Abstract
In classical proteome analyses, final experimental data are (a) images of 2D protein separations obtained by gel electrophoresis and (b) corresponding lists of proteins which were identified by mass spectrometry (MS). For data annotation, software tools were developed which allow the linking of protein identity data directly to 2D gels (“clickable gels”). GelMap is a new online software tool to annotate 2D protein maps. It allows (i) functional annotation of all identified proteins according to biological categories defined by the user, e.g., subcellular localization, metabolic pathway, or assignment to a protein complex and (ii) annotation of several proteins per analyzed protein “spot” according to MS primary data. Options to differentially display proteins of functional categories offer new opportunities for data evaluation. For instance, if used for the annotation of 2D Blue native/SDS gels, GelMap allows the identification of protein complexes of low abundance. A web portal has been established for presentation and evaluation of protein identity data related to 2D gels and is freely accessible at http://www.gelmap.de/.
Keywords: proteomics, two-dimensional gel electrophoresis, mass spectrometry, data annotation, data visualization, web portal, software
Introduction
Proteomics aims to identify large sets of proteins in defined biological fractions, e.g., tissue or cell extracts, isolated organelles, or protein fractions generated by biochemical preparation procedures. As a first step, the different proteins of the various fractions have to be separated. Protein separations can be achieved by gel electrophoresis or by chromatographic procedures. Next, the proteins need to be identified. Protein identifications today are nearly exclusively carried out by mass spectrometry (MS). However, since the mass of a native protein normally does not allow any conclusion on its identity, proteins have to be fragmented by specific endopeptidases before mass analyses to generate defined peptides. These peptide fragments are then searched against a database of theoretically digested proteins using a software package such as MASCOT, SEQUEST, or X! Tandem.
Many proteome projects use one- or two-dimensional (2D) gel electrophoresis for protein separation. Following this experimental strategy, proteins are first separated, then individually “picked” from gels, treated with an endopeptidase (trypsin) and finally identified by MS and database interrogation. As a result, “proteome reference maps” are generated which consist of gel images and linked information of protein identity. Proteome maps were developed in several fields of biology and are greatly appreciated by the scientific community (see for instance Gallardo et al., 2001; Komatsu et al., 2004; Giavalisco et al., 2005).
How can protein reference maps be published? Originally, maps were exclusively presented in scientific publications in the form of images and long lists of identified proteins which are assigned to the spots on the image using arrows and numbers. About a decade ago, the first web-based resources were developed which offer interactive features in the maps: upon “clicking” on a spot, information on a protein is presented in a small pop-up window which sometimes is linked to further background information in databases (examples at http://seed.proteome.free.fr/, http://gabi.rzpd.de/projects/Arabidopsis_Proteomics/, http://gene64.dna.affrc.go.jp/RPD/).
Recently, the GelMap software package for annotating gel-based proteome data was developed. It offers several new possibilities for the web-based publication of reference maps, e.g., functional annotation of proteins and assignment of multiple proteins to individual “spots” on a gel (Rode et al., 2011a). In the following sections, we summarize the characteristics of GelMap and introduce novel features which were newly implemented into the GelMap software package (GelMap 2.0).
The GelMap Software Package
GelMap is offered as a web tool free of charge at www.gelmap.de. Minimal requirement for building a new map are (a) an image file and (b) a protein table which includes the coordinates of the spots on the gel. The latter table can be produced “by hand” (by reading the spot’s coordinates from graphics software like Photoshop, GIMP, or even MS Paint) but is usually generated automatically by specialized proteomics software tools like Delta2D (Decodon, Greifswald, Germany), DeCyder (GE Healthcare, Munich, Germany), or Progenesis SameSpots (Non-linear Dynamics, Newcastle, UK). To use GelMap to full capacity, the table should be extended with information on (1) the spot identification number (corresponding to the number on the gel image), (2) the Mascot probability score (or the score of another matching software package), (3) coverage of a protein by identified peptides, (4) the accession number of a protein, (5) the database used for protein identification, (6) assignment of a protein to functional categories like a protein complex, a physiological area or subcellular localization, (7) molecular mass (calculated and/or apparent mass on the gel), etc. (Table 1). To ensure that GelMap recognizes IDs, coordinates, scores, accession numbers, and filters, a second line beneath the line with the column names has to include special keywords (Table 1). The third line can contain commentaries for the tooltips in GelMaps spreadsheet view (Figure 1). To mark the columns of high importance, the fourth line can be filled with the keyword “show” and the data will be shown in the information windows. Since GelMap provides a three-level filter tree, the fifth line can define if this column should be used as a (1) root level, (2) second level, or (3) third level filter. This control information is required for a fully functional map and is documented with examples on www.gelmap.de/howto.
Table 1.
Column names and information included in the “protein spreadsheet” file.
| Column name/label (first line) | GelMap keyword (second line) | General description/commentary (third line) | Show (forth line) | Filter tree (fifth line) | GelMap interpretation (how the keyword is realized by GelMap) |
|---|---|---|---|---|---|
| ID | ID | Spot number | ID for spot identification | ||
| X | X | Coordinate on x axis | X/Y-coordinates are used to generate the spot circles on the image. Automatically scaled to | ||
| Y | Y | Coordinate on y axis | 800*800 pixels | ||
| MS score | SCORE | Mascot probability score | Rank for the order of multiple hits per spot | ||
| Accession | ACC | Accession numbers according to database | Unique ID in the used database to generate an external link | ||
| Database | ACCSRC | Database for protein identification | Used to generate the base URL for the external link | ||
| Name | TITLE | Name of protein | Header of the quick info box when hovering a spot | ||
| Protein complex | Free text | Assigned mitochondrial protein complex | Show | 1 | Free text columns can be shown in the info box These fields have no special purpose like linking to external sites or sort algorithms. All fields are displayed in the supplemental “Protein spreadsheet” |
| Physiological function | Free text | Categorization by physiological function | 2 | ||
| Subcellular localization | Free text | Subcellular localization according to SUBA II | 3 | ||
| Coverage | Free text | Sequence coverage of a protein | Show | ||
| # Peptides | Free text | No. of unique peptides matching a protein hit | Show | ||
| MM (calculated) | Free text | Calculated molecular mass | Show | ||
| App mass 2D | Free text | Apparent mol. mass (second dimension) | Show | ||
| App mass 1D | Free text | Apparent mol. mass (first dimension) | Show |
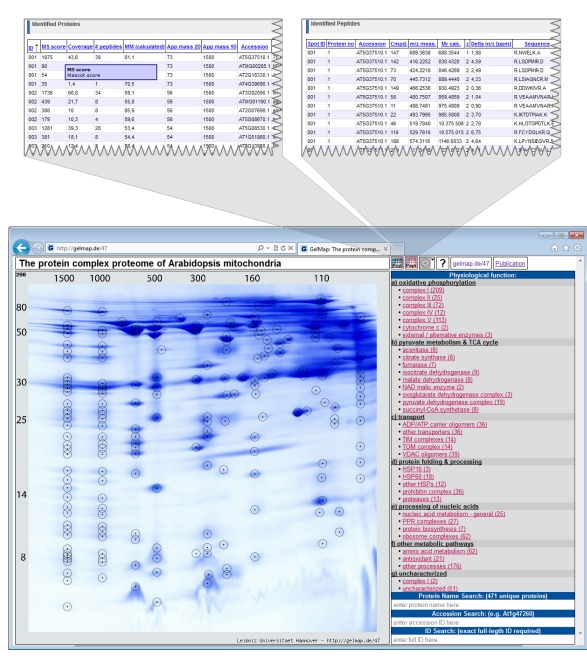
Figure 1.
Reference map of the mitochondrial proteome of Arabidopsis thaliana (Klodmann et al., 2011; http://www.gelmap.de/47). Proteins were separated by 2D Blue native/SDS PAGE. The two icons given in the header allow access to the “protein spreadsheet” and the “peptide spreadsheet.”
The resulting table is designated “protein spreadsheet” in GelMap 2.0. The image file and the “protein spreadsheet” file can be directly uploaded at www.gelmap.de/create.
The resulting map consists of the gel image, a side menu for functional categories of the identified proteins (right side), and frames for search options (Figure 1). Spots analyzed by MS are circled. On the map, information automatically is displayed in the following way: (i) upon hovering over a spot, all proteins identified within this spot are displayed within a tooltip. Proteins are ordered according to their Mascot probability scores (this, on a semi-quantitative level, reflects abundance of the identified proteins within a protein spot). (ii) After clicking a spot, the protein names included in the tooltip are transformed into links, which can be used to get protein-specific information within a second info frame. In addition to the information on the protein accession and the calculated molecular mass of the protein this frame can offer links to an external database (via the accession number) and to the “protein spreadsheet” (link “more protein details”). The “protein spreadsheet” also can be accessed directly by using an icon in the header (Figure 1). (iii) Proteins assigned to functional categories are accessible via the three-level tree in the side menu. Upon clicking onto a topic included in this menu, all proteins forming part of this category automatically become highlighted by circles on the gel image. At the same time, subcategories become visible for many topics of the main menu, which allow to differentially visualize proteins assigned to the next functional level. For details we recommend visiting one of the established GelMaps, e.g., http://gelmap.de/47.
To display primary MS data for all identified proteins, a second table called “peptide spreadsheet” was introduced in GelMap 2.0 (Figure 1). This table may include information on all identified peptides within a spot. In the case of the Arabidopsis thaliana BN/SDS map1, the table includes information on the amino acid sequences of the peptides, peptide modifications, and positions of peptides with respect to the complete protein sequences (column “range”: number of the first and the last amino acid of a peptide with respect to the N-terminal methionine). The latter information is essential for integration of gel maps into proteomics “meta portals” which are realized on the basis of peptide position data (see Integration of GelMap into the MASCP Gator). The linked icon for the “peptide spreadsheet” is located next to the “protein spreadsheet” icon at the top (Figure 1). Alternatively, the “peptide spreadsheet” is accessible through a text link in the information frames for individual proteins on the gel image. The content of this table can be freely defined as long as the first column contains the hit’s ID to link this information to the spots (see http://www.gelmap.de/47 for example).
In summary, GelMap offers the following features: Proteins of interest can easily be found using the different search options or by browsing/navigating through/using the categories in the side menu. Since all proteins are annotated according to functional criteria, sets of proteins involved in metabolic processes or other functional categories are easily visualized. Detailed information on the identified proteins is provided by the information frames which are accessible via the gel image. Background information and raw data on all proteins is given in the “protein spreadsheet” and the “peptide spreadsheet” accessible directly above every map or via the information frames for individual proteins. GelMap allows complete annotation of MS data for all analyzed protein spots: not only the protein with the highest Mascot probability score is given, but all identified proteins above a lower boundary score to be defined by the user. This feature excludes any loss of data during map building.
The GelMap Portal
The GelMap portal was established for creation and presentation of reference maps. Upon finishing a new map, a user can decide to make it accessible via the official GelMap site and/or via a direct link which can be presented in publications. GelMap also allows private (password protected) projects to share proteome data in a closed group or analyzing results without sharing them. Currently (February 28th, 2012), four projects are publicly available at the GelMap portal:
The Cyclamen persicum seed proteome (Rode et al., 2011b)2. In the frame of this project, the proteomes of zygotic and somatic embryos were compared. Identified proteins are assigned to 30 different metabolic pathways within 10 metabolic divisions. Furthermore. In addition to the features mentioned above, this GelMap allows to differentially display proteins of changed abundance between the two compared protein fractions. This illustrates the various extensions which easily can be realized based on the GelMap software package.
The A. thaliana mitochondrial proteome separated by 2D Blue native/SDS PAGE (Klodmann et al., 2011; see text footnote 1). This map is based on a special protein separation procedure: protein complexes are separated under native conditions on the first gel dimension and the subunits of the protein complexes under denaturing conditions on the second dimension, which is carried out in the presence of SDS. On the resulting gels, proteins belonging to the same protein complex form a vertical row of spots. For 2D Blue native/SDS PAGE, GelMap offers special advantages: selective display of functional categories allows identifying vertical positioning of proteins of low abundance. This led to the discovery of new protein complexes in Arabidopsis mitochondria. In the frame of this study, 471 non-redundant proteins were identified and assigned to more than 35 protein complexes.
The A. thaliana mitochondrial proteome separated by 2D IEF/SDS PAGE (Taylor et al., 2011)3. More than 250 proteins were identified and annotated on three functional levels.
The Oryza sativa mitochondrial proteome (Huang et al., 2009)4. 146 non-redundant proteins were identified and annotated on two functional levels.
Several further projects will be made available shortly.
Integration of GelMap Into MASCP Gator
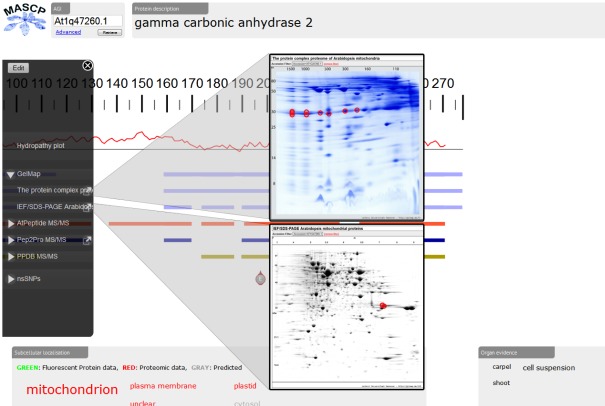
Recently, the proteomic aggregation utility “MASCP Gator” has been established to link proteome databases for the model plant A. thaliana (Joshi et al., 2011)5. In this platform, protein data is simultaneous displayed from many different international repositories to allow assessment of the global knowledge of a protein or several proteins. So far, projects integrated in the MASCP Gator exclusively represented shot-gun approaches not based on 2D PAGE. Since September 2011, GelMap offers a limited application programming interface (“API”) which allows other projects (such as Gator) to run automated search queries on the GelMap database and collect the results. The two GelMaps dedicated to the mitochondrial proteome of Arabidopsis very recently were the first gel-based projects integrated into the MASCP Gator. Upon submission of a protein accession, the user of the MASCP Gator directly can access the reference maps at www.gelmap.de (Figure 2). Information on peptides available at GelMap is graphically displayed at the MASCP Gator site together with peptide information of other databases. In the future, GelMap will open its database for seamless integration into other software products.
Figure 2.
Integration of the mitochondrial reference maps at GelMap into MASCP Gator (http://gator.masc-proteomics.org/). Upon submission of a protein accession (here: At2g47260), matching peptides from GelMap are graphically displayed at the Gator site. Direct access to GelMap is provided by links in the left side menu of the Gator.
Perspective
The GelMap software package allows complete annotation of MS data corresponding to 2D protein separations. Based on its central features – functional annotation of proteins and at the same time annotation of complete sets of proteins identified within each spot – it allows the comprehensive evaluation of gel-based proteome data sets. In the future, the GelMap platform will be used to annotate further projects. Specifically, annotation of additional organellar proteomes in Arabidopsis is planned. Integration of these projects into the MASCP Gator might soon allow extensive coverage of the Arabidopsis proteome by gel-based analyses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Deutsche Forschungsgemeinschaft (Grant Br 1829/10-1).
Footnotes
References
- Gallardo K., Job C., Groot S. P. C., Puype M., Demol H., Vanderkerckhove J., Job D. (2001). Proteomic analysis of Arabidopsis thaliana seed germination and priming. Plant Physiol. 126, 835–849 10.1104/pp.126.2.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P., Nordhoff E., Kreitler T., Klöppel K. D., Lehrach H., Klose J., Gobom J. (2005). Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5, 1902–1913 10.1002/pmic.200401062 [DOI] [PubMed] [Google Scholar]
- Huang S., Taylor N. L., Narsai R., Eubel H., Whelan J., Millar A. H. (2009). Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol. 149, 719–734 10.1104/pp.108.131300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi H. J., Hirsch-Hoffmann M., Baerenfaller K., Gruissem W., Baginsky S., Schmidt R., Schulze W. X., Sun Q., van Wijk K. J., Egelhofer V., Wienkoop S., Weckwerth W., Bruley C., Rolland N., Toyoda T., Nakagami H., Jones A. M., Briggs S. P., Castleden I., Tanz S. K., Millar A. H., Heazlewood J. L. (2011). MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 155, 259–270 10.1104/pp.110.168195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J., Senkler M., Rode C., Braun H. P. (2011). The protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598 10.1104/pp.111.182352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S., Kojima K., Suzuki K., Ozaki K., Higo K. (2004). Rice Proteome Database based on two-dimensional polyacrylamide gel electrophoresis: its status in 2003. Nucleic Acids Res. 32, D388–D392 10.1093/nar/gkh020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode C., Senkler M., Klodmann J., Winkelmann T., Braun H. P. (2011a). GelMap – a novel software tool for building and presenting proteome reference maps. J. Proteomics 74, 2214–2219 10.1016/j.jprot.2011.06.017 [DOI] [PubMed] [Google Scholar]
- Rode C., Gallien S., Heintz D., Van Dorsselaer A., Braun H. P., Winkelmann T. (2011b). Enolases: storage compounds in seeds? Evidence from a proteomic comparison of zygotic and somatic embryos of Cyclamen persicum Mill. Plant Mol. Biol. 75, 305–319 10.1007/s11103-010-9729-x [DOI] [PubMed] [Google Scholar]
- Taylor N. L., Heazlewood J. L., Millar A. H. (2011). The Arabidopsis thaliana 2-D gel mitochondrial proteome: refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 11, 1720–1733 10.1002/pmic.201000620 [DOI] [PubMed] [Google Scholar]