Abstract
Major intrinsic proteins (MIPs) also called aquaporins form pores in membranes to facilitate the permeation of water and certain small polar solutes across membranes. MIPs are present in virtually every organism but are uniquely abundant in land plants. To elucidate the evolution and function of MIPs in terrestrial plants, the MIPs encoded in the genome of the spikemoss Selaginella moellendorffii were identified and analyzed. In total 19 MIPs were found in S. moellendorffii belonging to 6 of the 7 MIP subfamilies previously identified in the moss Physcomitrella patens. Only three of the MIPs were classified as members of the conserved water specific plasma membrane intrinsic protein (PIP) subfamily whereas almost half were found to belong to the diverse NOD26-like intrinsic protein (NIP) subfamily permeating various solutes. The small number of PIPs in S. moellendorffii is striking compared to all other land plants and no other species has more NIPs than PIPs. Similar to moss, S. moellendorffii only has one type of tonoplast intrinsic protein (TIP). Based on ESTs from non-angiosperms we conclude that the specialized groups of TIPs present in higher plants are not found in primitive vascular plants but evolved later in a common ancestor of seed plants. We also note that the silicic acid permeable NIP2 group that has been reported from angiosperms appears at the same time. We suggest that the expansion of the number MIP isoforms in higher plants is primarily associated with an increase in the different types of specialized tissues rather than the emergence of vascular tissue per se and that the loss of subfamilies has been possible due to a functional overlap between some subfamilies.
Keywords: water channels, AQP, SIP, XIP, HIP, GIP, phylogeny
Introduction
The major intrinsic proteins (MIPs) constitute an ancient protein family and representatives can be found in virtually all types of organisms (Preston et al., 1992; Heymann and Engel, 1999). These proteins form pores in biological membranes and facilitate the passive transport of a range of small polar molecules, most notably water and glycerol. Several structural features are conserved throughout the protein family such as the six transmembrane helices (H1–H6) connected by five loops (LA–LE) as well as two conserved NPA motifs (Asn-Pro-Ala) at the N-terminal end of two short helices in LB and LE (Fu et al., 2000; Sui et al., 2001). These oppositely oriented short helices connected by the two NPA-motifs facing each other in the middle of the membrane form a seventh helical transmembrane structure. The dipole moment of each of the half transmembrane helices creates partial positive charges centered at the NPA motifs and these charges prevent the passage of protons through the pore (de Groot et al., 2003). Four amino acid residues, known as the aromatic/arginine-filter (ar/R filter), located toward the extracellular entrance form the narrowest part of the pore and are thought to determine the substrate selectivity (Fu et al., 2000; Beitz et al., 2006).
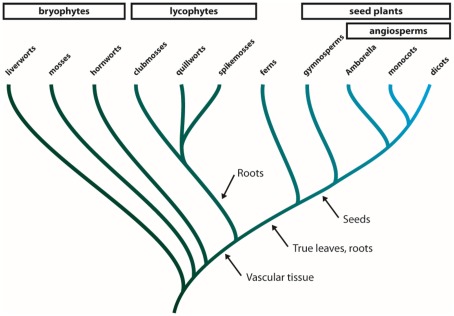
The MIPs are especially abundant and multifaceted in terrestrial plants suggesting that the expansion of this protein family has contributed to the adaptation of green plants to life on land (Danielson and Johanson, 2010; Anderberg et al., 2011). The green plants (viridiplantae) consist of two major clades, the chlorophytes, including the majority of all green algae, and the streptophytes (Leliaert et al., 2011). The streptophytes comprise a number of classes of freshwater algae, collectively known as the charophytes, and the land plants (embryophyta, Figure A1 in Appendix). The earliest diverging groups of embryophytes, the liverworts, mosses, and hornworts are all primitive in the sense that they are non-vascular plants (Shaw et al., 2011). Following the evolution of vascular tissue in plants the lycophytes emerged and came to dominate the flora during the Carboniferous period with some species growing to heights of 30 m (Banks, 2009). Even though all members of this group have leaf-like structures, these structures are not considered to be true leaves and only have a central vein (Banks, 2009). True leaves evolved in the lineage leading to ferns and spermatophytes and by the time angiosperms split from gymnosperms the ability to produce seeds had emerged in spermatophytes.
The MIPs of terrestrial plants can be divided into seven subfamilies, the plasma membrane intrinsic proteins (PIPs), the tonoplast intrinsic proteins (TIPs), the nodulin 26-like intrinsic proteins (NIPs), the small basic intrinsic proteins (SIPs), the X intrinsic proteins (XIPs), the hybrid intrinsic proteins (HIPs), and the GlpF-like intrinsic proteins (GIPs). While the moss Physcomitrella patens contains representatives from all seven groups the dicots at most have five of these subfamilies, the PIPs, TIPs, NIPs, SIPs, and XIPs and monocots only four, having lost also the XIPs (Danielson and Johanson, 2008). Whereas the number of subfamilies has decreased during the evolution of land plants the number of MIP isoforms present in each species seems to have increased. In P. patens only 23 isoforms are present while in higher angiosperms like A. thaliana and O. sativa 35 and 31 isoforms have been identified respectively (Johanson et al., 2001; Sakurai et al., 2005). This difference is mainly due to an expansion of the PIP, TIP, and NIP subfamilies which is also manifested by the appearance of new subgroups within the two latter subfamilies. The current understanding of the evolution of the MIP family in terrestrial plants is based on analyses of MIPs in a moss and in higher angiosperms, representing lineages which split more than 440 million years ago. In order to get a more comprehensive picture of the evolution of the plant MIP family it is necessary to investigate MIPs encoded in genomes of species belonging to lineages diverging later than mosses but before the emergence of higher angiosperms.
In the present study we have classified and annotated the MIPs encoded in the recently sequenced genome of the lycophyte Selaginella moellendorffii and complemented the analysis by MIPs encoded in ESTs of gymnosperms and basal angiosperms to provide a more detailed picture of the evolution of MIPs in terrestrial plants.
Results
Identification and classification of S. moellendorffii MIPs
The diploid genome of S. moellendorffii available at Joint Genome Institute was searched using the complete set of Physcomitrella patens MIPs as queries. Out of 20 unique hits one was deemed to be a pseudo NIP gene after manual inspection, containing just one NPA-box and only parts of helix 1–4. No MIP encoding sequence was found at this locus on the homologous chromosome. Of the remaining 19 sequences approximately half already had a satisfactory gene model, defining the coding sequence and exon/intron boarders, whereas new models encoding more typical MIP features were created for the rest (Table 1; Table A1 in Appendix).
Table 1.
Selectivity filters and NPA boxes of S. moellendorffii MIPs.
| Subfamily | Proteina | ar/R selectivity filterb |
NPA motifsc |
||||
|---|---|---|---|---|---|---|---|
| H2 | H5 | LE1 | LE2 | Loop B | Loop E | ||
| PIP | SmPIP1;1 | F | H | T | R | NPA | NPA |
| SmPIP2;1 | F | H | S | R | NPA | NPA | |
| SmPIP2;2 | F | H | T | R | NPA | NPA | |
| PpPIP1;1 | F | H | T | R | NPA | NPA | |
| TIP | SmTIP6;1 | H | I | G | R | NPA | NPA |
| SmTIP6;2 | H | I | G | R | NPA | NPA | |
| PpTIP6;1 | H | I | A | R | NPA | NPG | |
| NIP | SmNIP3;1 | S | I | A | R | NPS | NPV |
| SmNIP3;2 | P | N | A | R | NPS | NPA | |
| SmNIP3;3 | A | N | A | R | NPA | NPC | |
| SmNIP3;4 | A | N | A | R | NPA | NPI | |
| SmNIP3;5 | S | I | A | R | NPA | NPV | |
| PpNIP3;1 | A | I | A | R | NPA | NPV | |
| SmNIP5;1 | F | A | A | R | NPA | NPA | |
| SmNIP5;2 | N | N | A | R | NPA | NPA | |
| PpNIP5;1 | F | A | A | R | NPA | NPA | |
| SmNIP7;1 | A | V | G | R | NPS | NPA | |
| PpNIP6;1 | G | V | A | R | NPA | NPM | |
| SIP | SmSIP1;1 | I | I | P | N | NPT | NPA |
| PpSIP1;1 | V | V | P | N | NPT | NPA | |
| XIP | SmXIP1;1 | H | V | A | R | NPC | NPA |
| SmXIP1;2 | H | I | A | R | NPC | NPA | |
| SmXIP1;3 | L | T | A | R | NPI | NPA | |
| PpXIP1;1 | Q | I | A | R | NPC | NPG | |
| HIP | SmHIP1;1 | H | H | A | R | NPA | NPA |
| SmHIP1;2 | H | H | A | R | NPA | NPA | |
| PpHIP1;1 | H | H | A | R | NPA | NPA | |
aMIPs from P. patens are included as a reference and are written in bold.
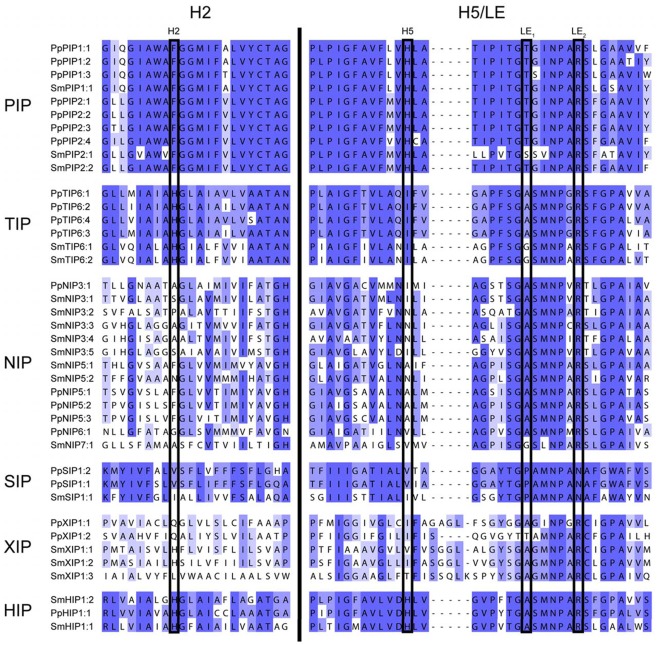
bThe ar/R filter is defined by one amino acid residue in helix 2, one in helix 5, and two in loop E. See Figure A2 in Appendix.
cThe sequences corresponding to the NPA motifs in loops B and E.
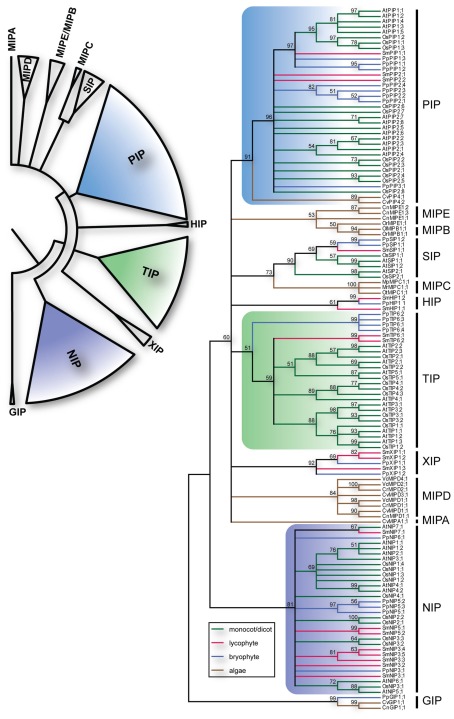
The identified S. moellendorffii MIPs (SmMIPs) were then subjected to phylogenetic analyses (Figure 1). Of the 19 SmMIPs, one clearly grouped with the SIP1 isoforms (SIP1s), three with the XIPs, and two with the HIPs. Hence, no further analysis was required for the classification of these MIPs and they were named SmSIP1;1, SmXIP1;1–SmXIP1;3, SmHIP1;1, and SmHIP1;2. Three of the SmMIPs clustered with the PIPs, whereof one was firmly nested within the PIP1s whereas the remaining two together with the PIP2s and PpPIP3;1 ended up basal to the PIP1s with an unresolved internal relationship. Two sequences clustered with the TIPs and eight with the NIPs. Although the classification into the seven different subfamilies was straightforward, the phylogenies within the PIPs, TIPs, and NIPs were largely unresolved. In an attempt to achieve a more precise classification of these MIPs, individual phylogenetic analyses based on separate alignments, were performed. These analyses also included the available relevant MIPs from plants lacking sequenced genomes but since these are likely to represent only a fraction of the MIPs encoded in these species, no names were suggested to leave the classification of these MIPs open for future more comprehensive analyses.
Figure 1.
Phylogeny of MIPs. The upper left panel summarize the different subfamilies of plant MIPs. The right panel depicts the bootstrap consensus tree of the complete set of MIPs from O. sativa (Os), A. thaliana (At), S. moellendorffii (Sm), P. patens (Pp) and the chlorophyte algae Chlamydomonas reinhardtii (Cr), Volvox carteri (Vc), Coccomyxa sp. C-169 (Cc), Chlorella sp. NC64A (Cn), Micromonas pusilla CCMP1545 (Mp), Micromonas sp. RCC299 (Mr), Ostreococcus lucimarinus (Ol), Ostreococcus sp. RCC809 (Or), and Ostreococcus tauri (Ot) using the Maximum Likelihood method. The branches are colored according to from what phyla the sequences are derived. The numbers by the nodes are bootstrap support in percentage and nodes with less the 50% support are collapsed. The vertical lines to the right delimit the different subfamilies.
Phylogeny of PIPs
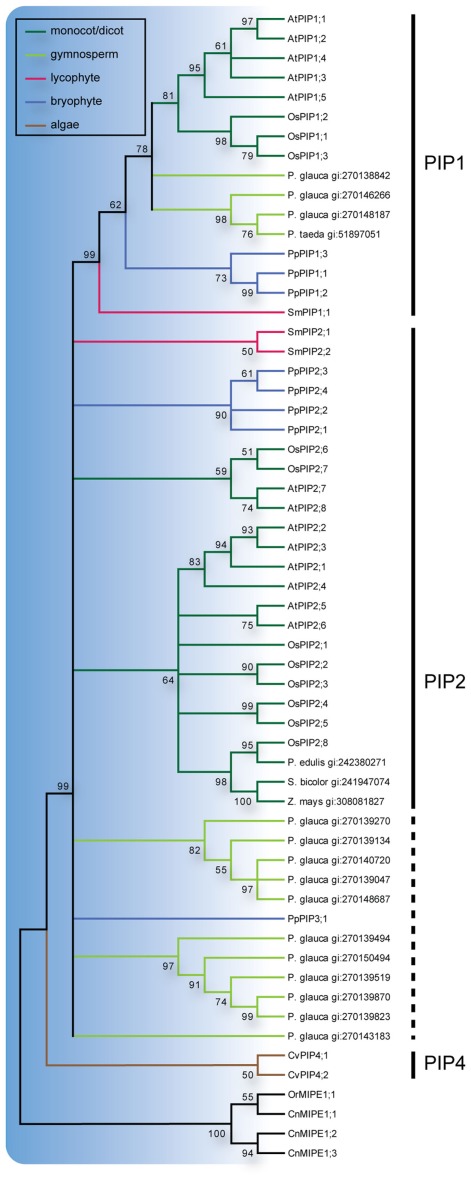
All three S. moellendorffii PIPs have ar/R filters identical or very similar to those in other PIPs and are likely to be specific for water (Table 1; Figure A2 in Appendix). The PIP consensus tree (Figure 2) shows a clearly defined PIP1 clade with a bootstrap support of 99% and include representatives from the gymnosperms P. glauca, P. taeda and one sequence from S. moellendorffii. This is consistent with the initial analysis (Figure 1), alas the phylogeny of the other PIPs in part remained unresolved. On the node basal to the PIP1s, the PIP2s of A. thaliana, O. sativa, and P. patens, as well as 2 sequences from S. moellendorffii and 11 from P. glauca clustered into 7 separate clades. Based on the phylogenetic analysis and a manual comparison to the reference PIPs, these S. moellendorffii PIPs were clearly not PIP1s and in contrast to the PpPIP3;1 they had the N- and C-terminal lengths characteristic of PIP2s, containing the among PIP2s conserved C-terminal phosphorylation site SFRS (Johansson et al., 2000). They were therefore classified as PIP2s and hence the S. moellendorffii PIPs were named SmPIP1;1, SmPIP2;1, and SmPIP2;2. In accordance with earlier analyses the two PIPs from the algae Coccomyxa C-169 were basal to all other PIPs (Anderberg et al., 2011).
Figure 2.
Phylogeny of PIPs. Maximum Likelihood bootstrap consensus tree of PIPs from plant species with sequenced genomes together with PIP-like sequences retrieved from the NCBI EST database. EST sequences are named with species name followed by their GenBank gi numbers. The PIP branches are color coded according to from what phyla the sequences are derived. Subgroups are delimited by the vertical lines to the left where the dashed line indicates sequences for which classification is uncertain. The robustness of nodes is denoted with bootstrap support in percentage and nodes with less than 50% support are collapsed.
Phylogeny of TIPs
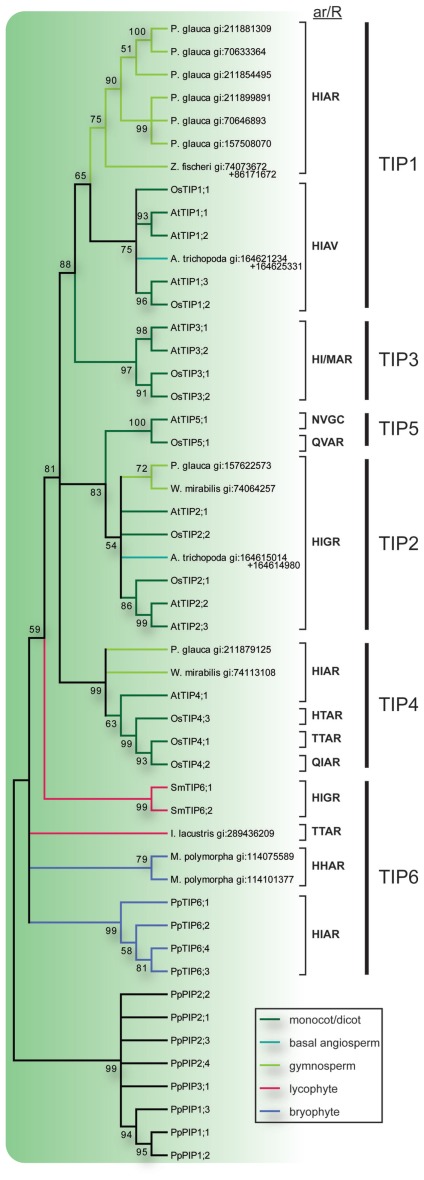
The TIP6s of P. patens together with two sequences from the liverwort Marchantia polymorpha and one from the quillwort Isoetes lacustris formed three separate groups on the most basal node of the TIP consensus tree (Figure 3). On the next node two sequences from S. moellendorffii formed a sister clade to the TIPs of seed plants and to indicate an orthologous relationship with the TIP6s in P. patens, the SmTIPs were named TIP6;1 and TIP6;2. The TIPs of seed plants formed five stable clades, TIP1 to TIP5. The clades of TIP1s and TIP2s included sequences from gymnosperms, basal angiosperms, monocots, and dicots, and the TIP4 clade sequences from gymnosperms, monocots, and dicots. In contrast, the TIP3s and TIP5s contained monocot and dicot sequences only. As shown by the bootstrap support the TIP3s and the TIP5s are clearly associated with the TIP1s and the TIP2s, respectively. To facilitate a comparison between phylogenetic relationship and possible substrate specificity the ar/R filters of the different subgroups are shown in Figure 3.
Figure 3.
Phylogeny of TIPs. The bootstrap consensus tree resulting from a Maximum Likelihood analysis of the TIPs of species with sequenced genome along with full length TIP-like EST sequences retrieved from the NCBI EST database. EST sequences are named with species name and GenBank gi number. In the case where two ESTs were used to compile a full length MIP sequence both GenBank gi numbers are provided. Selectivity filters (ar/R) are displayed next to the sequence names and the vertical lines to the right indicate the subgroup to which they belong. The TIP branches are color coded according to from what phyla the sequences are derived. Nodes with less than 50% bootstrap support are collapsed.
Phylogeny of NIPs
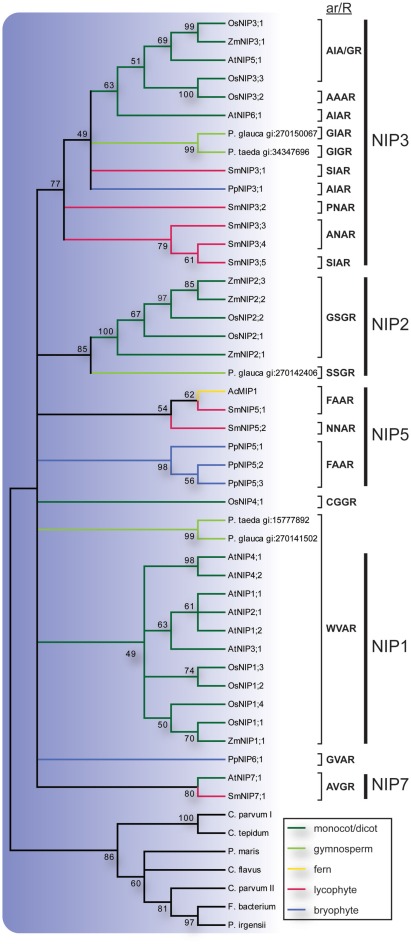
Previous studies of NIPs in higher plants have established at least three major groups, NIP1, NIP2, and NIP3 (Sakurai et al., 2005; Zardoya, 2005; Gupta and Sankararamakrishnan, 2009; Danielson and Johanson, 2010). In our analysis the NIP2s and NIP3s were well supported but to resolve the NIP1 group in the consensus tree it was necessary to lower the cut off value for collapsing nodes slightly from the standard 50% to 49% (Figure 4). Of these three groups the NIP3s included sequences from P. patens, S. moellendorffii, gymnosperms and angiosperms, the NIP2s sequences from gymnosperms and angiosperms, and the NIP1s sequences from angiosperms only. The S. moellendorffii sequences in the NIP3 group were named SmNIP3;1 to SmNIP3;5. In addition to the three major clades, two sequences from S. moellendorffii and one from the fern Adiantum capillus-veneris formed a separate clade as did the P. patens NIP5s. The ar/R filters suggest that at least one of these SmNIPs and the fern sequence AcMIP1 are functionally equivalent to the PpNIP5s and in accordance with the phylogenetic analysis, the SmNIPs of this group were both classified as NIP5s and were hence named SmNIP5;1 and SmNIP5;2. However it should be noted that these two sequences are not very similar and the constriction region of the latter deviates from other NIP5s and instead show some resemblance to SmNIP3;2–3;4. Finally, a single sequence from S. moellendorffii was named SmNIP7;1 since it clustered with NIP7;1 from A. thaliana and also share the same ar/R filter. In contrast to the SmNIPs, PpNIP6;1, OsNIP4;1 and two gymnosperm sequences showed no apparent association with any other sequence.
Figure 4.
Phylogeny of NIPs. The bootstrap consensus tree resulting from a Maximum Likelihood analysis of the NIPs of species with sequenced genome along with full length NIP-like EST sequences retrieved from the NCBI EST database. EST sequences are named with species name and GenBank gi number. Selectivity filters (ar/R) are displayed next to the sequence names and the vertical lines to the right indicate the subgroup to which they belong. The NIP branches are color coded according to from what phyla the sequences are derived. Nodes with less than 49% bootstrap support are collapsed.
Discussion
Selaginella moellendorffii constitute the smallest genome of a sequenced land plant but nevertheless this spikemoss overall encode more protein families than the about twofold larger genome of the moss P. patens (Banks et al., 2011), consistent with having a more intricate physiology. Despite this general increase in the number of protein families the number of MIPs in P. patens and in S. moellendorffii are very similar, 23 and 19 respectively. Thus there is no evidence of an overall expansion of the MIP family coinciding with the evolution of primitive vascular plants like the lycophytes. This suggests that the number of encoded MIPs instead increased at a later stage in the evolution of terrestrial plants with the emergence of spermatophytes (Table 2). In our analysis six of the seven MIP subfamilies previously identified in the moss P. patens are confirmed in S. moellendorffii. This indicates that the seventh subfamily, the GIPs, was lost early on in the lineage leading to vascular plants, already before the appearance of the lycophytes. Interestingly, also the number of members within some of the subfamilies differs substantially between P. patens and S. moellendorffii. Each of the six subfamilies found in S. moellendorffii is discussed in detail below.
Table 2.
Overview of MIP isoforms in terrestrial plants.
| Number of isoforms in subfamilies |
Total isoforms | Total subfamilies | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GIP | HIP | XIP | SIP | PIP | TIP | NIP | ||||
| Physcomitrella patens | 1 (4.3)b | 1 (4.3) | 2 (8.7) | 2 (8.7) | 8 (34.8) | 4 (17.4) | 5 (21.7) | 23 | 7 | Danielson and Johanson (2008) |
| Selaginella moellendorffii | – | 2 (10.5) | 3 (15.8) | 1 (5.3) | 3 (15.8) | 2 (10.5) | 8 (42.1) | 19 | 6 | This work |
| Solanum lycopersicuma | 1 (2.7) | 3 (8.1) | 18 (48.6) | 9 (24.3) | 6 (16.2) | 37 | 5 | Sade et al. (2009) | ||
| Populus trichocarpa | – | – | 6 (10.9) | 6 (10.9) | 15 (27.3) | 17 (30.9) | 11 (20.0) | 55 | 5 | Gupta and Sankararamakrishnan (2009) |
| Gossypium hirsutum | – | – | 1 (1.4) | 7 (9.9) | 28 (39.4) | 23 (32.4) | 12 (16.9) | 71 | 5 | Park et al. (2010) |
| Arabidopsis thaliana | – | – | – | 3 (8.6) | 13 (37.1) | 10 (28.6) | 9 (25.7) | 35 | 4 | Johanson et al. (2001) |
| Oryza sativa | – | – | – | 2 (6.1) | 11 (33.3) | 10 (30.3) | 10 (30.3) | 33 | 4 | Sakurai et al. (2005) |
aThe occurrence of MIPs in S. lycopersicum is based on an extensive analysis of ESTs.
bRelative size of subfamily in the species (%).
Surprisingly few PIPs in S. moellendorffii and unresolved PIP2 groups in seed plants
The PIPs are the most conserved plant MIPs and form a monophyletic clade including the basal algal PIP4s. Within this clade the PIP1 group is well defined and has a congruent topology with the exception of SmPIP1;1 which appear more basal than the moss PIP1s. Despite this small inconsistency with the phylogeny of the species, the present result suggests that all terrestrial plants have PIP1 orthologs, implying a fundamental function in land plants. Unexpectedly, compared to the three PIP1 genes found in P. patens, the S. moellendorffii genome only encode one and may therefore provide a useful naturally non-redundant system for addressing the physiological function of PIP1s. In addition to PIP1s it is clear that all of the terrestrial plants have at least one other type of PIPs, even though the phylogeny of these PIPs is not fully resolved. In P. patens PIP2s and a PIP3 have earlier been defined as separate groups although the functional integrity of the latter has been questioned (Danielson and Johanson, 2008) and therefore the remaining two Selaginella sequences were annotated as PIP2s. Interestingly, both the angiosperm and gymnosperm non-PIP1s form two distinct clades with unresolved relationship and it is conceivable that these clades correspond to two distinct isoforms of PIP2s with specialized functions which evolved already in a common ancestor to gymnosperms and angiosperms. All the PIPs appear to be classical water channels when the ar/R filter is considered; the algal PIP4s have the residues FHCR at the filter, the PpPIPs and PIPs in higher plants FHTR, and the SmPIPs FHTR or FHSR. Thus, any functional difference between different clades of PIPs is more likely to relate to regulatory features rather than substrate specificity. However, it should be noted that all PIPs in terrestrial plants are likely to share some regulatory features as key amino acid residues of the proposed gating mechanism are conserved (D28, E31, S115, H193 in SoPIP2;1; Tornroth-Horsefield et al., 2006; Anderberg et al., 2011).
TIPs diversified after the emergence of primitive vascular plants
In higher plants like Arabidopsis and rice five distinct subgroups of TIPs, TIP1 to TIP5, are discerned, whereas in more primitive plants like mosses and spikemosses only one type of TIP, TIP6, is found (Danielson and Johanson, 2008). Although there are several isoforms of TIP6s in the latter species, they are all very similar within each species, indicative of a specific distinct physiological role of the TIP6s at least within the species. The SmTIP6 genes share the same gene structure with two introns which is also conserved in TIPs of higher plants (Johanson et al., 2001), whereas only one of these introns is present in PpTIP6s, suggesting that the conserved TIP gene structure evolved in early vascular plants. Interestingly, ESTs corresponding to TIP6s in the drought resistant spikemoss S. lepidophylla were the seventh most abundant transcript, estimated to make up 0.5% of the total ESTs after 2.5 h of dehydration (Iturriaga et al., 2006). This implies that the TIP6s might have a direct role in water relations.
The presented phylogenetic analysis suggests that the first subgroups of TIPs appeared after the lycophytes had diverged from other vascular plants, when the TIP1s, TIP2s, and TIP4s evolved in the lineage leading to higher plants. This diversification happened before the angiosperm/gymnosperm split and these three groups are thus expected to be present in all seed plants. Furthermore, the well supported clustering of TIP1s with TIP3s and TIP2s with TIP5s suggest that each pair evolved from a common ancestor. Since TIP3s and TIP5s are basal to TIP1s and TIP2s, respectively, this implies that TIP3s and TIP5s were also present in the lineage leading to the gymnosperms but later lost. However, it should be noted that the apparent absence of TIP3s and TIP5s in gymnosperms is based on searches in EST libraries and the picture might change as genomic sequences become available for analyses.
Typically the ar/R filters of TIPs have a characteristic histidine at the first position followed by a large aliphatic residue (I/M/V) at the second, a small amino acid residue (A/G) at the third and the among MIPs conserved large charged arginine at the fourth position. Based on the frequencies of occurrence, the original filter in the last common ancestor of all TIPs was suggested to be HIAR (Danielson and Johanson, 2008). There are however some deviations from this consensus, e.g., the TIP6s from M. polymorpha similar to the HIPs have a histidine also at the second position of the filter, TIP5s and OsTIP4;2 have an asparagine or a glutamine at the first position, and one of the TIP4s from rice together with a TIP from quillwort have a threonine at the first and second positions. Most notably the TIP1s in angiosperms lack the arginine at the fourth position and instead have a valine (HIAV). Functional studies suggest that TIPs in general are permeable for both water and ammonia, whereas the TIPs missing the arginine have a wider range of substrates including hydrogen peroxide and urea (Soto et al., 2008; Azad et al., 2011). Based on the ar/R filter we speculate that the gymnosperm TIP1s might functionally correspond to the TIP3s but only partly to the TIP1s in angiosperms since they are not expected to permeate either hydrogen peroxide or urea. A similar physiological function is supported by the fact that the expression of the TIP3s have been reported to be seed specific (Maurel et al., 1997; Gattolin et al., 2011) and at least one gymnosperm TIP1-like MIP (MIPFG) is also expressed in the protein storage vacuoles of seeds (Hakman and Oliviusson, 2002).
NIPs constitute the most numerous subfamily in S. moellendorffii
Similar to the TIPs, the NIPs form a highly divergent subfamily with large variation in ar/R filters (Figure 4). Based on the ar/R residues at least three functional groups have been recognized (Wallace and Roberts, 2005; Mitani et al., 2008; Rouge and Barre, 2008; Ali et al., 2009) and substrate specificities have been reviewed extensively (Ma et al., 2008; Ali et al., 2009; Bienert and Jahn, 2010). It has been noted that the NIPs are surprisingly numerous in primitive plants and at least one of the phylogenetical subgroups (NIP3) had evolved already in a common ancestor of mosses and higher plants (Danielson and Johanson, 2008). The analyzed SmMIPs support this finding and almost half (8) of the 19 SmMIPs are NIPs, where of 5 are classified as NIP3s. Two of the SmNIPs are proposed to be orthologous to PpNIP5s based on the common ar/R-filter FAAR. This filter is also present in bacterial NIPs supporting that this represents the ancestral specificity of the plant NIPs (Danielson and Johanson, 2010). Interestingly, no NIP5 has yet been identified in higher plants. The somewhat similar ar/R filter of the NIP1s (WVAR) suggests that they have a similar substrate specificity and could therefore have replaced the NIP5s in higher plants. A single SmNIP grouped with AtNIP7;1 and both are likely to correspond to PpNIP6;1 since the ar/R filters are related (AVGR and GVAR). Thus similar to NIP3s, the NIP6/7 function might have evolved already in a common ancestor of mosses and higher plants or at the latest in a common ancestor of vascular plants.
The low expression of NIPs in Arabidopsis suggests that transcripts encoding NIPs may be rare also among ESTs from other species (Alexandersson et al., 2005). This indicates that many NIPs are yet to be identified in species for which a complete genomic sequence is not available. Nevertheless, we do find gymnosperm ESTs encoding NIPs that firmly cluster with NIP3s and NIP2s. Although the NIP3 group is well supported there is a large variation in the ar/R filters. The SmNIP3s can be divided into three types based on the ar/R filters (SIAR, PNAR, ANAR). Whether these also translate into differences in substrate specificity and if they differ in specificity from the other NIP3s remain to be seen. The NIP2 group contains the best characterized NIPs regarding their physiological function and have been shown to have a role in the uptake and distribution of silicic acid within the plant (Ma et al., 2006). NIP2 homologs have previously only been reported from monocots and dicots (Ma et al., 2008) but the, in this study identified, gymnosperm NIP2 suggest that this physiological function was present in a common ancestor of gymnosperms and angiosperms. Two other gymnosperm NIPs appear to correspond to the NIP1s judging from a strictly conserved ar/R filter (WVAR), although the bootstrap support for such an association is weak. Hence, both NIP1s and NIP2s may be present in many seed plants although most dicots seem to have lost the NIP2s.
SIP2s evolved later
The SIPs cluster together with the algal MIPC with high bootstrap support which may indicate an orthologous relationship. Intriguingly, previous analyses have indicated that MIPCs are actually more related to AQP11/12 in mammals than SIPs and this have hampered the classification of the MIPCs as SIPs (Anderberg et al., 2011). The two SIPs of P. patens and the single SIP of S. moellendorffii all clearly belong to the SIP1 group whereas the angiosperms also have a SIP2 group. The hydrophobic ar/R filter of the SIPs from moss and spikemoss also supports the SIP1 classification. Our result is consistent with previous analyses of ESTs (Johanson and Gustavsson, 2002) suggesting that SIP2s have only evolved in the angiosperms whereas SIP1s go deeper and are present in all terrestrial plants sharing a common ancestor with the mosses.
XIPs – alternative alignments and functional redundancy
There are three XIPs encoded in the genome of S. moellendorffii compared to two in P. patens. Previous analyses have shown that XIPs from moss and spikemoss are basal to all XIPs from higher plants (Gupta and Sankararamakrishnan, 2009). So far XIPs from higher plants have only been found in dicot plants and these cluster into XIP1s and XIP2s, although the latter have only been identified in two species (Ricinus communis and Populus trichocarpa). There have been several alternative suggestions for how H5 should be aligned to reference sequences and this has resulted in alternative ar/R filters, deviating at the second position. Here we suggest yet another alignment of H5, based on hydrophobicity plots (data not shown), where the predicted transmembrane region is shifted four residues toward the N-terminus of XIPs. Like earlier alignments this would preserve a glycine or an alanine at the position corresponding to G203 in SoPIP2;1, which is important for the close packing of H5 and H2 (Bansal and Sankararamakrishnan, 2007). More importantly, all XIPs will now have a conserved glycine also at the position corresponding to F207 in SoPIP2;1, which will release the structural constraint in H2 at the position corresponding to G82 in SoPIP2;1. This is consistent with the large size variation of amino acids (G/A/S/V/F) found at this position in the XIPs. This new alignment results in a slightly more hydrophobic filter with valine at the second position in the dicot XIPs. At this position SmXIP1;3 has a threonine whereas the PpXIP1;1 and SmXIP1;1–1;2 have isoleucine or valine similar to dicot XIPs. Nevertheless, the PpXIP1;1 and SmXIP1;1–1;2 deviate from other XIPs in having a TIP-like ar/R filter with histidine in the first position. Interestingly, the XIPs have recently been lost in both monocots and in Arabidopsis. These evolutionary events as well as the change of ar/R filters in higher plants may have been possible due to a functional redundancy with TIPs. However, with the present understanding of MIP subcellular localization this scenario is unlikely since the XIPs of higher plants have been shown to be targeted to the plasma membrane (Bienert et al., 2011) whereas TIPs reside in the tonoplast (Jauh et al., 1999).
HIPs and TIPs – shared ancestry and overlapping function?
So far HIPs have only been identified in moss and spikemoss. The ar/R filter in all three HIPs is conserved (HHAR) and appears to be a hybrid of the PIP (FHTR) and TIP (HIAR) filters, having histidine at both the first and second position (Danielson and Johanson, 2008). To the best of our knowledge the substrate specificity of such a filter has not been tested. However, we note that it is identical to the ar/R filter in a TIP6 from Marchantia polymorpha, leading us to speculate that it is functionally equivalent to the filter in other basal TIPs (HIA/GR). Although the subcellular localization of HIPs is not known, a redundant function with the TIPs would explain why the HIPs were lost in higher plants and would fit in time with the expansion of the TIP subfamily. We also note that the lengths of the two first exons of SmHIP1;1 are identical to SmTIP6s, and if an alternative gene model is used for this position in SmHIP1;2 and PpHIP1;1 (data not shown) the first intron position of SmTIP6s and TIPs of higher plants is conserved in both the SmHIPs and PpHIP1;1. This is a strong indication of a shared ancestry of this region in HIPs and TIPs where the first position of the ar/R filter is encoded. The precise details of this common evolutionary history are difficult to reconstruct from our current data.
General remarks and future perspectives
Land plants encode more MIP isoforms than any other type of organism. It has been suggested that the major expansion of MIPs occurred as an adaption to life on land, enabling the plant to exploit concentration gradients as well as to tolerate drought and hypo-osmotic stress (Anderberg et al., 2011). Here we have identified and analyzed MIPs encoded in the genome of the primitive vascular plant S. moellendorffii to investigate how the MIP family has evolved in terrestrial plants. The total number of SmMIPs is similar to that of moss and six of the seven MIP subfamilies in moss are also found in S. moellendorffii. Of the six subfamilies, the NIP subfamily is dominating in S. moellendorffii whereas both the PIPs and TIPs are uniquely few compared to all other land plants. Based on EST data a second expansion of the MIP family appears to coincide with the evolution of spermatophytes. It seems likely that this later expansion was a consequence of the development of more specialized types of tissue such as those present in the seed and the flower organs, requiring a more complex regulation of water and solute transport. Future analyses of sequenced genomes will hopefully corroborate the more precise timing of this event and provide more information on the transition from fresh water to life on land.
Materials and Methods
Identification and annotation of S. moellendorffii MIPs
The diploid S. moellendorffii genome, available at Joint Genome Institute1, was searched for MIPs using tblastn with P. patens MIPs as queries. In subsequent rounds of searches identified S. moellendorffii MIPs were included until no more MIPs could be found. The allele to be included in subsequent analyses was chosen randomly.
The genomic sequence around the hits was then checked for existing gene models. The models were evaluated and kept if they were found to accurately represent a MIP. New gene models were created if they were found to correspond poorly to the sequences of known MIPs with respect to conserved residues and lengths of predicted loops and transmembrane helices as well as conserved intron positions (Table 2). If no satisfactory gene model existed or could be created for a hit and no allelic variant could be found it was deemed to be a pseudo-gene and was excluded from further analyses.
Sequence alignments
The alignments used for the phylogenetic analyses was created in the program MEGA5 (Tamura et al., 2011) and was based on a structural alignment of BtAQP1 (1J4N, Sui et al., 2001), EcGLPF (1LDA, Fu et al., 2000), EcAQPZ (1RC2, Savage et al., 2003), SoPIP2;1 (1Z98, Tornroth-Horsefield et al., 2006), RnAQP4 (2D57, Hiroaki et al., 2006), MmAQPM (2F2B, Lee et al., 2005), PfAQP (3C02, Newby et al., 2008), HsAQP5 (3D9S, Horsefield et al., 2008), and BtAQP0 (2B6P, Gonen et al., 2005) made in DeepView/Swiss-PdbViewer v4.0.1. All MIP sequences from Arabidopsis thaliana, Physcomitrella patens and from the chlorophyte algae species Chlamydomonas reinhardtii, Volvox carteri, Coccomyxa sp. C-169, Chlorella NC64A, Micromonas pusilla CCMP1545, Micromonas RCC299, Ostreococcus lucimarinus, Ostreococcus RCC809, and Ostreococcus tauri (Anderberg et al., 2011) as well as those of Oryza sativa were then added and manually aligned to the initial structural alignment. Accession numbers are given in Table A2 in Appendix. Finally the MIP sequences identified in S. moellendorffii were added and aligned. Since the N- and C-terminal regions were not included in the subsequent phylogenetic analyses no effort was put into aligning these.
Subset alignments
Based on the resulting alignment three subsets including only PIPs, TIPs, or NIPs were created. The PIP alignment included all PIP sequences from the original alignment with the addition of PIPs encoded by full length ESTs from the gymnosperms Picea glauca and Pinus taeda. In preliminary analyses OsPIP2;8 associated with PpPIP3;1 however this was not observed in the final analysis when PIP2;8-like sequences from the monocots Phyllostachys edulis, Sorghum bicolor, and Zea mays were included. The algal MIPEs were included as an out-group. The NIP alignment, including all the NIP sequences from the original alignment, was supplemented with NIPs encoded by full length EST sequences identified in blast searches with selected NIPs against the NCBI database2. The searches were restricted to species belonging to viridiplantae, excluding monocots and dicots. To this alignment bacterial NIPs (bNIPs) which are the closest homologs to plant NIPs (Danielson and Johanson, 2010) were added as out-group. The TIP alignment was created in the same way but here the P. patens PIPs were included as out-group. If two ESTs from the same organism were found to overlap in such a way that they covered a whole coding sequence, the encoded MIP was also included.
Phylogenetic analyses
The phylogenetic analyses were performed using the Maximum Likelihood algorithm in MEGA5. The best substitution model for each alignment was determined within the MEGA 5 program and were rtREV + G + I + F for the alignment including all subfamilies, JTT + G for the PIP alignment and WAG + G + I + F for both the TIP and NIP alignments. For all alignments the number of discrete gamma categories was set to 5, all sites were used, the heuristic method was set to Nearest-Neighbor-Interchange and the initial tree was made automatically. For all analyses the robustness of the resulting best trees were assessed by 1000 bootstrap replications.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the U.S. Department of Energy Joint Genome Institute for sequencing the genome of S. moellendorffii and making the sequences available to the public. We would also like to thank Jonas Danielson for sharing valuable insights on the evaluation of gene models. This work was supported by funding from the Swedish Research Council.
Appendix
Figure A1.
Phylogeny of land plants. The land plants (embryophytes) can be divided into four groups. The bryophytes are a paraphyletic group comprised of the three earliest diverging lineages of land plants, i.e., the liverworts, mosses, and hornworts. The next group to diverge is the monophyletic lycophytes that consists of clubmosses, quillworts, and spikemosses and further up the tree the ferns represent a lineage diverging just before the emergence of seed plants. The seed plants are divided into two groups, the gymnosperms and the angiosperms (flowering plants). Within the angiosperms Amborella is thought to be the earliest diverging genus. Arrows indicate important morphological changes during the evolution of land plants.
Table A1.
Gene structure of S. moellendorffii MIPs.
| Name | Sca | Str.b | Start posc | E1d | I1d | E2 | I2 | E3 | I3 | E4 | I4 | E5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SmPIP1;1 | sc 43 | − | 543709 | 322 | 206 | 296 | 195 | 141 | 79 | 87 | ||
| SmPIP2;1 | sc 266 | + | 14192 | 292 | 244 | 296 | 255 | 141 | 69 | 126 | ||
| SmPIP2;2 | sc 17 | − | 1553601 | 289 | 71 | 302 | 107 | 141 | 76 | 141 | ||
| SmTIP6;1 | sc 1 | + | 4467056 | 127 | 83 | 248 | 61 | 360 | ||||
| SmTIP6;2 | sc 1 | − | 4470607 | 127 | 58 | 248 | 104 | 360 | ||||
| SmNIP3;1 | sc 23 | + | 391409 | 156 | 145 | 225 | 61 | 195 | 79 | 62 | 68 | 193 |
| SmNIP3;2 | sc 21 | − | 95220 | 96 | 64 | 222 | 63 | 195 | 56 | 62 | 50 | 196 |
| SmNIP3;3 | sc 30 | + | 1231132 | 162 | 49 | 225 | 2408 | 195 | 59 | 62 | 64 | 181 |
| SmNIP3;4 | sc 3 | + | 1364278 | 120 | 78 | 225 | 68 | 263 | 129 | 151 | ||
| SmNIP3;5 | sc 7 | − | 3041227 | 108 | 70 | 225 | 59 | 198 | 54 | 62 | 56 | 187 |
| SmNIP5;1 | sc 2 | + | 2404270 | 180 | 70 | 225 | 159 | 195 | 59 | 62 | 88 | 193 |
| SmNIP5;2 | sc 75 | − | 422849 | 162 | 60 | 225 | 95 | 198 | 50 | 62 | 196 | 226 |
| SmNIP7;1 | sc 13 | − | 1722259 | 147 | 62 | 228 | 56 | 198 | 59 | 62 | 52 | 175 |
| SmSIP1;1 | sc 51 | + | 562196 | 288 | 127 | 273 | 61 | 150 | ||||
| SmXIP1;1 | sc 1 | − | 2139783 | 123 | 58 | 828 | ||||||
| SmXIP1;2 | sc 1 | − | 2093139 | 102 | 74 | 792 | ||||||
| SmXIP1;3 | sc 4 | − | 630059 | 111 | 56 | 936 | ||||||
| SmHIP1;1 | sc 3 | − | 3858367 | 127 | 57 | 248 | 56 | 417 | ||||
| SmHIP1;2 | sc 9 | + | 2872669 | 48 | 61 | 118 | 113 | 191 | 55 | 187 | 58 | 197 |
aScaffold on which the gene is located.
bEncoding strand (+ or −).
cPosition of first nucleotide on scaffold.
dLength of exons (E) and introns (I).
Figure A2.
Alignment of regions determining the ar/R filter. Two regions of the alignment, helix 2 (H2) and helix 5/loop E (H5/LE) separated by a black bar, containing the four residues of the ar/R selectivity filter (boxed). All MIPs of S. moellendorffii and P. patens are included in the alignment. The blue shading reflects the degree of conservation within each subfamily.
Table A2.
Major intrinsic protein accession numbers.
| Accession No. |
|||
|---|---|---|---|
| Taxa name | Species | GenBanka | Phytozomeb |
| PpPIP1;1 | Physcomitrella patens | Pp1s1_535V6.4 | |
| PpPIP1;2 | Pp1s102_107V6.1 | ||
| PpPIP1;3 | Pp1s305_12V6.1 | ||
| PpPIP2;1 | Pp1s8_151V6.1 | ||
| PpPIP2;2 | Pp1s55_301V6.2 | ||
| PpPIP2;3 | Pp1s267_61V6.1 | ||
| PpPIP2;4 | Pp1s118_199V6.1 | ||
| PpPIP3;1 | Pp1s17_281V6.1c | ||
| PpTIP6;1 | Pp1s44_31V6.1 | ||
| PpTIP6;2 | Pp1s156_153V6.1 | ||
| PpTIP6;3 | Pp1s101_226V6.1 | ||
| PpTIP6;4 | Pp1s184_96V6.1 | ||
| PpNIP3;1 | Pp1s258_69V6.1 | ||
| PpNIP5;1 | Pp1s13_445V6.1 | ||
| PpNIP5;2 | Pp1s91_35V6.1 | ||
| PpNIP5;3 | Pp1s37_249V6.1 | ||
| PpNIP6;1 | d | ||
| PpSIP1;1 | Pp1s3_429V6.1 | ||
| PpSIP1;2 | Pp1s475_9V6.3 | ||
| PpXIP1;1 | Pp1s31_73V6.1 | ||
| PpXIP1;2 | Pp1s32_353V6.1 | ||
| PpHIP1;1 | d | ||
| PpGIP1;1 | Pp1s283_16V6.1 | ||
| AtPIP1;1 | Arabidopsis thaliana | CAB71073 | |
| AtPIP1;2 | AAC28529 | ||
| AtPIP1;3 | AAF81320 | ||
| AtPIP1;4 | AAF02782 | ||
| AtPIP1;5 | CAA20461 | ||
| AtPIP2;1 | CAB67649 | ||
| AtPIP2;2 | AAD18142 | ||
| AtPIP2;3 | AAD18141 | ||
| AtPIP2;4 | BAB09839 | ||
| AtPIP2;5 | CAB41102 | ||
| AtPIP2;6 | AAC79629 | ||
| AtPIP2;7 | CAA17774 | ||
| AtPIP2;8 | AAC64216 | ||
| AtTIP1;1 | AAD31569 | ||
| AtTIP1;2 | BAB01832 | ||
| AtTIP1;3 | AAC62778 | ||
| AtTIP2;1 | BAB01264 | ||
| AtTIP2;2 | CAB10515 | ||
| AtTIP2;3 | BAB09071 | ||
| AtTIP3;1 | AAG52132 | ||
| AtTIP3;2 | AAF97261 | ||
| AtTIP4;1 | AAC42249 | ||
| AtTIP5;1 | CAB51216 | ||
| AtNIP1;1 | CAA16760 | ||
| AtNIP1;2 | CAA16748 | ||
| AtNIP2;1 | AAC26712 | ||
| AtNIP3;1 | AAG50717 | ||
| AtNIP4;1 | BAB10360 | ||
| AtNIP4;2 | BAB10361 | ||
| AtNIP5;1 | CAB39791 | ||
| AtNIP6;1 | AAF14664 | ||
| AtNIP7;1 | AAF30303 | ||
| AtSIP1;1 | AAF26804 | ||
| AtSIP1;2 | BAB09487 | ||
| AtSIP2;1 | CAB72165 | ||
| OsPIP1;1 | Oryza sativa | BAD28398 | |
| OsPIP1;2 | EAZ31617 | ||
| OsPIP1;3 | BAD22920 | ||
| OsPIP2;1 | BAC15868 | ||
| OsPIP2;2 | BAD23735 | ||
| OsPIP2;3 | CAD41442 | ||
| OsPIP2;4 | BAC16113 | ||
| OsPIP2;5 | BAC16116 | ||
| OsPIP2;6 | CAE05002 | ||
| OsPIP2;7 | BAD46581 | ||
| OsPIP2;8 | AAP44741 | ||
| OsTIP1;1 | AAK98737 | ||
| OsTIP1;2 | BAB63833 | ||
| OsTIP2;1 | BAD25765 | ||
| OsTIP2;2 | BAD61899 | ||
| OsTIP3;1 | AAG13544 | ||
| OsTIP3;2 | CAE05657 | ||
| OsTIP4;1 | AAS98488 | ||
| OsTIP4;2 | BAA92993 | ||
| OsTIP4;3 | BAA92991 | ||
| OsTIP5;1 | BAF15407 | ||
| OsNIP1;1 | BAD27715 | ||
| OsNIP1;2 | BAD73177 | ||
| OsNIP1;3 | AAV44140 | ||
| OsNIP1;4 | BAD53665 | ||
| OsNIP2;1 | BAD16128 | ||
| OsNIP2;2 | BAD37471 | ||
| OsNIP3;1 | AAG13499 | ||
| OsNIP3;2 | BAC99758 | ||
| OsNIP3;3 | BAC65382 | ||
| OsNIP4;1 | BAB61180 | ||
| OsSIP1;1 | BAB32914 | ||
| OsSIP2;1 | ABF95655 | ||
| CcMIPA1;1 | Coccomyxa C-169 | e | |
| CcMIPD1;1 | |||
| CcMIPD3;1 | |||
| CcPIP4;1 | |||
| CcPIP4;2 | |||
| CcGIP1;1 | |||
| CnMIPD1;1 | Chlorella NC64A | e | |
| CnMIPE1;1 | |||
| CnMIPE1;2 | |||
| CnMIPE1;3 | |||
| CnGIP1;1 | |||
| CrMIPD1;1 | Chlamydomonas reinhardtii | e | |
| CrMIPD2;1 | |||
| MpMIPC1;1 | Micromonas pusilla CCMP1545 | e | |
| MrMIPC1;1 | Micromonas RCC299 | e | |
| OlMIPB1;1 | Ostreococcus lucimarinus | e | |
| OrMIPB1;1 | Ostreococcus RCC809 | e | |
| OrMIPE1;1 | |||
| OtMIPC1;1 | Ostreococcus tauri | e | |
| C. parvum I | Chlorobaculum parvum | ACF10864 | |
| F. bacterium | Flavobacteriales bacterium | EDP70495 | |
| P. irgensii | Polaribacter irgensii | EAR12182 | |
| C. flavus | Chthoniobacter flavus | EDY20715 | |
| P. maris | Planctomyces maris | EDL59061 | |
| C. parvum II | Chlorobaculum parvum | ACF11962 | |
| C. tepidum | Chlorobium tepidum | AAM72699 | |
| ZmNIP1;1 | Zea mays | AAK26750 | |
| ZmNIP2;1 | AAK26751 | ||
| ZmNIP2;2 | AAK26752 | ||
| ZmNIP2;3 | AAK26849 | ||
| ZmNIP3;1 | AAK26753 | ||
| AcMIP1 | Adiantum capillus-veneris | BAB12437 | |
Footnotes
References
- Alexandersson E., Fraysse L., Sjovall-Larsen S., Gustavsson S., Fellert M., Karlsson M., Johanson U., Kjellbom P. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484 10.1007/s11103-005-0352-1 [DOI] [PubMed] [Google Scholar]
- Ali W., Isayenkov S. V., Zhao F. J., Maathuis F. J. (2009). Arsenite transport in plants. Cell. Mol. Life Sci. 66, 2329–2339 10.1007/s00018-009-0021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg H. I., Danielson J. A., Johanson U. (2011). Algal MIPs, high diversity and conserved motifs. BMC Evol. Biol. 11, 110. 10.1186/1471-2148-11-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. K., Yoshikawa N., Ishikawa T., Sawa Y., Shibata H. (2011). Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochim. Biophys. Acta 1818, 1–11 [DOI] [PubMed] [Google Scholar]
- Banks J. A. (2009). Selaginella and 400 million years of separation. Annu. Rev. Plant Biol. 60, 223–238 10.1146/annurev.arplant.59.032607.092851 [DOI] [PubMed] [Google Scholar]
- Banks J. A., Nishiyama T., Hasebe M., Bowman J. L., Gribskov M., Depamphilis C., Albert V. A., Aono N., Aoyama T., Ambrose B. A., Ashton N. W., Axtell M. J., Barker E., Barker M. S., Bennetzen J. L., Bonawitz N. D., Chapple C., Cheng C., Correa L. G., Dacre M., Debarry J., Dreyer I., Elias M., Engstrom E. M., Estelle M., Feng L., Finet C., Floyd S. K., Frommer W. B., Fujita T., Gramzow L., Gutensohn M., Harholt J., Hattori M., Heyl A., Hirai T., Hiwatashi Y., Ishikawa M., Iwata M., Karol K. G., Koehler B., Kolukisaoglu U., Kubo M., Kurata T., Lalonde S., Li K., Li Y., Litt A., Lyons E., Manning G., Maruyama T., Michael T. P., Mikami K., Miyazaki S., Morinaga S., Murata T., Mueller-Roeber B., Nelson D. R., Obara M., Oguri Y., Olmstead R. G., Onodera N., Petersen B. L., Pils B., Prigge M., Rensing S. A., Riano-Pachon D. M., Roberts A. W., Sato Y., Scheller H. V., Schulz B., Schulz C., Shakirov E. V., Shibagaki N., Shinohara N., Shippen D. E., Sorensen I., Sotooka R., Sugimoto N., Sugita M., Sumikawa N., Tanurdzic M., Theissen G., Ulvskov P., Wakazuki S., Weng J. K., Willats W. W., Wipf D., Wolf P. G., Yang L., Zimmer A. D., Zhu Q., Mitros T., Hellsten U., Loque D., Otillar R., Salamov A., Schmutz J., Shapiro H., Lindquist E., Lucas S., Rokhsar D., Grigoriev I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 10.1126/science.1203810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A., Sankararamakrishnan R. (2007). Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Struct. Biol. 7, 27. 10.1186/1472-6807-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E., Wu B., Holm L. M., Schultz J. E., Zeuthen T. (2006). Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia, and protons. Proc. Natl. Acad. Sci. U.S.A. 103, 269–274 10.1073/pnas.0507225103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert G. P., Bienert M. D., Jahn T. P., Boutry M., Chaumont F. (2011). Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 66, 306–317 10.1111/j.1365-313X.2011.04496.x [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Jahn T. P. (2010). Major intrinsic proteins and arsenic transport in plants: new players and their potential role. Adv. Exp. Med. Biol. 679, 111–125 10.1007/978-1-4419-6315-4_9 [DOI] [PubMed] [Google Scholar]
- Danielson J. A., Johanson U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8, 45. 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson J. A., Johanson U. (2010). Phylogeny of major intrinsic proteins. Adv. Exp. Med. Biol. 679, 19–31 10.1007/978-1-4419-6315-4_2 [DOI] [PubMed] [Google Scholar]
- de Groot B. L., Frigato T., Helms V., Grubmuller H. (2003). The mechanism of proton exclusion in the aquaporin-1 water channel. J. Mol. Biol. 333, 279–293 10.1016/j.jmb.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Fu D., Libson A., Miercke L. J., Weitzman C., Nollert P., Krucinski J., Stroud R. M. (2000). Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290, 481–486 10.1126/science.290.5491.481 [DOI] [PubMed] [Google Scholar]
- Gattolin S., Sorieul M., Frigerio L. (2011). Mapping of tonoplast intrinsic proteins in maturing and germinating Arabidopsis seeds reveals dual localization of embryonic TIPs to the tonoplast and plasma membrane. Mol. Plant 4, 180–189 10.1093/mp/ssq051 [DOI] [PubMed] [Google Scholar]
- Gonen T., Cheng Y., Sliz P., Hiroaki Y., Fujiyoshi Y., Harrison S. C., Walz T. (2005). Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638 10.1038/nature04321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. B., Sankararamakrishnan R. (2009). Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 9, 134. 10.1186/1471-2229-9-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakman I., Oliviusson P. (2002). High expression of putative aquaporin genes in cells with transporting and nutritive functions during seed development in Norway spruce (Picea abies). J. Exp. Bot. 53, 639–649 10.1093/jexbot/53.369.639 [DOI] [PubMed] [Google Scholar]
- Heymann J. B., Engel A. (1999). Aquaporins: phylogeny, structure, and physiology of water channels. News Physiol. Sci. 14, 187–193 [DOI] [PubMed] [Google Scholar]
- Hiroaki Y., Tani K., Kamegawa A., Gyobu N., Nishikawa K., Suzuki H., Walz T., Sasaki S., Mitsuoka K., Kimura K., Mizoguchi A., Fujiyoshi Y. (2006). Implications of the aquaporin-4 structure on array formation and cell adhesion. J. Mol. Biol. 355, 628–639 10.1016/j.jmb.2005.10.081 [DOI] [PubMed] [Google Scholar]
- Horsefield R., Norden K., Fellert M., Backmark A., Tornroth-Horsefield S., Terwisscha Van Scheltinga A. C., Kvassman J., Kjellbom P., Johanson U., Neutze R. (2008). High-resolution x-ray structure of human aquaporin 5. Proc. Natl. Acad. Sci. U.S.A. 105, 13327–13332 10.1073/pnas.0801466105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga G., Cushman M. A. F., Cushman J. C. (2006). An EST catalogue from the resurrection plant Selaginella lepidophylla reveals abiotic stress-adaptive genes. Plant Sci. 170, 1173–1184 10.1016/j.plantsci.2006.02.004 [DOI] [Google Scholar]
- Jauh G. Y., Phillips T. E., Rogers J. C. (1999). Tonoplast intrinsic protein isoforms as markers for vacuolar functions. Plant Cell 11, 1867–1882 10.1105/tpc.11.10.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., Gustavsson S. (2002). A new subfamily of major intrinsic proteins in plants. Mol. Biol. Evol. 19, 456–461 10.1093/oxfordjournals.molbev.a004101 [DOI] [PubMed] [Google Scholar]
- Johanson U., Karlsson M., Johansson I., Gustavsson S., Sjovall S., Fraysse L., Weig A. R., Kjellbom P. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369 10.1104/pp.126.4.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I., Karlsson M., Johanson U., Larsson C., Kjellbom P. (2000). The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465, 324–342 10.1016/S0005-2736(00)00147-4 [DOI] [PubMed] [Google Scholar]
- Lee J. K., Kozono D., Remis J., Kitagawa Y., Agre P., Stroud R. M. (2005). Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc. Natl. Acad. Sci. U.S.A. 102, 18932–18937 10.1073/pnas.0506144102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F., Verbruggen H., Zechman F. W. (2011). Into the deep: new discoveries at the base of the green plant phylogeny. Bioessays 33, 683–692 10.1002/bies.201100035 [DOI] [PubMed] [Google Scholar]
- Ma J. F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M., Ishiguro M., Murata Y., Yano M. (2006). A silicon transporter in rice. Nature 440, 688–691 10.1038/nature04590 [DOI] [PubMed] [Google Scholar]
- Ma J. F., Yamaji N., Mitani N., Xu X. Y., Su Y. H., Mcgrath S. P., Zhao F. J. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U.S.A. 105, 9931–9935 10.1073/pnas.0802361105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C., Chrispeels M., Lurin C., Tacnet F., Geelen D., Ripoche P., Guern J. (1997). Function and regulation of seed aquaporins. J. Exp. Bot. 48, 421–430 10.1093/jxb/48.Special_Issue.421 [DOI] [PubMed] [Google Scholar]
- Mitani N., Yamaji N., Ma J. F. (2008). Characterization of substrate specificity of a rice silicon transporter, Lsi1. Pflugers Arch. 456, 679–686 10.1007/s00424-007-0408-y [DOI] [PubMed] [Google Scholar]
- Newby Z. E., O’Connell J., III, Robles-Colmenares Y., Khademi S., Miercke L. J., Stroud R. M. (2008). Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat. Struct. Mol. Biol. 15, 619–625 10.1038/nsmb.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Scheffler B. E., Bauer P. J., Campbell B. T. (2010). Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 10, 142. 10.1186/1471-2229-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston G. M., Carroll T. P., Guggino W. B., Agre P. (1992). Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256, 385–387 10.1126/science.256.5055.385 [DOI] [PubMed] [Google Scholar]
- Rouge P., Barre A. (2008). A molecular modeling approach defines a new group of Nodulin 26-like aquaporins in plants. Biochem. Biophys. Res. Commun. 367, 60–66 10.1016/j.bbrc.2007.12.079 [DOI] [PubMed] [Google Scholar]
- Sade N., Vinocur B. J., Diber A., Shatil A., Ronen G., Nissan H., Wallach R., Karchi H., Moshelion M. (2009). Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2;2 a key to isohydric to anisohydric conversion? New Phytol. 181, 651–661 10.1111/j.1469-8137.2008.02689.x [DOI] [PubMed] [Google Scholar]
- Sakurai J., Ishikawa F., Yamaguchi T., Uemura M., Maeshima M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577 10.1093/pcp/pci172 [DOI] [PubMed] [Google Scholar]
- Savage D. F., Egea P. F., Robles-Colmenares Y., O’Connell J. D., III, Stroud R. M. (2003). Architecture and selectivity in aquaporins: 2.5 Å X-ray structure of aquaporin Z. PLoS Biol. 1, e72. 10.1371/journal.pbio.0000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. J., Szovenyi P., Shaw B. (2011). Bryophyte diversity and evolution: windows into the early evolution of land plants. Am. J. Bot. 98, 352–369 10.3732/ajb.1000316 [DOI] [PubMed] [Google Scholar]
- Soto G., Alleva K., Mazzella M. A., Amodeo G., Muschietti J. P. (2008). AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582, 4077–4082 10.1016/j.febslet.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Sui H., Han B. G., Lee J. K., Walian P., Jap B. K. (2001). Structural basis of water-specific transport through the AQP1 water channel. Nature 414, 872–878 10.1038/414872a [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., Neutze R., Kjellbom P. (2006). Structural mechanism of plant aquaporin gating. Nature 439, 688–694 10.1038/nature04316 [DOI] [PubMed] [Google Scholar]
- Wallace I. S., Roberts D. M. (2005). Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44, 16826–16834 10.1021/bi0511888 [DOI] [PubMed] [Google Scholar]
- Zardoya R. (2005). Phylogeny and evolution of the major intrinsic protein family. Biol. Cell 97, 397–414 10.1042/BC20040134 [DOI] [PubMed] [Google Scholar]