Abstract
Glycosphingolipids (GSLs) are information-bearing biomolecules that play critical roles in embryonic development, signal transduction and carcinogenesis. Previous studies indicate that certain GSLs are associated with differentiation in acute myeloid leukemia (AML) cells. In this study, we collected bone marrow samples from healthy donors and AML patients and analyzed the GSL expression profiles comprehensively using electrospray ionization linear ion-trap mass spectrometry. The results showed that AML patients had higher expression of the GSL lactotriaosylceramide (Lc3), GM3 and neolactotetraosylceramide (nLc4) in their bone marrow than did the healthy donors (P < 0.05), especially the M1 subtype of AML. To further explore the molecular mechanisms of Lc3, we examined the expression of the Lc3 synthase β1,3-N-acetylglucosaminyltransferase5 (β3Gn-T5) and found that the bone marrow samples of AML patients had 16-fold higher expression of β3Gn-T5 than those of healthy donors (P < 0.05). Our results suggest that AML-associated GSLs Lc3, GM3 and nLc4 are possibly involved in initiation and differentiation of AML.
Keywords: acute myeloid leukemia, glycosphingolipidomics, lactotriaosylceramide, Lc3 synthase, mass spectrometry
Introduction
Glycosphingolipids (GSLs) are essential biomolecules present in the cell membranes of all animal cells. They are composed of carbohydrates covalently linked to a ceramide (N-acylsphingosine) lipid tail that anchors the molecule within the cell membrane. Given the differences between carbohydrate and lipid moieties, more than 300 GSL series have been identified to date (mostly in carbohydrates) on the basis of sequencing and anomeric linkages (Ichikawa and Hirabayashi 1998). GSLs in mammalian tissues can be generally divided into three major groups: ganglio-, lacto- and globo-GSLs. The biosynthesis and degradation of these GSLs involves numerous enzymes that act at various subcellular locations (Hakomori 2008; Wennekes et al. 2009).
Biologically, GSLs play critical roles in cell signaling, apoptosis, adhesion, receptor modulation, growth and differentiation (Jarvis et al. 1996; Hakomori 1998; Kasahara and Sanai 1999). Cellular differentiation and malignant transformation are often accompanied by dramatic changes in GSL expression. Many GSLs are capable of inducing differentiation, apoptosis, bone marrow suppression and metastasis (Sietsma et al. 1999; Liang et al. 2010; Ogasawara et al. 2011). Aberrant glycosylation occurs in essentially all types of human cancers. Numerous glycosyl epitopes constitute tumor-associated antigens. However, there is a long-standing debate over whether aberrant glycosylation is a result or a cause of cancer (Hakomori 2002).
Previous studies indicate that certain GSLs are associated with differentiation in acute myeloid leukemia (AML) cells. AML is the most common acute leukemia affecting adults, and its incidence increases with age. The symptoms of AML are caused by the replacement of normal bone marrow with leukemic cells. Several risk factors and chromosomal abnormalities have been identified, but the specific cause of AML is not clear. It is suspected, however, that cell-surface carbohydrates play roles in myelopoiesis. Carbohydrate antigens have been shown to facilitate vascular adhesion, cell activation and phagocytosis of granulocytes and monocytes. Neutrophilic and monocytic differentiations are also often accompanied by changes in N-glycan, ganglioside and neutral GSL expression (Saito 1993; McEver et al. 1995; Hinkovska-Galcheva et al. 1998; Liu et al. 1998; Cooling et al. 2003). In leukemic cells, GSLs have been found to be biomarkers of successive maturation stages (Yu 1994; Smolenska-Sym et al. 2004). For example, in HL-60 cells, granulocytic and monocytic differentiations are accompanied by distinct changes in ganglioside synthesis and expression (Nakamura et al. 1992).
Over the past decade, mass spectrometry (MS) technologies have been applied to the characterization and quantification of GSLs (Costello and Vath 1990; Peter-Katalinic and Egge 1990; Samuelsson et al. 1990; Levery 2005). We have used the ion-trap MS (MSn) technology for the analysis of complex mixtures of cellular GSLs, on the basis of our previous finding that signature ion fragments can be generated for each specific GSL after multiple rounds of fragmentation (Li, Teneberg et al. 2008; Li, Zhou et al. 2008). Here, we report the first cellular glycosphingolipidomics study of primary AML tumor cells using the MSn technology. We collected bone marrow samples from healthy donors and AML patients and analyzed the GSL expression profiles comprehensively to determine which GSLs are associated with AML.
Results
Major GSL expression pattern in human bone marrow samples
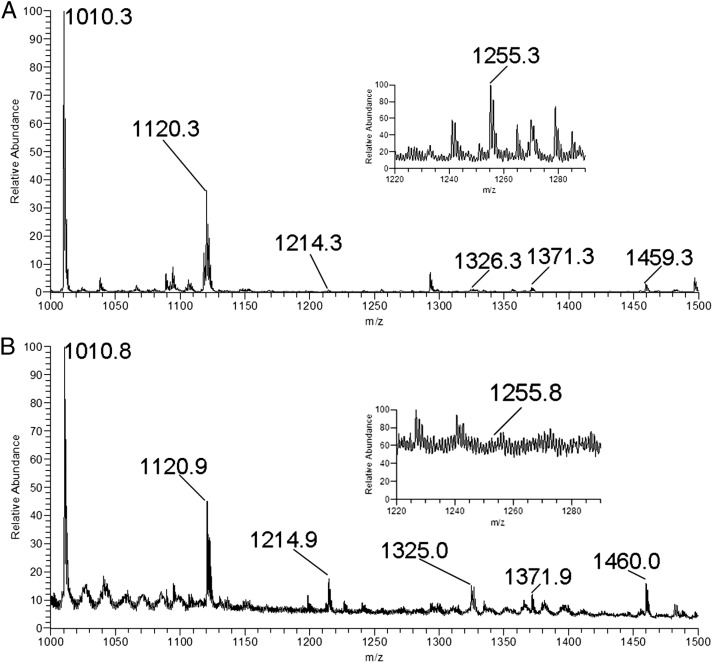
We used electrospray ionization linear MSn (ESI-LIT-MS) to analyze GSLs extracted from the human bone marrow samples. We found a number of potential GSL molecular ions (as Na+ adducts) in the ESI-LIT-MS1 profile spectrum of the permethylated GSL fraction isolated from the samples (Figure 1A and B). Subsequently, we systematically acquired MS2 spectra for every molecular precursor ion in the MS1 spectrum from the mass-to-charge ratio (m/z) 1000–1500, covering the range of dihexosylceramides and trihexosylceramides with ceramide compositions of d18:1 sphingosine and C16–C26 fatty acids. Major GSLs have the C16 or C24 fatty acid ceramide.
Fig. 1.
Positive electrospray ionization linear MSn analysis of bone marrow samples. (A) MS1 profile of GSLs isolated from an AML patient bone marrow sample; the expression of Lc3 (1255) was at a high level. (B) MS1 profile of GSLs isolated from a healthy control bone marrow sample; the expression of Lc3 (1255) was at a low level.
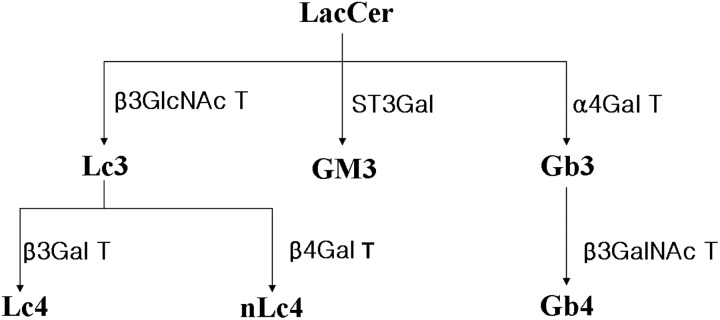
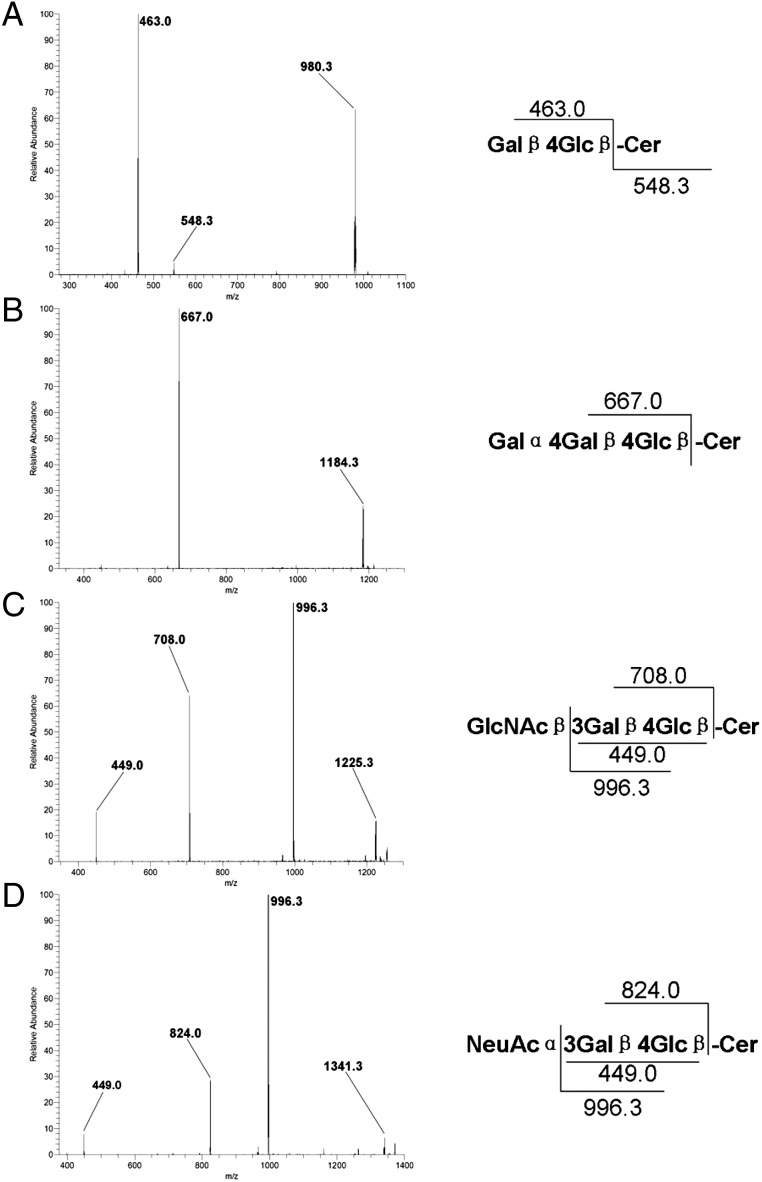
The relationship among major species of GSLs that exist in mammals is shown in Scheme 1. After multiple rounds of fragmentation, signature ion fragments for each specific GSL can be generated. Using the MSn technology, we can generally distinguish one series of GSLs from another. We analyzed the MS2 spectrum of the major molecular ion m/z 1010 in the MS1 spectrum and found that m/z 463 and 547 were the main fragments in the MS2 spectrum (Figure 2A). Using the fragmentation rules shown in Figure 2A (right), we identified the m/z 463 molecular ion as the Hex-Hex fragment (Galβ4Glc-OH; Li et al. 2009) and the m/z 547 molecular ion as the C16 fatty acid ceramide. The MS2 spectrum of the major molecular ion m/z 1120 in the MS1 spectrum (Supplementary data, Figure S1A) was similar to that of m/z 1010: we identified the m/z 463 molecular ion as the Hex-Hex fragment (Galβ4Glc-OH) and the m/z 657 molecular ion as the C24 fatty acid ceramide. We concluded that m/z 1120 was lactosylceramide (LacCer) with the C24 fatty acid ceramide (Supplementary data, Figure S1A). These results indicated that both m/z 1010 and 1120 were LacCer, though with different lengths of fatty acyl chains in ceramide.
Scheme 1.
Biosynthetic relationship of major GSL families in humans.
Fig. 2.
Qualitative analysis of major GSLs in the human bone marrow samples. MS2 spectrum, fragmentation pathway and characteristic fragmentation products for major permethylated GSLs in the human bone marrow samples in positive-mode MSn are shown. (A) MS2 spectrum and characteristic decomposition products of permethylated LacCer (1010) in the positive mode. (B) MS2 spectrum and characteristic decomposition products of permethylated Gb3 (1214) in the positive mode. (C) MS2 spectrum and characteristic decomposition products of permethylated Lc3 (1255) in the positive mode. (D) MS2 spectrum and characteristic decomposition products of permethylated GM3 (1371) in the positive mode.
We analyzed the MS2 spectra of other major molecular ions in the MS1 spectra, including m/z 1214, 1255 and 1371 as well as m/z 1326, 1368 and 1483 (Figure 2B–D and Supplementary data, Figure S1B–D, respectively). The MS2 spectra were compared with our previous study (Li et al. 2009), which revealed m/z 1214 and 1326 to be globotrihexosylceramide (Gb3) with C16 and C24 fatty acid ceramides, respectively; m/z 1255 and 1368 to be lactotriaosylceramide (Lc3) with C16 and C24 fatty acid ceramides, respectively; and m/z 1371 and 1483 to be GM3 with C16 and C24 fatty acid ceramides, respectively.
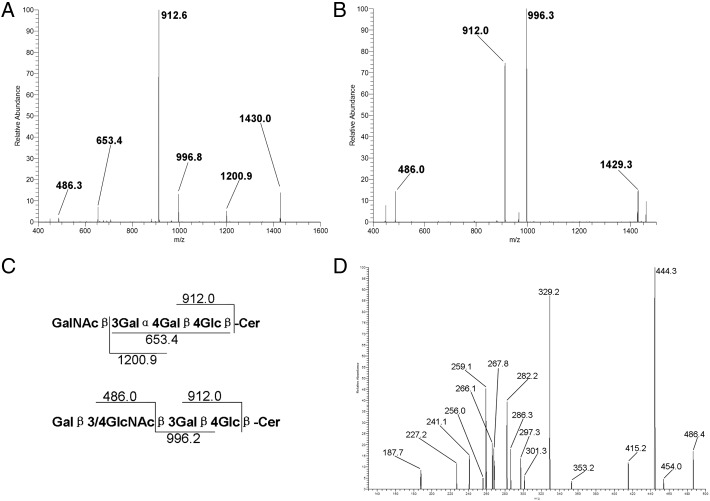
LacCer, Gb3, Lc3 and GM3 were present in the bone marrow samples from both AML patients and healthy donors. However, the isomers for m/z 1460, which represented GSL containing three hexose and one HexNAc residues, in healthy donors were significantly different from those in AML patients (Figure 3A and B). Healthy control had one major fragment, m/z 912, representing globotetraglycosylceramide (Gb4), whereas AML patients had m/z 912 plus another major fragment, m/z 996, representing lactotetraosylceramide (Lc4) or neolactotetraosylceramide (nLc4; Figure 3C). We hypothesized that m/z 1460 was a mixture comprising mainly Gb4 in healthy donors and primarily Lc4 or nLc4 in AML patents. To test this hypothesis, we generated an MS3 spectrum (m/z 1460 → 486→; Figure 3D) and compared it with that published by Ashline et al. (2005); our comparison suggested that Galβ4GlcNAc-ene was the terminal linkage. Therefore, we speculated that the m/z 1460 mixture in AML patients was mainly nLc4 with the C16 fatty acid ceramide. Similarly, we found that the m/z 1572 mixture in the AML patients was mainly nLc4 with the C24 fatty acid ceramide, whereas the mixture in the healthy controls was mainly Gb4 with the C24 fatty acid ceramide.
Fig. 3.
Qualitative analysis of Gb4 in the bone marrow samples of healthy donors and nLc4 in the bone marrow samples of AML patients. (A) MS2 spectrum of m/z 1460 precursor in the bone marrow samples of healthy donors. (B) MS2 spectrum of m/z 1460 precursor in the bone marrow samples of AML patients. (C) Fragmentation pathway and characteristic decomposition products for Gb4 (top) and (n) Lc4 (bottom) in the positive-mode MSn of permethylated GSLs. (D) MS3 spectrum (m/z 1460 → 486→) in the bone marrow samples of AML patients.
Differential expression of major GSLs in the bone marrow samples of healthy donors and AML patients
ESI-LIT-MS revealed four major GSLs in the human bone marrow samples: LacCer, Gb3, Lc3 and GM3. We used a method of relative quantification to analyze these GSLs. Among these GSLs, LacCer was the precursor to the other GSLs and much more abundant in than the other GSLs in the bone marrows of both healthy donors and AML patients. We used differential isotope labeling to compare the amount of LacCer in bone marrow cells of healthy donors with that in the bone marrow cells of AML patients. In bone marrow cells from AML patients, GSLs were permethylated with deuterated iodomethane and mixed with GSLs from the same number of iodomethane-permethylated bone marrow cells (as determined by hemocytometer cell count) from healthy donors. We found the levels of LacCer in the bone marrow samples from AML patients to be similar to those in the bone marrow samples from healthy donors (Supplementary data, Figure S2), so we chose LacCer as the internal standard, with m/z 1010 and 1120 as major LacCer ion fragments, with C16 and C24 fatty acid ceramides, respectively.
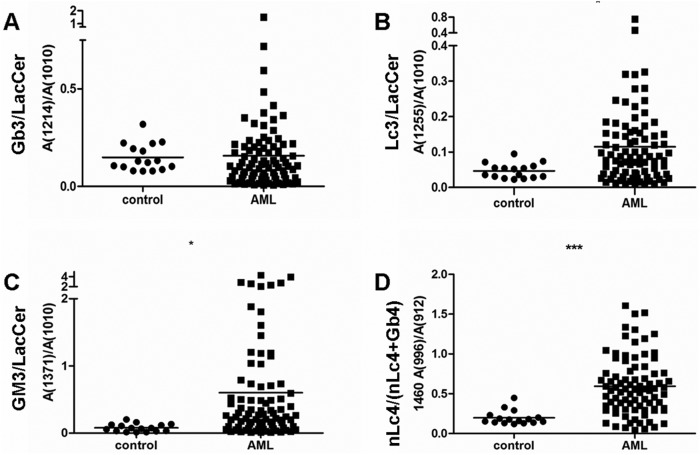
We used a method of relative quantification to determine the ratios of Gb3 (Figure 4A and Supplementary data, Figure S3A), Lc3 (Figure 4B and Supplementary data, Figure S3B) and GM3 (Figure 4C and Supplementary data, Figure S3C) to LacCer. The abundance of m/z 1460 and 1572 indicates the presence of a Gb4-and-nLc4 mixture. Although the relative quantities of Gb4 and nLc4 could not be determined from the MS1 spectrum, they could be determined from the MS2 spectrum, which revealed m/z 912 to be the common fragment of Gb4 and nLc4 and m/z 996/1108 to be the specific fragment of nLc4 (1460/1572). Gb4 had only a small m/z 996/1108 fragment. Therefore, the ratio of the abundance of m/z 996/1108 to that of m/z 912 represented the relative proportion of nLc4 (1460/1572) in the mixture (Figure 4D and Supplementary data, Figure S3D). These results suggested that Lc3, GM3 and nLc4 expression are higher in the bone marrow samples from AML patients than in the bone marrow samples from healthy donors. However, Gb3 expression in healthy donors and AML patients did not differ significantly (Figure 4A).
Fig. 4.
Relative quantitative analysis of the bone marrow samples in healthy donors and AML patients. With LacCer (1010) as the internal standard, relative quantity was represented by the ratios of abundance of Gb3, Lc3 and GM3 to LacCer. (A) Relative quantification of Gb3 (1214) in the bone marrow samples of healthy donors and AML patients. (B) Relative quantification of Lc3 (1255) in the bone marrow samples of healthy donors and AML patients. (C) Relative quantification of GM3 (1371) in the bone marrow samples of healthy donors and AML patients. (D) Relative quantification of nLc4 (1460) in the bone marrow samples of healthy donors and AML patients. The ratios of m/z 996 abundance to m/z 912 in the MS2 spectrum of m/z 1460 were chosen to represent the relative proportion of nLc4 (1460) in the mixture. Data represent the bone marrow samples from healthy donors (n = 16) and AML patients (n = 92). Statistical significance of differences between AML patients and controls was analyzed using the two-tailed Student's t-test of means. Compared with control, *P < 0.05, ***P < 0.001.
Differential GSL expression in the bone marrow samples of different subtypes of AML patients
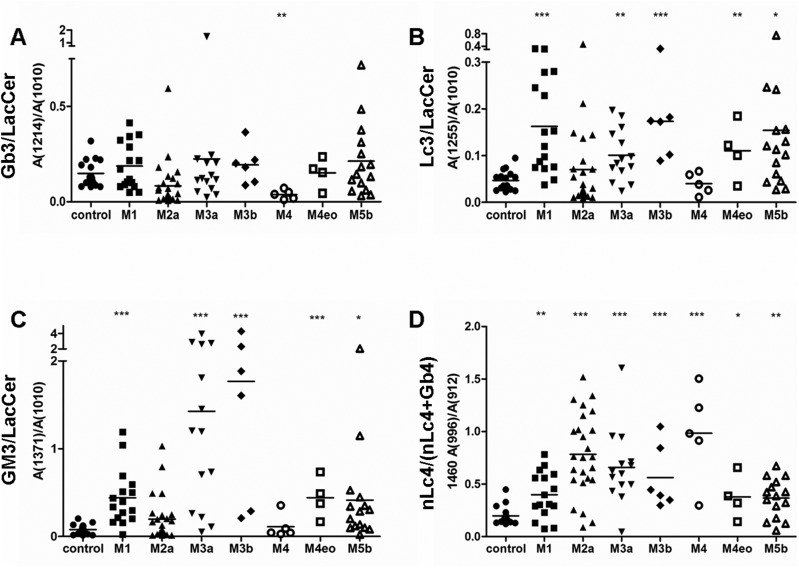
The French–American–British (FAB) classification system (Bennett et al. 1976) categorizes AML into eight subtypes, M0–M7, based on the type of cells from which the leukemia developed and the maturity of cells. Based on the clinical information, we identified different subtypes of AML and analyzed the expression of different GSLs in the subtypes. Differences in the relative quantities of Gb3 (Figure 5A and Supplementary data, Figure S4A), Lc3 (Figure 5B and Supplementary data, Figure S4B), GM3 (Figure 5C and Supplementary data, Figure S4C) and nLc4 (Figure 5D and Supplementary data, Figure S4D) were found in different subtypes of AML. In the M1 subtype (acute myeloblastic leukemia without maturation), Gb3, Lc3, GM3 and nLc4 were all more highly expressed than in the control group. However, in the M2a subtype (acute myeloblastic leukemia with granulocytic maturation), only nLc4 expression was higher than in healthy donors, whereas Gb3, Lc3 and GM3 expression did not differ significantly from the control. In the M3a group, both Lc3 and nLc4 had higher expression than in healthy donors. In the M4 subtype (acute myelomonocytic leukemia), only nLc4 expression was higher than in the healthy donors. In the M5 subtype, Gb3, Lc3, GM3 and nLc4 were all higher than in the healthy donors (Table I).
Fig. 5.
Relative quantitative analysis of different AML subtypes. With LacCer (1010) as the internal standard, relative quantification was represented by the ratios of abundance of Gb3, Lc3 and GM3 to LacCer. (A) Relative quantification of Gb3 (1214) in the bone marrow samples of healthy donors and patients with different subtypes of AML. (B) Relative quantification of Lc3 (1255) in the bone marrow samples of healthy donors and patients with different subtypes of AML. (C) Relative quantification of GM3 (1371) in the bone marrow samples of healthy donors and patients with different subtypes of AML. (D) Relative quantification of nLc4 (1460) in the bone marrow samples of healthy donors and patients with different subtypes of AML. The ratios of m/z 996 abundance to m/z 912 abundance in the MS2 spectrum of m/z 1460 were taken to represent the relative proportion of nLc4 (1460) in the mixture. Data represent the bone marrow samples of healthy donors (n = 16) and AML patients with the M1 (n = 16), M2a (n = 25), M3a (n = 14), M3b (n = 6), M4 (n = 5), M4eo (n = 4) and M5b (n = 15) subtypes. Statistical significance in differences between AML patient subtypes and controls was analyzed using the two-tailed Student's t-test of means. Compared with control, *P < 0.05, **P < 0.01, ***P < 0.001.
Table I.
The amounts of major GSLs in patients with different subtypes of AML when compared with healthy controls
| Control | M1 | M2a | M3a | M3b | M4 | M4eo | M5b | |
|---|---|---|---|---|---|---|---|---|
| Gb3 (1214) | 1.00 | 1.26 | 0.56 | 1.51 | 1.30 | 0.26 | 1.02 | 1.43 |
| Lc3 (1255) | 1.00 | 3.47 | 1.49 | 2.15 | 3.70 | 0.86 | 2.36 | 3.29 |
| GM3 (1371) | 1.00 | 5.61 | 2.51 | 18.19 | 22.54 | 1.43 | 5.62 | 5.28 |
| nLc4 (1460) | 1.00 | 2.02 | 3.97 | 3.34 | 2.85 | 4.99 | 1.91 | 1.86 |
Higher expression levels of Lc3 synthase β1,3-N-acetylglucosaminyltransferase5 in the bone marrow samples of AML patients versus healthy donors
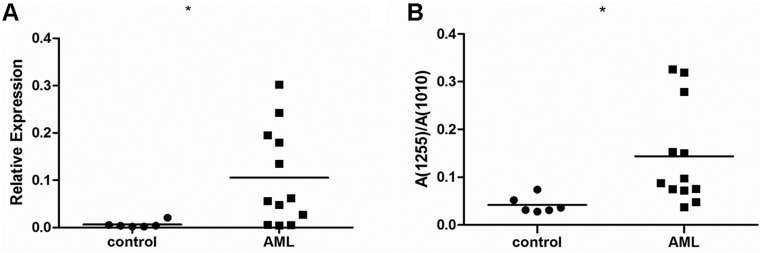
In our previous studies (Henion et al. 2001; Biellmann et al. 2008), β1,3-N-acetylglucosaminyltransferase5 (β3Gn-T5) was demonstrated to be Lc3 synthase that synthesizes the precursor structure for the lacto/neolacto-series GSLs in both in vitro and in vivo experiments. β3Gn-T5 is, in fact, the key enzyme responsible for the synthesis of lacto- and neolacto-series GSLs. Therefore, we compared the mRNA levels of β3Gn-T5 in the bone marrow samples from healthy donors and AML patients. Figure 6A shows that the expression of β3Gn-T5 was significantly higher in AML patients than in the healthy donors, which was consistent with MS analysis results, as shown in Figure 6B. Table II shows the β3Gn-T5 and Lc3 expression levels in each bone marrow sample of the healthy donors and the AML patients.
Fig. 6.
Expression level of Lc3 synthase β3Gn-T5 and corresponding relative quantification of Lc3 (1255) in the bone marrow samples of healthy donors and AML patients. (A) The expression of Lc3 synthase β3Gn-T5 in bone marrow samples was analyzed using the real-time PCR method. (B) Corresponding relative quantification of Lc3 (1255) in the bone marrow samples of healthy donors and AML patients was analyzed as in Fig. 4. Data represent the bone marrow samples of healthy donors (n = 6) and AML patients (n = 12). Statistical significance in differences between AML patients and controls was analyzed using the two-tailed Student's t-test of means. Compared with control, *P < 0.05.
Table II.
β3Gn-T5 and Lc3 levels in each of the healthy donors (No. 1–6) and AML patients (No. 7–18, randomly selected from 104 patients)
| No. | β3Gn-T5a | Lc3b | No. | β3Gn-T5 | Lc3 | No. | β3Gn-T5 | Lc3 |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.0055 | 0.0519 | 7 | 0.0056 | 0.0723 | 13 | 0.0622 | 0.0877 |
| 2 | 0.0022 | 0.0309 | 8 | 0.1953 | 0.2787 | 14 | 0.1801 | 0.1505 |
| 3 | 0.0039 | 0.0312 | 9 | 0.0481 | 0.0759 | 15 | 0.0059 | 0.0483 |
| 4 | 0.0209 | 0.0739 | 10 | 0.0561 | 0.0976 | 16 | 0.2426 | 0.3195 |
| 5 | 0.0042 | 0.0358 | 11 | 0.0049 | 0.0375 | 17 | 0.0275 | 0.0749 |
| 6 | 0.0023 | 0.0279 | 12 | 0.3022 | 0.3263 | 18 | 0.1353 | 0.1530 |
aThe expression of Lc3 synthase β3Gn-T5 in bone marrow samples was analyzed using real-time PCR.
bCorresponding relative quantitation of Lc3 (1255) in healthy donors' bone marrow samples and AML patients' bone marrow samples were analyzed as in Fig. 4.
Discussion
In the present study, we used a specific, sensitive and generally applicable method to analyze GSLs in tumor cells (Li, Teneberg et al. 2008; Li, Zhou et al. 2008). We detected Gb3, Lc3, GM3 and nLc4 as major GSLs expressed in bone marrow cells. Furthermore, significant differences in GSL expression between healthy donors and AML patients were found. Our findings confirm previous reports that AML cells have high expression of GM3. We found that Lc3 and nLc4 were expressed at higher levels in AML patients than in healthy donors. We also found small amounts of isoglobotriosylceramide (iGb3; Zhou et al. 2004) in Gb3/iGb3 isomers in AML patients (Wang et al., unpublished data).
Our study was made possible by the use of MSn, which has greatly improved the applicability of the MS technology for analyzing patient samples. The application of ESI and matrix-assisted laser desorption ionization, in conjunction with tandem collision-induced dissociation, time of flight and quadrupole ion-trap techniques, has revolutionized the sensitivity of MS (Reinhold et al. 1995; Reinhold and Sheeley 1998; Costello 1999; Harvey 1999; Dell and Morris 2001; Levery 2005). The selectivity of our method is based on the characteristic fragmentations of the sugar chain of permethylated GSLs, which differ from those of other GSL series when isolated and analyzed by MSn using the MSn analysis. Relative proportions of GSLs could be determined from the abundance ratios of selected characteristic fragmentation products of the GSLs.
In this study, the major GSL species in human bone marrow, including Gb3, Lc3, GM3 and nLc4, were analyzed in a semi-quantitative manner. Through statistical analysis, we minimized the deviation between the donors and the AML patients. Gb3 did not differ significantly between the control and AML groups, whereas Lc3, GM3 and nLc4 had significantly higher expression in the bone marrow samples of AML patients. GM3 serves as a precursor for most of the more complex ganglioside species, and ganglioside participates in the regulation of various cellular functions, including cell proliferation, apoptosis, migration and invasion. Hence, the finding of a large amount of GM3 in the bone marrow samples of AML patients is very valuable for further investigation.
Lc3 is composed of glucose, galactose and N-acetylglucosamine as carbohydrate chain constituents. This GSL is a precursor structure, which is elongated to the lacto- and neolacto-series GSLs. Lc3 is highly expressed on the cell surface of HL-60 cells (Nozaki et al. 2010; Togayachi et al. 2010). The glycosyltransferase β3Gn-T5 (Henion et al. 2001; Togayachi et al. 2001), which synthesizes Lc3, plays critical roles in embryonic development and differentiation. Its expression in leukemia cell lines has been reported previously (Nozaki et al. 2010). Our finding that Lc3 is highly expressed in primary AML suggests that it may serve as a potential biomarker and a target for therapy.
We detected high expression of Lc3 in the AML cell line HL-60, whereas little Lc3 was detected in the acute lymphoblastic leukemia (ALL) cell line Jurkat (Togayachi et al. 2010). We also detected only a small amount of Lc3 using MS in Jurkat cells (data not shown), and we did not find high expression of Lc3 in the bone marrow samples of T-ALL or B-ALL patients (data not shown). In K-562 cells, a chronic myeloid leukemia cell line, the expressions of Lc3 and structurally related GSL components were very low and could not even be detected by flow cytometry (Nozaki et al. 2010). In AML, both cell lines and clinical samples showed significantly higher expression of Lc3. Interestingly, the expression level of Lc3 varies in different subtypes of AML, indicating that the expression of Lc3 and/or nLc4 is closely related to cell differentiation. The expression of Lc3 in the M1 subtype (acute myeloblastic leukemia without maturation) was significantly higher than that in healthy controls. However, no significant difference was found in Lc3 expression between healthy controls and the M2a subtype (acute myeloblastic leukemia with granulocytic maturation). It indicates that the expression of Lc3 is closely related to cell differentiation in human bone marrow. Moreover, there may have differences in GSLs metabolism between the various leukemia subtypes that would be worthy of further investigation.
Inducing differentiation is currently one of the most important therapies for AML, especially for the M3 subtype, also known as acute promyelocytic leukemia (APL). Differentiation therapy with all-trans retinoic acid (ATRA) has been proven to be very effective and has become an established APL treatment (Degos and Wang 2001; Bolanos-Meade et al. 2003; Vitoux et al. 2007). In vitro studies have shown that the human leukemia cell lines HL-60 and NB4 can be induced to enter differentiation along the neutrophil lineage upon stimulation with ATRA or along the monocyte lineage after phorbol 12-myristate 13-acetate (PMA) treatment, resulting in morphological, functional and drug sensitivity changes accompanied by a loss of proliferative capacity. Thin-layer chromatography has been applied to analyze the changes in GSLs in the differentiated cells (Nakamura et al. 1992; Jasek et al. 2008). Thus, we used the MS method to analyze the GSLs in HL-60 and NB4 cells treated with ATRA and PMA and found that their Lc3 expression was lower than that of the untreated cells (Supplementary data, Figure S5).
The biosynthesis and degradation of GSLs is a complex process involving numerous enzymes that act at various subcellular locations. Among these enzymes, only one or a few key enzymes control the metabolism of GSLs. As Lc3 is a precursor structure of lacto- and neolacto-series GSLs, β3Gn-T5 also affects lacto- and neolacto-series GSLs (Togayachi et al. 2010). In this study, Lc3 is identified as a potential biomarker of AML. However, whether the presence of Lc3 is a result or a cause of AML is still unknown. This question may be resolved by silencing the Lc3 synthase β3Gn-T5 to reduce the expression of Lc3 and examining the effect of lacto-series GSLs on AML carcinogenesis.
Our results clearly indicate that Lc3 is abnormally expressed by AML cells, and the underlying mechanism is the overexpression of the Lc3 synthase. However, due to the semi-quantitative nature of MS assays, the exact level of Lc3 expression in AML patients must be further quantified by other independent methods, such as staining by specific monoclonal antibodies. Similarly, the high expression of GM3 and nLc4 in the bone marrow of AML patients also need to be quantified by other independent methods. The subcellular localization of Lc3 in AML cells remain to be studied, although its expression on cell surface has been reported by Nozaki et al. (2010).
In conclusion, we show for the first time in clinical samples that the increased expression of Lc3, GM3 and nLc4 is characteristic in AML patients and Lc3 is related to cell differentiation. AML-associated GSLs Lc3, GM3 and nLc4 are possibly involved in initiation and differentiation of AML. Future studies will be focused on the transcriptional regulation of Lc3 synthase, the receptors for Lc3 and the alterations of Lc3 in patients after induction therapy, as well as developing Lc3-specific antibodies for diagnosis and therapies. Similarly, all these studies are necessary for GM3 and nLc4.
Materials and methods
Tissues and cultured cells
Bone marrow samples were obtained from AML patients and healthy donors at the First Affiliated Hospital of Soochow University (Suzhou, China) under institutional guidelines. A total of 16 healthy donors and 104 patients were enrolled (Supplementary data, Table S1 and Supplementary data, Figure S2). All the bone marrow samples were obtained at initial diagnosis without chemotherapy. Human AML cell lines HL-60 (ATTC CCL 240) and NB4 (DSM ACC 207) were cultured in RPMI 1640 (Life Technologies, Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum (Life Technologies), 2 mM l-glutamine (Life Technologies) and 50 μg/mL of gentamicin. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Total lipid extraction from leukemia cells
Cultured leukemia cells (5 × 107 cell aliquots) were stored in 16 × 100 mm glass tubes at −80°C. Lipids were extracted by extensive sonication four times with mixed polarity solvents (1 mL each). The first and last solvent used was chloroform–methanol 1:1 (v/v). The second and third solvent used was isopropanol–hexane–water 55:25:20 (v/v/v, upper phase removed by aspiration before use). The supernatants were pooled and dried under a nitrogen stream at 40°C.
Per-N,O-methylation of GSLs
A modification of the method of Ciucanu and Costello (2003) was employed for per-N,O-methylation of GSLs. GSLs (1–20 µg) were introduced into a conical glass vial, and dimethyl sulfoxide (150 µL) was added without using special drying conditions or an inert gas atmosphere. Powdered sodium hydroxide (40–60 mg) was then added to the sample solution and stirred at room temperature until completely dissolved. Iodomethane or deuterated iodomethane (80 µL) was added with a syringe, and the mixture was shaken at room temperature for 1 h. The methylation reaction was quenched with water (2 mL). The permethylated products were extracted three times by the addition of dichloromethane (2 mL). The combined dichloromethane extracts were then washed three times with water (2 mL each). Following the final wash, the samples were transferred to a new tube and dried under a nitrogen stream at 35–40°C.
Electrospray ionization MSn of permethylated GSLs
Electrospray ionization MS (MS and MSn) was carried out in a positive ion mode on a linear ion-trap mass spectrometer (LTQ, Thermo Finnigan, San Jose, CA) using a electrospray source for direct infusion of samples dissolved in methanol, with a flow rate of 0.30 µL/min at a capillary temperature of 230°C, a sheath gas flow rate of 2 arb, a spray voltage of 3.50 kV, a capillary voltage of 28.00 kV, a tube lens of 120.00 V, an injection time of 100.00 ms, an activation time of 30 ms, an activation Q-value of 0.250 and an isolation width of m/z 1.5. Normalized collision energies were set to leave a minimal residual abundance of precursor ion; in this case, 30% was used for all product ion scans. All ions were detected as sodium adducts.
Quantitative real-time polymerase chain reaction assays
The RNA from each sample was reverse-transcribed into first-strand cDNA with an oligo-dT primer. The quantitative reverse transcriptase–polymerase chain reaction (PCR) analysis was performed using the ABI 7500 system (Life Technologies) with a Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific, Glen Burnie, MA) according to the manufacturer's protocol. The amount of β-actin mRNA was measured as an internal standard. PCR was performed using primers specific to each gene through 40 cycles at 95°C for 15 s and 60°C for 60 s after a preincubation at 50°C for 2 min and 95°C for 10 min. The specific primers used for amplification were as follows: β3Gn-T5-F: 5′-TTTTGGATTGGTCGTGTTCAT-3′; β3Gn-T5-R: 5′-CGGCTGTTAGTCAGGGTAAG-3′; β-actin-F: 5′-CACCATTGGCAATGAGCGGTTCC-3′; β-actin-R: 5′-GTAGTTTCGTGGATGCCACAGG-3′.
Statistical analysis
Statistical significance in differences between AML patient and control samples was analyzed using the two-tailed Student's t-test of means. For all tests, P < 0.05 was considered as significant.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The work was supported by the National Natural Science Foundation of China (31000370), the Natural Science Foundation of Jiangsu Province (BK2011288), the Priority Academic Program Development of Jiangsu Higher Education Institutions, Program for Changjiang Scholars and Innovative Research Team in University (IRT1075), the Suzhou Key Laboratories (SWG0904) and the National Institutes of Health through MD Anderson's Cancer Center Support grant (CA16672) and other grants (AI079232, AI078898, P30-AI36211).
Conflict of interest
D.Z. is the President of NanoCruise Suzhou Pharmaceutical Ltd.
Abbreviations
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; ATRA, all-trans retinoic acid; β3Gn-T5, β1,3-N-acetylglucosaminyltransferase5; ESI-LIT-MS, electrospray ionization linear ion-trap mass spectrometry; FAB, French–American–British; Gb4, globotetraglycosylceramide; Gb3, globotrihexosylceramide; GSL, glycosphingolipid; iGb3, isoglobotriosylceramide; LacCer, lactosylceramide; Lc3, lactotriaosylceramide; Lc4, lactotetraosylceramide; MS, mass spectrometry; MSn, ion trap mass spectrometry; nLc4, neolactotetraosylceramide; PCR, polymerase chain reaction; PMA, phorbol 12-myristate 13-acetate.
Supplementary Material
Acknowledgements
We thank Joe Munch and Dawn Chalaire for critical reading of the manuscript.
References
- Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. doi:10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33(4):451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. doi:10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- Biellmann F, Hülsmeier AJ, Zhou D, Cinelli P, Hennet T. The Lc3-synthase gene B3gnt5 is essential to pre-implantation development of the murine embryo. BMC Dev Biol. 2008;8:109. doi: 10.1186/1471-213X-8-109. doi:10.1186/1471-213X-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Meade J, Karp JE, Guo C, Sarkodee-Adoo CB, Rapoport AP, Tidwell ML, Buddharaju LN, Chen TT. Timed sequential therapy of acute myelogenous leukemia in adults: A phase II study of retinoids in combination with the sequential administration of cytosine arabinoside, idarubicin and etoposide. Leuk Res. 2003;27:313–321. doi: 10.1016/s0145-2126(02)00177-7. doi:10.1016/S0145-2126(02)00177-7. [DOI] [PubMed] [Google Scholar]
- Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125:16213–16219. doi: 10.1021/ja035660t. doi:10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- Cooling LL, Zhang DS, Naides SJ, Koerner TA. Glycosphingolipid expression in acute nonlymphocytic leukemia: Common expression of Shiga toxin and parvovirus B19 receptors on early myeloblasts. Blood. 2003;101:711–721. doi: 10.1182/blood-2002-03-0718. doi:10.1182/blood-2002-03-0718. [DOI] [PubMed] [Google Scholar]
- Costello CE. Bioanalytic applications of mass spectrometry. Curr Opin Biotechnol. 1999;10:22–28. doi: 10.1016/s0958-1669(99)80005-6. doi:10.1016/S0958-1669(99)80005-6. [DOI] [PubMed] [Google Scholar]
- Costello CE, Vath JE. Tandem mass spectrometry of glycolipids. Methods Enzymol. 1990;193:738–768. doi: 10.1016/0076-6879(90)93448-t. doi:10.1016/0076-6879(90)93448-T. [DOI] [PubMed] [Google Scholar]
- Degos L, Wang ZY. All trans retinoic acid in acute promyelocytic leukemia. Oncogene. 2001;20:7140–7145. doi: 10.1038/sj.onc.1204763. doi:10.1038/sj.onc.1204763. [DOI] [PubMed] [Google Scholar]
- Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291:2351–2356. doi: 10.1126/science.1058890. doi:10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Cancer-associated glycosphingolipid antigens: Their structure, organization, and function. Acta Anat (Basel) 1998;161:79–90. doi: 10.1159/000046451. doi:10.1159/000046451. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc Natl Acad Sci USA. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. doi:10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakomori SI. Structure and function of glycosphingolipids and sphingolipids: Recollections and future trends. Biochim Biophys Acta. 2008;1780:325–346. doi: 10.1016/j.bbagen.2007.08.015. doi:10.1016/j.bbagen.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom Rev. 1999;18:349–450. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. doi:10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Henion TR, Zhou D, Wolfer DP, Jungalwala FB, Hennet T. Cloning of a mouse β1,3N-acetylglucosaminyltransferase GlcNAc(β1,3)Gal(β1,4)Glc-ceramide synthase gene encoding the key regulator of lacto-series glycolipid biosynthesis. J Biol Chem. 2001;276:30261–30269. doi: 10.1074/jbc.M102979200. doi:10.1074/jbc.M102979200. [DOI] [PubMed] [Google Scholar]
- Hinkovska-Galcheva VT, Boxer LA, Mansfield PJ, Harsh D, Blackwood A, Shayman JA. The formation of ceramide-1-phosphate during neutrophil phagocytosis and its role in liposome fusion. J Biol Chem. 1998;273:33203–33209. doi: 10.1074/jbc.273.50.33203. doi:10.1074/jbc.273.50.33203. [DOI] [PubMed] [Google Scholar]
- Ichikawa S, Hirabayashi Y. Glucosylceramide synthase and glycosphingolipid synthesis. Trends Cell Biol. 1998;8:198–202. doi: 10.1016/s0962-8924(98)01249-5. doi:10.1016/S0962-8924(98)01249-5. [DOI] [PubMed] [Google Scholar]
- Jarvis WD, Grant S, Kolesnick RN. Ceramide and the induction of apoptosis. Clin Cancer Res. 1996;2:1–6. [PubMed] [Google Scholar]
- Jasek E, Mirecka J, Litwin JA. Effect of differentiating agents (all-trans retinoic acid and phorbol 12-myristate 13-acetate) on drug sensitivity of HL60 and NB4 cells in vitro. Folia Histochem Cytobiol. 2008;46:323–330. doi: 10.2478/v10042-008-0080-x. doi:10.2478/v10042-008-0080-x. [DOI] [PubMed] [Google Scholar]
- Kasahara K, Sanai Y. Possible roles of glycosphingolipids in lipid rafts. Biophys Chem. 1999;82:121–127. doi: 10.1016/s0301-4622(99)00111-8. doi:10.1016/S0301-4622(99)00111-8. [DOI] [PubMed] [Google Scholar]
- Levery SB. Glycosphingolipid structural analysis and glycosphingolipidomics. Methods Enzymol. 2005;405:300–369. doi: 10.1016/S0076-6879(05)05012-3. doi:10.1016/S0076-6879(05)05012-3. [DOI] [PubMed] [Google Scholar]
- Li Y, Teneberg S, Thapa P, Bendelac A, Levery SB, Zhou D. Sensitive detection of isoglobo and globo series tetraglycosylceramides in human thymus by ion trap mass spectrometry. Glycobiology. 2008;18:158–165. doi: 10.1093/glycob/cwm129. doi:10.1093/glycob/cwm129. [DOI] [PubMed] [Google Scholar]
- Li Y, Thapa P, Hawke D, Kondo Y, Furukawa K, Hsu FF, Adlercreutz D, Weadge J, Palcic MM, Wang PG, et al. Immunologic glycosphingolipidomics and NKT cell development in mouse thymus. J Proteome Res. 2009;8:2740–2751. doi: 10.1021/pr801040h. doi:10.1021/pr801040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou D, Xia C, Wang PG, Levery SB. Sensitive quantitation of isoglobotriaosylceramide in the presence of isobaric components using electrospray ionization-ion trap mass spectrometry. Glycobiology. 2008;18:166–176. doi: 10.1093/glycob/cwm127. doi:10.1093/glycob/cwm127. [DOI] [PubMed] [Google Scholar]
- Liang YJ, Kuo HH, Lin CH, Chen YY, Yang BC, Cheng YY, Yu AL, Khoo KH, Yu J. Switching of the core structures of glycosphingolipids from globo- and lacto- to ganglio-series upon human embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:22564–22569. doi: 10.1073/pnas.1007290108. doi:10.1073/pnas.1007290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AH, Liu F, Li Z, Gu JX, Chen HL. Alterations in glycosyltransferases during myeloid and monocytoid differentiation of HL-60 cells. Cell Biol Int. 1998;22:545–550. doi: 10.1006/cbir.1998.0293. doi:10.1006/cbir.1998.0293. [DOI] [PubMed] [Google Scholar]
- McEver RP, Moore KL, Cummings RD. Leukocyte trafficking mediated by selectin-carbohydrate interactions. J Biol Chem. 1995;270:11025–11028. doi: 10.1074/jbc.270.19.11025. doi:10.1074/jbc.270.19.11025. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Tsunoda A, Sakoe K, Gu J, Nishikawa A, Taniguchi N, Saito M. Total metabolic flow of glycosphingolipid biosynthesis is regulated by UDP-GlcNAc:lactosylceramide beta 1–>3N-acetylglucosaminyltransferase and CMP-NeuAc:lactosylceramide alpha 2–>3 sialyltransferase in human hematopoietic cell line HL-60 during differentiation. J Biol Chem. 1992;267:23507–23514. [PubMed] [Google Scholar]
- Nozaki H, Yanagida M, Koide K, Shiotani K, Kinoshita M, Kobayashi Y, Watarai S, Nakamura K, Suzuki A, Ariga T, et al. Production and characterization of monoclonal antibodies specific to lactotriaosylceramide. Glycobiology. 2010;20:1631–1642. doi: 10.1093/glycob/cwq117. doi:10.1093/glycob/cwq117. [DOI] [PubMed] [Google Scholar]
- Ogasawara N, Katagiri YU, Kiyokawa N, Kaneko T, Sato B, Nakajima H, Miyagawa Y, Kushi Y, Ishida H, Kiso M, et al. Accelerated biosynthesis of neolacto-series glycosphingolipids in differentiated mouse embryonal carcinoma F9 cells detected by using dodecyl N-acetylglucosaminide as a saccharide primer. J Biochem. 2011;149:321–330. doi: 10.1093/jb/mvq142. doi:10.1093/jb/mvq142. [DOI] [PubMed] [Google Scholar]
- Peter-Katalinic J, Egge H. Desorption mass spectrometry of glycosphingolipids. Methods Enzymol. 1990;193:713–733. doi: 10.1016/0076-6879(90)93446-r. doi:10.1016/0076-6879(90)93446-R. [DOI] [PubMed] [Google Scholar]
- Reinhold VN, Reinhold BB, Costello CE. Carbohydrate molecular weight profiling, sequence, linkage, and branching data: ES-MS and CID. Anal Chem. 1995;67:1772–1784. doi: 10.1021/ac00107a005. doi:10.1021/ac00107a005. [DOI] [PubMed] [Google Scholar]
- Reinhold VN, Sheeley DM. Detailed characterization of carbohydrate linkage and sequence in an ion trap mass spectrometer: Glycosphingolipids. Anal Biochem. 1998;259:28–33. doi: 10.1006/abio.1998.2619. doi:10.1006/abio.1998.2619. [DOI] [PubMed] [Google Scholar]
- Saito M. Bioactive gangliosides: Differentiation inducers for hematopoietic cells and their mechanism(s) of actions. Adv Lipid Res. 1993;25:303–327. [PubMed] [Google Scholar]
- Samuelsson BE, Pimlott W, Karlsson KA. Mass spectrometry of mixtures of intact glycosphingolipids. Methods Enzymol. 1990;193:623–646. doi: 10.1016/0076-6879(90)93442-n. doi:10.1016/0076-6879(90)93442-N. [DOI] [PubMed] [Google Scholar]
- Sietsma H, Kamps WA, Dontje B, Hendriks D, Kok JW, Vellenga E, Nijhof W. Leukemia-induced bone marrow depression: Effects of gangliosides on erythroid cell production. Int J Cancer. 1999;82:92–97. doi: 10.1002/(sici)1097-0215(19990702)82:1<92::aid-ijc16>3.0.co;2-j. doi:10.1002/(SICI)1097-0215(19990702)82:1<92::AID-IJC16>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Smolenska-Sym G, Spychalska J, Zdebska E, Wozniak J, Traczyk Z, Pszenna E, Maj S, Danikiewicz W, Bienkowski T, Koscielak J. Ceramides and glycosphingolipids in maturation process: Leukemic cells as an experimental model. Blood Cells Mol Dis. 2004;33:68–76. doi: 10.1016/j.bcmd.2004.04.002. doi:10.1016/j.bcmd.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Akashima T, Ookubo R, Kudo T, Nishihara S, Iwasaki H, Natsume A, Mio H, Inokuchi J, Irimura T, et al. Molecular cloning and characterization of UDP-GlcNAc: Lactosylceramide β1,3-N-acetylglucosaminyltransferase (β3Gn-T5), an essential enzyme for the expression of HNK-1 and Lewis X epitopes on glycolipids. J Biol Chem. 2001;276:22032–22040. doi: 10.1074/jbc.M011369200. doi:10.1074/jbc.M011369200. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Kozono Y, Ikehara Y, Ito H, Suzuki N, Tsunoda Y, Abe S, Sato T, Nakamura K, Suzuki M, et al. Lack of lacto/neolacto-glycolipids enhances the formation of glycolipid-enriched microdomains, facilitating B cell activation. Proc Natl Acad Sci USA. 2010;107:11900–11905. doi: 10.1073/pnas.0914298107. doi:10.1073/pnas.0914298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitoux D, Nasr R, de The H. Acute promyelocytic leukemia: New issues on pathogenesis and treatment response. Int J Biochem Cell Biol. 2007;39:1063–1070. doi: 10.1016/j.biocel.2007.01.028. doi:10.1016/j.biocel.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Wennekes T, van den Berg RJ, Boot RG, van der Marel GA, Overkleeft HS, Aerts JM. Glycosphingolipids—nature, function, and pharmacological modulation. Angew Chem Int Ed Engl. 2009;48:8848–8869. doi: 10.1002/anie.200902620. doi:10.1002/anie.200902620. [DOI] [PubMed] [Google Scholar]
- Yu RK. Development regulation of ganglioside metabolism. Prog Brain Res. 1994;101:31–44. doi: 10.1016/s0079-6123(08)61938-x. doi:10.1016/S0079-6123(08)61938-X. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306(5702):1786–1789. doi: 10.1126/science.1103440. doi:10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.