Abstract
We demonstrate that the unique green algal iron assimilatory protein, FEA1, is able to complement the Arabidopsis iron-transporter mutant, irt1, as well as enhance iron accumulation in FEA1 expressing wild-type plants. Expression of the FEA1 protein reduced iron-deficient growth phenotypes when plants were grown under iron limiting conditions and enhanced iron accumulation up to fivefold relative to wild-type plants when grown in iron sufficient media. Using yeast iron-uptake mutants, we demonstrate that the FEA1 protein specifically facilitates the uptake of the ferrous form of iron. Significantly, the FEA1 protein does not increase sensitivity to toxic concentrations of competing, non-ferrous metals nor facilitate their (cadmium) accumulation. These results indicate that the FEA1 protein is iron specific consistent with the observation the FEA1 protein is overexpressed in cadmium stressed algae presumably to facilitate iron uptake. We propose that the FEA1 iron assimilatory protein has ideal characteristics for the iron biofortification of crops and/or for facilitated iron uptake in plants when they are grown in low iron, high pH soils, or soils that may be contaminated with heavy metals.
Keywords: FEA1, iron uptake, yeast, Arabidopsis thaliana
Introduction
Iron is essential for all living organisms and its deficiency is among the most widespread human nutritional problems in the world. Among the two billion anemic people worldwide approximately 50% of all anemia cases are attributed to Fe deficiency (Mason et al., 2001). Globally, approximately 1.9 million disability adjusted life years and over 100,000 deaths are attributed to iron malnutrition (Caulfield et al., 2006). Often, subsistence farmers must rely primarily on plant-based sources of iron in their diets. The iron content of many plant-based foods may be insufficient to meet dietary requirements, however.
Even though iron is one of the most abundant elements in the earth’s crust, due to the low solubility of ferric iron it is considered as the third most limiting nutrient for plant growth (Grotz and Guerinot, 2006). This poses a problem for plants since iron is essential for a variety of metabolic processes (Hell and Stephan, 2003). Consequently, plants grown on calcareous (high pH) or low iron soils have reduced growth (Marschner, 1995). Excess iron can also be toxic to plants. The ferrous form of iron can react spontaneously with hydrogen peroxide to produce damaging oxygen radicals (Halliwell and Gutteridge, 1999). Since many plants reduce ferric iron to ferrous iron to increase its solubility, the uptake, and redox chemistry of iron must be highly regulated to meet the metabolic demand for iron as well as limit iron-induced damage to cells (Eide, 2000; Connolly and Guerinot, 2002; Curie and Briat, 2003).
Plants have evolved two strategies to obtain iron from the soil (Schmidt, 2003). The Strategy I mechanism used by all plants except graminaceous plants includes: (1) ATP-dependent proton extrusion to solubilize Fe(III), (2) reduction of the solubilized Fe(III) to Fe(II) by membrane-bound ferric reductases, and (3) transport of Fe(II) into the plant root cell by iron transporters following oxidation to Fe(III). It is well known that all these activities are upregulated in roots under iron deficiency (Eide et al., 1996; Robinson et al., 1999). Strategy II is a chelation-based approach limited to graminaceous plants (Von Wiren et al., 1994; Mori, 1999; Curie et al., 2001). It has been found that rice plants utilize both strategies for iron uptake in submerged growth conditions (Ishimaru et al., 2006).
Among single celled eukaryotes, iron uptake is perhaps best characterized in yeast (Saccharomyces cerevisiae; Askwith and Kaplan, 1998). In yeast, iron is solubilized by reduction of ferric to ferrous iron by ferric reductases, ferrous iron is then transported across the plasma membrane by either high affinity transporters (Fet3p/Ftr1p complex) or by low-affinity and low-specificity transporters (Fet4). The high affinity iron transporter complex includes the Fet3p protein, a multi-copper oxidase that oxidizes Fe(II) to Fe(III). Ferric iron is then transported across the membrane by the trivalent cation-specific permease, Ftr1p (De Silva et al., 1995; Stearman et al., 1996). Similar iron transport strategies are observed in fungi, bacteria, mammals, Chlamydomonas reinhardtii, and plants (Askwith and Kaplan, 1997; Herbik et al., 2002; Huston et al., 2002; La Fontaine et al., 2002; Hoopes and Dean, 2004). Yeast also utilizes low-affinity metal transporters such as FET4 which transports ferrous iron (Dix et al., 1997). In contrast to Ftr1p-mediated uptake, however, the low-affinity iron transport pathway is Fet3p-independent. Thus, this pathway can supply iron to the cell when Fet3p activity is absent (Hassett et al., 2000).
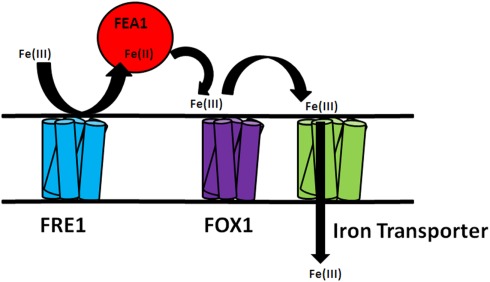
More recently, iron uptake and homeostasis has been characterized in single celled algae (Eckhardt and Buckhout, 1998; Lynnes et al., 1998; Weger, 1999; Weger and Espie, 2000; Allen et al., 2007; Chen et al., 2008; Long et al., 2008) and the associated iron transporters and periplasmic ferric reductases have been cataloged in Chlamydomonas (Merchant et al., 2007). The availability of a Chlamydomonas genome sequence has led to the identification of a number of metal transporters known from other organisms (La Fontaine et al., 2002; Rosakis and Köster, 2004; Merchant et al., 2007). Functional analysis based on the expression pattern of these genes is underway (Rubinelli et al., 2002; Hanikenne et al., 2005; Allen et al., 2007). One of the unique algal gene products involved in iron uptake is the H43 or FEA1 protein. The H43 protein (FEA1 is the homolog in Chlamydomonas reinhardtii) was shown to be secreted into the periplasm of the green alga, Chlorococcum littorale, during iron-deficient growth conditions (Sasaki et al., 1998). Subsequently, a gene encoding a similar protein was discovered in Chlamydomonas during a functional genomics analysis of genes whose expression was upregulated by cadmium exposure (Rubinelli et al., 2002). These studies demonstrated that FEA1 gene encodes an iron assimilation protein presumably functioning as an iron chaperonin that delivers iron to metal transporters (Figure 1).
Figure 1.
Hypothetical model of FEA1 iron assimilation in Chlamydomonas reinhardtii. Available ferric iron is reduced to ferrous iron by reductases (FRE1). FEA1 protein binds ferrous iron and facilitates uptake by iron transporters.
Recently, it was shown that the FEA1 protein is the major protein secreted into the periplasm by iron-deficient Chlamydomonas and is expressed coordinately with the FRE (encoding a ferrireductase) and FOX1 (encoding a multi-copper oxidase) genes (Allen et al., 2007). In this work, we demonstrate that the FEA1 protein complements the Arabidopsis irt1, iron-uptake mutant indicating that it is able to function in a variety of organisms. Additionally, roots of transgenic Arabidopsis plants (complemented wild-type plants) expressing the FEA1 protein show increased iron uptake even when grown in iron-deficient media or at high pH, conditions under which iron is very insoluble. We also show that the FEA1 protein facilitates only ferrous iron uptake and does not facilitate the uptake of other divalent metals even when present in toxic concentrations indicating the FEA1 protein is iron specific.
Materials and Methods
Construction of TI-plasmid binary vector and plant transformation
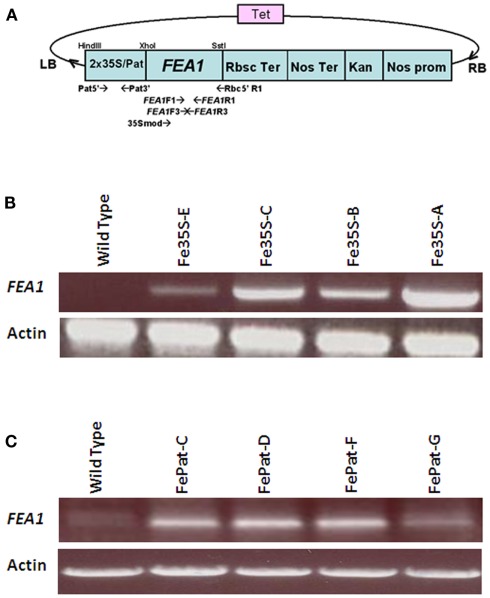
Plasmids used for transformation of wild-type Arabidopsis plants, were made by cloning the Chlamydomonas reinhardtii FEA1 as XhoI and SstI fragments behind 2 × 35S (enhanced Cauliflower mosaic virus) or patatin (potato, Solanum tuberosum L.) promoter in pKYLX plasmid backbone. Agrobacterium-mediated transformation of wild-type Arabidopsis plants (Columbia) was accomplished using floral-dip method (Clough and Bent, 1998) with Agrobacteria suspensions carrying the plasmid of choice. T1 seeds obtained from self-fertilization of the primary transformants were surface-sterilized and grown on Murashige and Skoog (MS) medium supplemented with kanamycin (40 mg/L) and vancomycin (500 mg/L). The antibiotic resistant plants were transferred to soil and self-fertilized to obtain the T2 seeds and was repeated to obtain T3 and T4 seeds. For transformation of the Arabidopsis irt1 mutant (Arabidopsis Biological Resource Center, Columbus, OH, USA), the FEA1 gene was cloned as SmaI and SstI fragments behind patatin promoter using vector pBI121 that carried the kanamycin resistant gene.
Plant growth conditions
The seeds of wild-type Arabidopsis thaliana (ecotype Columbia) were surface-sterilized, placed in the dark at 4°C for 2 days, and then sown on plates of MS medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 2% sucrose, 1 mM MES, and 0.7% agar, pH 5.7. Transgenic plants were selected on plates supplemented with kanamycin (40 μg/mL) and vancomycin (500 μg/mL). Plates were incubated at 23°C under constant illumination for 10–14 days until they reached the four- to six-true-leaf stage. Seedlings were then transferred to Metro-mix or Fafard #2 potting soil (Conrad Fafard, Inc., Agawam, MA, USA) after selection. T4 seedlings were transferred to either iron sufficient [50 μM Fe(III)-EDTA] or iron-deficient {0 μM Fe(III)-EDTA or 300 μM FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]; Sigma-Aldrich, St. Louis, MO, USA} standard MS plates.
RNA analysis of transgenic plants
Total RNA from leaves and roots of wild-type and FEA1 transgenic plants (both 35S and patatin promoter) was isolated using the RNA-easy kit from Qiagen (Qiagen Inc., Valencia, CA, USA) according to the manufacturer’s instructions. To remove contaminating genomic DNA, RNAs were treated with the DNAase I (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The concentrations of RNAs were assessed using a Nanodrop-2000C (Thermo-scientific, Wilmington, DE, USA) according to the manufacturer’s instructions. DNase-treated RNA samples (0.5 μg) were reverse transcribed with an anchored oligo (dT) primer and 200 units superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) in a volume of 20 μl according to the manufacturer’s instructions. The primers used for FEA1 detection were FEA1-F1 (5′-CAAGCCCGTCGCACAGTTAAC-3′) and FEA1-R1, (5′-GCCTTGAAGTTGCGCAGCTTG-3′) amplifying an 850 bp fragment. Actin was used as an internal control.
Cotyledon emergence
Forty seeds each from wild-type and transgenic plants were surfaced-sterilized and sown on plates that were either iron sufficient [50 μM Fe(III)-EDTA] or iron-deficient [0 μM Fe(III)-EDTA]. Cotyledon emergence at 23°C under constant illumination was monitored for 10 days. Each value is the mean of three experiments.
Root growth analysis
Seeds from wild-type and transgenic plants were surfaced-sterilized, placed in the dark at 4°C for 2 days, and sown on MS medium. After 9 days incubation at 23°C under constant illumination, plants were transferred to large plates that were either iron sufficient [50 μM Fe(III)-EDTA] or iron-deficient [0 μM Fe(III)-EDTA]. Plates were placed in the growth chamber vertically so that the roots grew down along the surface of the agar. Pictures were taken at day 10.
pH dependent growth assay
The plants had been germinated and grown to four- to six-true-leaf stage on MS medium, and transferred to either iron sufficient [50 μM Fe(III)-EDTA] or iron-deficient {300 μM FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]} plates for 3 days before the pH assay was performed. The localized pattern of pH change around wild-type and transgenic roots was visualized by placing seedlings on medium containing 0.2 mM CaSO4 and the pH indicator Bromocresol Purple (0.006%, (w/v), solidified with 0.7% (w/v) agar. The pH of the medium was adjusted to 6.0 with NaOH.
Chlorophyll fluorescence analysis
Wild-type, IRT1, and transgenic seeds were surfaced-sterilized and sown on plates that were either iron sufficient [50 μM Fe(III)-EDTA] or iron-deficient [0 μM Fe(III)-EDTA]. After 2 weeks, seedlings were subjected to 2 s of actinic light (sensitivity 70%, irradiance 30%, electronic shutter at 500−1 or 30000−1 s) and the chlorophyll fluorescence intensity was measured every 0.04 s for 5 s using a kinetic fluorescence CCD camera (Handy FluorCam FC 1000-H; Photon system Instruments, Czech Republic). Each curve represents a general pattern shared by multiple areas on several seedlings of the same plant.
Growth and mineral analysis in Arabidopsis
Both wild-type and transgenic Arabidopsis (35S: FEA1 and patatin: FEA1, three independent lines for each) were grown with 0.072 M Fe (Sprint 330) for 48 days in hydroponics (as described by Gibeaut et al., 1997). For pH experiments, the plants were also grown in hydroponics and the pH was adjusted to 5, 7, and/or 8.5. The pH was checked and adjusted as needed each day during the entire course of experiment. To remove extraplasmic Fe, roots were washed with 5 mM sodium dithionite and 1.0 M magnesium sulfate for 7 min, followed by a 5 min wash with deionized water. Plant tissues were collected and dried for 48 h in a 60°C oven. Samples were sent to The Ohio State University, Wooster (http://oardc.osu.edu/starlab/default.asp) for Inductively Coupled Plasma-Mass Spectrometry analysis (ICP-MS).
Cloning into Δftr1 strain
Yeast strain BY4743Δftr1 (MATa/MATα, his3Δ1/his3Δ1, ura3Δ0/ura3Δ0, leu2Δ0/leu2Δ0, lys2Δ0/ + , met15Δ0/+) was obtained from Open Biosystems (Hunsville, AL, USA). The FEA1–Flag gene fusion (in the pYES2 yeast expression vector) was constructed as described in Rubinelli et al., 2002. The pYES2 vector containing FEA1 was transformed into the Δftr1 Saccharomyces cerevisiae strain (BY4743) using a standard lithium acetate/heat shock protocol (Gietz and Schiestl, 1991; Adams et al., 1998). Transformants were selected on synthetic complete–uracil (SC–URA) medium supplemented with 2% (w/v) glucose at pH 3.5 (Q-BIOgene, Inc.), and 100 μM FeSO4.Colonies that appeared after 3 days were verified for the presence of the FEA1 gene. FEA1 expression was induced by 2% (w/v) galactose.
Assessment of growth of the Δftr1 strain
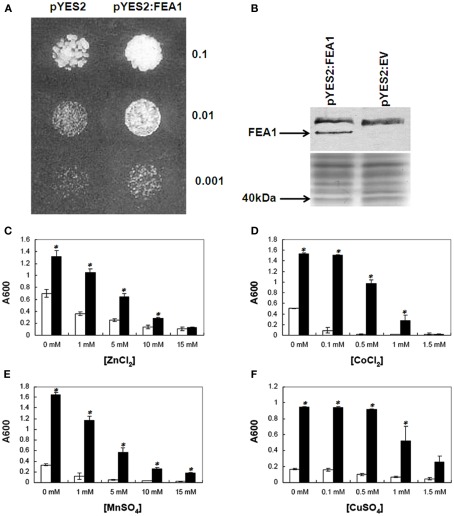
Δftr1 was transformed with either pYES2 or pYES2–FEA1 and grown overnight in SC–URA media supplemented with 2% (w/v) galactose at pH 3.5. Yeast transformations were generated by the lithium acetate-based method (Gietz and Schiestl, 1991). Twenty microliters of cells corresponding to a final OD600/mL of 0.1, 0.01, or 0.001 was spotted on SC–URA medium supplemented with 2% (w/v) galactose at pH 3.5.
Immunoblot analysis
Transformed yeast cells were grown overnight in SC–URA media supplemented with 2% (w/v) galactose at pH 3.5. Equal amounts of culture based on OD at 600 nm were harvested by centrifugation, washed once with water and resuspended in 100 μL of sample buffer (0.06 M Tris–HCl, pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 0.0025% (w/v) bromophenol blue). Solubilized cells were heated at 95°C for 5 min. Samples were centrifuged for 30 s at 15,000 g to remove debris and 50 μl of the sample was then separated by SDS-PAGE using 10% ready cast gels (Bio-Rad, Hercules, CA, USA) at 20 mA for 3 h. Immunoblots were performed according to the method of Rajamani (2006). Membrane was immuno-detected with anti-FLAG M2-alkaline phosphatase antibody.
Assessment of metal sensitivity/toxicity
The sensitivity of Δftr1 yeast, transformed with either the pYES2 empty vector or pYES2:FEA1, to transition metals was tested in SC–URA media containing 2% (w/v) galactose at pH 3.5 supplemented with various concentrations of cobalt, copper, manganese, or zinc. Yeast cultures were grown in 25 mL of medium for 18 h at 28 C. The cultures were then diluted to a starting concentration of 10 cells/mL in 12 mL of liquid medium supplemented with the appropriate amount of transition metal. The cultures were grown for 24 h at 28 C, and growth rates were monitored spectrophotometrically at A600 nm.
Iron dependent growth assay in yeast iron uptake mutants
Δfer1fer2 reductase mutant yeast strains were kindly provided by Caroline C. Philpott, NIH, MD, USA. The ferric reductase mutant strain was transformed with either the pESC–LEU–FEA1 or the empty vector pESC–LEU (EV). Yeast transformations were performed by the lithium acetate-based method (Gietz and Schiestl, 1991). Cultures were grown overnight on SD–LEU + 50 μM FeCl3 and induced with SG–LEU (no iron added) for 16–18 h. The optical density of the culture was adjusted to 0.1 and diluted 10 and 100 times. A small volume (5 μL) of each dilution was spotted on plates containing synthetic defined SG–LEU media containing 50 μM FeCl3 and SG–LEU media containing 50 μM FeCl3 + 1.0 mM sodium ascorbate. Plates were grown at 28°C for 2–3 days.
RNA extraction and semi-quantitative RT-PCR analysis
Total RNA was extracted from yeast cells using MasterPure™ yeast RNA Purification Kit (Epicenter Biotechnologies., Madison, WI, USA) according to manufacturer’s instructions. cDNA was synthesized from 1–2 μg total RNA using the first-strand synthesis protocol with M-muLV reverse transcriptase (New England BioLabs., MA, USA) according to manufacturer’s instructions. Yeast actin gene Act1p (Accession number: NP_116614) was used as internal control. RT-PCR primers for FEA1 (FEA1-F: 5′ GAGAGCGGCCGCAATGTCGGTCGGATTTC 3′ and FEA1-R: 5′ GAGAGAGCTC CGCAATGCTGCGCAGGGTCT 3′) and Act1p (Actin-F: 5′ TCGAACAAGAAAT GCAAACCG 3′ and Actin-R: 5′ GGCAGATTCCAAACCCAAAAC 3′) were used. The reaction mixture containing template, primers, buffer, dNTPs, and Taq DNA polymerase was subjected to initial denaturation (94°C) for 4 min, followed by repeated denaturation (94°C) for 30 s, annealing (53°C) for 30 s, and elongation (72°C) for 1 min for a total of 35 cycles. Final elongation step was carried out at (72°C) for 10 min.
Iron–cadmium competition experiment
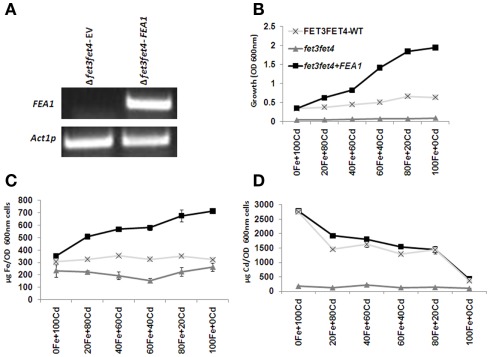
For iron-cadmium competition experiments, wild-type FET3FET4 (DY150; MATa/MATa ade2/+ can1his3 leu2 trp1 ura3), mutant Δfet3fet4 (DEY1453; MATa/MATa ade2/+ can1 his3 leu2 trp1 ura3 fet3–2::HIS3 fet4–1::LEU2), and Δfet3fet4 yeast mutants transformed with the pYES2:FEA1 were grown in SC–URA media containing 2% (w/v) galactose at various concentrations of cadmium and iron. Yeast cultures were grown in 25 mL of medium for 24 h at 28 C, and growth rates were monitored spectrophotometrically at A600 nm. Yeast cells were washed with 5.0 mM sodium dithionite and 1.0 mM EDTA, pH 8.0, followed by three washes of distilled water. Samples are lyophilized and sent to The Ohio State University, Wooster (http://oardc.osu.edu/starlab/default.asp) for ICP-MS analysis.
Results
FEA1 complements the Arabidopsis irt1 mutant
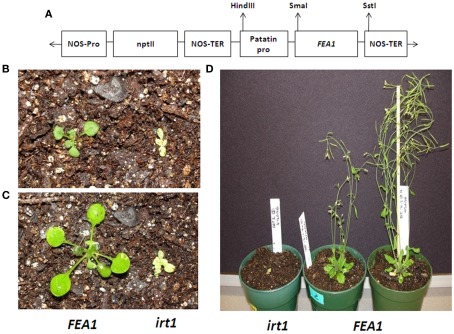
The IRT1 protein is one of the essential metal transporters required for iron transport into plant root hairs (Vert et al., 2002). To determine if the FEA1 protein facilitates iron assimilation in plants, the Arabidopsis irt1 mutant (irt1-1) was transformed with the Chlamydomonas FEA1 gene. The FEA1 gene was expressed under control of the patatin promoter (Figure 2A). As shown in Figures 2B,C, the FEA1 gene was able to rescue the lethal irt1 mutant phenotype. At the four- to six-true-leaf stage, FEA1 complemented irt1 plants showed normal seedling size relative to wild-type plants while the non-complemented irt1 mutant was stunted and chlorotic (Figures 2B,C). At 5 weeks post germination, the FEA1 complemented plants produced flowers and fully fertile siliques while the non-complemented irt1 mutant failed to develop beyond the four- to six-true-leaf stage (Figure 2D).
Figure 2.
FEA1 complements Arabidopsis irt1 mutant. (A) The T-DNA region pBI121 containing patatin promoter and FEA1 gene used in the transformation of Arabidopsis mutant line irt1 (B) Growth of irt1 mutant and irt1 mutant and wild-type complemented with the FEA1 at the four- to six-true-leaf stage (day 1), (C) day 5, (D) day 35.
FEA1 transgenic plants show enhanced cotyledon emergence
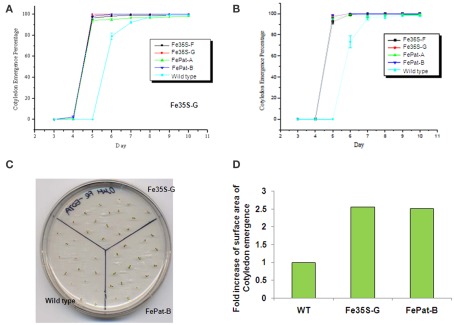
To examine further FEA1’s ability to facilitate iron uptake in planta, we determined the phenotypic effects of expressing the FEA1 gene in wild-type Arabidopsis (ecotype Columbia) driven by the patatin and the 2 × 35S promoter (Figure 3A). RT-PCR experiments confirmed the expression of the FEA1 gene in leaf and root tissues of transgenic plants (Figures 3B,C). Initially, germination rate and cotyledon emergence were evaluated. Seeds from wild-type and transgenic FEA1 plants were sown on iron-deficient [0 μM Fe(III)-EDTA] and iron sufficient [50 μM Fe(III)-EDTA] medium. In both media, over 90% of the transgenic seeds had full cotyledon emergence by day 5. However, cotyledon emergence was not observed on day 5 for wild-type seeds and it was below 80% on day 6 when they were sown on iron-deficient media. All plants had completed cotyledon emergence, however, by day 8 (Figures 4A–D).
Figure 3.
RT-PCR analysis of expression in FEA1 transgenic plants. (A) Modified pKYLX–FEA1 vector used for transformation of wild-type Arabidopsis thaliana. Two separate plasmids were used for transforming plants with the FEA1 gene driven by the 35S promoter (constitutive) and patatin (storage organ) promoter. (B) RT-PCR analysis of FEA1 expression in leaves of independent 2 × 35S–FEA1 transgenic plants. (C) RT-PCR analysis of FEA1 expression in roots of independent patatin-FEA1 transgenic plants.
Figure 4.
FEA1 enhances cotyledon emergence from seeds. Time of cotyledon emergence was scored on wild-type and transgenic seeds sown on plates that were (A) iron-deficient [0 μM Fe(III)-EDTA] or (B) iron sufficient [50 μM Fe(III)-EDTA]. (C) Visual representation of a plate showing cotyledon emergence grown under iron-deficient [0 μM Fe(III)-EDTA] media at day 5. (D) ImageJ based measurement of Arabidopsis cotyledon emergence grown under iron sufficient [50 μM Fe(III)-EDTA] media at day 14. Surface area was measured for 20 plants for each transgenic and wild-type line. Values for wild-type were normalized to 1 and the fold increase for the transgenic plants is shown.
FEA1 transgenic plants expresses strategy I responses under iron-deficient growth conditions
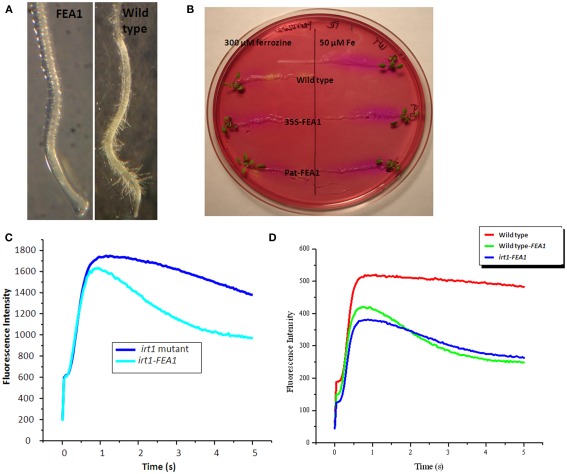
One indicator of iron deficiency is a proliferation of root hair growth (Vert et al., 2002). Root hairs are the primary structures through which iron uptake takes place. To determine if FEA1 transgenic wild-type plants were more efficient at iron uptake, root hair growth of wild-type and FEA1 transgenic wild-type plants was compared on iron sufficient or deficient media. Wild-type plants grown under Fe-deficient conditions developed more lateral root branches and root hairs than transgenic plants (Figure 5A). At the root tip, extensive root hair formation was observed for wild-type plants while transgenic plants had few root hairs (Figure 5A). These results suggest that FEA1 transgenic plants are more iron sufficient than wild-type plants grown under limiting iron concentrations.
Figure 5.
FEA1 transgenic plants expresses strategy I responses under iron-deficient growth conditions. (A) Lateral root branches of wild-type and FEA1 transgenic plants grown in iron-deficient media. Pictures were taken at day 10. (B) Iron deficiency induced acidification of the rhizosphere in wild-type and FEA1 transgenic roots. (C) Chlorophyll fluorescence induction kinetics of irt1 mutant and irt1 complemented FEA1 seedlings sown on iron sufficient media for 2 weeks. (D) Chlorophyll fluorescence induction kinetics of wild-type, irt1 complemented FEA1, and FEA1 transgenic seeds grown on iron-deficient media for 2 weeks.
One of the major responses of Strategy I plants to iron deficiency is acidification of the rhizosphere to solubilize iron (Yi et al., 1994). To determine whether FEA1 transgenic wild-type plants exhibited rhizosphere acidification plants were transferred to iron-deficient media followed by transfer to iron-free media containing the pH indicator Bromocresol purple. Roots of iron-deficient wild-type plants reduced the pH of the medium to below 5.2, as indicated by the pH indicator color change from red to yellow. In contrast, iron sufficient plants and transgenic plants grown on iron-deficient media did not acidify the medium (Figure 5B) indicating that transgenic plants were iron sufficient. These results suggest that expression of the FEA1 protein in wild-type plants enhances their ability to take up iron.
Since iron is an essential cofactor of the photosynthetic electron transfer chain, we hypothesized that FEA1 expression could complement impairments in photosynthetic electron transfer when wild-type plants were grown under iron-deficient growth conditions. Seeds from FEA1 complemented irt1 plants and the non-complemented irt1 mutants were grown on iron sufficient medium for 2 weeks prior to measuring chlorophyll fluorescence induction kinetics. These studies allow us to monitor potential impairment in whole chain electron transfer processes including non-photochemical quenching (NPQ) of chlorophyll fluorescence associated with the generation of a sufficient proton gradient (Müller et al., 2001). As shown in Figure 5C, the irt1 mutant exhibited normal variable fluorescence (Fv = Fmax − Fo), however, after reaching Fmax the subsequent decrease in fluorescence associated with NPQ was diminished in the irt1 mutant relative to the FEA1 complemented mutant. These results indicate impairment in the ability to generate a proton gradient sufficient to induce NPQ in the irt1 mutant. We also measured chlorophyll fluorescence kinetics in wild-type, FEA1 complemented irt1mutants, and patatin-FEA1 transgenic plants after 2 weeks growth on iron-deficient [0 μM Fe(III)-EDTA] medium. Iron-deficient wild-type plants exhibited a dramatic rise in chlorophyll fluorescence intensity with no subsequent decrease (Figure 5D). In contrast, the FEA1 transgenic wild-type and FEA1 complimented irt1 mutant had near normal chlorophyll fluorescence kinetics demonstrating the typical Kautsky curve (Figure 5D). These results demonstrate that wild-type and irt1 mutants expressing the FEA1 protein had enhanced photosynthetic ability when grown under iron-deficient conditions unlike wild-type plants. The reduced ability to quench maximum chlorophyll fluorescence may result from impairment of electron transfer in the cytochrome b6f complex which has multiple iron-containing redox-active cofactors.
FEA1 transgenic plants show increased iron concentrations
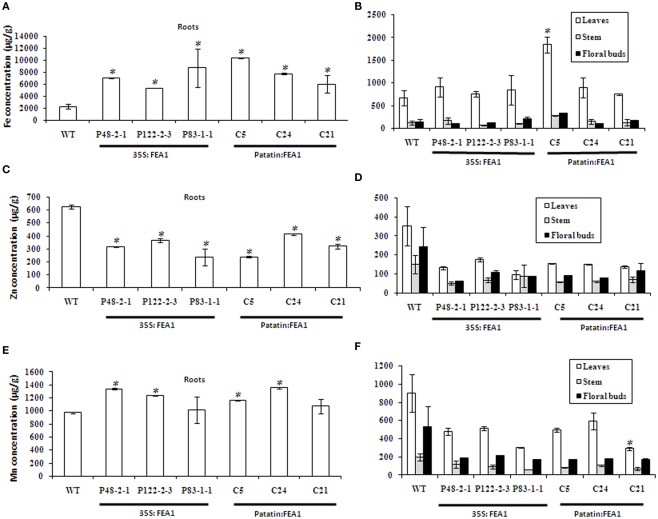
To compare iron accumulation in wild-type and transgenic FEA1 lines, plants were grown hydroponically for 48 days. Roots, stem, leaves, and floral buds were then harvested and used for ICP-MS elemental analysis. Three independent transgenic lines for each construct (35S:FEA1 and patatin: FEA1) were included in the study. Results revealed a three to fivefold increase of Fe levels in transgenic roots compared to wild-type roots (Figure 6A). No significant differences were observed in the iron levels of leaves between the transgenic and wild-type plants except for the C5 (patatin: FEA1) line which had a two-threefold increase in leaf iron levels (Figure 6B). Similarly, no significant differences were observed in the iron levels of stems and floral buds between the FEA1 transgenics and wild-type plants (Figure 6B). Interestingly, Zn concentrations in the roots were reduced in both 35S and patatin-FEA1 transgenics relative to wild-type (Figure 6C). Zinc concentrations in the leaves, stems, and floral buds were slightly reduced in the transgenic plants when compared with the wild-type, however, the differences were not significant (Figure 6D). Manganese concentrations in transgenic roots were slightly elevated when compared with the wild-type (Figure 6E), however, manganese concentrations in the leaves, stems and floral buds were reduced in the FEA1 transgenic plants when compared with the wild-type (Figure 6F). Again, however, manganese levels were not significantly different between transgenic and wild-type plants. These results indicate that the impact of FEA1 expression is most pronounced in roots actively involved in iron uptake.
Figure 6.
FEA1 specifically increases root iron levels in transgenic plants. (A) Iron levels in roots; (B) Iron levels in leaves, stems, and floral buds; (C) Zinc levels in roots; (D) Zinc levels in leaves, stems, and floral buds; (E) Manganese levels in roots; (F) Manganese levels in leaves, stems, and floral buds. Values are the average of triplicates. Error bars represent SE. The asterisk (*) indicates a statistically significant differences between wild-type and transgenics, determined by Student’s t-test, with P < 0.05.
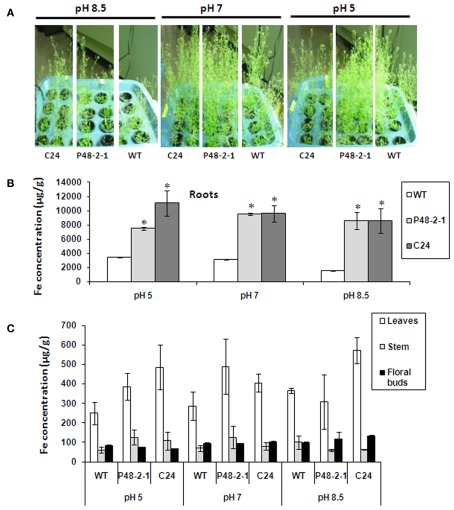
Crops grown in high soil pH often have substantially impaired growth due to iron deficiency (Marschner, 1995). We compared the growth of wild-type and transgenic FEA1 plants grown at pHs 5, 7, and 8.5. As expected, all the plants grown at pH 5 and 7 were normal and healthy. Significantly, patatin-FEA1 transgenic plants grew much better than the wild-type plants at high pH 8.5 (Figure 7A). There was also three to fourfold increase in iron levels in the roots of 35S- and patatin-FEA1 transgenic plants relative to wild-type plants at pH 8.5 (Figure 7B). Similar results were observed for roots at other pH levels tested (Figure 7C). There were no significant differences in leaf, stem and floral bud iron concentrations between the transgenics and wild-type at different pH levels (Figure 7C). These results indicate that the FEA1 iron assimilatory protein functions well at high pH conditions and its expression does not alter iron steady-state levels between different plant organs.
Figure 7.
FEA1 facilitates iron uptake in Arabidopsis at high pH. Wild-type and transgenic Arabidopsis plants were grown hydroponically on 0.072 M Fe (Sprint 330) until day 48. (A) Image of wild-type (WT) and transgenic Arabidopsis plants grown at different pH levels (pH 5, 7, and 8.5). (B) Iron concentration of roots. (C) Iron concentrations of stems, leaves, and floral buds. Values are average of triplicates. Error bars represent SE. The asterisk (*) indicates statistically significant differences between wild-type and transgenics, determined by Student’s t-test, with P < 0.05.
FEA1 protein facilitates uptake of ferrous not ferric iron in yeast
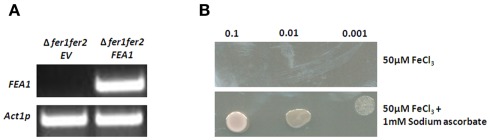
To increase iron solubility strategy I plants and yeast reduce ferric iron to more soluble ferrous iron. To determine which iron valence state is utilized by FEA1, ferric, and ferrous iron dependent growth in yeast ferric reductase mutants (Δfer1fer2) complemented with the FEA1 gene was assessed. RT-PCR analysis for the FEA1 transcript in yeast iron uptake mutants (Δfer1fer2) transformed with pESC–LEU–FEA1 confirmed expression of the transgene (Figure 8A). As shown in Figure 8B, the FEA1 transformants did not grow on media lacking sodium ascorbate [reduces Fe(III) to Fe(II)] indicating the FEA1 protein does not facilitate ferric iron uptake. On the contrary, yeast transformed with the FEA1 gene grew well with sodium ascorbate and ferric iron (Figure 8B).
Figure 8.
FEA1 protein facilitates uptake of ferrous iron in yeast. The yeast ferric reductase (Δfer1fer2) mutant was transformed with the pESC–LEU–FEA1 vector. (A) RT-PCR expression of FEA1 transgenic yeast. Actin was used as an internal expression control. (B) Growth of yeast on synthetic defined SG-LEU media containing 50 μM FeCl3 or SG-LEU media containing 50 μM FeCl3 + 1 mM sodium ascorbate. Plates were grown at 28°C for 2–3 days.
FEA1 does not transport non-ferrous metals
Iron-deficient yeast strains (ftr1 and fet3 mutants) are particularly sensitive to high concentrations of non-ferrous transitions metals due to their facilitated uptake by the facultative over-expression of the low-specificity iron transporter, Fet4p (Li and Kaplan, 1998). When the ftr1mutant strain is grown under limiting iron conditions the expression of the low-affinity, low-specificity iron transporter, FET4, is induced. Under these conditions, ftr1 mutants become more sensitive to high concentrations of competing metals including zinc, manganese, cobalt, or copper ions due to their non-specific transport and accumulation in cells mediated by the FET4 transporter. As shown in Figures 9A,B, expression of the FEA1 gene in the yeast ftr1 iron permease mutant (BY4743–Δftr1) complemented growth. To determine if expression of FEA1 protein ftr1 mutants altered sensitivity to competing non-ferrous metals, iron-deficient cells were grown in the presence of elevated concentrations of non-ferrous metals. We observed no increased sensitivity to cadmium, cobalt, copper, zinc, or manganese in transgenic cells expressing the FEA1 protein relative to cells transformed with the empty vector. These results indicated that the FEA1 protein did not facilitate transport of non-ferrous metals under iron-deficient growth conditions (Figures 9C–F). In fact, in some cases, growth inhibition by competing non-ferrous metals was relieved FEA1 transgenics (see Figure 9D, for example) presumably due to increased iron uptake. Overall, these results indicate that the FEA1 protein facilitates uptake of only ferrous iron.
Figure 9.
FEA1 complements yeast Δ ftr1 mutant and is iron specific. (A) Cells transformed with either the pYES2: vector, or with pYES2:FEA1 were grown in liquid SC-URA galactose (pH 3.5) medium overnight. Cells equivalent to the indicated absorbance values at 600 nm were spotted on SC-URA galactose (pH 3.5) plates. OD for the grown cultures was adjusted to 0.1 and diluted 10 and 100 times. The picture was taken after 72 h incubation at 28 C. (B) Western blot of Flag-tagged FEA1 immuno-detected with anti-FLAG M2-alkaline phosphatase antibody. The Coomassie-blue stained gel is shown below the western blot to show equal loading of the protein. Sensitivity to toxic metal concentrations: Yeast transformed with the pYES2 empty vector (unfilled columns) or the pYES2:FEA1 (filled columns) vector were grown in SC-URA galactose (pH 3.5) for 18 h at 28 C and diluted to 10 cells/mL in fresh SC-URA galactose media (pH 3.5) supplemented with various concentrations of (C) ZnCl2, (D) CoCl2, (E) MnSO4, and (F) CuSO4. Growth was monitored after 24 h by measuring the absorbance of the culture at 600 nm. Data are means + SE of three replicate experiments. The asterisk (*) indicates statistically significant differences between pYES2 empty vector and pYES2:FEA1 yeast as determined by Student’s t-test, with P < 0.05.
The FEA1 protein does not facilitate the uptake of cadmium
To further test this hypothesis, we measured intracellular metal concentrations in cells grown in the presence of toxic concentrations of cadmium. Previously, we observed that the expression of the FEA1 gene was induced in Chlamydomonas cells exposed to high concentrations of cadmium (Rubinelli et al., 2002). We hypothesized that FEA1 expression under these conditions facilitated iron uptake to repair proteins and enzymes poisoned by cadmium. This hypothesis also implies that the FEA1 protein does not facilitate transport of metals other than iron. To determine whether the FEA1 protein facilitated cadmium uptake, we compared growth rates and metal content of the following yeast strains grown at various ratios of iron and cadmium; (1) yeast iron uptake mutants (Δfet3fet4), (2) FEA1 complemented Δfet3fet4 mutants, and (3) the wild-type yeast. RT-PCR analysis of Δfet3fet4 mutants transformed with pYES:FEA1 or with empty vector indicated that the FEA1 transgene was actively transcribed (Figure 10A). Significantly, the growth rate of the FEA1 complemented Δfet3fet4 mutants was enhanced at all iron concentrations tested relative to the empty vector transformed mutant and wild-type (Figure 10B). Analyses of the iron content of FEA1 complemented mutants indicated that there was greater iron accumulation in FEA1 expressing cell lines relative to wild-type and Δfet3fet4 mutants (Figure 10C). Importantly, there was no significant difference in cadmium content between the FEA1 complemented lines and the wild-type (Figure 10D). These results provide further evidence that the FEA1 protein is iron specific.
Figure 10.
FEA1 does not transport cadmium in transgenic yeast. (A) RT-PCR analysis of FEA1 expression in Δfet3fet4 mutant yeast cells transformed with pYES2:FEA1. Actin expression was used as an internal control. (B) Growth rates of wild-type, iron uptake mutants (Δfet3fet4), and mutants complemented with pYES2:FEA1. Cell were grown in SC-URA medium containing 2% (w/v) galactose with various amounts (μM) of cadmium and iron (C) ICP-MS analysis of iron concentrations normalized for cell number. (D) ICP analysis of cadmium concentrations normalized for cell number. Data are means + SE of three replicate experiments.
Discussion
FEA1 is a novel iron specific assimilatory protein
One of the unique features of the FEA1 protein is its high specificity for iron. We observed that the growth of yeast ftr1 mutants complemented with FEA1 was not altered relative to empty vector transformants by potentially toxic concentrations of copper, cobalt, zinc, or manganese (Figures 9C–F). These results suggest that FEA1 protein does not facilitate transport of other transition metals. In addition, analyses of the cadmium content of wild-type and FEA1 expressing transgenic yeast grown at various iron and cadmium ratios revealed no difference in cadmium content between strains expressing or not expressing the FEA1 protein (Figure 10D). These results are consistent with the previous observations that cadmium induces FEA1 expression in Chlamydomonas presumably to facilitate iron uptake in the presence of potentially competing toxic metals (Rubinelli et al., 2002).
In oxygenated aqueous solutions, iron exists primarily in the ferric form and significant proportion is chelated by organic acids in different organisms (Morel et al., 2008). The redox state of iron transported through the plasma membrane is still a major dispute (Koropatkin et al., 2007; Badarau et al., 2008). Our results demonstrate that FEA1 transports ferrous iron in yeast (Figure 8B) These results are consistent with the observed co-expression of ferric reductase (FRE) and FEA1 during iron-deficient growth in Chlamydomonas (Allen et al., 2007).
FEA1 functions in dicotyledonous plants
Significantly, the FEA1 iron assimilatory protein complements Arabidopsis mutants impaired in iron uptake. The Arabidopsis irt1 mutant was fully rescued by complementation with the FEA1 gene (Figure 2). In addition, full fertility was recovered in FEA1 complemented irt1 mutants. FEA1 expressing plants produced normal flowers, siliques, and seeds when production of these structures has shown to be arrested in wild-type plants grown in low iron (Waters et al., 2006). Earlier studies have shown that iron is essential in germination of nramp3 nramp4 double mutant plants. The germination arrest of this mutant under low-Fe supply was rescued either by expression of the AtNRAMP3 or AtNRAMP4 genes, or by supplying high Fe levels to the seedlings (Lanquar et al., 2005). Both 35S and patatin-FEA1 transgenic plants also exhibited rapid cotyledon emergence under both iron sufficient and deficient growth conditions compared to wild-type plants suggesting greater iron storage reserves or iron uptake efficiency in FEA1 transgenics (Figures 4A,B). During the first 2 days of seed germination, iron is mobilized from vacuoles. By the third day IRT1 expression increases rapidly (Lanquar et al., 2005). In the Arabidopsis transgenics, FEA1 expression may have increased both the initial iron supply as well as enhanced iron uptake by IRT1 (Figure 4C).
FEA1 transgenic plants shows iron sufficient response
One typical response to iron deficiency in plants is increased root hair growth (Schmidt et al., 2000). Previous studies have clearly indicated that lack of nutrients may increase in lateral root branching in plants (Linkohr et al., 2002). Moreover, IRT1 expression is shown to be localized in the root hairs and epidermis of iron-deficient plants (Vert et al., 2002). Lateral root branching and root hair formation thus facilitates iron uptake, a default iron stress response. The dense and extensive root hair formation in wild-type roots grown under iron-deficient [0 μM Fe(III)-EDTA] medium clearly demonstrated this iron stress response (Figure 5A). The fact that FEA1 transgenic plants showed normal lateral root growth and reduced root hair formation compared to wild-type plants (Figure 5A) suggests that FEA1 protein facilitates iron uptake and supplies adequate amounts of iron even under iron-deficient conditions.
In addition to increasing lateral root branching and root hair formation under low iron conditions, Strategy I plants also acidify the rhizosphere through activation of aspecific plasma membrane H+-ATPase in root epidermal cells (Vert et al., 2002; Curie and Briat, 2003). Iron-deficient wild-type plants reduced the pH in the medium whereas iron-deficient FEA1 transgenic plants did not, a phenotype consistent with growth under iron sufficient conditions (Figure 5B). The lack of a rhizosphere acidification response under iron limited growth in FEA1 transgenic plants indicated that the transgenic plants were able to take up iron more efficiently than wild-type plants. When wild-type plants were exhibiting iron stress responses, both 35S and patatin-FEA1 transgenic plants were still able to utilize trace amounts of iron from the medium presumably due to facilitated iron assimilation by the FEA1 protein (Figure 5B).
One of the physiological processes most sensitive to iron deficiency is photosynthesis. The high abundance of redox-active iron cofactors in the cytochrome b6f complex relative to other electron transfer complexes may impart a greater susceptibility to iron depletion (Okegawa et al., 2005). Regulation of NPQ under high light conditions requires a functional cytochrome b6f complex to regulate chlorophyll fluorescence quenching by carotenoids including zeaxanthin whose abundance increases at low-luminal pH. The low luminal pH is generated in part by proton pumping across the thylakoid membrane associated with an active cytb6f complex. We have shown that the FEA1 complemented irt1 plants grown on iron sufficient medium exhibited normal variable chlorophyll fluorescence and a typical decline in fluorescence after reaching Fmax indicative of functional NPQ-mediated energy quenching processes (Figure 5C). Significantly, when FEA1 complemented irt1 (irt1–FEA1) and FEA1 transgenic wild-type plants (WT-FEA1) were grown on iron-deficient medium only the FEA1 expressing wild-type plants exhibited normal chlorophyll fluorescence kinetics indicating that the FEA1 transgenic wild-type plants had greater Cytb6f activity consistent with an elevated iron status relative to wild-type plants when grown under low iron conditions (Figure 5D).
FEA1 transgenic plants have increased iron concentrations in roots
Mineral analysis of wild-type and transgenic FEA1 plants revealed a three to fivefold increase in iron concentrations in both 35S:FEA1 and patatin: FEA1 roots relative to wild-type (Figure 6A) indicating that the FEA1 facilitates iron uptake in plants Interestingly, there was a slight reduction of zinc concentrations in roots of the transgenic plants expressing the FEA1 protein driven by the 35S promoter relative to wild-type. In contrast, when FEA1 was expressed in cassava roots using the patatin promoter, there were no significant differences in zinc concentrations in the roots between the wild-type and transgenic plants (unpublished data).
Importantly, FEA1 expressing Arabidopsis plants exhibited increased iron levels even when grown at high pHs, conditions where iron is very insoluble (Figure 7B). It is well known that most dicot plants reduce ferric iron to ferrous iron (Schmidt, 2003). Rice plants usually utilize strategy II mechanisms, but recent reports indicate that transgenic rice plants expressing the refre1/372 gene (yeast Fe3+ chelate-reductase gene) display tolerance to low-Fe availability in calcareous soils by enhancing Fe3+ chelate-reductase activity (Ishimaru et al., 2006). These transgenic rice plants took up more ferrous iron than control plants under Fe-deficient conditions, indicating that the transformants successfully reduced chelated Fe3+ to Fe2+ and took up Fe2+ by a Fe2+ transporter. We demonstrate that FEA1 transgenic plants exhibit enhanced tolerance to high pH environments presumably by an analogous mechanism of facilitated ferrous iron uptake.
In summary, we have shown that the FEA1 assimilatory protein is functional in a diverse group of organisms including yeast and Arabidopsis. FEA1 transgenic plants had three to fivefold higher iron levels than wild-type plants and grew well under iron-deficient conditions that impair the growth of wild-type Arabidopsis. The high metal specificity and the ability to facilitate iron uptake at high pH suggest that expression of the FEA1 protein in plants may be an ideal strategy to increase iron uptake even in soils with reduced iron levels or soils contaminated with toxic heavy metals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Drs. Narayanan and Ihemere contributed equally to this work. We are grateful to Dr. Caroline Philpott, NIH, MD, USA for all the mutant yeast strains. Technical assistance from Richard A. Gallenstein is greatly appreciated. This research was supported by Ohio Sea Grant (NOAA) and the Bill and Melinda Gates Foundation for BioCassava Plus Project.
References
- Adams A., Gottschling D., Kaiser C., Stearns T. (1998). Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Allen M. D., del Campo J. A., Kropat J., Merchant S. S. (2007). FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6, 1841–1852 10.1128/EC.00205-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C., Kaplan J. (1997). An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272, 401–405 10.1074/jbc.272.1.401 [DOI] [PubMed] [Google Scholar]
- Askwith C., Kaplan J. (1998). Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23, 135–138 10.1016/S0968-0004(98)01192-X [DOI] [PubMed] [Google Scholar]
- Badarau A., Firbank S. J., Waldron K. J., Yanagisawa S., Robinson N. J., Banfield M. J., Dennison C. (2008). FutA2 is a ferric binding protein from Synechocystis PCC 6803. J. Biol. Chem. 283, 12520–12527 10.1074/jbc.M709907200 [DOI] [PubMed] [Google Scholar]
- Caulfield L. E., Richard S. A., Rivera J. A., Musgrove P., Black R. E. (2006). ‘Stunting, wasting, and micronutrient deficiency disorders,” in Disease Control Priorities in Developing Countries, eds Jamison D., Breman J., Measham A. R., Alleyne G., Claeson M., Evans D. B., Jha P., Mills A., Musgrove P. (Washington, DC: Oxford University Press; ), 551–567 [Google Scholar]
- Chen J. C., Hsieh S. I., Kropat J., Merchant S. (2008). A Ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell 7, 541–545 10.1128/EC.00463-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Plant responses to metal toxicity. C. R. Acad. Sci. Paris 322, 43–54 [DOI] [PubMed] [Google Scholar]
- Connolly E. L., Guerinot M. (2002). Iron stress in plants. Genome Biol. 3, 1024–1030 10.1186/gb-2002-3-8-reviews1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Briat J.-F. (2003). Iron Transport and Signaling in Plants. Annu. Rev. Plant Biol. 54, 183–206 10.1146/annurev.arplant.54.031902.135018 [DOI] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- De Silva D. M., Askwith C. C., Eide D., Kaplan J. (1995). The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270, 1098–1101 10.1074/jbc.270.7.2974 [DOI] [PubMed] [Google Scholar]
- Dix D., Bridgham J., Broderius M., Eide D. (1997). Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J. Biol. Chem. 272, 11770–11777 10.1074/jbc.272.18.11770 [DOI] [PubMed] [Google Scholar]
- Eckhardt U., Buckhout T. J. (1998). Iron assimilation in Chlamydomonas reinhardtii involves ferric reduction and is similar to strategy I higher plants. J. Exp. Bot. 49, 1219–1226 10.1093/jexbot/49.324.1219 [DOI] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 5624–5628 10.1073/pnas.93.20.10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D. J. (2000). Metal ion transport in eukaryotic microorganisms: insights from Saccharomyces cerevisiae. Adv. Microb. Physiol. 43, 1–38 10.1016/S0065-2911(00)43001-8 [DOI] [PubMed] [Google Scholar]
- Gibeaut D. M., Hulett J., Cramer G. R., Seemann J. R. (1997). Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115, 317–319 10.1104/pp.115.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H. (1991). Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast 7, 253–263 10.1002/yea.320070307 [DOI] [PubMed] [Google Scholar]
- Grotz N., Guerinot M. L. (2006). Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochem. Biophys. Acta 763, 595–608 [DOI] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. M. C. (1999). Free Radicals in Biology and Medicine, 3rd Edn, Oxford: Oxford University Press [Google Scholar]
- Hanikenne M., Krämer U., Demoulin V., Baurain D. (2005). A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschyzon merolae. Plant Physiol. 137, 428–446 10.1104/pp.104.054189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett R., Dix D. R., Eide D. J., Kosman D. J. (2000). The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 351, 477–484 10.1042/0264-6021:3510477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R., Stephan U. W. (2003). Iron uptake, trafficking and homeostasis in plants. Planta 216, 541–551 [DOI] [PubMed] [Google Scholar]
- Herbik A., Bolling C., Buckhout T. J. (2002). The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 130, 2039–2048 10.1104/pp.013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes J. T., Dean J. F. (2004). Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol. Biochem. 42, 27–33 10.1016/j.plaphy.2003.10.011 [DOI] [PubMed] [Google Scholar]
- Huston W. M., Jennings M. P., McEwan A. G. (2002). The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45, 1741–1750 10.1046/j.1365-2958.2002.03132.x [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Suzuki M., Tsukamoto T., Suzuki K., Nakazono M., Kobayashi T., Wada Y., Watanabe S., Matsuhashi S., Takahashi M., Nakanishi H., Mori S., Nishizawa N. K. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346 10.1111/j.1365-313X.2005.02624.x [DOI] [PubMed] [Google Scholar]
- Koropatkin N., Randich A. M., Bhattacharyya-Pakrasi M., Pakrasi H. B., Smith T. J. (2007). The structure of the iron-binding protein, FutA1, from Synechocystis 6803. J. Biol. Chem. 282, 27468–27477 10.1074/jbc.M704136200 [DOI] [PubMed] [Google Scholar]
- La Fontaine S., Quinn J. M., Nakamoto S. S., Page M. D., Göhre V., Moseley J. L., Kropat J., Merchant S. (2002). Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1, 736–757 10.1128/EC.1.5.736-757.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V., Lelievre F., Bolte S., Hames C., Alcon C., Neumann D., Vansuyt G., Curie C., Schroder A., Kramer U., Barbier-Brygoo H., Thomine S. (2005). Mobilization of vacuolar iron by AtNRAMP3 and AtNAMP4 is essential for seed germination on low iron. EMBO J. 24, 4041–4051 10.1038/sj.emboj.7600864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Kaplan J. (1998). Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273, 22181–22187 10.1074/jbc.273.35.22181 [DOI] [PubMed] [Google Scholar]
- Linkohr B. I., Williamson L. C., Fitter A. H., Leyser O. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29, 751–760 10.1046/j.1365-313X.2002.01251.x [DOI] [PubMed] [Google Scholar]
- Long J., Sommer F., Allen M., Lu S., Merchant S. S. (2008). FER1 and FER2 encoding two ferritin complexes in Chlamydomonas reinhardtii chloroplasts are regulated by iron. Genetics 179, 137–147 10.1534/genetics.107.083824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynnes J. A., Derzaph T. L. M., Weger H. G. (1998). Iron limitation results in induction of ferricyanide reductase and ferric chelate reductase activities in Chlamydomonas reinhardtii. Planta 204, 360–365 10.1007/s004250050267 [DOI] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants, 2nd Edn, London: Academic press [Google Scholar]
- Mason J. B., Lotfi M., Dalmiya N., Sethuraman K., Deitchler M., Geibel S., Gillenwater K., Gilman A., Mason K., Mock N. (2001). The Micronutrient Report: Current Progress and Trends in the Control of Vitamin A, Iodine, and Iron Deficiencies. Ottawa, Canada: The Micronutrient Initiative/International Development Research Center, 2001 [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal-Drouard L., Marshall W. F., Qu L.-H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Grimwood J., Schmutz J., Lucas S., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel F. M. M., Kustka A. B., Shaked Y. (2008). The role of unchelated Fe in the iron nutrition of phytoplankton. Limnol. Oceanogr. 53, 400–404 10.4319/lo.2008.53.1.0400 [DOI] [Google Scholar]
- Mori S. (1999). Iron acquisition by plants. Curr. Opin. Plant Biol. 2, 250–253 10.1016/S1369-5266(99)80043-0 [DOI] [PubMed] [Google Scholar]
- Müller P., Li X. P., Niyogi K. K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566 10.1104/pp.125.2.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okegawa Y., Tsuyama M., Kobayashi Y., Shikanai T. (2005). The pgr1 mutation in the rieske subunit of the cytochrome b6f complex does not affect PGR5-dependent cyclic electron transport around photosystem I. J. Biol. Chem. 280, 28332–28336 10.1074/jbc.M505703200 [DOI] [PubMed] [Google Scholar]
- Rajamani S. (2006). Small Molecule Signaling and Detection Systems in Protists and Bacteria. Thesis, The Ohio State University, Columbus [Google Scholar]
- Robinson N. J., Proctor C. M., Connolly E. L., Guerinot M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697 10.1038/17800 [DOI] [PubMed] [Google Scholar]
- Rosakis A., Köster W. (2004). Transition metal transport in the green microalga Chlamydomonas reinhardtii-genomic sequence analysis. Res. Microbiol. 155, 201–210 10.1016/j.resmic.2003.12.004 [DOI] [PubMed] [Google Scholar]
- Rubinelli P., Siripornadulsil S., Gao-Rubinelli F., Sayre R. T. (2002). Cadmium- and iron-stress-inducible gene expression in the green alga Chlamydomonas reinhardtii: evidence for H43 protein function in iron assimilation. Planta 215, 1–13 10.1007/s00425-001-0711-3 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kurano N., Miyachi S. (1998). Cloning and characterization of high-CO2-specific cDNAs from a marine microalga, Chlorococcum littorale, and effect of CO2 concentration and iron deficiency on the gene expression. Plant Cell Physiol. 39, 131–138 [DOI] [PubMed] [Google Scholar]
- Schmidt W. (2003). Iron solutions: acquisition strategies and signaling pathways in plants. Trends Plant Sci. 8, 188–193 10.1016/S1360-1385(03)00048-7 [DOI] [PubMed] [Google Scholar]
- Schmidt W., Tittel J., Schikora A. (2000). Role of hormones in the induction of iron deficiency responses in Arabidopsis roots. Plant Physiol. 122, 1109–1118 10.1104/pp.122.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearman R., Yuan D. S., Yamaguchi-Iwai Y., Klausner R. D., Dancis A. (1996). A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271, 1552–1557 10.1126/science.271.5255.1552 [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M. L., Briat J. F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233 10.1105/tpc.001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wiren N., Mori S., Marschner H., Romheld V. (1994). Iron inefficiency in maize mutant ys1 (Zea mays L. cv yellow-stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiol. 106, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters B. M., Chu H., Didonato R. J., Roberts L. A., Eisley R. B., Lahner B., Salt D. E., Walker E. L. (2006). Mutations in Arabidopsis yellow-stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458 10.1104/pp.106.082586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger H. G. (1999). Ferric and cupric reductase activities in the green alga Chlamydomonas reinhardtii: experiments using iron-limited chemostats. Planta 207, 377–384 10.1007/s004250050495 [DOI] [Google Scholar]
- Weger H. G., Espie G. S. (2000). Ferric reduction by iron-limited Chlamydomonas cells interacts with both photosynthesis and respiration. Planta 210, 775–781 10.1007/s004250050679 [DOI] [PubMed] [Google Scholar]
- Yi Y., Saleeba J. A., Guerinot M. L. (1994). “Iron uptake in Arabidopsis thaliana,” in Biochemistry of Metal Micronutrients in the Rhizosphere, eds Manthey J., Luster D., Crowley D. E. (Chelsea, MI: Lewis Publishers, Inc.), 295–307 [Google Scholar]