Abstract
Background/Aims
Recessive mutations in the LHX3 ho-meodomain transcription factor gene are associated with developmental disorders affecting the pituitary and nervous system. We describe pediatric patients with combined pituitary hormone deficiency (CPHD) who harbor a novel mutation in LHX3.
Methods
Two female siblings from related parents were examined. Both patients had neonatal complications. The index patient had CPHD featuring deficiencies of GH, LH, FSH, PRL, and TSH, with later onset of ACTH deficiency. She also had a hypoplastic anterior pituitary, respiratory distress, hearing impairment, and limited neck rotation. The LHX3 gene was sequenced and the biochemical properties of the predicted altered proteins were characterized.
Results
A novel homozygous mutation predicted to change amino acid 194 from threonine to arginine (T194R) was detected in both patients. This amino acid is conserved in the DNA-binding homeodomain. Computer modeling predicted that the T194R change would alter the homeodomain structure. The T194R protein did not bind tested LHX3 DNA recognition sites and did not activate the α-glycoprotein and PRL target genes.
Conclusion
The T194R mutation affects a critical residue in the LHX3 protein. This study extends our understanding of the phenotypic features, molecular mechanism, and developmental course associated with mutations in the LHX3 gene.
Key Words: Growth, Transcription, LIM, Development, Pediatric patients
Introduction
Development of the hormone-secreting cells of the human anterior pituitary gland requires the coordinated actions of regulatory genes that encode signaling pathway components and transcription factors leading to the subsequent expression of specialized genes that establish the differentiated character of the cells [1, 2, 3, 4]. Within these regulatory genes, inherited and de novo mutations in those encoding transcription factors have been associated with pediatric diseases including both isolated hormone deficiencies and combined pituitary hormone deficiency (CPHD) diseases. Critical pituitary transcription factors include the HESX1, ISL1, LHX3, LHX4, PAX6, PIT-1 (POU1F1 gene), OTX1, PITX1, PITX2, PROP1, SIX3, and SIX6 proteins that possess structurally related homeodomain tertiary structures that interact with target cis DNA elements in the promoters and enhancers of pituitary genes [1, 2, 3, 4].
LHX3 is a member of the LIM-homeodomain (LIM-HD) subfamily of transcription factor proteins that are characterized by two amino terminal LIM domains (cysteine-rich structures that are important for protein-protein interactions) and a centrally-located DNA-binding homeodomain [5]. Biochemical studies have shown that LHX3 can activate pituitary genes encoding transcription factors, hypothalamic signal receptors, and hormones [e.g. [5, 6, 7]], placing the LHX3 gene in an early regulatory position involved in the determination and differentiation of all five pituitary cell types [reviewed in [4]]. During embryogenesis, the LHX3 gene is expressed in the embryonic brain, spinal cord and Rathke's pouch, an early structure in pituitary development; expression is also detectable in the adult pituitary gland [5, 8, 9, 10]. Mice with inactive Lhx3 genes die shortly following birth of presumptive nervous system deficiencies and have poor structural pituitary development with an associated absence of four of the differentiated cell types and only a minimal population of ACTH-producing corticotropes [11]. By contrast, mice lacking only the carboxyl terminus of the protein are viable but have anterior pituitary deficits causing dwarfism, female infertility, and thyroid insufficiency [12].
Patients who are homozygous for the known rare, recessive mutations in the LHX3 gene have CPHD with deficiencies in GH, PRL, TSH, LH, and FSH. Other syndromic symptoms associated with these patients can include normal, hypoplastic, or enlarged anterior pituitaries, a rigid spine with limited neck rotation, hearing loss, and ACTH deficiency [13, 14, 15, 16, 17, 18]. The endocrine deficits associated with CPHD can result in short stature, metabolic defects, failure of puberty, and other symptoms resulting from pituitary gland inaction. There appears to be a structure/function relationship between the predicted changes that the observed mutations cause, and the resulting disease. Mutations that compromise the overall function of the gene or protein and mutations affecting the LIM domains or the homeodomain appear to result in syndromes involving both the endocrine (CPHD) and nervous (limited neck rotation, possible deafness) systems. By contrast, the carboxyl terminus of LHX3 proteins appears to have roles that are limited to pituitary development and function: mutations affecting only this domain in both human patients and engineered animal models are associated with pituitary diseases without any apparent nervous system deficits [12, 17].
In this study, we describe CPHD patients with a syndrome including GH, LH, FSH, PRL, and TSH deficiency, evolving onset of ACTH deficiency, hypoplastic anterior pituitary morphology, respiratory distress, hearing impairment, and limited neck rotation. These patients harbor a novel LHX3 mutation that causes substitution of a conserved, critical amino acid in the homeodomain (T194R) that abrogates DNA-binding and pituitary gene activation in molecular analyses.
Patients and Methods
Patients
Two female siblings were examined at the University Children's Hospital in Munich, Germany. Both developed respiratory distress combined with hypoglycemia shortly after birth. The first child of the family (patient IV2, fig. 1a) was born full term at 41 + 1 weeks to a 29-year-old mother originating from India. The family reported that there is a common connection four or five generations back (fig. 1a). She was delivered by caesarean section for breech presentation and pathological cardiotocography. Apgar was 8/9/10, umbilical cord pH was 7.35. Birth weight and length corresponded to the 3rd percentile (table 1). At the age of 20 h, she started to breathe irregularly, and a cardiopulmonary resuscitation was necessary 1 h later. After intubation, a low blood glucose was present. Neonatal screening (i.e. detection of more than 30 inborn errors of metabolism, TSH, 17-hydroxyprogesterone) was normal; however, TSH levels were retrospectively not detectable. On echocardiography she had a large patent ductus artery and small atrial septal defect. She developed a severe edema of the brain, recurrent seizures, and no spontaneous movements. She died at the age of 20 days of cardiorespiratory insufficiency. With consent, fibroblasts were preserved, and genomic DNA was later extracted.
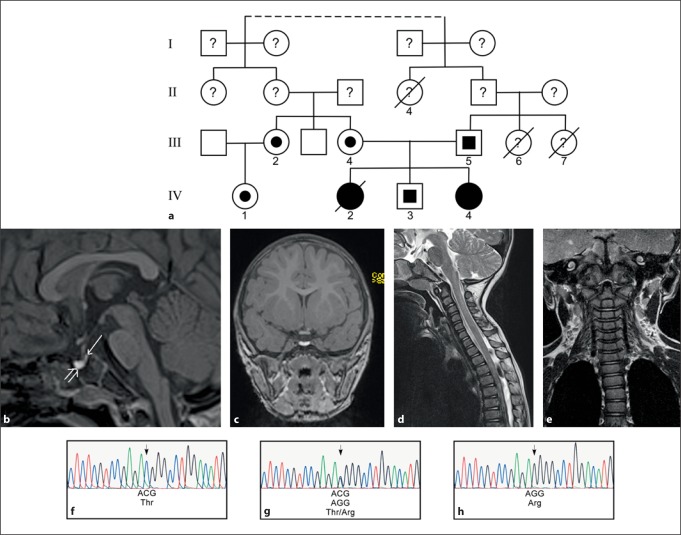
Fig. 1.
DNA analysis reveals a homozygous mutation in the LHX3 gene in patients with CPHD. a Pedigree of the affected family. Filled symbols indicate patients with homozygous genotype. Half-filled symbols denote heterozygous individuals. A line through a symbol indicates that the subject is deceased. The dotted line indicates a family connection four or five generations back from the index patient (IV4). b A 3-Tesla MRI of patient IV4 reveals an absent or severely hypoplastic anterior pituitary gland. The anterior part of the sella is filled with a cystic structure with high signal intensity in non-enhanced T1-weighted sequence (open arrow) and a reduced signal intensity in T2-weighted sequences (not shown), most likely corresponding to a cyst with high protein content. The posterior pituitary gland is found in an orthotopic location (arrow). c Coronal view. d, e A T2-weighted sequence of the cervical spine demonstrates a reduced cervical lordosis with slight kyphosis of the vertebral bodies in segment C2 to C5. f-h DNA sequencing reveals a C to G mutation in the LHX3 gene altering a threonine (Thr) to an arginine (Arg) (f control; g III4 (mother of patient IV4 with heterozygous genotype); h patient IV4).
Table 1.
Clinical data for patients with the T194R LHX3 mutation at newborn age
| Patient | Gestational age weeks | Birth length cm (SD) | Birth weight g (SD) | Age at manifestation, h | Basal PRL nmol/l [44–213] | Basal TSH* μU/ml [0.7–15.2] | Basal GH ng/ml [>7] | Basal cortisol μmol/l [0.14–0.55] | Basal ACTH pmol/ml [2.2–11] | Basal LH mU/ml [0.1–7.7] | Basal FSH mU/ml [0.1–13] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IV2 | 41+1 | 48 (−1.7) | 2,800 (−2.0) | 20 | N/A | 0 | N/A | N/A | N/A | N/A | N/A |
| IV4 | 37+1 | 45 (−2.1) | 2,501 (−1.7) | 48 | 17.5 | 0 | <0.1 | 0.075 | 7.68 | <0.1 | 0.2 |
Age-adjusted normative data are in square brackets.
SD = Standard deviation (according to the normative Swiss data of Prader et al. [33]); N/A = not available.
TSH levels from neonatal screening.
The index patient (patient IV4, fig. 1a) was a full-term infant of 37 + 1 weeks, born by caesarean section due to premature labor. By that time, her mother was 32 years old (G3P3). At birth, her Apgar was 9/8/9, umbilical cord was pH 7.29, and weight was 2,501 g (9th percentile). Postnatally, patient IV4 showed signs of respiratory distress. After 48 h, an intubation was required. A chest X-ray revealed bilateral alveolar infiltrates, a grade 3 respiratory distress syndrome. Surfactant was administrated to the patient, and she began to improve. Four days later, she was extubated without any difficulties. After 3 more days, no further artificial respiration was required. Her examination showed a large anterior and posterior fontanel, prominent eyes, short neck, rigid spine, failure to thrive, sucking weakness, hypoglycemia, and neonatal jaundice. She was completely tube fed until the 9th day of life. Afterwards, additional tube feeding was still necessary due to sucking weakness. She developed recurrent hypoglycemia without symptoms despite nutritional supplements such as maltodextrin. The overall weight gain was poor.
Endocrine diagnostics of patient IV4 on day 3 showed a TSH below the detection limit consistent with central hypothyroidism (table 1), thyroxin levels were also low (0.005 nmol/l), therefore thyroid replacement was started on day 5 of life. Further tests of the hypothalamic-pituitary axis were performed on day 14 and revealed GH (<0.1 ng/ml), IGF-I (<25 μg/l), PRL (17.5 nmol/l), LH (<0.1 mU/ml) and FSH (0.2 mU/ml) deficiencies together with a cortisol (0.075-0.44 μmol/l) in the low normal range (table 1). ACTH levels were within normal limits. She was started on GH treatment on day 17. At the age of 4 months, LH and FSH were re-evaluated and showed the same results as in the newborn age. At the age of 1 year, the patient developed an addisonian crisis, and detected ACTH levels were low by that time (1.32 pmol/l) as well as cortisol (<0.006 μmol/l).
A recent brain MRI at the age of 1.5 years revealed the presence of an absent or severely hypoplastic anterior pituitary and orthotopic posterior pituitary that may be enlarged with a lipoid cyst (fig. 1b). MRI of the cervical spine showed normal alignment and configuration of the cervical spine without any apparent structural abnormalities with a loss of cervical lordosis. In addition, there was impaired cervical rotation limiting movement of the patient's neck to approximately 35° to each side. A brainstem-evoked response audiometry test was negative for both ears. Hearing impairment was confirmed recently at the age of 1.5 years.
DNA Analysis
The array of hormone deficiencies and the associated impaired cervical rotation suggested that patient IV4 may have a mutation in the LHX3 transcription factor gene. After obtaining informed consent, blood samples were collected, including also from first-degree relatives whenever possible. The seven exons of LHX3 were amplified by PCR and sequenced with the ABI PRISM Big Dye Terminator v3.1 Ready Reaction Cycle Sequencing kit (Applied Biosystems, Foster City, Calif., USA). Electrophoresis was performed on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems).
Plasmid Construction and in vitro Transcription/Translation
Human LHX3 cDNA expression plasmids have been described [7, 19]. The T194R mutation was introduced using the QuikChange® kit (Stratagene/Agilent, La Jolla, Calif., USA) with the following oligonucleotide primers: 5′-agctctcgtccgagaggggcctgg-3′, 5′-ccaggcccctctcggacgagagct-3′. Radiolabeled wild-type (WT) and mutant proteins were synthesized in vitro from transcribed cDNAs using TNT rabbit reticulocyte lysate reagents (Promega, Madison, Wisc., USA) and 35S-methionine as described [20]. Proteins were analyzed by electrophoresis in 12% acrylamide gels followed by fluorography as described [21]. Dried gels were imaged with a Storm phosphorimager (Amersham Biosciences, Piscataway, N.J., USA) or BioMax MR X-ray film (Kodak, Rochester, N.Y., USA).
Homology Structural Modeling
Homology structural modeling of the LHX3 homeodomain was performed following methodology described previously [21]. Briefly, the LHX3 homeodomain amino acid sequence was screened against the PDB database using BLAST-p. Among all the possible templates, the 1HDD file corresponding to an engrailed homeodomain DNA complex at 2.8 Å resolution [22] was selected as the best suitable (BLAST score 1e-8) displaying homology higher than 50% (46% identity). The raw LHX3 homeodomain sequence was structurally aligned to the 1HDD, subjected to energy minimization and evaluated with ANOLEA, as described [21]. The accepted model of each homeodomain was docked into the DNA structure of the 1HDD protein-DNA complex structure. The T194 residue of the LHX3 model was changed to arginine (R). All the rotamers for this residue were tested to find putative contacts with other amino acids in the homeodomain structure. The resultant 3D PDB files were exported to pov format and rendered using PovRay 3.62 to obtain high-quality pictures.
Electrophoretic Mobility Shift Analysis
Electrophoretic mobility shift assays (EMSA) were performed using radiolabeled DNA probes representing the pituitary glycoprotein basal element of the α-glycoprotein subunit (αGSU) gene promoter or the LHX3 consensus-binding site as described [23]. Proteins for EMSA were generated by in vitro translation as described above except that cold amino acids were substituted for 35S-methionine. Results were visualized by autoradiography or using a phosphorimager.
Luciferase Assays
Mouse pituitary GHFT1 cells were a gift from Dr. Pamela Mellon (University of California, San Diego, Calif., USA). GHFT1 cells were cultured and transfected as described [20]. Luciferase activity was measured 48 h after transfection [20, 24]. All assay points were performed in triplicate. Total cell protein was determined by the Bradford method (BioRad, Hercules, Calif., USA), and luciferase activity was normalized to protein concentration.
Results
Patient Gene Analysis
Genomic sequencing of patients IV2 and IV4 revealed a homozygous C→G transversion mutation in the fourth coding exon of the LHX3 gene (fig. 1a, h). The clinically unaffected parents (III4 and III5), a brother (IV3), an aunt (mother's sister, III2) and a first cousin (IV1) were heterozygous for the mutation (fig. 1a, g).
The Mutation Causes Alteration of a Conserved Amino Acid in the Homeodomain
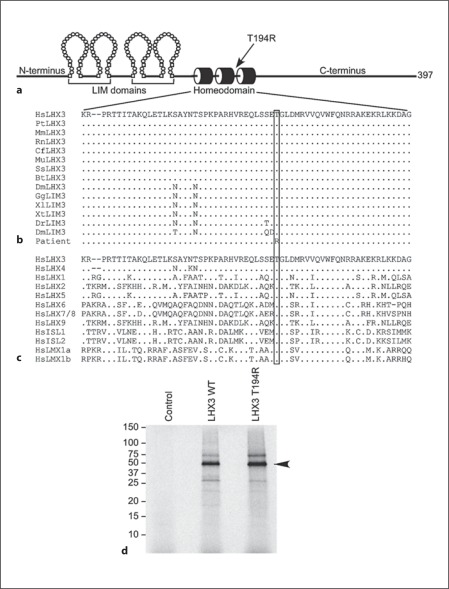
The detected mutation changes a threonine amino acid at position 194 (T194) of the LHX3a protein to an arginine residue, a non-conservative biochemical substitution (T194R). The threonine at position 194 is located in the homeodomain which mediates LHX3 transcription factor interactions with target DNA sequences (fig. 2a). This threonine is entirely conserved in LHX3/LIM3 proteins in mammals, birds, amphibians, fish, and insects (fig. 2b). Further, the equivalent position is also a threonine in each of the human LIM-HD transcription factor proteins (fig. 2c), suggesting that it plays a critical role in the homeodomain of this class of developmental regulators. In vitro transcription/translation of radiolabeled WT and T194R human LHX3a proteins produced full-length protein products that migrated in electrophoresis in accord with their predicted molecular sizes (fig. 2d).
Fig. 2.
A point mutation in the LHX3 gene causes a T194R amino acid substitution in the homeodomain. a Diagram of the key domains of the 397-amino-acid human LHX3a protein. N-terminus = Amino terminus; C-terminus = carboxyl terminus. b Sequence analyses demonstrate that T194 is conserved in LHX3/LIM3 proteins. Alignments of the homeodomain primary amino acid sequence are shown. Dashes denote gaps introduced to optimize alignments; dots denote amino acid identity at that position; the box shows the equivalent amino acids for T194. Comparisons are of human (HsLHX3) to chimpanzee (PtLHX3), rhesus macaque (MmLHX3), rat (RnLHX3), dog (CfLHX3), mouse (MuLHX3), pig (SsLHX3), cow (BtLHX3), opossum (DmLHX3), chicken (GgLIM3), Xenopus laevis (XlLIM3), Xenopus tropicalis (XtLIM3), zebrafish (DrLIM3), Drosophila melanogaster (DmLIM3), and the patient. c The threonine at position 194 in LHX3 is conserved in human LIM-HD class transcription factor proteins. Alignments of the homeodomain primary amino acid sequence are shown. Dashes denote gaps introduced to optimize alignments; dots denote amino acid identity at that position; the box shows the equivalent amino acids for T194. d Generation of radiolabeled WT and T194R LHX3 proteins from cDNA expression vectors by in vitro transcription/translation using rabbit reticulocyte lysates in the presence of 35S-methionine. Labeled products were separated by SDS electrophoresis and dried gels were visualized by fluorography. The migration positions of protein standards (in kDa) are shown. Control = Reaction from a lysate programmed with empty vector (negative control).
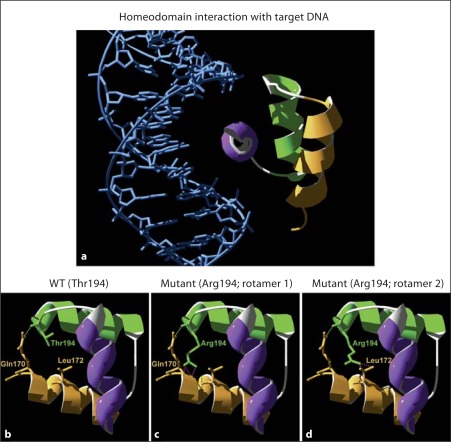
In order to explore the possible effect of the mutation-caused amino acid substitution in the LHX3 homeodomain, we first made a bioinformatic structural prediction using the LHX3 homeodomain amino acid sequence. A model predicting LHX3 interaction with target DNA was performed, using the engrailed homeodomain/DNA complex structure [22] as a template. The LHX3 homeo-domain is composed of three predicted α-helix motifs in a helix-turn-helix structure (fig. 3a). In the 3D model predicted for the WT protein, T194 is located at the end of the second α-helix (green in fig. 3), and it contributes to stabilize the tertiary structure of the homeodomain by establishing a predicted hydrogen bond with a glutamine (Q170) residue located at the beginning of the first α-helix (fig. 3b). When T194 is mutated to arginine, the hydrogen bond with the glutamine 170 amino acid is maintained. However, from the 26 possible predicted rotamer structures for an arginine residue at position 194 (194R), 25 predictions result in steric hindrances, mainly with Q170 and a nearby leucine (L172) (fig. 3c, d), suggesting that these steric hindrances may destabilize the helix-turn-helix tertiary structure, thereby reducing DNA-binding and gene activation.
Fig. 3.
Structural prediction of LHX3 homeodomain/DNA interaction. A model of LHX3 interaction with target DNA was performed using the engrailed homeodomain/DNA complex as a template [22]. a Ribbon model of WT LHX3 homeodomain/DNA showing the three α-helices in orange (helix 1), green (helix 2) and purple (helix 3). b Predicted WT LHX3 homeo-domain with a threonine at position 194 (Thr194). c, d Predicted T194R LHX3 homeodomain with an arginine at position 194 (Arg194). Twenty-six different rotamers for this residue are possible. In all cases, except rotamer 21, these result in steric hindrances with other amino acids (mainly with glutamine 170 [Gln170] and leucine 172 [Leu172]). The figure shows two of the R194 rotamers (green side chains) that destabilize the tertiary structure by causing steric hindrance (purple dotted lines) with glutamine 170 and leucine 172 (orange side chains).
DNA-Binding Properties of T194R LHX3
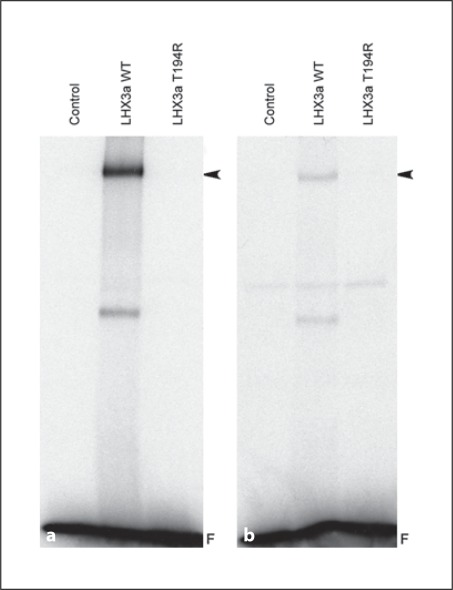
To test DNA binding, WT and T194R LHX3 proteins were generated in vitro and DNA binding was studied by EMSA analysis using radiolabeled LHX3-binding sites. Whereas WT LHX3 bound to probes representing the LHX3 consensus-binding site [23] or theαGSU promoter pituitary glycoprotein-binding element, T194R LHX3 did not interact with these sites (fig. 4a, b).
Fig. 4.
The LHX3 T194R protein does not bind to LHX3 DNA recognition elements. EMSA experiments were performed using the WT or T194R LHX3 proteins translated in vitro in rabbit reticulocyte lysates and radiolabeled probes representing the LHX3 consensus-binding site [23] (a) or theαGSU promoter pituitary glycoprotein-binding element (b). Unprogrammed lysates were used as negative controls (Control). The LHX3/DNA complexes are denoted by arrowheads. F = Free probe.
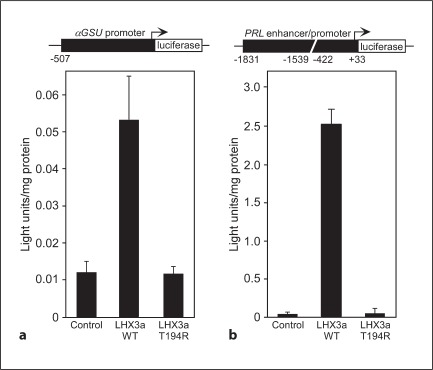
T194R LHX3 Does Not Activate Tested Pituitary Target Genes
To examine the ability of the T194R LHX3 protein to activate pituitary genes, LHX3 expression vectors were cotransfected with a αGSU promoter reporter gene into pituitary GHFT1 cells. WT LHX3a activated the αGSU promoter, while T194R LHX3 did not (fig. 5a). To test a different type of pituitary promoter, the prolactin (PRL) promoter/enhancer also was investigated. LHX3 alone is a moderate activator of PRL transcription, but in combination with the PIT-1 pituitary transcription factor, the effects are synergistic [5, 7]. The GHFT1 cells contain some endogenous PIT-1. As for αGSU, WT LHX3 activated the PRL reporter gene, but T194R LHX3 was inactive (fig. 5b).
Fig. 5.
The LHX3 T194R protein is unable to activate pituitary target genes. a Expression vectors for WT and T194R LHX3 proteins were transiently cotransfected into mouse pituitary GHFT1 cells with a luciferase reporter gene under the control of theαGSU promoter. Promoter activity was assayed by measuring luciferase activity 48 h after transfection. Negative controls (Control) received equivalent amounts of empty expression vector plasmid. Activities are mean (light units/10 s/μg total protein) of triplicate assays ± SEM. A representative experiment of at least three experiments is depicted. b Similar experiments to those depicted in a were performed using a prolactin (PRL) promoter/enhancer reporter gene.
Discussion
We have characterized a novel mutation (T194R) in the human LHX3 transcription factor gene that is associated with a syndrome featuring CPHD (GH, PRL, FSH, LH deficiency and later onset of ACTH deficiency), limited neck rotation, pituitary structural defects including a hypoplastic anterior lobe and a cystic structure within the anterior sella, respiratory distress, and sensorineural hearing loss. In addition, patient IV2 had cardiac defects including a patent ductus artery and an atrial septal defect. It is hard to determine whether these cardiac problems are associated with the T194R LHX3 mutation; or because the parents are distantly related, the cardiac defects could represent a second genetic defect. The observation of cystic structure within the anterior part of the sella is a new finding for LHX3 patients but may be related to other observations of uncharacterized pituitary masses in LHX3 patients [e.g. [13, 15]].
LHX3 gene mutations are comparatively rare [17], and their phenotypic outcomes are likely dependent on several parameters including the nature of the result of the mutation and the genetic background of the affected families. The family described in this report has several reported cases of infant deaths (patients II4, III6, III7 in fig. 1a), suggesting that if these patients carried homozygous T194R mutations, in the absence of successful medical intervention, the T194R mutation is a relatively severe mutation that can be lethal without patient treatment. Perinatal mortality has been reported in other families with LHX3 gene mutations that are predicted to disable protein function [e.g. [13, 17]].
Respiratory distress was observed in both patients characterized with LHX3 T194R mutations. This has been seen before with some LHX3 patients [14]. Mutations in genes encoding other pituitary transcription factors such as LHX4 and PROP1 have been shown to cause defects in lung development and respiratory distress syndrome symptoms in mouse models [25, 26] and in human patients with PIT-1 gene mutations [e.g. [27]]. In a mouse PROP1 deficiency model, pituitary hormone deficiencies (especially perhaps the thyroid axis hormones) lead to lower levels of the NKX2.1/TITF1 transcription factor, atelectasis of the lungs, and reduced lung surfactant, resulting in respiratory distress syndrome [26]. Interestingly, these phenotypes vary significantly depending on the genetic background [26]. In the patients described here, the genetic background may perhaps have promoted or exacerbated the respiratory distress associated with the LHX3 mutation.
The index patient (IV4) described here initially presented with a deficiency of all anterior pituitary hormones except ACTH. However, within about 1 year, ACTH deficiency developed. Some other forms of LHX3 mutations have also been associated with ACTH deficiencies, whereas others have not [13, 14, 15, 16, 17, 18]. It is clear therefore that patients with LHX3-associated CPHD should be monitored for evolving ACTH axis deficiencies during childhood and as adults, as have been similarly described for mutations in the gene encoding the PROP1 homeodomain transcription factor [e.g. [28]].
The striking conservation of the threonine at position 194 in human LHX3a in all known LHX3/LIM3 proteins and in other human LIM-HD factors (fig. 2) suggests that it is critical to the function of LIM-HD class transcription factors. Indeed, it is part of the amino acid sequence ‘signature’ that defines the LIM subclass of homeodomain proteins [29]. This residue is predicted to lie at the end of the second helix of the homeodomain and the equivalent amino acid in many other homeodomains is often also a threonine (e.g. in PITX2, PROP1, PAX4) or an isoleucine or leucine residue (e.g. in POU factors such as PIT1/POU1F1, in NK class homeodomains, and in many of the HOX proteins) [29]. Intriguingly, several homeodomain proteins have been experimentally altered at this equivalent position (e.g. by changing a leucine to a proline) to generate non-DNA-binding derivatives that can act as dominant negative proteins in overexpression experiments to probe the role of the endogenous homeodomain factor. Such approaches have been especially pursued with other developmental proteins such as NKX2.5 and MIX1 [30, 31]. In these experiments, it is thought that overexpression of the altered protein can titrate essential cofactor proteins away from the endogenous factor and thereby interfere with its normal activity. In the case of the patients described in this report, it appears that the T194R LHX3 protein has lost DNA-binding ability, consistent with observations of alteration of this position in other homeodomain proteins such as MIX1 [31]. However, the ‘T194R’ mutation in LHX3 appears to act in a simple recessive fashion, rather than in a dominant negative mode, with heterozygous individuals in the family not demonstrating affected symptoms. This is consistent with the previously described phenotypes of heterozygous and homozygous LHX3 patients and experimental Lhx3 model animals.
This report describes the first example of a missense mutation in exon IV of the LHX3 gene. The alteration of an essential amino acid in the DNA-binding domain provides a clear example of structure/function consequences of LHX3 mutations. To date, a range of mutations affecting the LHX3 gene and its encoded protein have been described (table 2). These include deletions that remove all or most of the gene, splice site mutations that are predicted to block gene function, missense point mutations that cause critical amino acid substitutions, or nonsense mutations that cause inactivating truncations of the protein, and nonsense mutations that result in loss of the carboxyl terminus (W224Ter) [13, 14, 15, 16, 17, 18]. Mutations that impact the overall function of the gene or protein appear to result in diseases affecting both pituitary gland development and physiology and the nervous systems (causing limited neck rotation, deafness, developmental delay, etc.). The T194R mutation described here falls into this class. Similarly, the A210V missense mutation apparently also results in an inactive LHX3 protein; however, unlike the T194R form, this substitution does retain some slight DNA-binding function [17]. The A210V patients were not all described to have ACTH axis deficiencies [17], but whether this reflects the nature of the mutation or an influence of genetic background is unclear. These phenotypes suggest that the LIM domains and homeodomain are required for all LHX3 functions. However, the carboxyl terminus of LHX3 contains an activation domain that is required for pituitary gene activation [32], and patients and mouse models of CPHD with mutations causing specific loss of the carboxyl terminus of the LHX3 protein have pituitary disease but not the limited neck rotation or other nervous system symptoms, demonstrating that the roles in the pituitary and nervous system are molecularly separable [12, 17].
Table 2.
Recessive mutations in the human LHX3 gene causing CPHD
| p.Y111C (Netchine et al. [16], Rajab et al. [18]) | 23-bp deletion (Netchine et al. [16], Rajab et al. [18]) | g.159delT (Bhangoo et al. [13]) | p.A210V (Pfäffle et al. [17]) | p.E173Ter (Pfäffle et al. [17]) | p.W224Ter (Pfäffle et al. [17]) | LHX3 gene deletion (Pfäffle et al. [17]) | p.K50Ter (Rajab et al. [18]) | Intragenic deletion of 3,088 bp (Rajab et al. [18]) | A→G splice mutation in intron 3 (Kristrom et al. [15]) | p.R77Ter (Bonfig et al. [14]) | p.T194R (this study) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hormone deficiencies reported | GH, TSH, PRL, LH, FSH | GH, TSH, PRL, LH, FSH | GH, TSH, PRL, LH, FSH | GH, TSH, PRL, LH, FSH (ACTH sometimes low) | GH, TSH, PRL, LH, FSH (ACTH low) | GH, TSH, PRL, LH, FSH | GH, TSH, PRL, LH, FSH | GH, TSH, PRL, LH, FSH, ACTH | GH, TSH, PRL, LH, FSH, ACTH | GH, TSH, PRL, LH, FSH, ACTH | GH, TSH, PRL, LH, FSH, ACTH | GH, TSH, PRL, LH, FSH, ACTH |
| Anterior pituitary morphology | Hypoplastic | Enlarged | Hypointense pituitary lesion | Enlarged | Hypoplastic | Normal | Severely hypoplastic/aplastic | Hypoplastic | Hypoplastic | Aplastic/hypoplastic (sometimes cysts) | Hypoplastic | Hypoplastic (possible cyst) |
| Neck rotation | Limited | Limited | Limited | Limited | Limited | Normal | Limited | Limited | Limited | Limited | Limited | Limited |
| Hearing deficiencies | Moderate to mild sensorineural hearing defect | Completely deaf | Not reported | Not reported | Not reported | Not reported | Not reported | Sensorineural deafness | Sensorineural hearing defect | Sensorineural hearing defect | Sensorineural hearing defect | Sensorineural hearing defect |
| Mental/learning deficiencies | Not reported | Extreme mental retardation | Some mental retardation | Not reported | Not reported | Not reported | Psychomotor development is severely retarded | Not reported | Some learning difficulties | Not reported | Not reported | Not reported |
| Location of mutation | Missense mutation within exon 3 | Deletion affecting exon 3 and splice-donor site | Deletion in exon 2 | Missense mutation in exon 5 | Complex mutation in exon 3 | Nonsense mutation in exon 5 | Complete gene deletion | Nonsense mutation in exon 2 | Deletion of exons 2–5 | A→G splice acceptor mutation in intron 3 | Nonsense mutation in exon 2 | Missense mutation in exon 4 |
| Protein domain(s) affected | Second LIM domain | Multiple | Multiple | HD | Multiple | Loss of carboxyl terminus | Multiple | Multiple | Multiple | Multiple | Multiple | HD |
Protein domains: Multiple = multiple domains affected or deleted resulting in an inactivated gene function; HD = homeodomain.
In summary, this study expands the range of known molecular defects in the LHX3 gene and extends our understanding of the phenotypic features and developmental disease course associated with mutations in this gene.
Acknowledgements
We thank the patients for participation in this study. We thank Dr. Pamela Mellon for the GHFT1 cells and Dr. Birgit Ertl-Wagner for the MRI pictures and their interpretation. Supported by a grant to S.J.R. from the National Institutes of Health (HD42024) and a National Science Foundation predoctoral fellowship to R.E.M.
References
- 1.Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH, Brinkmeier ML, Mendonça BB, Brue T, Camper SA. Molecular mechanisms of pituitary organogenesis: in search of novel regulatory genes. Mol Cell Endocrinol. 2010;323:4–19. doi: 10.1016/j.mce.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelberman D, Dattani MT. Role of transcription factors in midline central nervous system and pituitary defects. Endocr Dev. 2009;14:67–82. doi: 10.1159/000207478. [DOI] [PubMed] [Google Scholar]
- 3.Pfäffle R, Klammt J. Pituitary transcription factors in the aetiology of combined pituitary hormone deficiency. Best Pract Res Clin Endocrinol Metab. 2011;25:43–60. doi: 10.1016/j.beem.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Prince KL, Walvoord EC, Rhodes SJ. The role of homeodomain transcription factors in heritable pituitary disease. Nat Rev Endocrinol. 2011;7:727–737. doi: 10.1038/nrendo.2011.119. [DOI] [PubMed] [Google Scholar]
- 5.Bach I, Rhodes SJ, Pearse RV, 2nd, Heinzel T, Gloss B, Scully KM, Sawchenko PE, Rosenfeld MG. P-lim, a lim homeodomain factor, is expressed during pituitary organ and cell commitment and synergizes with pit-1. Proc Natl Acad Sci USA. 1995;92:2720–2724. doi: 10.1073/pnas.92.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granger A, Bleux C, Kottler ML, Rhodes SJ, Counis R, Laverriere JN. The lim-homeodomain proteins isl-1 and lhx3 act with steroidogenic factor 1 to enhance gonadotrope-specific activity of the gonadotropin-releasing hormone receptor gene promoter. Mol Endocrinol. 2006;20:2093–2108. doi: 10.1210/me.2005-0184. [DOI] [PubMed] [Google Scholar]
- 7.Sloop KW, Meier BC, Bridwell JL, Parker GE, Schiller AM, Rhodes SJ. Differential activation of pituitary hormone genes by human lhx3 isoforms with distinct DNA binding properties. Mol Endocrinol. 1999;13:2212–2225. doi: 10.1210/mend.13.12.0395. [DOI] [PubMed] [Google Scholar]
- 8.Seidah NG, Barale JC, Marcinkiewicz M, Mattei MG, Day R, Chretien M. The mouse homeoprotein mlim-3 is expressed early in cells derived from the neuroepithelium and persists in adult pituitary. DNA Cell Biol. 1994;13:1163–1180. doi: 10.1089/dna.1994.13.1163. [DOI] [PubMed] [Google Scholar]
- 9.Sheng HZ, Moriyama K, Yamashita T, Li H, Potter SS, Mahon KA, Westphal H. Multistep control of pituitary organogenesis. Science. 1997;278:1809–1812. doi: 10.1126/science.278.5344.1809. [DOI] [PubMed] [Google Scholar]
- 10.Sobrier ML, Attie-Bitach T, Netchine I, Encha-Razavi F, Vekemans M, Amselem S. Pathophysiology of syndromic combined pituitary hormone deficiency due to a lhx3 defect in light of lhx3 and lhx4 expression during early human development. Gene Expr Patterns. 2004;5:279–284. doi: 10.1016/j.modgep.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Sheng HZ, Zhadanov AB, Mosinger B, Jr, Fujii T, Bertuzzi S, Grinberg A, Lee EJ, Huang SP, Mahon KA, Westphal H. Specification of pituitary cell lineages by the lim homeobox gene lhx3. Science. 1996;272:1004–1007. doi: 10.1126/science.272.5264.1004. [DOI] [PubMed] [Google Scholar]
- 12.Colvin SC, Malik RE, Showalter AD, Sloop KW, Rhodes SJ. Model of pediatric pituitary hormone deficiency separates the endocrine and neural functions of the lhx3 transcription factor in vivo. Proc Natl Acad Sci USA. 2011;108:173–178. doi: 10.1073/pnas.1009501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhangoo AP, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, Walvoord EC, Ten S, Rhodes SJ. Clinical case seminar: a novel lhx3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab. 2006;91:747–753. doi: 10.1210/jc.2005-2360. [DOI] [PubMed] [Google Scholar]
- 14.Bonfig W, Krude H, Schmidt H. A novel mutation of lhx3 is associated with combined pituitary hormone deficiency including ACTH deficiency, sensorineural hearing loss, and short neck - a case report and review of the literature. Eur J Pediatr. 2011;170:1017–1021. doi: 10.1007/s00431-011-1393-x. [DOI] [PubMed] [Google Scholar]
- 15.Kristrom B, Zdunek AM, Rydh A, Jonsson H, Sehlin P, Escher SA. A novel mutation in the lim homeobox 3 gene is responsible for combined pituitary hormone deficiency, hearing impairment, and vertebral malformations. J Clin Endocrinol Metab. 2009;94:1154–1161. doi: 10.1210/jc.2008-0325. [DOI] [PubMed] [Google Scholar]
- 16.Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, Marcos E, Duriez B, Cacheux V, Moers A, Goossens M, Gruters A, Amselem S. Mutations in lhx3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet. 2000;25:182–186. doi: 10.1038/76041. [DOI] [PubMed] [Google Scholar]
- 17.Pfäffle RW, Savage JJ, Hunter CS, Palme C, Ahlmann M, Kumar P, Bellone J, Schoenau E, Korsch E, Bramswig JH, Stobbe HM, Blum WF, Rhodes SJ. Four novel mutations of the lhx3 gene cause combined pituitary hormone deficiencies with or without limited neck rotation. J Clin Endocrinol Metab. 2007;92:1909–1919. doi: 10.1210/jc.2006-2177. [DOI] [PubMed] [Google Scholar]
- 18.Rajab A, Kelberman D, de Castro SC, Biebermann H, Shaikh H, Pearce K, Hall CM, Shaikh G, Gerrelli D, Grueters A, Krude H, Dattani MT. Novel mutations in lhx3 are associated with hypopituitarism and sensorineural hearing loss. Hum Mol Genet. 2008;17:2150–2159. doi: 10.1093/hmg/ddn114. [DOI] [PubMed] [Google Scholar]
- 19.Sloop KW, Dwyer CJ, Rhodes SJ. An isoform-specific inhibitory domain regulates the lhx3 lim homeodomain factor holoprotein and the production of a functional alternate translation form. J Biol Chem. 2001;276:36311–36319. doi: 10.1074/jbc.M103888200. [DOI] [PubMed] [Google Scholar]
- 20.Sloop KW, Parker GE, Hanna KR, Wright HA, Rhodes SJ. Lhx3 transcription factor mutations associated with combined pituitary hormone deficiency impair the activation of pituitary target genes. Gene. 2001;265:61–69. doi: 10.1016/s0378-1119(01)00369-9. [DOI] [PubMed] [Google Scholar]
- 21.Pfäffle RW, Hunter CS, Savage JJ, Duran-Prado M, Mullen RD, Neeb ZP, Eiholzer U, Hesse V, Haddad NG, Stobbe HM, Blum WF, Weigel JF, Rhodes SJ. Three novel missense mutations within the lhx4 gene are associated with variable pituitary hormone deficiencies. J Clin Endocrinol Metab. 2008;93:1062–1071. doi: 10.1210/jc.2007-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 23.Bridwell JA, Price JR, Parker GE, McCutchan Schiller A, Sloop KW, Rhodes SJ. Role of the lim domains in DNA recognition by the lhx3 neuroendocrine transcription factor. Gene. 2001;277:239–250. doi: 10.1016/s0378-1119(01)00704-1. [DOI] [PubMed] [Google Scholar]
- 24.Savage JJ, Hunter CS, Clark-Sturm SL, Jacob TM, Pfäffle RW, Rhodes SJ. Mutations in the lhx3 gene cause dysregulation of pituitary and neural target genes that reflect patient phenotypes. Gene. 2007;400:44–51. doi: 10.1016/j.gene.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Witte DP, Branford WW, Aronow BJ, Weinstein M, Kaur S, Wert S, Singh G, Schreiner CM, Whitsett JA, et al. Gsh-4 encodes a lim-type homeodomain, is expressed in the developing central nervous system and is required for early postnatal survival. EMBO J. 1994;13:2876–2885. doi: 10.1002/j.1460-2075.1994.tb06582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasonkin IO, Ward RD, Raetzman LT, Seasholtz AF, Saunders TL, Gillespie PJ, Camper SA. Pituitary hypoplasia and respiratory distress syndrome in prop1 knockout mice. Hum Mol Genet. 2004;13:2727–2735. doi: 10.1093/hmg/ddh311. [DOI] [PubMed] [Google Scholar]
- 27.De Zegher F, Pernasetti F, Vanhole C, De-vlieger H, Van den Berghe G, Martial JA. The prenatal role of thyroid hormone evidenced by fetomaternal pit-1 deficiency. J Clin Endocrinol Metab. 1995;80:3127–3130. doi: 10.1210/jcem.80.11.7593413. [DOI] [PubMed] [Google Scholar]
- 28.Böttner A, Keller E, Kratzsch J, Stobbe H, Weigel JF, Keller A, Hirsch W, Kiess W, Blum WF, Pfäffle RW. Prop1 mutations cause progressive deterioration of anterior pituitary function including adrenal insufficiency: a longitudinal analysis. J Clin Endocrinol Metab. 2004;89:5256–5265. doi: 10.1210/jc.2004-0661. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee-Basu S, Baxevanis AD. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29:3258–3269. doi: 10.1093/nar/29.15.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, xnkx2-3 and xnkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- 31.Mead PE, Brivanlou IH, Kelley CM, Zon LI. Bmp-4-responsive regulation of dorsal-ventral patterning by the homeobox protein mix.1. Nature. 1996;382:357–360. doi: 10.1038/382357a0. [DOI] [PubMed] [Google Scholar]
- 32.Sloop KW, Dwyer CJ, Rhodes SJ. An isoform-specific inhibitory domain regulates the lhx3 lim homeodomain factor holoprotein and the production of a functional alternate translation form. J Biol Chem. 2001;276:36311–36319. doi: 10.1074/jbc.M103888200. [DOI] [PubMed] [Google Scholar]
- 33.Prader A, Largo RH, Molinari L, Issler C. Physical growth of Swiss children from birth to 20 years of age. First Zurich longitudinal study of growth and development. Helv Paediatr Acta Suppl. 1989;52:1–125. [PubMed] [Google Scholar]