 16 carbons or odd-carbon-number are not typically present in plant membranes and, thus, these fatty acids appear to be specific to the plant phloem. Yet their function remains unknown. Though they could simply be transported in the phloem, a role in pathogen response, or signaling has been discussed in the literature (Sanz et al., 1998; Hamberg et al., 1999). We showed that these fatty acids are intrinsic to phloem exudates and suggest that they are either transported in the phloem as one form of photoassimilate and cell building blocks destined for sink tissues or, more likely, as developmental signals. The question is: are there more complex lipids in the plant phloem and could they also play a role in long-distance signaling?
Thin-layer chromatography confirmed the presence of polar lipids in the phloem exudate, several of which have retention times different from typical leaf/membrane lipids. This suggests that they are indeed not contaminants but may instead perform separate functions. LC–MS was used to confirm the occurrence of lipids and other hydrophobic compounds in exudates. Mass over charge-based characterization of ions in conjunction with detection of at least two predicted MS/MS fragments using lipid profiler and alternative manual interpretation of mass spectra suggests the presence of species of phosphatidic acid (PA), lysophosphatidic acid (LPA), phosphatidylglycerol, phosphatidylinositol (PI) plus its phosphates (PIPs), di- and triacylglycerols, and phosphatidylcholine (PC) in the phloem exudates (Guelette et al., 2012).
Inositol-3-phosphate and PIP2 are important second messengers in animals as well as in plants (Ananieva and Gillaspy, 2009; Munnik and Vermeer, 2010); in plants phophatidic acid (PA) and lysophosphatidic acid (LPA) also appear to play an important role (Wang, 2004; Katagiri et al., 2005; Munnik and Testerink, 2009). PA has been proposed to be involved in intracellular abscisic acid (ABA), wound, and pathogen-related signaling/response (Katagiri et al., 2005; Testerink and Munnik, 2005; Lee and Lee, 2008). So far, there is no evidence about their long-distance transport and action, which could potentially be via the phloem as representatives of all three lipid groups (PIPs, PA, LPA) were identified in our preliminary characterization of phloem lipids.
A second lipid group we identified were phosphatidyl cholines (PC). In addition to their function as membrane components, PCs may play a role in signaling through the generation of diacylglycerols by PC-specific phospholipases. The resulting diacylglycerols would be more saturated and have different activities compared to PI-derived diacylglycerols. It has been suggested that PC may play a role in intracellular signaling in prokaryotes, humans, and possibly even plants (Hunt, 2006). Our findings raise the possibility that phloem lipids could play a similar role in plant long-distance signaling.
Examples of Long-Distance Lipid Transport and Signaling in Other Biological Systems
The presence of hydrophobic compounds in an aqueous system like the sieve elements, though unusual, is not unheard of in biological systems: other aqueous biological systems like the human blood contain a variety of lipids, many of which play a crucial role in human health. They are typically bound to proteins which can serve multiple functions such as transport to other tissues for storage, use, modification, or degradation. The best-known example would be the lipid cholesterol. It is either bound to low-density lipoproteins (LDL) and transported throughout the bloodstream for uptake into cells and incorporation into membranes; or it is bound to high-density lipoproteins (HDL) or within chylomicrons and moved to the liver for degradation (for a summary see Nelson and Cox, 2008). Thus, the nature of the protein to which the lipid is bound not only determines its direction of transport but also its fate. Other proteins transport essential vitamins or play a role in signaling (Blaner, 1989; Glatz et al., 1995; Charbonneau et al., 2009). In several cases lipid–protein complexes serve as messengers and affect transcription factor activity (Tontonoz et al., 1994; Nagy and Szanto, 2005). These mechanisms are generally important in mammalian systems but their possible importance in plants is virtually unexplored. They raise the possibility that lipids and the respective lipid-binding proteins in the phloem serve similar functions in plants and play an important role in stress and developmental signaling. Many lipids (phosphatidic acid, phosphoinositides) are already known to function as second messengers. However, virtually nothing is known about their possible long-distance transport. It is conceivable that these lipids are transported in the phloem in the form of protein–lipid complexes. This mechanism of lipid signaling in the plant through phloem transport is a new area of research which is expected to contribute new concepts to plant development. We hypothesize that plant phloem lipids are bound to and transported by lipid-binding proteins using mechanisms similar to those in the human blood. Consequently plant phloem lipid–protein complexes may have regulatory and transport functions similar to those in mammals and may play an important role in stress related and developmental signaling beyond what is already known about hormones like JA. The question remains: Are there any phloem proteins which bind lipids and could serve as good candidates for this function?
Putative Lipid-Binding Proteins in the Phloem and Their Possible Function in Relation to Signaling
Lipid-binding proteins in the phloem could have several roles:
-
(i)
they could mediate the release of the lipid into sieve element either by participating in the transport of lipids into the sieve element or in its release from the membrane,
-
(ii)
they could bind specific phloem lipids, thus facilitating their solubilization in the exudate as well as their (targeted) transport,
-
(iii)
they could be part of a receptor which senses the lipid (-signal) and transfers it out of the sieve element, or,
-
(iv)
the protein could be the signal itself, with its activity modulated by the bound lipid.
These possibilities remain to be tested, however, three known examples of phloem lipid-binding proteins and predictions based on homologies to known proteins support these options.
While many lipid-binding proteins have been identified in different plant species, only three have previously been described in phloem exudates: DIR1, a lipid transfer protein (LTP) was shown to play a role in systemic acquired resistance in Arabidopsis and tomato (Maldonado et al., 2002; Mitton et al., 2009). Similarly, an Acyl-CoA-binding protein has been identified in phloem exudates from rice (Suzui et al., 2006). However, as of yet, no connection has been made to phloem lipids. Representatives of both protein classes have been found in the phloem as well as in other plant tissues like the leaves. In both cases, the mechanism of their function in the phloem remains unknown. A third example of a predicted lipid-binding protein in the phloem is Flowering locus t (FT). It belongs to the family of Phosphatidylethanolamine-binding proteins (PEBPs), which also have a low affinity for Phosphatidylinositol1. In animals, PEBPs play a role in neuronal development, Alzheimer’s disease, and in the regulation of signaling pathways like the MAP kinase pathway (Vallée et al., 2003; Zhao et al., 2011). The PEBP homolog in the plant Antirrhinum appears to play a role as a kinase regulator (Banfield and Brady, 2000). Whether FT functions in a similar fashion and what its lipid-binding partners are remains to be shown (Kikuchi et al., 2009; Danilevskaya et al., 2011; Karlgren et al., 2011).
In our analysis of Arabidopsis phloem exudates we were able to detect 14 putative lipid-binding proteins, 10 of which have characteristics which could allow a function in lipid-based long-distance signaling and are discussed below. Expression in the phloem/companion cells for all but two of them has been shown (Table 1; Mustroph et al., 2009; Guelette et al., 2012).
Table 1.
Lipid-binding proteins identified in Arabidopsis thaliana phloem exudates.
| Identified proteins | Accession number | MW (kDa) | mRNA in CCs | Tryptic fragments |
|---|---|---|---|---|
| Lipase class 3 family protein | At4g16820 | 58 | No | VPGIFADNDK |
| VDMKMSPYLK | ||||
| Aspartic protease | At4g04460 | 56 | Yes | LNDENADMVPLK |
| Glycine-rich protein (oleosin) GRP17 | At5g07530 | 53 | Yes | KKKCMSGGMSGSEEGMSGSEGGMSSGGGSK |
| DNPPPAGLPPNSGAGAGGAQSLIKKSK | ||||
| KSMSGGMSGSEEGMSGSEGGMSSGGGSK | ||||
| Put. PIG-P | At2g39435 | 50 | No | YDQQNFKSK |
| GDSL motiv Lipase/hydrolase | At1g29660 | 40 | Yes | ITFSGQVENYK |
| Annexin* | At1g35720 | 36 | Yes | TGTDEGALTR |
| YGDDHGEEILK | ||||
| TLDKELSNDFER | ||||
| TSTQLLHAR | ||||
| 14-3-3-like protein GF14 epsilon psi | At1g22300 | 28 | Yes | ILSSIEQK |
| At2g10450 | 9 | DSTLIMQLLP | ||
| DSTLIMKILR | ||||
| Lipid-associated family protein | At4g39730 | 20 | Yes | IYDKDGDYIGIK |
| Major latex protein/Bet v I like protein* | At1g70890 | 18 | Yes | SFAFTLQVTPK |
| VLEGDLMNEYK | ||||
| NKIEAVDPEK | ||||
| IEAVDPEK | ||||
| EIDEHLLAEE | ||||
| Bet v 1 allergen-like protein | At1g23130 | 18 | Yes | ERIEAVDQEK |
| VGSVIFWNYAIDGQPK |
Two of the proteins identified in our phloem exudates are characterized as lipases. Proteins are often included in that category based on sequence similarity with known lipases. Lipases are lipolytic enzymes which can function in membrane synthesis and turnover or in signaling (Beisson et al., 2003). Lipases have been implicated in salicylic acid signaling (Feys et al., 2001) and wound response (Guan and Nothnagel, 2004). The first lipase-like protein (At4g16820) is annotated as a phospholipase A (PLA-Iβ2), a triacylglycerol lipase or DAD1-like acyl hydrolase. It acts in a redundant manner together with other lipases in salt stress as well as in wound and pathogen-induced production of Jasmonic acid (Beisson et al., 2003; Ellinger and Kubigsteltig, 2010; Ellinger et al., 2010)2. It contains one transmembrane domain suggesting that it is anchored in the membrane. Thus, rather than participating in long-distance transport of (signaling) lipids, it may assist in the release of those lipids into the sieve element.
The second lipase-like protein, At1g29660 is annotated as a GDSL lipase/fatty acyl transferase. It has been found in the nucleus, ER, peroxisomes, and extracellular space and appears to be reduced in response to drought (Huang et al., 2008; Ding et al., 2009). Its function is unknown and may range from hydrolysis of triacylglycerols to the fatty acid transfer between phosphatidylcholine and sterols (cd01846)3. As for the DAD1-like lipase, the exact enzymatic/lipolytic activity has yet to be characterized.
The phloem exudate also contains an aspartic protease. Some aspartic proteases have the ability to cleave membrane proteins. Their activity can sometimes be modulated by association with certain membrane lipids. It has been proposed that apoplastic aspartic proteases may cleave lipids off acyl-CoA-binding proteins to act (in conjunction with DIR1) in pathogen response/signaling (Xiao and Chye, 2011). Hence it appears more likely that they serve in a localized process, for example in the release of lipids for binding to DIR1 rather than in long-distance transport.
GRP17 has previously been identified in Arabidopsis pollen and in seeds (Mayfield et al., 2001). It contains an oleosin domain. The role of oleosins in the seed is to prevent lipid aggregation and control oil-body size (Mayfield and Preuss, 2000). It is conceivable that it plays a similar role in the phloem sap, functioning to solubilize lipids in an otherwise aqueous environment.
Another of the proteins in phloem exudates is the subunit P of a phosphatidylinositol N-acetylglucosaminyltransferase (PIG-P), which catalyzes the transfer of N-acetyl glucosamine to phosphatidyl inositol in the first step of the glycosylphosphatidylinositol (GPI)-anchor synthesis. GPI anchors are glycolipids which anchor proteins to membranes (Watanabe et al., 2000). In animals, GPI anchors may interact directly with receptor proteins in the signaling of neural and brain development (Peles et al., 1997; Ferrando-Miguel et al., 2004). Only few GPI-anchored proteins have been found in plants. They include arabinogalactan proteins (AGPs), which have been shown to contain a classical C-terminal GPI anchor signal sequence (Schultz et al., 1998): upon synthesis in the ER they are secreted into the apoplast, where the AGPs could be cleaved by phospholipases and act as signal molecules. It is possible that GPI-anchored proteins participate in cell-to-cell or even long-distance interactions.
Annexins have been shown to bind phospholipids/membranes and are involved in Ca2+-signaling as well as callose formation (Andrawis et al., 1993; Mortimer et al., 2008). Expression of Arabidopsis annexin 1 is upregulated in response to salt and water stress as well as salicylic acid and ABA (Konopka-Postupolska et al., 2009). Similarly, a Medicago truncatula annexin-like protein AnxMs2 is induced under stress conditions (salt, osmotic, drought) and localizes in the nucleolus even though it contains no typical nuclear localization signal. This suggests that it could play a role in membrane trafficking and intracellular signal transduction (Kovács et al., 1998). Annexin transcript levels vary depending on tissue type and age, suggesting specific purposes during different developmental stages. They possibly play a role in tip growth, gravitropic response, mechanical stress, Ca2+-signaling as well as callose formation and cold, oxidative, saline, and ABA stress responses (Andrawis et al., 1993; Mortimer et al., 2008). Plant annexins have been identified in the plant phloem before (Barnes et al., 2004; Giavalisco et al., 2006). Yet their role in long-distance transport is not clear. It is conceivable that they function in phospholipid-based long-distance signaling processes.
14-3-3 proteins are small conserved proteins, which play an important role in regulatory processes and signal transduction. In animal systems, free Raf kinases appear to be bound to 14-3-3 proteins, however, once they bind to liposomes/membranes, the 14-3-3 protein dissociates (Hekman et al., 2006). This suggests that 14-3-3 proteins may modulate binding of proteins to lipids. In plants they activate transcription by binding to activators in the nucleus (Ferl, 1996; Fu et al., 2000; Emi et al., 2001). Taoka et al. (2011) have shown that they can act as receptors for rice Hd3a. Hd3a is the rice homolog to FT, a putative phosphatidyl ethanolamine-binding protein (PEBP) which travels in sieve elements and is essential for the induction of flowering (Corbesier et al., 2007; Lin et al., 2007; Tamaki et al., 2007). Many PEBPs act as kinase regulators in signaling pathways in both animals and plants (Banfield and Brady, 2000; Zhao et al., 2011). This suggests that rather than transporting lipids/lipid signals 14-3-3 proteins may act as receptors for lipid/protein signals.
The major latex protein-like protein and the Bet v 1 allergen-like protein belong to the Bet v 1 like superfamily. Members of this superfamily can be major plant allergens but also play a role in the response to biotic and abiotic stress and in development (Mogensen et al., 2002). The Bet v 1 superfamily contains proteins with low sequence similarity but a common hydrophobic fold. This fold has high structural similarity with the cholesterol-binding fold of the human MLN64 protein suggesting the possibility of a role in phytosteroid transport (Neudecker et al., 2001). Members of this superfamily include ceramide transfer proteins, phosphatidylcholine transfer proteins, phosphatidyinositol transfer proteins, acetyl-CoA hydrolases, and HD-Zip transcription factors from plants (Ariel et al., 2007; Radauer et al., 2008). The discovery of representatives of the proteins in phloem exudates suggests that they may not just play a role in transmembrane lipid transport but in long-distance lipid transport and signaling as well. While their lipid-partners remain to be identified it is intriguing that phosphatidylcholine and phosphatidylinositol transfer proteins belong into this group of proteins while the respective lipids are also present in phloem exudates.
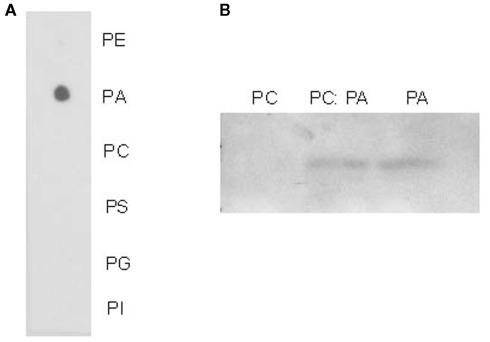
One of the smallest putative lipid-binding proteins we identified in phloem exudates is a hypothetical protein characterized as a lipid-associated family protein (PLAFP). It contains a PLAT/LH2 domain. The proposed function of this domain is to mediate interaction with lipids or membrane-bound proteins. While the function of this domain is yet unknown, proteins containing it appear to be stress-induced. To examine the lipid-binding properties of PLAFP, we cloned PLAFP into the pET15b expression vector (Novagen) and overexpressed it in E. coli host strain OrigamiB(DE3)pLysS (Novagen; see Appendix). The protein was purified and lipid-binding assessed using lipid overlay and lysosome-binding assays (Figure 1; Awai et al., 2006; Lu and Benning, 2009; see Appendix). Both tests showed that PLAFP binds phosphatidic acid (PA) but not the other tested phospholipids.
Figure 1.
Lipid-binding assays. (A) Protein–lipid overlay assay using purified PLAFP. Ten nanomole of selected phospholipids were spotted on a hybond-C membrane and overlaid it with 1 μl/ml purified PLAFP. PE, phosphatidyl ethanolamine; PA, phosphatidic acid; PC, phosphatidyl choline; PS, phosphatidyl serine; PG, phosphatidyl glycerol; PI, phosphatidyl inositol. (B) Liposome binding assay using purified PLAFP. Liposomes were prepared from PC, PA, or PC:PA (1:1, w/w). Bound protein was detected by antibody against the His-tag.
This is the first example where a phloem lipid-binding protein (PLAFP), and its respective lipid partner (PA) have both been found within phloem exudates and suggests that PA is indeed transported within or into the phloem while bound to a protein. Whether the function of this protein is that of loading/unloading the lipid into and out of the phloem, as a receptor or as a transport molecule, and whether it has a true signaling function remains to be shown.
Our detection of these lipids and a lipid-binding protein with specific PA-binding properties suggests that lipids and long-distance lipid signaling may play a larger and more complex role in the plant than previously thought and should be explored.
Conclusion
We and others have been able to detect several fatty acids and lipids in phloem exudates, some of which are known factors in intracellular signaling (PA, PIP, and PC). In addition, we find proteins with the predicted, or in some cases known, ability to bind and transport these lipids and with a role in intracellular signaling (i.e., Annexin; Andrawis et al., 1993).
Together, this suggests that phloem lipids may not only be involved in intracellular signaling but also play an important role in long-distance lipid signaling. We postulate that the lipids may be released, possibly from membranes, in response to an environmental stimulus (lipases, aspartic protease) followed by transport (PLAFP, annexin, Bet v 1-like proteins) into and throughout the phloem and binding to a receptor at the target tissue (PLAFP, 14-3-3 proteins).
It is clear that even though lipids and lipid-binding proteins in the phloem have been studied very little, those that have been studied in other plant tissues, have a significant impact in signaling. However, the mechanisms of lipid uptake, translocation, and action are largely unknown suggesting that this aspect of phloem transport needs to be studied further. Lipid signaling in the plant through phloem transport is a new area of research which is expected to contribute new concepts to plant development.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by funding through the Michigan State University – College of Natural Sciences and by the USDA National Institute of Food and Agriculture, Hatch project number MICL02233.
Appendix
Materials and methods
Harvest of phloem exudate
Phloem exudate from 6-week-old Arabidopsis plants using the EDTA-facilitated method described in Guelette et al. (2012). In short, leaves were cut at the base of petiole under 20 mM K2-EDTA, pH 7.0, and incubated in microtubes containing 1.2 ml of the same solution for 1 h. After 1 h the solution was discarded. The petioles were washed thoroughly and placed in a new microtube containing 1.2 ml deionized water. Exudates were collected for 6–8 h in a dark, humid atmosphere, immediately frozen in liquid nitrogen, lyophilized, and stored at −80°C.
Analysis of proteins in the phloem exudates
Phloem exudates from 20 leaves per replicate (three replicates total) were subjected to 15% SDS-polyacrylamide gel electrophoresis. Protein bands were excised and digested with trypsin according to Shevchenko et al. (1996). For liquid chromatography/mass spectrometry (LC–MS/MS) tryptic fragments were dissolved in 2% ACN/0.1% formic acid. Analysis was performed on a Capillary LC system (Waters Corp., Milford, MA, USA) coupled to a LCQ DECA ion trap mass spectrometer (Thermofinnigan, San Jose, CA, USA) equipped with a nanospray ionization source. The sample was trapped onto a Peptide Cap Trap (Michrom BioResources, Auburn, CA, USA) and flushed onto a 5 cm × 75 μm ID picofrit column packed with 5 μm ProteoPep C18 material (New Objective, Woburn, MA, USA), and eluted with a gradient of 2–95% ACN in 0.1% formic acid at a flow rate of 200 nl/min for 60 min. Peptides/proteins were identified using the programs SEQUEST, MASCOT, or gpm.org. Carbamidomethyl Cys was set as fixed modification and oxidation of Met was allowed. Up to two missed tryptic sites were permitted. Peptide tolerance was set to 2.5 Da and MS/MS tolerance was set to 0.8 Da. Positive identification required at least two unique peptides per proteins counting only peptides with significant scores (95% confidence per peptide; >2.5 for SEQUEST). Three proteins with a single peptide identification were included due to the fact that their mass corresponded to the predicted size from the gel and they were seen in at least two independent preparations. Database searches using individual tryptic fragments were performed using the BLAST searches at NCBI (http://www.ncbi.nlm.nih.gov/blast).
Analysis of lipids in phloem exudates
Phloem exudates were phase partitioned against chloroform:methanol (1:1, v/v), concentrated under N2 and submitted to LC–MS. Lipid analysis was performed using a Waters LCT Premier mass spectrometer (LC-TOF–MS) with multiplexed CID (collision-induced dissociation at 20, 35, 50, 65, and 80 V) and a gradient from 10% A (10 mM Ammonium formate) to 99% B (acetonitrile:isopropanol; 1:2) on the column described above using both positive and negative ion mode. Data are representatives of three biological replicates.
Protein expression and purification
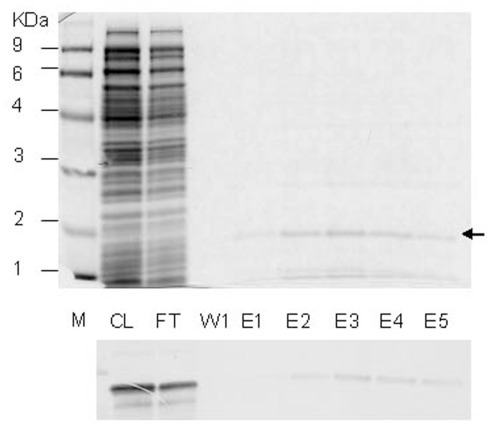
A cDNA clone for PLAFP (At4g39730), U21720, was obtained from Arabidopsis Biological Resource Centre, Ohio State University (Columbus, OH, USA). The coding region of PLAFP, excluding 69 nucleotide region encoding the 23 amino-acid predicted signal peptide, was PCR amplified using the forward primer, 5′-GCGCATATGGAAGATGATCCAGACTGTGTATACA-3′ and reverse primer, 5′-GCGCATATGTTAAACGACCCAAGAAAGCTTTTTCCG-3′ which introduced NdeI sites at both ends of the PCR product. The PCR product was cloned into pGEMT-easy vector (Promega), and subcloned into pET15b expression vector (Novagen) using NdeI site, to generate the expression clone, pET15b-PLAFP. E. coli host strain OrigamiB(DE3)pLysS (Novagen) was transformed with pET15b-PLAFP and the transformants were selected by Amp, Kan, Cm, and Tet resistance. Protein expression was induced by adding IPTG up to the final concentration of 0.5 mM. Protein was extracted and purified using the HisLink™ resin (Promega) using the HEPES buffers containing different concentrations of imidazole, following the manufacturer’s protocol. Purification steps are shown in Figure A1 below. The purified protein was exchanged into 10 mM KH2PO4 (Lu and Benning, 2009) using a PD10 column (GE healthcare).
Figure A1.
Purification of PLAFP. (Top) SDS-PAGE gel for different fractions of purification of PLAFP-His. M, molecular weight marker; CL, cleared lysate; FT, flow-through; W1, wash fraction; E1–E6, elution fractions. (Bottom) Western blot analysis for the above listed factions using Anti-His antibody to visualize PLAFP-His.
Protein–lipid overlay assay
A phospholipid strip was prepared by spotting 10 nmol of different phospholipids (Avanti Polar Lipids) onto a Hybond-C membrane (GE Healthcare). Protein–lipid overlay assay was performed as described (Awai et al., 2006). The lipid strip was blocked with the blocking buffer (3% BSA in TBST, 10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.2% Tween 20) for 1 h and then incubated in the blocking buffer containing purified PLAFP (1 μg/ml) at 4°C overnight. The membrane was washed with TBST twice and incubated in the blocking buffer with Anti-polyHis mouse monoclonal antibody (Sigma) at 1:1000 dilution for 2 h at room temperature. The membrane was washed twice with TBST and incubated in the blocking buffer with horseradish peroxidase-conjugated anti-mouse antibody (BioRad) at 1:10,000 dilution for 1 h at room temperature. The membrane was washed twice with TBST and detection was done using a chemiluminescence detection system (Thermo scientific).
Liposome binding assay
Liposomes were prepared using PC, PA, or a mixture of PC and PA, following the method described in (Awai et al., 2006). Liposomes (250 μg) were mixed with 1 μg of purified PLAFP and TBS (50 mM Tris–HCl, pH7.0, 0.1 M NaCl) to make 100 μl solution. The mixture was incubated at 30°C for 30 min, followed by centrifugation at 10,000 × g for 10 min at 4°C. The pellet was washed twice with ice-cold TBS and then mixed with SDS-PAGE sample buffer. Western blot analysis was done using the antibodies and detection system mentioned above.
Footnotes
References
- Ananieva E. A., Gillaspy G. E. (2009). Switches in nutrient and inositol signaling. Plant Signal. Behav. 4, 304–306 10.4161/psb.4.4.8063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrawis A., Solomon M., Delmer D. P. (1993). Cotton fiber annexins – a potential role in the regulation of callose synthase. Plant J. 3, 763–772 10.1111/j.1365-313X.1993.00763.x [DOI] [PubMed] [Google Scholar]
- Aoki K., Kragler F., Xoconostle-Cázares B., Lucas W. J. (2002). A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proc. Natl. Acad. Sci. U. S. A. 99, 16342–16347 10.1073/pnas.252427999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Yamaya T., Sakakibara H. (2005). CmPP16-1-interacting phloem sap proteins are involved in the regulation of CmPP16-1 long-distance movement. Plant Cell Physiol. 46, S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F. D., Manavella P. A., Dezar C. A., Chan R. L. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12, 419–426 10.1016/j.tplants.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Awai K., Xu C., Tamot B., Benning C. (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U.S.A. 103, 10817–10822 10.1073/pnas.0602754103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield M. J., Brady R. L. (2000). The structure of antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297, 1159–1170 10.1006/jmbi.2000.3619 [DOI] [PubMed] [Google Scholar]
- Barnes A., Bale J., Constantinidou C., Aston P., Jones A., Pritchard J. (2004). Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J. Exp. Bot. 55, 1473–1481 10.1093/jxb/erh161 [DOI] [PubMed] [Google Scholar]
- Behmer S. T., Grebenok R. J., Douglas A. E. (2011). Plant sterols and host plant suitability for a phloem feeding insect. Funct. Ecol. 25, 484–491 10.1111/j.1365-2435.2010.01810.x [DOI] [Google Scholar]
- Behnke H.-D. (1991). Distribution and evolution of forms and types of sieve-element plastids in the dicotyledons. Aliso 3, 167–182 [Google Scholar]
- Behnke H.-D., Schulz A. (1983). The development of specific sieve-element plastids in would phloem of Coleus blumei (S-type) and Pisum sativum (P-type), regenerated from amyloplast-containing parenchyma cells. Protoplasma 114, 125–132 10.1007/BF01279876 [DOI] [Google Scholar]
- Beisson F., Koo A. J., Ruuska S., Schwender J., Pollard M., Thelen J. J., Paddock T., Salas J. J., Savage L., Milcamps A., Mhaske V. B., Cho Y., Ohlrogge J. B. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132, 681–697 10.1104/pp.103.022988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaner W. S. (1989). Retinol-binding protein: the serum transport protein for vitamin A. Endocr. Rev. 10, 308–316 10.1210/edrv-10-3-308 [DOI] [PubMed] [Google Scholar]
- Burton R. A., Gibeaut D. M., Bacic A., Findlay K., Hamilton A., Boulcombe D. C., Fincher G. B. (2000). Virus-induced silencing of a plant cellulose synthase. Plant Cell 12, 691–706 10.2307/3870995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau D., Beauregard M., Tajmir-Riahi H.-A. (2009). Structural analysis of human serum albumin complexes with cationic lipids. J. Phys. Chem. B 113, 1777–1781 10.1021/jp8092012 [DOI] [PubMed] [Google Scholar]
- Chen S., Petersen B. L., Olsen C. E., Schulz A., Halkier B. A. (2001). Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 127, 194–201 10.1104/pp.127.1.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zambryski P. (2000). Systemic transport of RNA in plants. Trends Plant Sci. 5, 52–54 10.1016/S1360-1385(99)01540-X [DOI] [PubMed] [Google Scholar]
- Corbesier L., Coral V., Seonghoe J., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Prinsen E., Jacqmard A., Lejeune P., Van Onckelen H., Perilleux C., Bernier G. (2003). Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J. Exp. Bot. 54, 2511–2517 10.1093/jxb/erg276 [DOI] [PubMed] [Google Scholar]
- Cronshaw J. (1981). Phloem structure and function. Annu. Rev. Plant Physiol. 32, 465–484 10.1146/annurev.pp.32.060181.002341 [DOI] [Google Scholar]
- Danilevskaya O. N., Meng X., McGonigle B., Muszynski M. G. (2011). Beyond flowering time: pleiotropic function of the maize flowering hormone florigen. Plant Signal. Behav. 9, 1267–1270 10.4161/psb.6.9.16423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Itaya A., Qi Y. J. (2003). Symplasmic protein and RNA traffic: regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6, 596–602 10.1016/j.pbi.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Ding Y., Lapko H., Ndamukong I., Xia Y., Al-Abdallat A., Lalithambika S., Sadder M., Saleh A., Fromm M., Riethoven J.-J., Lu G., Avramova Z. (2009). The Arabidopsis chromatin modifier ATX1, the myotubularin-like AtMTM and the response to drought. Plant Signal. Behav. 4, 1049–1058 10.4161/psb.4.11.10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Kubigsteltig I. I. (2010). Involvement of DAD1-like lipases in response to salt and osmotic stress in Arabidopsis thaliana. Plant Signal. Behav. 5, 1269–1271 10.4161/psb.5.10.13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D., Stingl N., Kubigsteltig I. I., Bals T., Juenger M., Pollmann S., Berger S., Schuenemann D., Mueller M. J. (2010). Dongle and defective in anther dehiscence1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: redundant lipases contribute to jasmonate formation. Plant Physiol. 153, 114–127 10.1104/pp.110.155093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi T., Kinoshita T., Shimazaki K. (2001). Specific binding of vf14-3-3a isoform to the plasma membrane H+-ATPase in response to blue light and fusicoccin in guard cells of broad bean. Plant Physiol. 125, 1115–1125 10.1104/pp.125.2.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl R. J. (1996). 14-3-3 proteins and signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 49–73 10.1146/annurev.arplant.47.1.49 [DOI] [PubMed] [Google Scholar]
- Ferrando-Miguel R., Cheon M. S., Lubec G. (2004). Protein levels of genes encoded on chromosome 21 in fetal down syndrome brain (part V): overexpression of phosphatidyl-inositol-glycan class P protein (DSCR5). Amino Acids 26, 255–261 10.1007/s00726-004-0065-9 [DOI] [PubMed] [Google Scholar]
- Feys B. J., Moisan L. J., Newman M. A., Parker J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. Embo J. 20, 5400–5411 10.1093/emboj/20.19.5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. B., Wu Y., Ku M. S. B. (1992). Turnover of soluble-proteins in the wheat sieve tube. Plant Physiol. 10, 1433–1441 10.1104/pp.100.3.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. A., Subramanian R. R., Masters S. C. (2000). 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 10.1146/annurev.pharmtox.40.1.617 [DOI] [PubMed] [Google Scholar]
- Giavalisco P., Kapitza K., Kolasa A., Buhtz A., Kehr J. (2006). Towards the proteome of Brassica napus phloem sap. Proteomics 6, 896–909 10.1002/pmic.200500155 [DOI] [PubMed] [Google Scholar]
- Glatz J. F. C., Boerchers T., Spener F., van der Vusse G. J. (1995). Fatty acids in cell signaling: modulation by lipid binding proteins. Prostaglandins Leukot. Essent. Fatty Acids 52, 121–127 10.1016/0952-3278(95)90010-1 [DOI] [PubMed] [Google Scholar]
- Guan Y., Nothnagel E. A. (2004). Binding of arabinogalactan proteins by yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol. 135, 1346–1366 10.1104/pp.104.039370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelette B. S., Benning U. F., Hoffmann-Benning S. (2012). Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana. J. Exp. Bot. 10.1093/jxb/ERS028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelette B. S., Chamberlin B., Benning U. F., Hoffmann-Benning S. (2007). “Indications of lipids/lipid signaling in the phloem exudates of Arabidopsis thaliana and Perilla ocymoides,” in Proceedings of the 17th International Symposium of Plant Lipids, eds Benning C., Ohlrogge J. (East Lansing: Michigan State University; ), 92–95 [Google Scholar]
- Haebel S., Kehr J. (2001). Matrix-assisted laser desorption/ionization time of flight mass spectrometry peptide mass fingerprints and post source decay: a tool for the identification and analysis of phloem proteins from Cucurbita maxima Duch. separated by two-dimensional polyacrylamide gel electrophoresis. Planta 213, 586–593 10.1007/s004250100523 [DOI] [PubMed] [Google Scholar]
- Ham B. K., Brandom J. L., Xoconostle-Cázares B., Ringgold V., Lough T. J., Lucas W. J. (2009). Polypyrimidine tract binding protein, CmRBP50, forms the basis of a pumpkin phloem ribonucleoprotein complex. Plant Cell 21, 197–215 10.1105/tpc.108.061317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M., Sanz A., Castresana C. (1999). Alpha-oxidation of fatty acids in higher plants – Identification of a pathogen-inducible oxygenase (PIOX) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J. Biol. Chem. 274, 24503–24513 10.1074/jbc.274.35.24503 [DOI] [PubMed] [Google Scholar]
- Hause B., Hause G., Kutter C., Miersch O., Wasternack C. (2003). Enzymes of jasmonate biosynthesis occur in tomato sieve elements. Plant Cell Physiol. 44, 643–648 10.1093/pcp/pcg072 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Fukuda A., Suzui N., Fujimaki S. (2000). Proteins in the sieve element-companion cell complexes: their detection, localization and possible functions. Aust. J. Plant Physiol. 27, 489–496 [Google Scholar]
- Haywood V., Yu T. S., Huang N. C., Lucas W. J. (2005). Phloem long-distance trafficking of gibberellic acid-insensitive RNA regulates leaf development. Plant J. 42, 49–68 10.1111/j.1365-313X.2005.02351.x [DOI] [PubMed] [Google Scholar]
- Hekman M., Albert S., Galmiche A., Rennefahrt U. E., Fueller J., Fischer A., Puehringer D., Wiese S., Rapp U. R. (2006). Reversible membrane interaction of BAD requires two C-terminal lipid binding domains in conjunction with 14-3-3 protein binding. J. Biol. Chem. 281, 17321–17336 10.1074/jbc.M600292200 [DOI] [PubMed] [Google Scholar]
- Hoffmann-Benning S., Gage D. A., McIntosh L., Kende H., Zeevaart J. A. D. (2002). Comparison of peptides in the phloem sap of flowering and non-flowering Perilla and lupine plants using microbore HPLC followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Planta 216, 140–147 10.1007/s00425-002-0916-0 [DOI] [PubMed] [Google Scholar]
- Huang D., Wu W., Abrams S. R., Cutler A. J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007 10.1093/jxb/erm336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. N. (2006). Dynamic lipidomics of the nucleus. J. Cell. Biochem. 97, 244–251 10.1002/jcb.20691 [DOI] [PubMed] [Google Scholar]
- Karlgren A., Gyllenstrand N., Källman T., Sundström J. F., Moore D., Lascoux M., Lagercrantz U. (2011). Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol. 156, 1967–1977 10.1104/pp.111.176206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T., Ishiyama K., Kato T., Tabata S., Kobayashi M., Shinozaki K. (2005). An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. Plant J. 1, 107–117 10.1111/j.1365-313X.2005.02431.x [DOI] [PubMed] [Google Scholar]
- Kehr J. (2006). Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 57, 767–774 10.1093/jxb/erj087 [DOI] [PubMed] [Google Scholar]
- Kehr J., Butz A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85–92 10.1093/jxb/erm176 [DOI] [PubMed] [Google Scholar]
- Kehr J., Haebel S., Blechschmidt-Schneider S., Willmitzer L., Steup M., Fisahn J. (1999). Analysis of phloem protein patterns from different organs of Cucurbita maxima Duch. by matrix-assisted laser desorption/ionization time of flight mass spectroscopy combined with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Planta 207, 612–619 10.1007/s004250050525 [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Kawahigashi H., Ando T., Tonooka T., Handa H. (2009). Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149, 1341–1353 10.1104/pp.108.132134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka-Postupolska D., Clark G., Goch G., Debski J., Floras K., Cantero A., Fijolek B., Roux S., Hennig J. (2009). The role of annexin 1 in drought stress in Arabidopsis. Plant Physiol. 150, 1394–1410 10.1104/pp.109.135228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A. J. K., Howe G. A. (2009). The wound hormone jasmonate. Phytochemistry 70, 1571–1580 10.1016/j.phytochem.2009.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács I., Ayaydin F., Oberschall A., Ipacs I., Bottka S., Pongor S., Dudits D., Toth E. C. (1998). Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. Plant J. 15, 185–197 10.1046/j.1365-313X.1998.00194.x [DOI] [PubMed] [Google Scholar]
- Kühn C., Franceschi V. R., Schulz A., Lemoine R., Frommer W. B. (1997). Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275, 1298–1300 10.1126/science.275.5304.1298 [DOI] [PubMed] [Google Scholar]
- Lee Y., Lee Y. (2008). Roles of phosphoinositides in regulation of stomatal movements. Plant Signal. Behav. 3, 211–213 10.4161/psb.3.4.5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Taoka K., Yoo B.-C., Ben-Nissan G., Kim D.-J., Lucas W. J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17, 2817–2831 10.1105/tpc.105.034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-K., Belanger H., Lee Y.-J., Varkonyi-Gasic E., Taoka K.-I., Miura E., Xoconostle-Cazares B., Gendler K., Jorgensen R. A., Phinney B., Lough T. J., Lucas W. J. (2007). FT protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19, 1488–1506 10.1105/tpc.107.051920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.-K., Lee Y.-J., Lough T. J., Phinney B., Lucas W. J. (2009). Analysis of the pumpkin phloem proteome provides functional insights into angiosperm sieve tube function. Mol. Cell Proteomics 8, 343–356 [DOI] [PubMed] [Google Scholar]
- Lough T. J., Lucas W. J. (2006). Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57, 203–232 10.1146/annurev.arplant.56.032604.144145 [DOI] [PubMed] [Google Scholar]
- Lu B., Benning C. (2009). A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284, 17420–17427 10.1074/jbc.M109.008821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W. J., Bouche-Pillon S., Jackson D. P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983 10.1126/science.270.5244.1980 [DOI] [PubMed] [Google Scholar]
- Madey E., Nowack L. M., Thompson J. E. (2002). Isolation and characterization of lipid in phloem sap of canola. Planta 214, 625–634 10.1007/s004250100649 [DOI] [PubMed] [Google Scholar]
- Maeda H., Song W., Sage T. L., DellaPenna D. (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18, 2710–2732 10.1105/tpc.105.039404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado A. M., Doerner P., Dixon R. A., Lamb C. J., Cameron R. K. (2002). A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 419, 399–403 10.1038/nature00962 [DOI] [PubMed] [Google Scholar]
- Marentes E., Grusak M. A. (1998). Mass determination of low-molecular-weight proteins in phloem sap using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Exp. Bot. 49, 903–911 10.1093/jexbot/49.322.903 [DOI] [Google Scholar]
- Mayfield J. A., Fiebig A., Johnstone S. E., Preuss D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292, 2482–2485 10.1126/science.1060972 [DOI] [PubMed] [Google Scholar]
- Mayfield J. A., Preuss D. (2000). Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2, 128–130 10.1038/35017146 [DOI] [PubMed] [Google Scholar]
- Mitton F. M., Pinedo M. L., de la Canal L. (2009). Phloem sap of tomato plants contains a DIR1 putative ortholog. J. Plant Physiol. 166, 543–547 10.1016/j.jplph.2008.07.002 [DOI] [PubMed] [Google Scholar]
- Mogensen J. E., Wimmer R., Larsen J. N., Spangfort M. D., Otzen D. E. (2002). The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277, 23684–23692 10.1074/jbc.M202065200 [DOI] [PubMed] [Google Scholar]
- Mortimer J. C., Laohavisit A., Macpherson N., Webb A., Brownlee C., Battey N. H., Davies J. M. (2008). Annexins: multifunctional components of growth and adaptation. J. Exp. Bot. 59, 533–544 10.1093/jxb/erm344 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M. E., Jang C. J. H., Holtan H. E., Repetti P. P., Galbraith D. W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18843–18848 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Testerink C. (2009). Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. 50, S260–S265 10.1194/jlr.R800098-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Vermeer J. E. M. (2010). Osmotic stress-induced phosphoinositde and inositol signaling in plants. Plant Cell and Environ. 33, 655–669 10.1111/j.1365-3040.2009.02097.x [DOI] [PubMed] [Google Scholar]
- Nagy L., Szanto A. (2005). Roles for lipid activated transcription factors. Mol. Nutr. Food Res. 49, 1072–1074 10.1002/mnfr.200590042 [DOI] [PubMed] [Google Scholar]
- Nelson D. L., Cox M. M. (2008). Lehninger: Principles of Biochemistry, 5th Edn New York: W. H. Freeman and Company, 836–841 [Google Scholar]
- Neudecker P., Schweimer K., Nerkamp J., Scheurer S., Vieths S., Sticht H., Rosch P. (2001). Allergic cross-reactivity made visible: solution structure of the major cherry allergen Pru av 1. J. Biol. Chem. 276, 22756–22763 10.1074/jbc.M101657200 [DOI] [PubMed] [Google Scholar]
- Peles E., Nativ M., Lustig M., Grumet M., Schilling J., Martinez R., Plowman G. D., Schlessinger J. (1997). Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 16, 978–988 10.1093/emboj/16.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radauer C., Lackner P., Breiteneder H. (2008). The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286–304 10.1186/1471-2148-8-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J. A. (1991). Long-term functioning of enucleate sieve elements: possible mechanisms of damage avoidance and damage repair. Plant Cell Environ. 14, 139–146 10.1111/j.1365-3040.1991.tb01330.x [DOI] [Google Scholar]
- Ruiz-Medrano R., Xoconostle-Cázares B., Lucas W. J. (1999). Phloem long-distance transport of CmNACP mRNA: implications for supracellular regulation in plants. Development 126, 4405–4419 [DOI] [PubMed] [Google Scholar]
- Ryabov E. V., Robinson D. J., Taliansky M. E. (1999). A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. U.S.A. 96, 1212–1217 10.1073/pnas.96.4.1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuth T., Schobert C., Pecsvaradi A., Eichholz A., Komor E., Orlich G. (1993). Specific proteins in the sieve-tube exudate of Ricinus communis l seedlings – separation, characterization and in vivo labeling. Planta 191, 207–213 10.1007/BF00199751 [DOI] [Google Scholar]
- Sanz A., Moreno J. I., Castresana C. (1998). PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell 10, 1523–1537 10.1105/tpc.10.9.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A. L., Howe G. A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8, 369–377 10.1016/j.pbi.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Schobert C., Grossmann P., Gottschalk M., Komor E., Pecsvaradi A., Zurnieden U. (1995). Sieve-tube exudate from Ricinus communis l seedlings contains ubiquitin and chaperones. Planta 196, 205–210 10.1007/BF00201375 [DOI] [Google Scholar]
- Schultz C., Gilson P., Oxley D., Youl J., Bacic A. (1998). GPI-anchors on arabinogalactan-proteins: implication for signalling in plants. Trends Plant Sci. 3, 426–431 10.1016/S1360-1385(98)01328-4 [DOI] [Google Scholar]
- Seo H. S., Song J. T., Cheong J.-J., Lee Y.-H., Lee Y.-W., Hwang I., Lee J. S., Choi Y. D. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98, 4788–4793 10.1073/pnas.081557298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. J., Jacob R. (1969). Development and structure of phloem in the petiole of Langenaria siceraria (Mol.). Standl. and Momordica charantia L. Ann. Bot. 33, 855–863 [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 10.1021/ac950914h [DOI] [PubMed] [Google Scholar]
- Suárez-López P. (2005). Long-range signalling in plant reproductive development. Int. J. Dev. Biol. 49, 761–771 10.1387/ijdb.052002ps [DOI] [PubMed] [Google Scholar]
- Suzui N., Nakamura A., Fujiwara T., Hayashi T., Yoneyama T. (2006). A putative acyl-CoA-binding protein is a major phloem sap protein in rice (Oryza sativa L.). J. Exp. Bot. 57, 2571–2576 10.1093/jxb/erl021 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H. L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036 10.1126/science.1141753 [DOI] [PubMed] [Google Scholar]
- Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., Yamaguchi M., Nakashima C., Purwestri Y. A., Tamaki S., Ogaki Y., Shimada C., Nakagawa A., Kojima C., Shimamoto K. (2011). 0.14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335 10.1038/nature10272 [DOI] [PubMed] [Google Scholar]
- Testerink C., Munnik T. (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375 10.1016/j.tplants.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Thorpe M. R., Ferrieri A. P., Herth M. M., Ferrieri R. A. (2007). (11)C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226, 541–551 10.1007/s00425-007-0503-5 [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B. M. (1994). Stimulation of adipogenesis in fibroblasts by PPARg2, a lipid-activated transcription factor. Cell 79, 1147–1156 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- Truman W., Bennett M. H., Kubigsteltig I., Turnbull C., Grant M. (2007). Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. U.S.A. 104, 1075–1080 10.1073/pnas.0605423104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée B. S., Coadou G., Labbé H., Sy D., Vovelle F., Schoentgen F. (2003). Peptides corresponding to the N- and C-terminal parts of PEBP are well-structured in solution: new insights into their possible interaction with partners in vivo. J. Pept. Res. 61, 47–57 10.1034/j.1399-3011.2003.21039.x [DOI] [PubMed] [Google Scholar]
- van Bel A. J. E., Knoblauch M. (2000). Sieve element and companion cell: the story of the comatose patient and the hyperactive nurse. Aust. J. Plant Physiol. 27, 477–487 [Google Scholar]
- Wang X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7, 329–336 10.1016/j.pbi.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Watanabe R., Murakami Y., Marmor M. D., Inoue N., Maeda Y., Hino J., Kangawa K., Julius M., Kinoshita T. (2000). Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 19, 4402–4411 10.1093/emboj/19.16.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dinneny J. R., Crawford K. M., Rhee Y., Citovsky V., Zambryski P. C., Weigel D. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130, 3735–3745 10.1242/dev.00577 [DOI] [PubMed] [Google Scholar]
- Xiao S., Chye M. L. (2011). Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 156, 2069–2081 10.1104/pp.111.176933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xoconostle-Cazares B., Yu X., Ruiz-Medrano R., Wang H. L., Monzer J., Yoo B. C., McFarland K. C., Franceschi V. R., Lucas W. J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98 10.1126/science.283.5398.94 [DOI] [PubMed] [Google Scholar]
- Yoo B. C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y. M., Lough T. J., Lucas W. J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000 10.1105/tpc.104.023614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., O’Donnell V. B., Balzar S., St Croix C. M., Trudeau J. B., Wenzel S. E. (2011). 15-Lipoxygenase 1 interacts with phosphatidylethanolamine-binding protein to regulate MAPK signaling in human airway epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 14246–14251 10.1073/pnas.1017419108 [DOI] [PMC free article] [PubMed] [Google Scholar]