Abstract
Transposable elements (TEs) are capable of inducing heritable de novo genetic variation. The sequences capable of reactivation, and environmental factors that induce mobilization, remain poorly defined even in well-studied genomes such as maize. We treated maize tissue culture with the demethylating agent 5-aza-2-deoxcytidine and examined long-term tissue culture lines to discover silenced TEs that have the potential to induce heritable genetic variation. Through these screens we have identified a novel low copy number hAT transposon, Tissue Culture Up-Regulated (TCUP), which is transcribed at high levels in long-term maize black Mexican sweet (BMS) tissue culture and is transcribed in response to treatment with 5-aza-2-deoxycytidine. Analysis of the TIGR Maize Gene Index revealed that this element is the most frequently represented EST from the BMS cell culture library and is not represented in other tissue libraries, which is the basis for its name. A full-length sequence was assembled in inbred B73 that contains the putative functional motifs required for autonomous movement of a hAT transposon. Transposon display detected novel TCUP insertions in two long-term tissue-cultured cell lines of the genotype Hi-II A × B and BMS. This research implicates TCUP as a transposon that is capable of reactivation and which may also be particularly sensitive to the stress of the tissue culture environment. Our findings are consistent with the hypothesis that epigenetic alterations potentiate genomic responses to stress during clonal propagation of plants.
Keywords: transposable element, tissue culture, DNA methylation, genome stress, epigenetics, somaclonal variation
Introduction
Transposable elements (TEs) are ubiquitous members of all higher eukaryotic genomes and compose the majority of DNA present in the maize genome (Schnable et al., 2009). These mobile DNAs are critical elements in the creation of genetic variation that can drive the reorganization and evolution of genomes. Even though these repetitive elements make up an extensive proportion of higher eukaryotic genomes, they are normally maintained in a transcriptionally silent and immobile state (Chomet et al., 1987; Lippman et al., 2003, 2004; Dooner and Weil, 2007). TEs are known to become activated from their dormant state by both biotic (Johns et al., 1985; Grandbastien et al., 1997; Kashkush et al., 2003; Xu et al., 2004) and abiotic stresses (Neuffer, 1966; Carpenter et al., 1987; Peschke et al., 1987; Kalendar et al., 2000; Kimura et al., 2001; Jiang et al., 2003).
The transcription of TEs near or within genic regions can change the spatial and temporal expression of genes leading to altered phenotypes by both genetic and epigenetic mechanisms (Morgan et al., 1999; Kashkush et al., 2003). They may also be mutagenic when mobilized and transpose into or near genes. The mobilization of all types of transposons requires transcription from autonomous elements. Activation of dormant TEs may be a contributor to the phenomenon of somaclonal variation. Somaclonal variation is defined as genetic or phenotypic variation among clonally propagated plants derived from a single donor clone and results in a wide range of phenotypes displayed in the progenies of regenerated plants (Kaeppler et al., 2000).
Tissue culture induced activation of silenced TEs does not affect all elements equally, with some elements predictably activated by the culture process (Hirochika et al., 1996; Takeda et al., 1999), and most others remaining quiescent. For example, silenced Mutator elements are not reactivated through tissue culture and slightly active elements can become methylated and silenced in tissue-cultured lines (Planckaert and Walbot, 1989). Other TEs, such as Tto1, are predictably activated during tissue culture. Tto1 contains cis regulatory sequences sensitive to the auxins used in tissue culture, providing an explanation for the sensitivity of this element to culture stress (Takeda et al., 1999).
The cell culture transcriptome of maize is enriched with TE ESTs compared with other organ tissue, but not all TEs are equally transcribed. The hAT transposon Mx is transcribed while the related element Ac is not, highlighting the differential response of TE to the tissue culture process (Vicient, 2010). The maize miniature inverted repeat TEs (MITE) ZmTPApong is transcriptionally activated in cell culture and mobilized in the regenerated progeny (Barret et al., 2006) and the MITE related P instability factor (PIF) is enriched in cell culture compared to other tissues (Vicient, 2010). The activation of TEs has been associated with a general loss of DNA methylation in heterochromatic regions, but specific elements become hypomethylated and gain H3Kme2 in both heterochromatic and euchromatic chromosome locations (Tanurdzic et al., 2008). The unique sensitivity of different TEs to specific stresses underlies the types and frequency of genetic variation induced in specific environments.
The goal of this research was to characterize a novel maize class 2 hAT transposable element that was first identified based on high expression specifically in black Mexican sweet (BMS) long-term cell culture. Based on its initial in silico identification indicating EST evidence of high expression in BMS and no expression in other tissues, we named the element Tissue Culture Up-Regulated (TCUP). A subsequent pharmacological analysis using the DNA methylation inhibitor, 5-aza-2-deoxycytidine, was used to evaluate the effect of DNA methylation on TCUP transcription in newly initiated cultures. The mobility of TCUP was also evaluated in a series of long-term Hi-II A × B tissue culture lines.
Materials and Methods
Callus lines
The BMS cell line was initiated in the 1970s at the University of Minnesota and was most recently acquired for our studies from Charles Armstrong at Monsanto in 2001. Independent callus lines were created from individual Hi-II A × B embryos harvested 12 days after pollination. Callus lines were maintained on N6 media supplemented with 1.5 mg/l of 2,4-dichlorophenoxyacetic acid (Armstrong, 1994). The embryogenic cell cultures were transferred to fresh media monthly.
Plant material
The inbred stocks obtained from the Maize Genetics Cooperation Stock Center were BMS [Accession: B542B], Hi-II A [Accession: T0940A], and Hi-II B [Accession: T09040B] (Armstrong et al., 1991). All seeds were bulked and maintained using sib crosses in field nurseries. Hi-II A × B seeds were generated by crossing Hi-II B pollen onto Hi-II A ears. Embryos used for tissue culture initiation were acquired from green house grown ears of a self-pollinated Hi-II A × B plant.
Suspension culture treatments
Each culture line was initiated using 1.5 g of Hi-II A × B type II embryogenic callus broken into small clumps. The culture lines were each split into two flasks prior to initiation of the experiment. One flask within each of the three cell lines was treated with 25 μM 5-aza-2-deoxycytosine, and the other flask was used as a non-treated control. Liquid N6 medium was replaced with either treated or untreated medium every 3 days for 9 days to ensure an adequate treatment length.
Genomic DNA isolations from plant tissue
Genomic DNA was isolated from plant tissue using the CTAB method (Saghai-Maroof et al., 1984). The DNA was suspended in LTE (1 mM Tris–HCl pH 8.0, 0.1 mM EDTA pH 8.0). DNA was extracted from callus using the Plant DNAzol reagent (Invitrogen catalog # 10978-021).
PCR amplification of genomic DNA
PCR reactions contained the following components: 1× Taq DNA polymerase buffer, 2.0 mM Mg2Cl, 200 μM dNTPs, 0.6 μM each primer, 1 U Taq DNA polymerase, 100 ng of genomic DNA, and sterile distilled deionized water to a volume of 25 μl. Cycling parameters were generally as follows: 94°C 2 min, 30–35× (94°C for 30 s, 58°C for 45 s, 72°C 1 min per kilobase of amplicon) 72°C for 7 min. TCUP 5′ probe sequence corresponding to 9–875 bp of accession DQ324364.1 was amplified using primers TCUP5′F GCCAAATGGCACTAACACGAC and TCUP5′R GAGGAGAGTACCAGTGCCAGT. The TCUP internal probe sequence corresponding to 2203–3439 bp was amplified using primers InternalF GCTGGTGTGCTTGCTGATTATG and InternalR CGTCGATGATCCTGCCAGTTA. The TCUP 3′ probe sequence corresponding to 3313–4127 bp was amplified using primers TCUP3′F GGTGGCATCAGCACAAACTCA, TCUP3′R TATAGATGGCCAACCGGGCCGCACGGCACG. Reamplification of the excised and sequenced novel band from transposon display was performed using H6_band1 CACGGCGCGAACTTGAACATATAG and display TCUP3′-1 ACTGGTAGTGCCGTGCCTGG.
DNA sequencing
Sequencing reactions consisted of 1 μl of BigDye Terminator mix version 3.1 (Applied Biosystems), 1.5 μl of BigDye buffer version 3.1 (Applied Biosystems catalog # 4336697), 1 μM of primer, and 0.2 μg of plasmid DNA in a final reaction volume of 10 μl. The cycle parameters were 95°C for 3 min; 35× (96°C for 10 s; 55°C for 15 s; 60°C for 3 min) followed by 72°C for 7 min. Sequence was determined at the University of Wisconsin Biotech Center DNA Sequencing facility.
DNA gel blot analysis
Genomic DNA (5 μg) was digested with 20 U of restriction enzyme under manufacturer’s specified conditions for 16 h. Digested genomic DNA was electrophoresed through 1% agarose TBE gels and transferred using the neutral transfer protocol supplied with Hybond N+ membranes (Amersham catalog # rpn203b) according to manufacturer’s instructions. After transfer, blots were UV crosslinked and baked for 2 h at 80°C. Prehybridization was performed with hybridization buffer (5× SSPE, 5× Denhart’s, 1% SDS, 100 μg/ml sheared salmon sperm DNA) for 2–5 h at 65°C. Hybridizations were performed in hybridization buffer at 65°C for 16 h with denatured [α-32P] dCTP (Perkin Elmer catalog # BLU513H) labeled probes. After hybridization, blots were stringently washed with 0.2× SSC, 0.2% SDS three to four times at 65°C for 20 min each.
Probe generation and labeling for DNA gel blot analysis
Segments of TCUP were PCR amplified from B73 genomic DNA, cloned using the pGEM®-T Easy Vector System II kit (Promega catalog # A1380), and confirmed by sequencing. The clones used as probes for DNA gel blots are as follows: TCUP5′ bp 9–875; TCUP internal bp 2203–3439; TCUP 3′ bp 3313–4127. Random labeling was performed with 25 ng of purified probe using the Prime-a-Gene 5× labeling buffer (Promega catalog # U1151).
Informatic and phylogenetic analysis
The TCUP sequence (GenBank Accession DQ324364) was completed by manual assembly of PCR sequences and sequence from trace archives for BAC clone ZMMBBc0310A01 (GenBank AC148160). The translated protein sequence of TCUP was analyzed for known domains using the BLAST-based NCBI conserved domain database (Marchler-Bauer and Bryant, 2004; Marchler-Bauer et al., 2005). The default settings were used for all searches. The position of this sequence in version 2 of the B73 reference genome (AGPv2; http://ftp.maizesequence.org/current/assembly/; Schnable et al., 2009) was determined in February 2011 on chromosome 7 corresponding to contig AC195167 derived from BAC clone ZMMBBc0310A01. The location of the insertion was confirmed by identification of the target site duplication (TSD) sequence AAGCAAAG.
Protein sequences were aligned using ClustalX (Thompson et al., 1997). The phylogenetic analysis was performed on Clustal aligned transposase protein sequences according to Xu and Dooner (2005). All transposases used in the alignment were derived from putative full-length elements except for the sorghum sequence translated from TC93360 which appears to be a partial transposase. The phylogenetic tree was created with the MEGA3 program (Kumar et al., 2004) using the neighbor joining method by performing 1000 bootstraps. The pairwise deletion method was selected to handle gaps in the alignments.
The accession numbers used in alignment are as follows: DQ324364 TCUP Zea mays, S13518 snapdragon Tam3, P08770 Maize Ac ORFA, AAV32822.1 Maize Mx, AAB60236.1 House Fly Hermes, AAD24567.1 Arabidopsis thaliana Tag2, BAA36225.1 Ipomoea purpurea Tip100, CAA66182.1 N. tabacum Slide, AAL93203.1 Bactrocera dorsalis Hopper, CAA93759.1 Tolypocladium inflatum Restless, P12258 D. melanogaster Hobo, BAB01787.1 A. thaliana Tip100, TC93360 Sorghum bicolor TCUP-like, BAD93710.1 Oryza sativa TCUP-like, P0024G09 O. sativa TCUP-like S, AP002071 O. sativa TCUP-like L, 2021344A P. glaucum Pac2, CAB82966.1 A. thaliana, NP191811.1 A. thaliana, AAL31650.1 O. sativa, BAA96580.1 O. sativa, AAM97760.1 O. sativa, NP920480.1 O. sativa, NP188098.2 A. thaliana, AF283502 C. parasitica Crypt1, BAA32244 F. oxysporum Tfo1, AAA64851 L. cuprina Hermit, AAD03082 B. tryoni Homer, XP309253.2 A. gambiae, NP192758.1 A. thaliana.
RNA extractions
RNA was extracted using the TRIZOL reagent (Invitrogen catalog # 15596-026) following the manufacturer’s protocol with optional spin steps included for plant tissue. The callus RNA extraction protocol had the following modification: callus RNA was extracted using an extra partitioning with chloroform to reduce phytogel contamination in the RNA pellet.
Reverse transcriptase-PCR
Total RNA (5 μg) was DNAse treated [1× RQ1 RNase-Free DNase 10× buffer, 2 U RQ1 RNase-Free DNase (Promega catalog # M6101)] for 20 min at 37°C. The cDNA was primed with Oligo (dT)15 (Promega catalog # C1101). Reverse transcription was carried out using 1× M-MLV reaction buffer, 0.5 mM dNTPs (Promega catalog # U1330), 200 U M-MLV reverse transcriptase (Promega catalog # M1701), 20 U RNAsin (Promega catalog # N2511). The reaction was incubated at 42°C for 2 h and then heated to 70°C for 10 min to stop the reaction. PCR was performed using the following conditions [1× Tth Buffer (Epicentre catalog # TTH72250), 2 mM MgCl2, 0.4 μM primers, 3× Masteramp enhancer, 1 U Tth DNA polymerase] on 3–6 μl of cDNA and cycled 25 times (94°C for 35 s, 58°C for 45 s, 72°C for 60 s). The 2.15-kb fragment was amplified using primers TCUP3′RACE2 ACGATGAAACTGTTGAGATCGACTGC and TCUP3′rev.1 ACCACATCAGACTTCAATCTCCTCCTC.
5′ and 3′rapid amplification of cDNA ends
Rapid amplification of cDNA ends (RACE) was performed using materials and the protocol from the BD SMART™ RACE cDNA Amplification Kit (BD Biosciences catalog # 634914). The PCR amplicons from nested PCR were isolated using the QIAquick Gel Extraction Kit (Qiagen catalog # 28704) and cloned using the pGEM®-T Easy Vector System II kit (Promega catalog # A1380). The TCUP specific primers used for 5′ RACE were 5′ TCUP RACE GGTCTAAGCAACCTCATGGCAGCAGCA, TCUP2R.5′TCCACCTAACCTCCATGTCCAAC, TCUP5′RACE1 ACAAGTTAGGAGGGACAACCCTGTTCA, TCUP5′F-CL307541 CATGTCTCTGGTGGTGGTTTGC, and TCUP5′RACE3 CCTCTGATGTGAGTGTCATTAGCATAGTTC. The TCUP specific primers used for 3′ RACE were TCUPRACE3′ GTGAGTTTGTGCTGATGCCACCAGTGCAAG, TCUP3′RACE1 TCCTAGCTCCGGTCTCTCCTAGCTCT, TCUP3′RACE2 ACGATGAAACTGTTGAGATCGACTGC, and TCUP3′RACE3 CTTACTCCGTTGAGGAACCGGGACT.
Transposon display
Transposon display was performed following the protocol described by Casa et al. (2004) using genomic DNA isolated from long-term callus lines and inbred plant DNA. The TCUP specific primers used for transposon display were disTLE3′-1 ACTGGTAGTGCCGTGCCTGG, disTCUP3′-1nest AGTGCCGTGCCGTGCGGCCCGGTT, disTCUP5′-1 TGCCAGTGCCGTGCTTTTTAGTG, and disTCUP5′-1nest GTGTTAGTGCCATTTGGCCA.
Results
Identification and characterization of TCUP
Tissue Culture Up-Regulated was first identified as TC249207 by evaluation of EST abundance in BMS suspension cultures using the TIGR gene index. This was the most abundant transcript from BMS and there was no evidence of expression in any other tissue based on this gene index. This observation led to us to name the element as TCUP. Expression of TCUP in BMS, newly initiated Hi-II callus cultures, seedling leaf, seedling root, and developing endosperm was experimentally evaluated by RT-PCR and TCUP transcription was not detected in seedling, leaf, root, endosperm, and newly initiated untreated callus. These evaluations supported that at least one member of the TCUP family is actively transcribed in BMS callus cultures, and that TCUP is stably quiescent in the other plant tissues as evaluated by EST and RT-PCR.
A full-length TCUP sequence (GenBank accession DQ324364) was assembled and confirmed using informatics and targeted sequencing (Figure 1). The sequence is 4132 bp in length and contains a single intronless 738 amino acid open reading frame. The AGP version 2 position of this sequence is on chromosome 7, but bridges several contigs AC195167.2-contig69, AC195167.2-contig72, and AC195167.2-contig36 within the unordered BAC clone ZMMBBc0310A01. The assembly of these contigs does not produce a contiguous sequence suggesting that the assemblies utilized for AGP version 2 of this BAC clone may not be correct. An alternate assembly for BAC clone ZMMBBc0310A01 was identified (AC148160.3) that contains a contiguous sequence which is identical to the TCUP sequenceDQ324364 except for 4 bp.
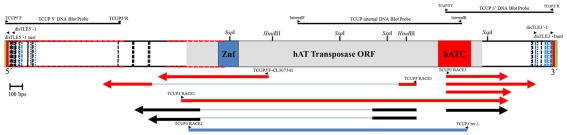
Figure 1.
Diagram of the of the TCUP sequence DQ324364.1 drawn to scale. The DNA sequence of the predicted hAT transposase ORF is represented as a gray box, the Znf-BED domain is represent by a blue box, and the hATC domain is represent with a red box. The TSD sequences (AAGCAAG) are presented as green bars at the 5′ and 3′ ends of the DNA sequence. The TIR sequence [TATAG(T/A)TGGCCAA] is represented by red bars at the 5′ and 3′ end of the sequence. The 3′ and 5′subterminal repeat regions containing 39 repeats of the sequence CGGCAC/CACGGC are represented by dotted lines with repeats in the sense orientation colored black and repeats in the antisense orientation colored light blue. The CpG islands are represent as red dotted lines along the outline of the DNA sequence. The cut sites of restriction enzymes utilized in the DNA gel blot analysis in Figure 2 are noted on the diagram (SwaI and EcoRI restriction enzyme do not cut within the TCUP sequence) and the location of the DNA gel blot probe sequences are represented as lines above the diagram. Primers used for RT-PCR related experiments and transposon display are represented as small arrowheads. RT-PCR and RACE related transcripts are represent below the DNA sequence with those derived from azadC treated callus cultures colored red while BMS derived sequences are colored black. Sequenced 5′ RACE clones are represented by arrows facing the 5′ direction and 3′ RACE clones are represented by arrows facing in the 3′ direction. The 2.15-kb predicted RT-PCR amplicon derived from the primers utilized in Figure 6 is represented as a blue bar underneath the sequenced RACE clones.
Protein alignments and conserved domain analysis were used to characterize the predicted translation products of the TCUP sequence. This search revealed homology to the hAT family of transposons. The specific domains contained in TCUP include a ZnF-BED domain, and a hATC domain. The ZnF-BED domain is a zinc finger motif responsible for DNA binding and is present in many types of TEs. This domain is required for terminal inverted repeat (TIR) and subterminal repeat binding and gives the unique target specificity to the transposase (Becker and Kunze, 1997). The hATC domain is present in all hAT transposases, and is involved in dimerization of the transposase. This domain is required for proper transposase function (Essers et al., 2000). These two domains, which are required for autonomous transposition, are present in the transposases Ac and Tam3 along with other transposases in the hAT family.
The TCUP sequence is flanked by an 8-bp TSD with perfect identity, which is a characteristic of hAT transposons. The sequence also contains 13 bp imperfect TIRs that vary in sequence at the sixth base pair where there is a T in the 5′ TIR and an A at the 3′ TIR. The TIR sequences are not homologous to any previously reported transposable element indicating that this is a member of a novel, previously uncharacterized family of TEs. Analysis of the GC content of TCUP revealed a 60–80% GC content at the 5′ UTR and regions proximal of TIRs containing two CpG islands at 47–890 and 1216–1659 bp that is similar to other characterized hAT transposons. GC rich regions in TEs have been shown to be important sites of DNA methylation-dependent transcriptional regulation (Chomet et al., 1987; Kunze and Starlinger, 1989; Brutnell and Dellaporta, 1994; Brutnell et al., 1997).
Subterminal repeats are repetitive sequence motifs that are required for transposase binding that follow the TIR and are found in autonomous and non-autonomous TEs. In most transposons, subterminal repeats range in size from 5 to 12 bp, are present within inverted repeats, and can occur five to more than 50 times in some elements (Kunze and Weil, 2002). We therefore sought to identify subterminal repeats in TCUP. A CGGCAC motif was identified which is present 21 times in both forward and reverse complement orientations in the subterminal regions. The reverse of this repeat, CACGGC, is also present 18 times in both the forward and reverse complement orientations. Based on its abundance and location in the element, the CGGCAC/CACGGC sequence is the most likely candidate for a transposase binding site.
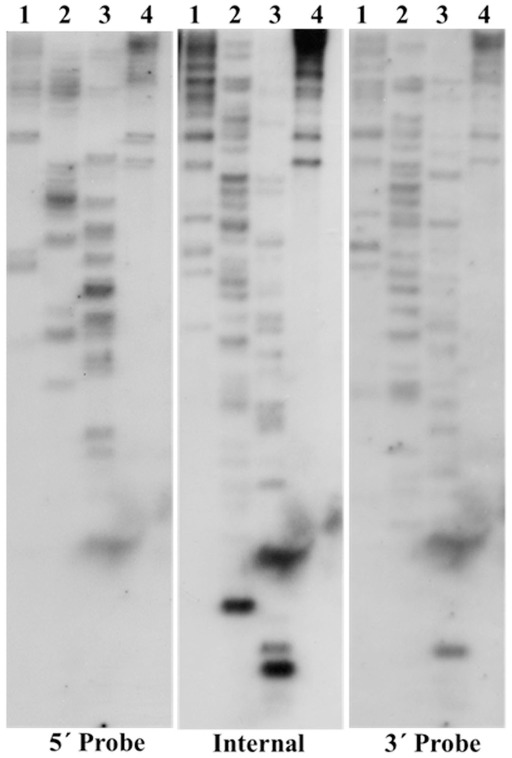
The copy number of TCUP was assessed by BAC library screening and DNA gel blot analysis using 5′, 3′, and internal segments of TCUP as probes, and by informatics analysis of the AGP version 2 genome assembly. The ZMMBBb BAC HindIII B73 library (Tomkins et al., 2002) covering 14 genome equivalents was screened by hybridization using the TCUP internal probe. The results from the BAC library screen suggest that an estimated 17 copies (233 positive BACs/14 genome equivalents) that hybridize to the TCUP internal sequence are present in the B73 genome. The banding pattern of DNA gel blots digested with the restriction enzymes EcoRI, HindIII, SspI, and SwaI detected between 13 and 16 partial copies using TCUP5′, TCUP internal, or TCUP3′ probes (Figure 2). Further analysis of fragment size and band intensity on the DNA gel blots provided an estimate of three to five putative full-length elements based on the presence of an equivalent sized band across all probes. Evaluation of the AGP version 2 assembly using a minimum e-value cut off of 1e-10 and minimum identity of 75 identified 70 (DQ324364.1) or 64 (TCUP cds) fragments in the genome that are greater than 500 bp without another fragment within 3000 bp of said fragment. Of these fragments, two putative autonomous elements on chromosomes 7 and 8, and 6 putative non-autonomous, but nearly full-length, elements on chromosomes 1,3,4, 9, and 10 were identified in the maize genome based on the presence of TIRs and TSD sequences. The putative autonomous elements contained uninterrupted TCUP transposase ORFs (Table 1). The number of putative autonomous elements identified by in silico analysis of the maize genome agrees with the number of sequences observed through DNA gel blot analysis (Figure 2).
Figure 2.
DNA gel blot investigating TCUP copy number. B73 genomic DNA was digested with four different restriction enzymes. Each panel is the same DNA gel blot hybridized with different TCUP probes (see Materials and Methods). Lanes 1, EcoRI, 2, HindIII, 3, SspI, 4, SwaI.
Table 1.
Summary of autonomous TCUP elements in the maize genome (AGPv2).
| Sequence | 5′ TIR | 3′ TIR | TSD | Chr | 5′TIR Start | 3′ TIR Stop | Length |
|---|---|---|---|---|---|---|---|
| DQ324364* | TATAGTTGGCCAA | TTGGCCATCTATA | AAGCAAAG | 7 | 4137 | ||
| TCUP-7a1 | TATAGTTGGCCAA | TTGGCCATCTATA | CCTCGCGG | 7 | 115988344 | 115992684 | 4341 |
| TCUP-8a1 | TATAGaTGGCCAA | TTGGCCAaCTATA | TCAGTGGG | 8 | 170041903 | 170046049 | 4147 |
*TCUP sequence determined by targeted sequencing and BAC information prior to whole genome assembly. Position in AGPv2 does not represent actual length due to assembly of BAC ZMMBBc0310A01 which is inconsistent with our sequence.
TIR, terminal inverted repeat; TSD, target site duplication; Chr, chromosome.
Phylogenetic analysis reveals related sorghum and rice TCUP homologs that are expressed in cultured cells and regenerated plants, respectively
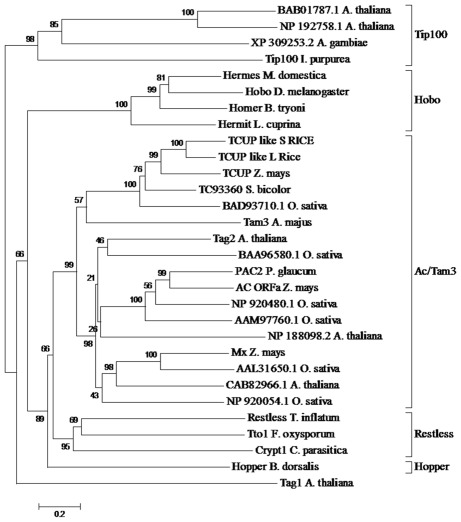
Phylogenetic analysis was performed on the putative TCUP transposase to examine its relationship to a diverse set of hAT transposases (Figure 3). Sequences included in this alignment were chosen to sample the diversity of hAT elements present in both plant and animal genomes (Xu and Dooner, 2005). An alignment of hAT transposases was used to determine the relationship of TCUP to groups of transposases within the hAT superfamily of elements. The interpretation of the grouping of the transposases was based on Xu and Dooner (2005) in which the authors surveyed a large sample of hAT transposases and categorized them into five groups (Figure 3). This analysis places TCUP within the Ac/Tam3 group of transposases in the hAT family. The TCUP transposase clusters with a group of uncharacterized sequences from rice and sorghum that are most closely related to Tam3.
Figure 3.
Phylogenetic analysis of hAT transposases from multiple genera. The accessions chosen for this tree were based on previous analysis performed by Xu and Dooner (2005). The different clades of hAT transposases are marked with brackets. The values in each bracket represent percentage of times the clade is present among 1000 bootstraps.
EST distribution for the most closely related sequences was evaluated in the TIGR Gene Indices (ZmGI release 15.0). Interestingly, the most closely related sorghum sequence TC93360 (SbGI release 8.0) is represented only by ESTs from tissue culture and the most closely related rice (O. sativa japonica) sequences TC253370 and TC267041 (OsGI release 16.0) are represented only by ESTs from regenerated plants. This suggests that this family of transposons may have a conserved sensitivity across grasses for activation under the conditions imposed by tissue culture.
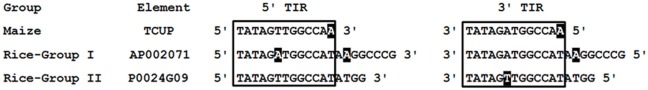
The rice (O. sativa japonica) genomic sequence at NCBI was queried to identify putative TCUP homologs. Seventeen different rice sequences with amino acid homology to the TCUP transposase were identified. The rice transposases, when compared to the maize TCUP element’s predicted hAT transposase ORF, had 53 and 54% identical and 67 and 72% conserved residues. The identified rice sequences were separated into two distinct groups based on protein and nucleotide sequences. The two groups of rice transposases were 71% identical and had 82% conserved amino acid residues relative to each other. Group I is comprised of six sequences 4450 bp in length encoding a 756 amino acid putative transposase. Group II contains 11 sequences 3916 bp in length encoding a 763 amino acid putative transposase. Analysis of the rice transposases found that several elements from each group still encode uninterrupted open reading frames containing Znf-BED and hATC domains. The majority of these sequences however appeared to be derived from immobile elements lacking the necessary sequence motifs for transposition or non-autonomous transposons that no longer contain transposase ORFs. The terminal regions of the Group I and Group II sequences are very similar with Group I possessing 20 bp perfect TIRs and Group II having 17 bp perfect TIRs. The TIRs from these two groups of elements are also most identical and vary at two base pairs along their regions of homology (Figure 4). A 12-bp core region of the TIRs is conserved among the rice and maize TCUP elements (Figure 4). This core region has an A/T variation at the sixth base pair between the different groups of rice elements that is at the same position as the A/T variation in the imperfect TIRs of TCUP.
Figure 4.
Comparison of maize TCUP and rice TCUP-like TIRs. Rice elements are grouped according to size. TIR sequences identical to the maize TCUP sixth position are in bold. The core 12 bp of similarity is boxed. Sequences not matching the consensus are shaded in black.
The TCUP transposase protein sequence was used to search the genome sequences of S. bicolor (sorgum), Setaria italica (foxtail millet), Populus trichocarpa (poplar) and A. thaliana for closely related sequences. If a protein sequence was identified with at least 40% homology to the maize TCUP transposase, the surrounding 10 kb of DNA sequence was evaluated to identify TIRs. TCUP-like sequences were identified in sorghum and millet, but not in poplar (JGI assembly release 1.0) and A. thaliana (TAIR9 assembly). TCUP sequences encoding putative transposase ORFs with up to 79% identity and 87% conserved amino acids was identified in the sorghum genome (Sbi1.4) assembly on chromosomes 5, 8, 9, and 10. These sequences were evaluated and contained a putative TIR sequence that was identical to the maize TCUP TIR. The foxtail millet genome assembly (JGI assembly release v1.0) also contained TCUP-like sequences encoding putative transposase ORFs with up to 79% identity and 88% conserved amino acids.
TCUP expression is increased by treatment with the DNA methylation inhibitor 5-aza-2-deoxycytidine and multiple isoforms are expressed
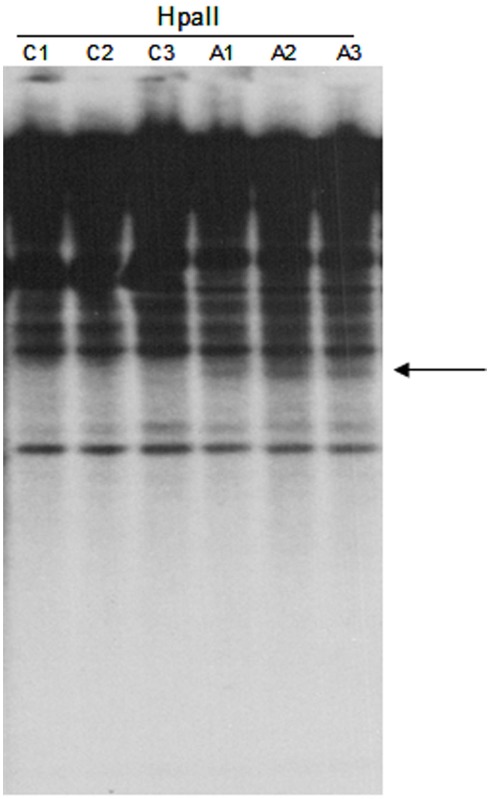
Epigenetic derepression of transposons has been implicated as a mechanism underlying transposon activation. We conducted an experiment using the DNA methyltransferase inhibitor, 5-aza-2-deoxycytidine (azadC) to test whether this treatment can activate TCUP. Previous studies across various organisms (maize by Pan and Peterson, 1989, snap dragon by Martin et al., 1989, and human cell lines by Davis et al., 1989; Neidhart et al., 2000) provide a precedent for using azadC to activate TEs through its inhibitory action on DNA methylation. Suspension cell cultures were prepared from newly initiated Hi-II callus cells. The suspension cultures were split into two groups, a control group and a 5-aza-2-deoxycytidine (azadC) treatment group with three replicates per treatment group (Figure 5). The effectiveness of the 5-aza-2-deoxycytidine treatment in reducing DNA methylation was verified using a DNA gel blot of HpaII digested DNA probed with a 5S rDNA probe (Figure A1 in Appendix). DNA gel blots containing genomic DNA of azadC treated callus cultures were used to assess changes in DNA methylation of TCUP sequences caused by the azadC treatments relative to untreated callus. A reduction in CpG DNA methylation at the 5S locus was observed in the HpaII digested DNA from the azadC treated cultures but not in the untreated control cultures. A reduction in methylation was detected in HpaII digested DNA but the band is faint indicating that either a portion of cells in the callus cultures were demethylated and/or that a specific copy of TCUP was demethylated (Figure A2 in Appendix).
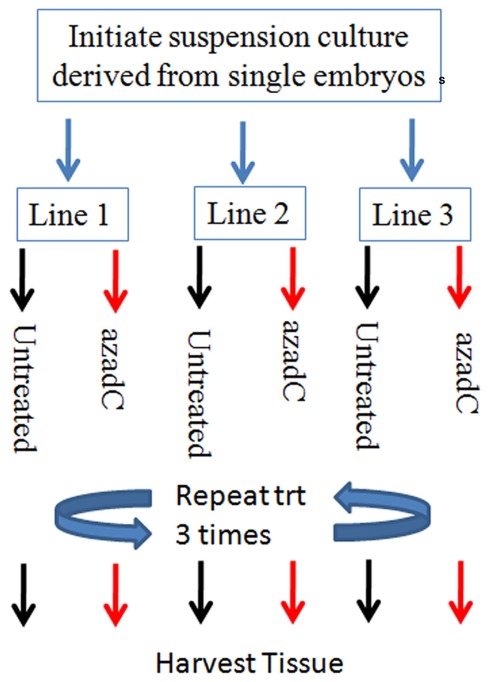
Figure 5.
Diagram of the experimental design used for the 25-μM 5-aza-2-deoxycytosine treatment experiments. Biological replicates consist of three suspension cultures initiated from three independent Hi-II A × B explants. Each suspension culture line was initiated with 1.5 g of Hi-II A × B type II embryogenic tissue and allowed to bulk. The three cultures were each split into two flasks, one treated 25 μM 5-aza-2-deoxycytosine and the other left untreated. The liquid medium was replaced with either media supplemented with 25 μM 5-aza-2-deoxycytosine or untreated medium every 3 days for 9 days to ensure an adequate length of treatment.
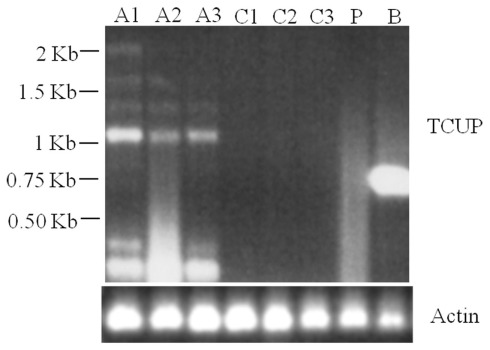
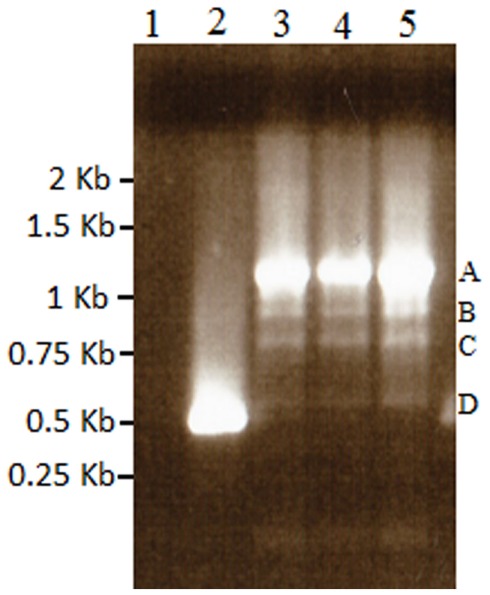
RT-PCR analysis of RNA isolated from suspension cultures treated with azadC, untreated suspension cultures, long-term cultured BMS callus, and whole plant seedlings was used to assess TCUP expression. The initial experiments to test for induction by azadC treatments yielded multiple transcripts in azadC treated cultures and a single transcript in BMS cultures. To further characterize these transcripts and identify if a transcript containing an intact ORF was produced, both 5′ and 3′ RACE were performed on treated suspension cultures and long-term BMS callus to identify transcription start sites and polyadenylation sites. DNA sequencing of cloned RACE PCR products identified the presence of multiple transcripts in azadC treated suspension cultures with at least three different polyadenylation sites at 3785, 3810, and 3994 bps, and an abundant BMS transcript with a polyadenylation site at 3776 bps of the TCUP transposon sequence DQ324364 was identified (Figure 1; Figure A3 in Appendix). Based on the results of the sequenced RACE PCR products, primers were designed to common sequences of the azadC and BMS RACE transcripts. RT-PCR was again performed on the suspension culture treated with azadC, untreated suspension cultures, long-term cultured BMS callus, and whole plant seedlings to confirm the observed TCUP expression pattern (Figure 6). These results indicate that some proportion of the transcripts produced in the azadC treated cell cultures are related to transcripts capable of encoding a functional transposase while the major transcript present in the long-term BMS cell culture contain an internal deletion and do appear capable of producing function TCUP transposase.
Figure 6.
RT-PCR demonstrating multiple TCUP transcripts present in azadC treated callus. Six separate transcripts are detected with a primer pair designed to amplify a 2.15-kb internal region of the TCUP coding sequence. Actin is shown as a control. Abbreviations: A1–A3, independent 25 μM azadC treated Hi-II A × B tissue culture cell lines; C1–C3, independent untreated Hi-II A × B tissue culture cell lines; P, Hi-II A × B seedling; B, BMS callus.
Evidence of TCUP transposition
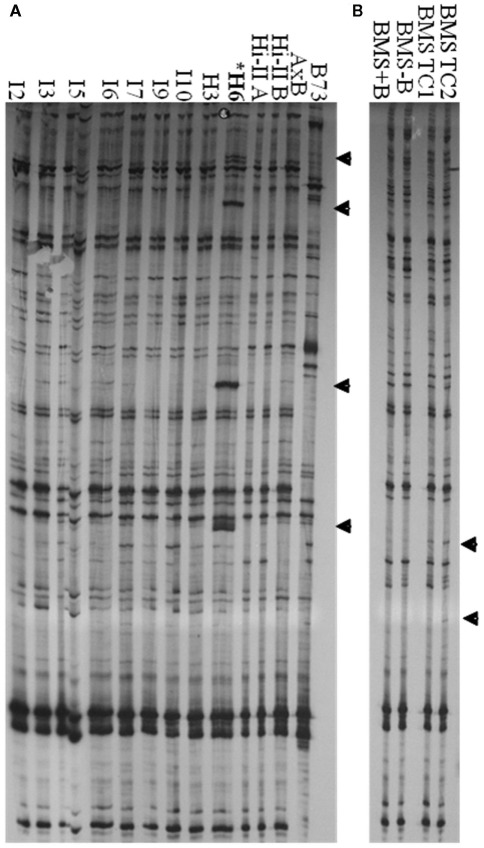
Tissue Culture Up-Regulated transcription was detected in long-term maize cell lines. We therefore sought to determine if TCUP also transposed in long-term cultured cells. Several independent Hi-II A × B cell lines ranging in age from 2 to 3 years following initiation and a 28-year long-term BMS cell line were analyzed for transposition using transposon display. Since cultured cells contain two homologous chromosomes and may be a mosaic of cell types, it is most likely that transposition events will appear as new bands without loss of existing bands. Transposon display using 3′ specific primers detected an increase of four copies of TCUP in line H6 and an increase in copy number in BMS tissue culture (Figure 7). New insertions were not detected in callus lines I2, I3, I5, I6, I9, I10, H3, and seedlings from genotypes Hi-II-A, Hi-II-B, Hi-II-A × B, B73, BMS + B chromosomes, BMS − B chromosomes. Transposon display using the 5′ specific primers did not detect any changes or mobility of TCUP. The new copies of TCUP were present in all three H6 culture biological replicates, indicating that the transposition likely occurred near the time of culture initiation. It is not possible to determine the timing of transposition in the BMS cell lines.
Figure 7.
3′ TCUP transposon display performed on genomic DNA from Hi-II A × B callus lines, B73 seedlings, BMS seedlings, and BMS callus. (A) Independent Hi-II A × B callus lines compared to Hi-II A × B and B73 seedlings. DNA from biological replicates was used to confirm the consistency of the transposon display results. Tissue culture lines are represented by I or H followed by the biological replicated number. Samples within each tissue line were: I2-1 and I2-5, I3-1 and I3-5, I5-2 and I5-6, I6-2, I6-3, and I6-4, I7-1 and I7-2, I9-2 and I9-3, I10-2 and I10-3, H3-3 and H3-6, H6-2, H6-3, and H6-4. (B) BMS seedling tissue from lines with and without B chromosomes compared to BMS callus lines. Novel bands are marked with an arrow and line H6 is marked with an asterisk.
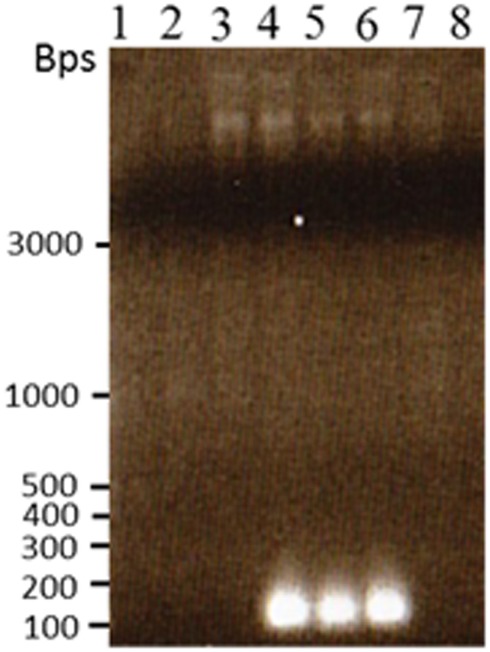
One of the novel bands in the H6 lines was confirmed to be novel TCUP insertions by excising the novel bands, reamplifying the PCR product, and sequencing the cloned PCR products. The sequenced clone contained 35 bps of TCUP sequence containing the primer sequence and 11 bps of TCUP sequence that corresponded to the TIR except for a C/T transversion at the fifth position in lines H6-2, H6-3, and H6-4 and 222 bp of genomic DNA sequence. Primers were designed from the flanking genomic sequence and PCR was performed using both the 3′ primary and nested TCUP specific primers. Amplification was detected in H6 lines but not in B73, Hi-A × B, BMS callus, and two independent Hi-II A × B callus lines with both primers indicating the uniqueness of this insertion (Figure 8).
Figure 8.
Reamplification of the novel TCUP transposon site that was cloned and sequenced from the transposon display gel fragments. PCR was performed on undigested genomic DNA to confirm the novel sequence was present in H6 genomic DNA and not in other callus or plant genomics DNA. Lane 1 B73 seedling, lane 2 Hi-II A × B seedling, lane 3 Hi-II A × B callus line H3-6, lane 4 Hi-II A × B callus line H6-2, lane 5 Hi-II A × B callus line H6-3, lane 6 Hi-II A × B callus line H6-4, lane 7 Hi-II A × B callus line I9-9, lane 8 BMS callus.
Discussion
TCUP is a novel maize hAT transposon
Tissue Culture Up-Regulated expands the list of TEs that are active in maize and is different from previously reported transposons as indicated by its novel TIRs. Activation of transposons by tissue culture is a common occurrence in plant cell culture, but does not affect all transposons equally. The reactivation of the Ac and Spm transposons occurred at a frequency of 3 and 1% in regenerated plants (Peschke et al., 1987, 1991). In rice, the Ping transposase is activated in anther culture leading to a 20% increase in the transposition frequency of the MITE mPing, which could be one of the sources leading to increased mutation rates in the rice haploid breeding program (Kikuchi et al., 2003). The rice retrotransposon, Tos17, is the transposon with highest known activity in tissue culture with almost every regenerated plant containing 5–30 new copies of the Tos17 element (Hirochika et al., 1996). The TCUP element was found to have transposed in one Hi-II A × B cell line out of the nine screened, and had also transposed in BMS callus indicating that it may be activated at a higher frequency in maize tissue culture than Ac or Spm elements.
Epigenetic changes associated with tissue culture may lead to TCUP activation
Epigenetic changes are associated with the tissue culture process and have been implicated as a significant source of somaclonal variation. Epigenetic changes are reversible alterations in phenotype or gene expression that are stably inherited through mitosis or meiosis and not due to changes in primary DNA sequence. The mechanisms that cause the epigenetic repression or cycling of gene expression have been attributed to DNA methylation, RNA interference, and modification of histone tails. These epigenetic pathways are critical in repressing the transcription of repetitive sequences such as TEs (Chomet et al., 1987; Banks et al., 1988; Lippman et al., 2003, 2004; Zilberman et al., 2003). DNA methylation and RNA interference pathways are linked in plants by RNA dependent DNA methylation that is involved with the regulation of heterochromatin and gene silencing (Mette et al., 2000). Transcriptional repression of TEs, such as class 2 transposons, is associated with the production of 24 nt sRNAs, DNA methylation of the corresponding genomic sequence, and H3K9me2 (Lippman et al., 2004; Zilberman et al., 2007; Tanurdzic et al., 2008; Cantu et al., 2010).
Changes in the pattern of DNA methylation are thought to be a major source of epigenetic variation leading to somaclonal variation (Kaeppler and Phillips, 1993; Phillips et al., 1994; Kaeppler et al., 2000). Tissue culture induction is known to cause a general shift toward a hypomethylated genome (Kaeppler and Phillips, 1993; Koukalova et al., 2005; Law and Suttle, 2005) likely through the loss of DNA methylation of repetitive sequences within heterochromatin rather than genic sequences (Tanurdzic et al., 2008). The age of a culture is also associated with increased somaclonal variation which may be due in part to a gradual overall demethylation of low copy sequences and subsequent activation of TEs (Fukui, 1983; Hirochika et al., 1996). Increased transpositional activation of Ac is associated with tissue culture induced demethylation and increased with tissue culture age (Dennis and Brettell, 1990). The observation of TCUP transcription in long-term BMS cell cultures and azadC treated cell cultures is consistent with the role of transposons in somaclonal variation.
Both class 1 retrotransposons and class 2 transposons are known to become active from biotic and abiotic stresses. Some of the class 2 transposons known to be activated by tissue culture are Ac (Peschke et al., 1987), Spm (Peschke et al., 1991), Slide (Grappin et al., 1996), and Ping (Kikuchi et al., 2003). Retrotransposons are also activated by tissue culture stress and the majority of TEs known to be activated by tissue culture fall into this group. These activation events are associated with changes in DNA methylation. They include tobacco elements Tnt1 (Grandbastien et al., 1989) and Tto1 (Hirochika, 1993), and the rice elements Tos17 (Hirochika et al., 1996) and Karma (Komatsu et al., 2003).
The activation of TEs could be one consequence of altered DNA methylation patterns. Important interactions occur between transposases and methylated cytosine residues in transposon regulatory sequences. This is true of both Ac/Ds and En/Spm elements, where transpositional activity is dependent on the level of DNA methylation (Chomet et al., 1987; Banks et al., 1988). The transpositional activity of Ac is strongly correlated with DNA methylation present at the 5′ TIR and several different Ac alleles exist because of the different cytosine residues methylated in the subterminal repeat region of the element (Chomet et al., 1987; Brutnell and Dellaporta, 1994; Brutnell et al., 1997). Changes in DNA methylation, often observed as a decrease, occur as a result of the tissue culture process (Kaeppler and Phillips, 1993). Since TCUP was first discovered as a transcribed element in long-term tissue-cultured cell lines, possibly by the demethylation associated with tissue culture age, we used pharmacological treatments to reduce DNA methylation in young Hi-II A × B callus and test if induction of TCUP transcription may occur within a hypomethylated genomic environment.
AzadC and its analog 5-azacytidine (azaC) have been used to activate TEs in maize (Pan and Peterson, 1989), snap dragon (Martin et al., 1989), and human cell lines (Davis et al., 1989; Neidhart et al., 2000) through its inhibitory action on DNA methylation. Treatment with azaC was associated with new excision, integration, and/or demethylation events of endogenous Tam3 element in snap dragon (Martin et al., 1989) and activation of transcript of L1 elements in human fibroblasts treated with azadC (Neidhart et al., 2000). We hypothesized that azadC treatment would cause a general decrease in DNA methylation mimicking the effect of tissue culture age, which is associated with hypomethylation, and activate TCUP in young cell cultures. AzadC treatments caused transcription of a quiescent TCUP in treated Hi-II A × B recently cultured suspension cell lines and not in untreated controls. Further research is necessary to characterize the role of DNA methylation in TCUP expression. However, it is well established that DNA methylation plays an important role in regulating expression and movement of other class II TEs including Ac (Dennis and Brettell, 1990), Spm (Banks et al., 1988), and Mu (Chandler and Walbot, 1986) and class I retrotransposons (Hsiao et al., 1986; Hirochika et al., 2000). At least four different transcripts are detected by RT-PCR, which could be related to transcriptional activation of both autonomous and non-autonomous TCUP elements through the genome wide demethylation caused by the azadC treatments. It has been observed that crossing an active Ac element into a background with silent Ds elements will induce transcription of the Ds elements (Kunze and Weil, 2002). Treatment with azadC may have activated a silenced TCUP element capable of trans activating other TCUP sequences creating the large number of different transcripts.
In summary, we have identified a novel hAT transposon with homology to Tam3 that exhibits differential transcription in long-term maize tissue-cultured cell lines. The element is also transcribed in newly initiated cultures in response to treatment with the DNA methylation inhibitor, 5-aza-2-deoxycytidine, when compared to untreated cultures. Mobility of TCUP was demonstrated in two independent long-term cell lines. TCUP has close relatives in rice and sorghum and EST evidence supports expression associated with tissue culture in these species. This research implicates TCUP and its related elements in rice and sorghum as a novel family of transposons that are highly sensitive to the tissue culture environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Nathan Springer for technical assistance and scientific discussions.
Appendix
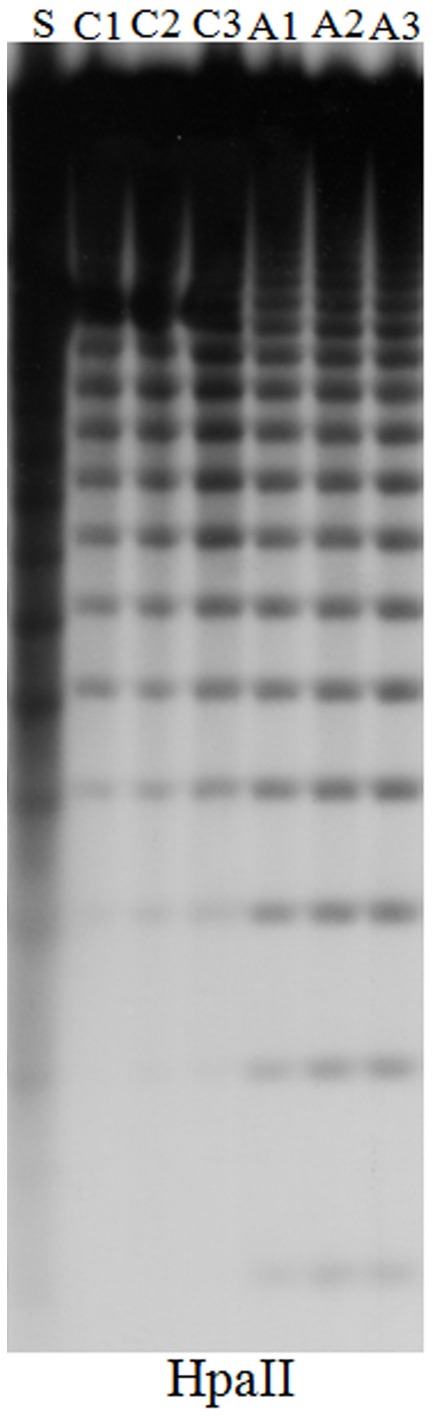
Figure A1.
DNA gel blot comparing DNA methylation at the 5S ribosomal DNA loci in untreated and 5-aza-2-deoxycytidine treated Hi-II A × B tissue-cultured cell lines. The plant and callus DNA was digested with HpaII to examine and CpG methylation respectively. An increase in number and intensity of low molecular weight bands in the HpaII digested DNA indicates a decrease in DNA methylation. Hi-II A × B genomic DNA isolated from 14-day-old seedlings (S), from independent untreated control cultures (C1–C3), from independent 5-aza-2-deoxycytidine cultures (A1–A3).
Figure A2.
DNA gel blot of untreated and 5-aza-2-deoxycytidine treated Hi-II A × B suspension cultures hybridized with the TCUP internal probe. HpaII digested genomic DNA of three independent untreated cultures (C1–C3) and three independent 5-aza-2-deoxycytidine treated cultures (A1–A3). An arrow marks the location of novel bands present in the HpaII digested DNA.
Figure A3.
Results of nested 3′ RACE demonstrating the single BMS transcript in lane 2, and azadC transcripts labeled A–D (lanes 3–5). Lane 1 is a ddH2O negative control. These bands were cloned and sequenced to identify the polyadenylation sites present in the TCUP transcripts. Sequencing of these PCR products along with 5′ RACE products were used to generate the primers used to amplify the transcripts in Figure 6.
References
- Armstrong C. L. (1994). “Regeneration of plants from somatic cell cultures: applications for in vitro genetic manipulation,” in The Maize Handbook, eds Freeling M., Walbot V. (New York, NY: Springer-Verlag; ), 663–671 [Google Scholar]
- Armstrong C. L., Green C. E., Phillips R. L. (1991). Development and availability of germplasm with high type II culture formation response. Maize Genet. Coop. Newslett. 65, 92–93 [Google Scholar]
- Banks J. A., Masson P., Fedoroff N. (1988). Molecular mechanisms in the developmental regulation of the maize suppressor-mutator transposable element. Genes Dev. 2, 1364–1380 10.1101/gad.2.11.1364 [DOI] [PubMed] [Google Scholar]
- Barret P., Brinkman M., Beckert M. (2006). A sequence related to rice Pong transposable element displays transcriptional activation by in vitro culture and reveals somaclonal variations in maize. Genome 49, 1399–1407 10.1139/g06-109 [DOI] [PubMed] [Google Scholar]
- Becker H. A., Kunze R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254, 219–230 10.1007/s004380050410 [DOI] [PubMed] [Google Scholar]
- Brutnell T. P., Dellaporta S. L. (1994). Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138, 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell T. P., May B. P., Dellaporta S. L. (1997). The Ac-st2 element of maize exhibits a positive dosage effect and epigenetic regulation. Genetics 147, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D., Vanzetti L. S., Sumner A., Dubcovsky M., Matvienko M., Distelfeld A., Michelmore R. W., Dubcovsky J. (2010). Small RNAs, DNA methylation and transposable elements in wheat. BMC Genomics 11, 408. 10.1186/1471-2164-11-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter R., Martin C., Coen E. (1987). Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol. Gen. Genet. 207, 82–89 10.1007/BF00331494 [DOI] [Google Scholar]
- Casa A. M., Nagel A., Wessler S. R. (2004). “MITE display,” in Mobile Genetic Elements: Protocols and Genomic Applications, eds Mille W., Capy P. (New York, NY: Humana Press; ), 175–188 [Google Scholar]
- Chandler V. L., Walbot V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. U.S.A. 83, 1767–1771 10.1073/pnas.83.6.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet P. S., Wessler S., Dellaporta S. L. (1987). Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6, 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. M., Constantinides P. G., van der Riet F., van Schalkwyk L., Gevers W., Parker M. I. (1989). Activation and demethylation of the intracisternal A particle genes by 5-azacytidine. Cell Differ. Dev. 27, 83–93 10.1016/0922-3371(89)90279-7 [DOI] [PubMed] [Google Scholar]
- Dennis E. S., Brettell R. I. (1990). DNA methylation of maize transposable elements is correlated with activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 217–229 10.1098/rstb.1990.0006 [DOI] [PubMed] [Google Scholar]
- Dooner H. K., Weil C. F. (2007). Give-and-take: interactions between DNA transposons and their host plant genomes. Curr. Opin. Genet Dev. 17, 486–492 10.1016/j.gde.2007.08.010 [DOI] [PubMed] [Google Scholar]
- Essers L., Adolphs R. H., Kunze R. (2000). A highly conserved domain of the maize activator transposase is involved in dimerization. Plant Cell 12, 211–224 10.1105/tpc.12.2.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui K. (1983). Sequential occurrence of mutations in a growing rice callus. Theor. Appl. Genet. 65, 225–230 10.1007/BF00308073 [DOI] [PubMed] [Google Scholar]
- Grandbastien M. A., Lucas H., Morel J. B., Mhiri C., Vernhettes S., Casacuberta J. M. (1997). The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100, 241–252 10.1023/A:1018302216927 [DOI] [PubMed] [Google Scholar]
- Grandbastien M. A., Spielmann A., Caboche M. (1989). Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337, 376–380 10.1038/337376a0 [DOI] [PubMed] [Google Scholar]
- Grappin P., Audeon C., Chupeau M. C., Grandbastien M. A. (1996). Molecular and functional characterization of slide, an Ac-like autonomous transposable element from tobacco. Mol. Gen. Genet. 252, 386–397 10.1007/s004380050242 [DOI] [PubMed] [Google Scholar]
- Hirochika H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Okamoto H., Kakutani T. (2000). Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12, 357–369 10.1105/tpc.12.3.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. U.S.A. 93, 7783–7788 10.1073/pnas.93.15.7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W. L., Gattoni-Celli S., Weinstein I. B. (1986). Effects of 5-azacytidine on expression of endogenous retrovirus-related sequences in C3H 10T1/2 cells. J. Virol. 57, 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Hirochika H., Eddy S. R., McCouch S. R., Wessler S. R. (2003). An active DNA transposon family in rice. Nature 421, 163–167 10.1038/nature01214 [DOI] [PubMed] [Google Scholar]
- Johns M. A., Mottinger J., Freeling M. (1985). A low copy number, copia-like transposon in maize. EMBO J. 4, 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler S. M., Kaeppler H. F., Rhee Y. (2000). Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43, 179–188 10.1023/A:1006423110134 [DOI] [PubMed] [Google Scholar]
- Kaeppler S. M., Phillips R. L. (1993). Tissue culture-induced DNA methylation variation in maize. Proc. Natl. Acad. Sci. U.S.A. 90, 8773–8776 10.1073/pnas.90.19.8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R., Tanskanen J., Immonen S., Nevo E., Schulman A. H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. U.S.A. 97, 6603–6607 10.1073/pnas.110587497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106 10.1038/ng1063 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Terauchi K., Wada M., Hirano H. Y. (2003). The plant MITE mPing is mobilized in anther culture. Nature 421, 167–170 10.1038/nature01218 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Tosa Y., Shimada S., Sogo R., Kusaba M., Sunaga T., Betsuyaku S., Eto Y., Nakayashiki H., Mayama S. (2001). OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 42, 1345–1354 10.1093/pcp/pce130 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Shimamoto K., Kyozuka J. (2003). Two-step regulation and continuous retrotransposition of the rice LINE-type retrotransposon Karma. Plant Cell 15, 1934–1944 10.1105/tpc.011809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukalova B., Fojtova M., Lim K. Y., Fulnecek J., Leitch A. R., Kovarik A. (2005). Dedifferentiation of tobacco cells is associated with ribosomal RNA gene hypomethylation, increased transcription, and chromatin alterations. Plant Physiol. 139, 275–286 10.1104/pp.105.061788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5, 150–163 10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Kunze R., Starlinger P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8, 3177–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze R., Weil C. (2002). “The hAT and CACTA superfamilies of plant transposons,” in Mobile DNA II, eds Craig N. L., Craigie R., Gellert M., Lambowitz A. (Herndon: VAASM Press; ), 997–1007 [Google Scholar]
- Law R. D., Suttle J. C. (2005). Chromatin remodeling in plant cell culture: patterns of DNA methylation and histone H3 and H4 acetylation vary during growth of asynchronous potato cell suspensions. Plant Physiol. Biochem. 43, 527–534 10.1016/j.plaphy.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A. V., Black M., Vaughn M. W., Dedhia N., McCombie W. R., Lavine K., Mittal V., May B., Kasschau K. D., Carrington J. C., Doerge R. W., Colot V., Martienssen R. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476 10.1038/nature02651 [DOI] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1, E67. 10.1371/journal.pbio.0000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., Cherukuri P. F., DeWeese-Scott C., Geer L. Y., Gwadz M., He S., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Marchler G. H., Mullokandov M., Shoemaker B. A., Simonyan V., Song J. S., Thiessen P. A., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196 10.1093/nar/gki069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 10.1093/nar/gkh454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Prescott A., Lister C., MacKay S. (1989). Activity of the transposon Tam3 in Antirrhinum and tobacco: possible role of DNA methylation. EMBO J. 8, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M. F., Aufsatz W., van der Winden J., Matzke M. A., Matzke A. J. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201 10.1093/emboj/19.19.5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H. D., Sutherland H. G., Martin D. I., Whitelaw E. (1999). Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314–318 10.1038/15490 [DOI] [PubMed] [Google Scholar]
- Neidhart M., Rethage J., Kuchen S., Kunzler P., Crowl R. M., Billingham M. E., Gay R. E., Gay S. (2000). Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 43, 2634–2647 [DOI] [PubMed] [Google Scholar]
- Neuffer M. G. (1966). Stability of the Suppressor element in two mutator systems at the A1 locus in maize. Genetics 53, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. B., Peterson P. A. (1989). Tagging of a maize gene involved in kernel development by an activated Uq transposable element. Mol. Gen. Genet. 219, 324–327 10.1007/BF00261195 [DOI] [PubMed] [Google Scholar]
- Peschke V. M., Phillips R. L., Gengenbach B. G. (1991). Genetic and molecular analysis of tissue-culture-derived Ac elements. Theor. Appl. Genet. 82, 121–129 10.1007/BF00226202 [DOI] [PubMed] [Google Scholar]
- Peschke V. M., Phillips R. L., Gengenbach B. G. (1987). Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science 238, 804–807 10.1126/science.238.4828.804 [DOI] [PubMed] [Google Scholar]
- Phillips R. L., Kaeppler S. M., Olhoft P. (1994). Genetic instability of plant tissue cultures: breakdown of normal controls. Proc. Natl. Acad. Sci. U.S.A. 91, 5222–5226 10.1073/pnas.91.12.5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planckaert F., Walbot V. (1989). Molecular and genetic characterization of Mu transposable elements in Zea mays: behavior in callus culture and regenerated plants. Genetics 123, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T. A., Minx P., Reily A. D., Courtney L., Kruchowski S. S., Tomlinson C., Strong C., Delehaunty K., Fronick C., Courtney B., Rock S. M., Belter E., Du F., Kim K., Abbott R. M., Cotton M., Levy A., Marchetto P., Ochoa K., Jackson S. M., Gillam B., Chen W., Yan L., Higginbotham J., Cardenas M., Waligorski J., Applebaum E., Phelps L., Falcone J., Kanchi K., Thane T., Scimone A., Thane N., Henke J., Wang T., Ruppert J., Shah N., Rotter K., Hodges J., Ingenthron E., Cordes M., Kohlberg S., Sgro J., Delgado B., Mead K., Chinwalla A., Leonard S., Crouse K., Collura K., Kudrna D., Currie J., He R., Angelova A., Rajasekar S., Mueller T., Lomeli R., Scara G., Ko A., Delaney K., Wissotski M., Lopez G., Campos D., Braidotti M., Ashley E., Golser W., Kim H., Lee S., Lin J., Dujmic Z., Kim W., Talag J., Zuccolo A., Fan C., Sebastian A., Kramer M., Spiegel L., Nascimento L., Zutavern T., Miller B., Ambroise C., Muller S., Spooner W., Narechania A., Ren L., Wei S., Kumari S., Faga B., Levy M. J., Mcmahan L., Van Buren P., Vaughn M. W., Ying K., Yeh C. T., Emrich S. J., Jia Y., Kalyanaraman A., Hsia A. P., Barbazuk W. B., Baucom R. S., Brutnell T. P., Carpita N. C., Chaparro C., Chia J. M., Deragon J. M., Estill J. C., Fu Y., Jeddeloh J. A., Han Y., Lee H., Li P., Lisch D. R., Liu S., Liu Z., Nagel D. H., McCann M. C., SanMiguel P., Myers A. M., Nettleton D., Nguyen J., Penning B. W., Ponnala L., Schneider K. L., Schwartz D. C., Sharma A., Soderlund C., Springer N. M., Sun Q., Wang H., Waterman M., Westerman R., Wolfgruber T. K., Yang L., Yu Y., Zhang L., Zhou S., Zhu Q., Bennetzen J. L., Dawe R. K., Jiang J., Jiang N., Presting G. G., Wessler S. R., Aluru S., Martienssen R. A., Clifton S. W., McCombie W. R., Wing R. A., Wilson R. K. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Takeda S., Sugimoto K., Otsuki H., Hirochika H. (1999). 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18, 383–393 10.1046/j.1365-313X.1999.00460.x [DOI] [PubMed] [Google Scholar]
- Tanurdzic M., Vaughn M. W., Jiang H., Lee T. J., Slotkin R. K., Sosinski B., Thompson W. F., Doerge R. W., Martienssen R. A. (2008). Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6, e302. 10.1371/journal.pbio.0060302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins J. P., Davis G., Main D., Duru N., Musket T., Goicoechea J. L., Frisch D. A., Coe E. H., Wing R. A. (2002). Construction and characterization of a deep-coverage bacterial artificial chromosome library for maize. Crop Sci. 42, 928–933 10.2135/cropsci2002.0928 [DOI] [Google Scholar]
- Vicient C. M. (2010). Transcriptional activity of transposable elements in maize. BMC Genomics 11, 601. 10.1186/1471-2164-11-601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Dooner H. K. (2005). Mx-rMx, a family of interacting transposons in the growing hAT superfamily of maize. Plant Cell 17, 375–388 10.1105/tpc.104.027797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Yan X., Maurais S., Fu H., O’Brien D. G., Mottinger J., Dooner H. K. (2004). Jittery, a Mutator distant relative with a paradoxical mobile behavior: excision without reinsertion. Plant Cell 16, 1105–1114 10.1105/tpc.019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S. E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 10.1126/science.1079695 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69 10.1038/ng1929 [DOI] [PubMed] [Google Scholar]