Abstract
The apical plasma membrane of pollen tubes contains different PI4P 5-kinases that all produce phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] but exert distinct cellular effects. In the present example, overexpression of Arabidopsis AtPIP5K5 or tobacco NtPIP5K6-1 caused growth defects previously attributed to increased pectin secretion. In contrast, overexpression of Arabidopsis AtPIP5K2 caused apical tip swelling implicated in altering actin fine structure in the pollen tube apex. AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 share identical domain structures. Domains required for correct membrane association of the enzymes were identified by systematic deletion of N-terminal domains and subsequent expression of fluorescence-tagged enzyme truncations in tobacco pollen tubes. A variable linker region (Lin) contained in all PI4P 5-kinase isoforms of subfamily B, but not conserved in sequence, was recognized to be necessary for correct subcellular localization of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2. Deletion of N-terminal domains including the Lin domain did not impair catalytic activity of recombinant AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 in vitro; however, the presence of the Lin domain was necessary for in vivo effects on pollen tube growth upon overexpression of truncated enzymes. Overexpression of catalytically inactive variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 did not influence pollen tube growth, indicating that PtdIns(4,5)P2 production rather than structural properties of PI4P 5-kinases was relevant for the manifestation of growth phenotypes. When Lin domains were swapped between NtPIP5K6-1 and AtPIP5K2 and the chimeric enzymes overexpressed in pollen tubes, the chimeras reciprocally gained the capabilities to invoke tip swelling or secretion phenotypes, respectively. The data indicate that the Lin domain directed the enzymes into different regulatory contexts, possibly contributing to channeling of PtdIns(4,5)P2 at the interface of secretion and actin cytoskeleton.
Keywords: PtdIns(4,5)P2; PI4P 5-kinase; alternative functions; membrane association; pollen tubes
Introduction
The minor phospholipid, PtdIns(4,5)P2, is involved in the control of various cellular processes in eukaryotic cells, including plants (Meijer and Munnik, 2003; Thole and Nielsen, 2008; Heilmann, 2009). Although the amounts of PtdIns(4,5)P2 in plant membranes are low and in vegetative tissues the lipid is hard to detect under standard conditions (König et al., 2007, 2008; van Leeuwen et al., 2007), specialized plant cells contain detectable amounts of PtdIns(4,5)P2 (Vincent et al., 2005; Ischebeck et al., 2008, 2011; Kusano et al., 2008; Sousa et al., 2008; Stenzel et al., 2008). An example is presented by pollen tubes, which are characterized by a distinct lipid microdomain in the apical plasma membrane that is enriched in PtdIns(4,5)P2 (Kost et al., 1999; Kost, 2008). Although the presence of this PtdIns(4,5)P2-containing membrane microdomain had been reported some time ago (Kost et al., 1999), its relevance for pollen tube growth has remained unclear until the PI4P 5-kinases were described that form PtdIns(4,5)P2 in the apical plasma membrane of pollen tubes (Ischebeck et al., 2008, 2011; Sousa et al., 2008; Zhao et al., 2010).
PI4P 5-kinases are cytosolic enzymes that associate peripherally with the plasma membrane to modify the headgroup of inositol-containing lipids (Anderson et al., 1999). Based on their domain structure, PI4P 5-kinases can be categorized into the subfamilies A and B (Mueller-Roeber and Pical, 2002). Experimental evidence (Ischebeck et al., 2008, 2011; Sousa et al., 2008; Zhao et al., 2010) and Genevestigator analysis (Zimmermann et al., 2004) indicate that at least 6 out of the 11 PI4P 5-kinase isoforms encoded in the Arabidopsis genome are expressed in pollen. Both subfamilies are represented in the group of pollen-expressed PI4P 5-kinases (subfamily A, AtPIP5K10 and AtPIP5K11; subfamily B, AtPIP5K2, AtPIP5K4, AtPIP5K5, and AtPIP5K6), suggesting a complex situation of PtdIns(4,5)P2 functions in these cells. By characterizing the effects of underexpression or overexpression of various PI4P 5-kinase isoforms on pollen tube growth and morphology, it has previously been reported that different cellular processes were affected by PtdIns(4,5)P2 originating from different PI4P 5-kinases. For instance, pollen tubes of Arabidopsis double mutants deficient in the type B PI4P 5-kinases, AtPIP5K4 or AtPIP5K5 exhibited reduced germination and growth. Overexpression of these enzymes in tobacco pollen tubes resulted in increased apical deposition of pectin and phenotypes ranging from wavy growth over tip branching to protoplast trapping and stunted growth (Ischebeck et al., 2008; Sousa et al., 2008). While not all of these phenotypes have previously been reported for overexpression of AtPIP5K6 in tobacco pollen tubes (Zhao et al., 2010), in our hands the overexpression phenotypes of AtPIP5K4, AtPIP5K5, and AtPIP5K6 were all similar. The range of these overexpression phenotypes has previously been jointly described as “secretion phenotypes” (Ischebeck et al., 2010b). In contrast, pollen tubes of Arabidopsis double mutants deficient in the type A PI4P 5-kinases, AtPIP5K10 and AtPIP5K11, exhibited increased sensitivity to the actin polymerization inhibitor, latrunculin B, while overexpression of these enzymes in tobacco pollen tubes resulted in aggregation of the apical actin fine structure and a tip-swelling phenotype (Ischebeck et al., 2011). When reviewing these previous data, a number of key points must be highlighted: (i) the manifestation of secretion phenotypes upon overexpression of AtPIP5K4 or AtPIP5K5 was not accompanied by notable changes in the actin cytoskeleton or by tip swelling (Ischebeck et al., 2008); (ii) overexpression of AtPIP5K10 or AtPIP5K11 did not result in increased deposition of pectin or the manifestation of secretion phenotypes (Ischebeck et al., 2011); and (iii) coexpressed AtPIP5K5:CFP and AtPIP5K11:EYFP exhibited localization patterns in growing and non-growing pollen tubes, that suggest discrete modes of recruitment to the plasma membrane (Ischebeck et al., 2011). The notion that PI4P 5-kinases must be recruited to the correct place in the membrane to enable functionality is supported by the previous finding that defects in root hair growth in an Arabidopsis mutant lacking the root-expressed PI4P 5-kinase, AtPIP5K3, were not complemented by a catalytically active but mistargeted truncated variant of AtPIP5K3 (Kusano et al., 2008; Stenzel et al., 2008).
Based on the available information it was the working hypothesis of this study that the different physiological effects of PI4P 5-kinases in pollen tubes were a consequence of alternative recruitment of the respective enzymes to different microdomains within the plasma membrane. Therefore, it was one goal of this study to define protein domains of different plant PI4P 5-kinases that are required for correct localization and membrane association in pollen tubes. The PI4P 5-kinases investigated in this study are all members of the subfamily B of PI4P 5-kinases and are characterized by a highly conserved pattern of protein domains (Mueller-Roeber and Pical, 2002). A short N-terminal domain (NT) is followed by the membrane occupation and recognition nexus (MORN) domain that contains multiple MORN-repeats. MORN-repeats are present in a number of proteins of animal or plant origin that are involved in mediating protein-membrane contacts, such as the Arabidopsis ARC3 protein involved in plastidial fission (Shimada et al., 2004; Maple et al., 2007), the Toxoplasma gondii protein, MORN1, involved in cell-division (Gubbels et al., 2006), or junctophilins, which mediate endomembrane-to-plasma membrane attachment in mammalian cells (Takeshima et al., 2000). The PIP5K1 protein from Arabidopsis has been reported to display enhanced activity upon deletion of its MORN domain, indicating an autoinhibitory function for the MORN domain of PI4P 5-kinases (Im et al., 2007). It has also been speculated that the MORN domain of PI4P 5-kinases might exert its regulatory effects on catalytic activity of the enzymes through interactions with the lipid environment of substrate membranes (Im et al., 2007). Both the NT and MORN domains are specific for the PI4P 5-kinase isoforms of the subfamily B, whereas all other domains discussed here are also present in isoforms of subfamily A, which more closely resemble PI4P 5-kinases of mammalian origin (Mueller-Roeber and Pical, 2002). The NT and MORN domains are joined to the rest of the protein by a linker (Lin) region, which is not conserved in sequence and to which, therefore, no particular in vivo functionality has been attributed to date. Based on the crystal structure of the related human HsPIPKIIβ, PI4P 5-kinases are thought to be dimers, and a dimerization (Dim) domain represents the postulated interface between the monomers (Rao et al., 1998). The catalytic (Cat) domain is highly conserved between PI4P 5-kinases, with the exception of a variable insert region close to the C-terminal end of the protein. The Cat domain also contains a feature termed the activation loop (AL; Mueller-Roeber and Pical, 2002), which has been demonstrated to be important in defining substrate specificity (Kunz et al., 2000, 2002).
It is a working hypothesis of this study that PI4P 5-kinases are recruited to their target membranes by specific protein–protein interactions. In this context it is relevant that interactions of AtPIP5K2 with other proteins have previously been demonstrated (Camacho et al., 2009). Specifically, it was shown that interaction of the MORN-repeat domain of PIP5K2 with small GTPases of the RabE family mediates recruitment of the enzyme and may stimulate temporally or spatially localized production of PtdIns(4,5)P2 at the plasma membrane (Camacho et al., 2009). An interaction of AtPIP5K9 with cytosolic invertases has also previously been reported (Lou et al., 2007). Considering the assumption that different PI4P 5-kinases are recruited to distinct sites of action by specific protein–protein interactions, it follows that likely none of the highly conserved domains of PI4P 5-kinases will be involved in mediating such differential recruitment. Instead, it was a working hypothesis of this work that non-conserved domains might be important for the specificity of protein–protein interactions. In brief, we show that the Arabidopsis type B PI4P 5-kinase, AtPIP5K2, exerts different effects on pollen tube growth than other type B enzymes, enabling the comparison of type B PI4P 5-kinases with distinct functionality. The dissimilar Lin domains of three type B PI4P 5-kinases, Arabidopsis AtPIP5K5, the newly isolated tobacco enzyme NtPIP5K6-1 and Arabidopsis AtPIP5K2, are identified as essential for correct membrane association and for physiological functionality of the enzymes in tobacco pollen tubes. The exchange of Lin domains between NtPIP5K6-1 and AtPIP5K2 resulted in reciprocally changed functionality of the chimeric enzymes, demonstrating that the Lin domains contribute to the specific membrane recruitment of PI4P 5-kinases.
Results
Pectin secretion-phenotypes and tip swelling upon overexpression of different type B PI4P 5-kinases
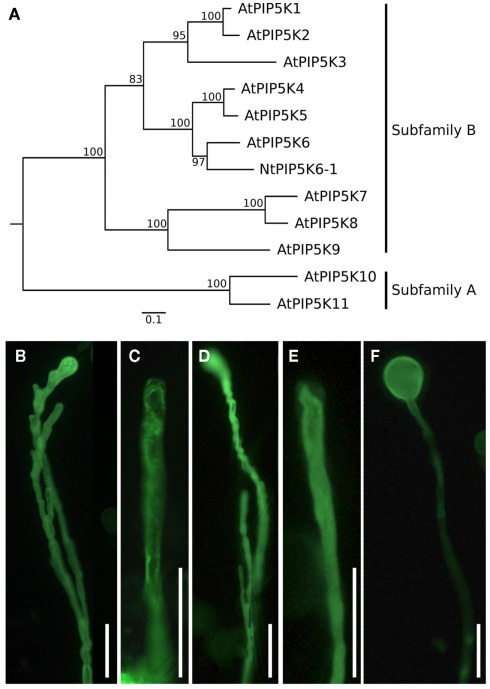
In order to document different regulatory effects of N-terminal domains on the in vivo functionality of PI4P 5-kinases, pollen-expressed PI4P 5-kinases of subfamily B (Figure 1A) were analyzed for effects their overexpression exerted on pollen tube growth. Besides the previously reported Arabidopsis AtPIP5K5 (Ischebeck et al., 2008; Sousa et al., 2008), the newly identified tobacco PI4P 5-kinase NtPIP5K6-1 was tested. A cDNA clone encoding this new PI4P 5-kinase was isolated from a tobacco pollen tube-specific cDNA library generously provided by Prof. Dr. Benedikt Kost (Friedrich-Alexander-University Erlangen–Nürnberg, Germany) and termed NtPIP5K6-1 in analogy to its closest Arabidopsis homolog, AtPIP5K6 (Figure 1A). Also included in further analyses was Arabidopsis AtPIP5K2, which is ubiquitously expressed in the plant and shows strong expression also in pollen according to Genevestigator analysis (Zimmermann et al., 2004). Overexpression of AtPIP5K5:EYFP and NtPIP5K6-1:EYFP in tobacco pollen tubes resulted in secretion phenotypes (Figures 1B–E) previously reported for type B PI4P 5-kinases (Ischebeck et al., 2008, 2010b; Sousa et al., 2008; Zhao et al., 2010). In contrast, overexpression of AtPIP5K2:EYFP resulted in severe tip swelling of the pollen tubes (Figure 1F), similar to that previously reported for overexpression of the type A PI4P 5-kinases, AtPIP5K10, or AtPIP5K11 (Ischebeck et al., 2011). In this context, please note that it has previously been proposed that besides the two PI4P 5-kinases of subfamily A, AtPIP5K10 and AtPIP5K11, at least one additional isoform of subfamily B must perform a similar role in pollen tubes (Ischebeck et al., 2011). The data presented indicate that AtPIP5K2 represents a candidate for this function. The observation of different phenotypes upon overexpression of AtPIP5K5 or NtPIP5K6-1 vs. AtPIP5K2 indicates that even PI4P 5-kinases with similar domain structures can perform different physiological roles. In subsequent experiments, the N-terminal domains of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 were investigated.
Figure 1.
Phylogenetic tree of plant PI4P 5-kinases and effects of PI4P 5-kinase overexpression on the morphology of tobacco pollen tubes. (A) Maximum likelihood (ML) tree created with the full-length protein sequences of A. thaliana PI4P 5-kinases and N. tabacum PIP6K6-1. Values on nodes represent% ML bootstrap support. Association of isoforms with PI4P 5-kinase subfamilies A and B according to Mueller-Roeber and Pical (2002), as indicated. (B–F) Selected PI4P 5-kinases of subfamily B were overexpressed in tobacco pollen tubes and the effects on growth were monitored. (B,C) Pollen tube branching (B) and protoplast trapping (C) upon expression of AtPIP5K5; (D, E) Pollen tube branching (D) and protoplast trapping (E) upon expression of NtPIP5K6-1; (F) Tip swelling upon expression of AtPIP5K2. Images are representative for ≥300 transgenic pollen tubes observed. Bars, 50 μm.
The Lin domain is required for correct subcellular localization of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2
First, experiments were initiated to determine whether an N-terminal domain is required for correct membrane association of selected PI4P 5-kinases of subfamily B. N-terminal domains were successively deleted and fluorescence-tagged truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 were expressed at low levels in tobacco pollen tubes (Figure 2). The exact truncations were as follows: AtPIP5K5ΔNT–MORN, AtPIP5K5253–772; AtPIP5K5ΔNT–MORN–Lin, AtPIP5K5370–772; NtPIP5K6-1ΔNT–MORN, NtPIP5K6-1241–775; NtPIP5K6-1ΔNT–MORN–Lin, NtPIP5K6-1366–775; AtPIP5K2ΔNT–MORN, AtPIP5K2260–754; AtPIP5K2ΔNT–MORN–Lin, AtPIP5K2344–754. The domain structures of the truncated variants are depicted in Figure 2A. Full-length AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 displayed plasma membrane association that was restricted to the apical plasma membrane PtdIns(4,5)P2-microdomain of pollen tubes (Figures 2B,E,H), as previously reported for AtPIP5K5 (Ischebeck et al., 2008; Sousa et al., 2008). The deletion of the NT-domain alone (not shown) or the combined NT and MORN domains (Figures 2C,F,I) did not result in altered localization of the fusion proteins; however, all three enzymes exhibited altered localization patterns when the truncations made included deletion of the Lin domain (Figures 2D,G,J). For AtPIP5K5 and NtPIP5K6-1 the deletion of N-terminal domains including the Lin domain resulted in a loss of membrane association and cytosolic localization (Figures 2D,G). The corresponding AtPIP5K2ΔNT–MORN–Lin still associated with the plasma membrane; however, the association pattern was more relaxed than that of the full-length enzyme and the localization of the truncated fusion protein basolaterally exceeded that of the apical plasma membrane microdomain (compare Figures 2H,J). The data define the Lin domains of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 as key factors directing correct membrane association.
Figure 2.
The Lin domain is required for correct localization of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2. The PI4P 5-kinases AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 belong to subfamily B and share the same domain structure. N-terminal domains of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 were systematically deleted, the truncated variants fused to EYFP-tags, the fusion proteins expressed in tobacco pollen tubes, and the fluorescence distribution was monitored. (A) Graphical representation of the domain structure of full-length AtPIP5K5, NtPIP5K6-1, or AtPIP5K2. NT, N-terminal domain; MORN, MORN domain; Lin, linker domain; Dim, dimerization domain; Cat, catalytic domain; AL, activation loop; EYFP, fluorescence-tag. ΔNT–MORN, graphical representation of the ΔNT–MORN truncations; ΔNT–MORN–Lin, graphical representation of the ΔNT–MORN–Lin truncations. (B–D) Subcellular distribution of AtPIP5K5 (B), AtPIP5K5ΔNT–MORN (C), and AtPIP5K5ΔNT–MORN–Lin (D). (E–G) Subcellular distribution of NtPIP5K6-1 (E), NtPIP5K6-1ΔNT–MORN (F), and NtPIP5K6-1ΔNT–MORN–Lin (G). (H–J) Subcellular distribution of AtPIP5K2 (H), AtPIP5K2ΔNT–MORN (I), and AtPIP5K2ΔNT–MORN–Lin (J). Images are representative for ≥300 transgenic pollen tubes observed. Bars, 10 μm.
Deletion of N-terminal domains does not impair catalytic activity of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2
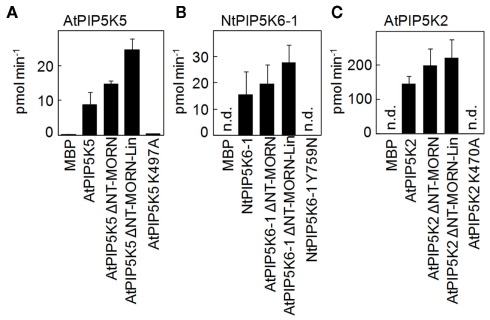
In order to determine whether the deletions of N-terminal domains of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 had detrimental effects on catalytic activity of the enzymes, the truncated variants were expressed in E. coli and the recombinant proteins tested in vitro (Figure 3). The data indicate that the N-terminal deletions did not impair catalytic activity against the substrate lipid, PtdIns4P, and even increased the activity of the enzymes (Figure 3). Maltose-binding protein (MBP) was used as a negative control. Furthermore, recombinant proteins carrying point mutations, PIP5K5 K497A, PIP5K6-1 Y759N, and PIP5K2 K470A, were generated to be used in further experiments. For AtPIP5K2 and AtPIP5K5 the point mutations targeted a conserved K in the ATP-binding site as was previously reported (Ishihara et al., 1998), rendering the recombinant mutant enzymes inactive. Because the equivalent exchange in NtPIP5K6-1 did not yield an inactive enzyme, we selected another site based on an analysis of the point mutation in a Neurospora crassa PIPK (Mss4) mutant (Seiler and Plamann, 2003). In the N. crassa mutant a conserved tyrosine close to the substrate-binding site of the enzyme, as determined based on the 3D-structure of the human HsPIPKIIβ (Rao et al., 1998), was altered; an exchange of the corresponding tyrosine of NtPIP5K6-1 (Y759A) rendered the tobacco enzyme inactive. PIP5K5 K497A, PIP5K6-1 Y759N, and PIP5K2 K470A were all found to be reduced in catalytic activity below the limit of detection (Figure 3). While the in vitro characterization demonstrated the catalytic capabilities under artificial conditions, it must be noted that the behavior of the enzymes in planta may deviate from the results obtained by in vitro tests.
Figure 3.
Deletion of N-terminal domains does not impair in vitro catalytic activity of recombinant truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2. PI4P 5-kinase variants used for localization experiments or to determine physiological effects on pollen tube growth were heterologously expressed in E. coli as fusions to N-terminal maltose-binding protein (MBP)-tags. AtPIP5K5, NtPIP5K6-1, AtPIP5K2, and the respective ΔNT–MORN and ΔNT–MORN–Lin truncations were included as well as the catalytically inactive variants, AtPIP5K5 K497A, NtPIP5K6-1 Y759N, and AtPIP5K2 K470A. The recombinant proteins were tested in vitro for catalytic activity against PtdIns4P. MBP was used as a negative control. Protein amounts were adjusted using immunodetection (not shown). (A) AtPIP5K5 and derived proteins; (B) NtPIP5K6-1 and derived proteins; (C) AtPIP5K2 and derived proteins. Data are means ± SD from three independent experiments. n.d., Not detected.
The Lin domain and catalytic activity are required for the manifestation of overexpression phenotypes in pollen tubes
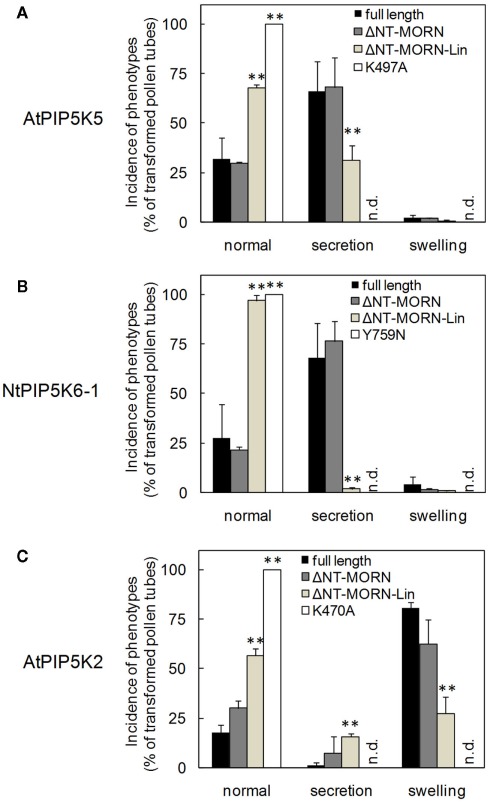
Next, the capability of the truncated variants of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 to influence pollen tube morphology (cf. Figure 1) was tested. Upon overexpression of full-length AtPIP5K5 or NtPIP5K6-1 at moderate to strong levels, pectin secretion phenotypes were observed (Figures 4A,B black bars), including tip branching and protoplast trapping (cf. Figures 1B–E), as has previously been reported for AtPIP5K5 (Ischebeck et al., 2008; Sousa et al., 2008). Approximately 25–30% of transformed pollen tubes exhibiting low fluorescence intensity did not exhibit altered growth, and only very few pollen tubes (<1%) displayed moderate tip swelling. Deletion of the NT and MORN domains of AtPIP5K5 or NtPIP5K6-1 did not significantly alter the phenotypic distribution in the overexpression experiments (Figures 4A,B, dark gray bars). When truncated variants of AtPIP5K5 or NtPIP5K6-1 were overexpressed, in which additionally the Lin domain was deleted, a substantial shift toward more morphologically unaltered pollen tubes was observed (Figures 4A,B, light gray bars) concomitant with fewer pollen tubes exhibiting secretion phenotypes. No altered pollen tubes were observed upon overexpression of the catalytically inactive variants AtPIP5K5 K497A or NtPIP5K6-1 Y759N (Figures 4A,B, white bars).
Figure 4.
Deletion of N-terminal domains abolishes effects of truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 on pollen tube growth. The effects of overexpression of truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 on pollen tube growth were analyzed (gray bars, as indicated). Phenotypes were categorized as normal, as secretion phenotypes (wavy growth, tip branching and protoplast trapping), or as tip swelling. Full-length enzymes (black bars) and catalytically inactive variants (white bars) were used as controls. (A) AtPIP5K5 and derived proteins; (B) NtPIP5K6-1 and derived proteins; (C) AtPIP5K2 and derived proteins. Data are means ± SD from four independent transformation experiments and represent ≥300 transgenic pollen tubes observed for each construct. n.d., Not detected. The asterisks indicate a significant difference from the full-length control within each category according to a Student’s t test (**p < 0.01).
Even though overexpression of AtPIP5K2 in tobacco pollen tubes resulted in a different phenotype (tip swelling, cf. Figure 1), the effects of successively deleting N-terminal domains on the swelling phenotype were similar to those observed with AtPIP5K5 or NtPIP5K6-1 and their associated secretion phenotype. Upon expression of full-length AtPIP5K2 around 20% of transformed pollen tubes exhibiting low expression levels did not display altered growth. Furthermore, only very few pollen tubes (<1%) displayed moderate secretion phenotypes (Figure 4C, black bars). Overexpression of AtPIP5K2ΔNT–MORN resulted in an only mildly altered phenotypic distribution (Figure 4C, dark gray bars). In contrast, a pronounced effect on the phenotypic distribution was observed when AtPIP5K2ΔNT–MORN–Lin was overexpressed, resulting in a substantial increase in the proportion of normal growing pollen tubes, concomitant with a decrease in the fraction of altered pollen tubes (Figures 4B,C, light gray bars). No altered pollen tubes were observed upon overexpression of the catalytically inactive AtPIP5K2 K470A (Figure 4C, white bars). The data indicate that despite of their dissimilar primary sequences the Lin domains of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 are required for the functionality of the enzymes, as indicated by the incidence of the respective pollen tube overexpression phenotypes.
In order to rule out that the different effects of the various truncations were a consequence of variable expression levels of the fluorescent proteins, the distribution of fluorescence intensities was scored individually for each set of experiments as previously described (Ischebeck et al., 2010b). The fluorescence distribution patterns (Figure 5) indicate that the reduced incidence of phenotypes observed upon expression of truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 (Figure 4) was not due to reduced expression levels of these fusion proteins (Figure 5). Quite contrary, the data indicate that PI4P 5-kinase variants that did not cause altered pollen tube growth upon overexpression (Figure 4) were tolerated by the cells at equal or even higher levels than variants exerting effects on growth (Figure 5).
Figure 5.
Similar fluorescence intensity patterns upon overexpression of truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 in pollen tubes. The fluorescence intensities during overexpression of truncated variants of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 on pollen tube growth were determined (gray bars, as indicated). Fluorescence intensities were also scored for full-length controls (black bars) and catalytically inactive variants (white bars). (A), AtPIP5K5 and derived proteins; (B), NtPIP5K6-1 and derived proteins; (C), AtPIP5K2 and derived proteins. Data are means ± SD from four independent transformation experiments and represent ≥300 transgenic pollen tubes observed for each construct. n.d., Not detected.
The Lin domain is not conserved in sequence between PI4P 5-kinase isoforms, but functionally corresponding domains share similarities in secondary structure
The Lin domain of type B PI4P 5-kinases is poorly conserved in sequence (Figure 6A) and therefore can only be recognized based on its position between the MORN and Dim domains. Computer-aided sequence analysis of the Lin-domains using blastp1 or psiblast2 did not reveal similarity to known sequences. While the dissimilarity of sequences between Lin domains of different PI4P 5-kinase isoforms escapes the definition of a protein “domain,” the presence of non-conserved sequence stretches provides PI4P 5-kinase isoforms with the capability to selectively interact with distinct partners. Despite their limited similarity in primary sequence, Lin domains serving equivalent functions, such as those of AtPIP5K5 and NtPIP5K6-1, display similar secondary structures that differ from that of the Lin domain of AtPIP5K2 (Figure 6B), which directs the enzyme toward other contexts (cf. Figures 4 and 7). Based on the data so far, it appears possible that certain secondary structures of Lin domains are recognized by partner proteins involved in specific membrane recruitment of PI4P 5-kinases.
Figure 6.
The Lin domain is not conserved in sequence between PI4P 5-kinase isoforms, but functionally corresponding domains share similarities in secondary structure. Lin domains of PI4P 5-kinases lack common sequence motifs and are defined by their position between the N-terminal MORN domain and the Dim and catalytic domains. (A) Local alignment of relevant parts of plant PI4P 5-kinases of subfamily B. Black, identical amino acids; gray, similar amino acids. The positions of MORN domain, Lin domain and Dim domain are indicated. (B), Secondary structure prediction for the Lin domains of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2. H, α-helix; E, β-sheet.
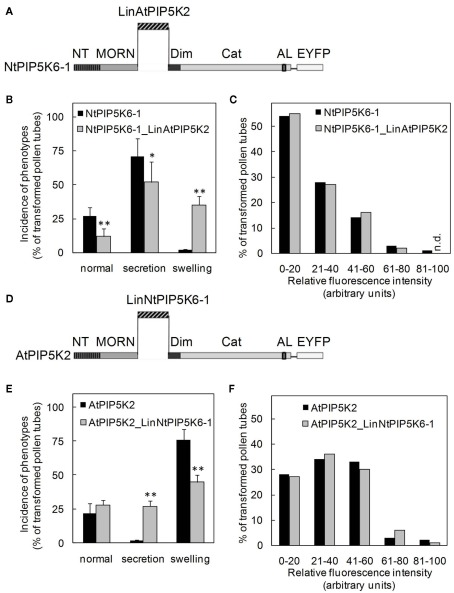
Figure 7.
Exchanging the Lin domains between AtPIP5K2 and NtPIP5K6-1 results in reciprocal gain of the capability to induce secretion-phenotypes or tip-swelling. The Lin domains of AtPIP5K2 and NtPIP5K6-1 were exchanged between the enzymes, yielding the chimeric proteins NtPIP5K6-1_LinAtPIP5K2 and AtPIP5K2_LinNtPIP5K6-1. The chimeric enzymes were overexpressed in tobacco pollen tubes as fusions to C-terminal EYFP-tags and the effects on pollen tube growth were monitored. The fluorescence intensity distributions of the expressed proteins were recorded as controls. (A–C), NtPIP5K6-1_LinAtPIP5K2. (A), Graphical representation of the chimeric fusion protein NtPIP5K6-1_LinAtPIP5K2; (B), Incidence of phenotypic categories observed upon overexpression of NtPIP5K6-1 (black bars) or NtPIP5K6-1_LinAtPIP5K2 (gray bars); (C), Fluorescence intensity distribution corresponding to overexpression of NtPIP5K6-1 (black bars) or NtPIP5K6-1_LinAtPIP5K2 (gray bars) shown in (B). (D), Graphical representation of the chimeric fusion protein AtPIP5K2_LinNtPIP5K6-1; (E), Incidence of phenotypic categories observed upon overexpression of AtPIP5K2 (black bars) or AtPIP5K2_LinNtPIP5K6-1 (gray bars); (F), Fluorescence intensity distribution corresponding to overexpression of AtPIP5K2 (black bars) or AtPIP5K2_LinNtPIP5K6-1 (gray bars) shown in (E). Data are means ± SD from four independent transformation experiments and represent ≥300 transgenic pollen tubes observed for each construct. n.d., Not detected. The asterisks indicate a significant difference from the full-length control within each category according to a Student’s t test (*p < 0.05; **p < 0.01).
Exchanging the Lin domains between AtPIP5K2 and NtPIP5K6-1 results in reciprocal gain of the capability to induce secretion-phenotypes or tip-swelling
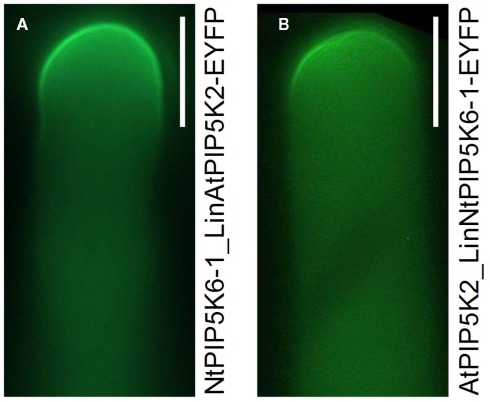
Based on the results so far, we speculated whether the Lin domains were sufficient to determine physiological functionality of PI4P 5-kinases of subfamily B. To test this hypothesis, two PI4P 5-kinases, NtPIP5K6-1 and AtPIP5K2 were chosen, which in previous experiments had exerted clear and different effects on pollen tube growth, and their Lin domains were reciprocally exchanged (Figures 7A,D). The domain structures of the chimeric proteins are depicted in Figure 7. The resulting chimeric enzymes were overexpressed in tobacco pollen tubes and the incidence of secretion vs. tip swelling phenotypes was scored (Figures 7B,E). When full-length NtPIP5K6-1 was overexpressed in tobacco pollen tubes, a typical phenotypic distribution was observed (cf. Figure 4B) with a large proportion of secretion phenotypes and close to no tip swelling (Figure 7B, black bars). In contrast, when the chimeric NtPIP5K6-1 containing the Lin domain of AtPIP5K2 (termed NtPIP5K6-1_LinAtPIP5K2) was overexpressed, a reduced proportion of pollen tubes displaying secretion phenotypes was observed concomitant with an increased proportion of cells exhibiting tip swelling (Figure 7B, gray bars). The difference in phenotypic effects was not due to variations in expression levels between the two fusion proteins (Figure 7C). Reciprocally, overexpression of the chimeric AtPIP5K2 containing the Lin domain of NtPIP5K6-1 (termed AtPIP5K2_LinNtPIP5K6-1) resulted in a reduced incidence of cells displaying tip swelling concurrent with the appearance of a substantial fraction of cells exhibiting secretion phenotypes (Figure 7E, gray bars). When full-length AtPIP5K2 was overexpressed as a control, a typical distribution of phenotypes was recorded (cf. Figure 4C) with close to no secretion phenotypes and a large proportion of cells exhibiting tip swelling (Figure 7E, black bars). Again, the difference in phenotypic effects was not due to variations in expression levels for the two fusion proteins (Figure 7F). The chimeric proteins NtPIP5K6-1_LinAtPIP5K2 and AtPIP5K2_LinNtPIP5K6-1 both localized correctly in tobacco pollen tubes (Figure 8) and the localization patterns were indistinguishable from those of their respective parental enzymes (cf. Figure 2). The data indicate that the exchange of Lin domains between NtPIP5K6-1 and AtPIP5K2 resulted in a reciprocal change in physiological functionality in vivo. The exchange of no other domains was needed for the effects observed, highlighting an important contribution of the Lin domains in specifying the signaling context of PI4P 5-kinases.
Figure 8.
Localization of the chimeric proteins, NtPIP5K6-1_LinAtPIP5K2-EYFP and AtPIP5K2_LinNtPIP5K6-1-EYFP in tobacco pollen tubes. The chimeric enzymes were overexpressed in tobacco pollen tubes as fusions to C-terminal EYFP-tags and the fluorescence distribution was monitored. (A), NtPIP5K6-1_LinAtPIP5K2-EYFP; (B), AtPIP5K2_LinNtPIP5K6-1-EYFP. Images are taken from experiments shown in Figure 7 and are representative for ≥300 transgenic pollen tubes observed for each construct. Bars, 10 μm.
Functional specification of PIP5K2 correlates with increased rate of mutation in DNA-regions encoding the PIP5K2-Lin-domain
The observation that AtPIP5K2 exerted pollen tube overexpression effects different from those of AtPIP5K5 or NtPIP5K6-1 (Figure 1) indicates that AtPIP5K2 might have functionally diverged from other type B PI4P 5-kinases of Arabidopsis. As the Lin-domain of AtPIP5K2 differs structurally from those of other type B PI4P 5-kinases (Figure 6) and the alternative functionality could partially be transferred by exchanging Lin-domains between PI4P 5-kinases (Figure 7), sequence divergence within the Lin-domains in type B PI4P 5-kinases was investigated in more detail. We assessed the rate of sequence evolution across the coding sequence of AtPIP5K1-9 by comparing the A. thaliana sequence to orthologous sequences from its completely sequenced sister species A. lyrata. Figure 9 represents a sliding window analysis depicting the ratio of the number of non-synonymous substitutions per non-synonymous site (KA) to the number of synonymous substitutions per synonymous site (KS). The KA/KS ratio is generally used as an indicator of selective pressure acting on a protein-coding gene. By successively performing the analysis for an entire coding sequence (“sliding window”-analysis), it is possible to assign regions of increased KA/KS ratios, thereby identifying regions undergoing increased rates of mutation at non-synonymous sites (i.e., positive selection). The analysis identifies numerous insignificant peaks of KA/KS ratios (<1) for all variable regions of PI4P 5-kinases, including sequence stretches encoding the NT-domains, the Lin-domains, and the variable inserts in the catalytic domains. The highest KA/KS ratios (>2) were observed for DNA-regions encoding the NT- and Lin-domains of AtPIP5K2, indicating a high rate of mutations accumulating in these domains. The observation of high KA/KS ratios (>1) and therefore signatures of positive or directional selection predominantly in AtPIP5K2 correlates well with the notion of an evolving new functionality of AtPIP5K2 arising from an altered Lin-domain and – possibly – an ensuing altered means of membrane recruitment.
Figure 9.
Rate of evolution of Arabidospis thaliana PI4P 5-kinase sequences according to sliding window analysis of KA/KS ratios comparing orthologous sequences of A. thaliana and A. lyrata. The rate of sequence evolution across the coding sequence of AtPIP5K1-9 was assessed by comparing the A. thaliana sequences to orthologous sequences from A. lyrata. The sliding window analysis depicts the ratios of the number of non-synonymous substitutions per non-synonymous site (KA) to the number of synonymous substitutions per synonymous site (KS). All plots are drawn to the same scale. Increased KA/KS ratios in sequence regions of AtPIP5K2 encoding the NT-domain, the Lin-domain and the variable insert region are annotated in the AtPIP5K2 plot, as indicated. The pairs of orthologous genes from A. thaliana and A. lyrata used for the analysis are given in the Section “Materials and Methods.”
Discussion
In the present study it was tested whether certain protein domains would direct PI4P 5-kinase isoforms into potentially different regulatory contexts underlying characteristic pollen tube overexpression phenotypes. In order to systematically compare the functionality of domains of candidate proteins, the domain structures of the candidates must be similar. Different functionality of pollen-expressed PI4P 5-kinase isoforms reported previously (Ischebeck et al., 2008, 2011; Sousa et al., 2008; Zhao et al., 2010) had been attributed to enzymes representing the subfamilies A and B of PI4P 5-kinases that differ notably in their domain structure, precluding a direct comparison of protein domains. However, from data previously published on the Arabidopsis pip5k10 pip5k11 double mutant it has been proposed that at least one PI4P 5-kinase of subfamily B would perform similar functions as the two enzymes of subfamily A (Ischebeck et al., 2011). In the present study, a corresponding type B enzyme was identified as AtPIP5K2 (Figure 1F), enabling the comparison of PI4P 5-kinases of subfamily B, AtPIP5K5, NtPIP5K6-1, and AtPIP5K2, which have similar domain structures while in part exerting different regulatory effects in pollen tubes (Figures 1B–F). Following the previous annotation of protein domains of plant PI4P 5-kinases (Mueller-Roeber and Pical, 2002), a systematic deletion approach was pursued to first identify domains involved in membrane association (Figure 2). The data indicate that all truncations in which the Lin-domain was deleted were cytosolic (Figures 2D,G) or were altered in the pattern of membrane association (Figure 2J), indicating a role for the Lin-domain in membrane recruitment. In experiments performed to reciprocally test whether N-terminal domains of AtPIP5K5, NtPIP5K6-1, or AtPIP5K2 would direct a fluorescent protein to the plasma membrane all fusion proteins tested localized to the cytosol (data not shown). This observation might be explained by the notion that other, so far unidentified regions of the PI4P 5-kinase proteins may be required for correct membrane recruitment of the enzyme. In this scenario the Lin-domain appears necessary (Figure 2) but not sufficient for membrane recruitment. Alternatively, it is possible that in particular the smaller NT-domains alone might not fold correctly when expressed as fusions to a fluorescent protein, thereby precluding correct targeting.
The positive observation that the Lin domains had key influence on membrane association of all three PI4P 5-kinases tested was surprising, as not the Lin-domain but rather the MORN domain has previously been implicated in protein–protein interactions (Camacho et al., 2009) and membrane association (Mueller-Roeber and Pical, 2002). This apparent conflict might be explained by the particular N-terminal deletions made to define effects of the MORN domain in previous experiments, where MORN and Lin domains were not always clearly distinguished. Based on the data presented here, the role of the MORN domain in membrane association and regulation of PI4P 5-kinases in pollen tubes remains unresolved.
Truncated variants of AtPIP5K5, NtPIP5K6-1, and AtPIP5K2 used in this study all exhibited full catalytic activity in vitro (Figure 3), and therefore, detrimental effects on their in vivo functionality (Figure 4) should be attributed to the interaction with the correct reaction partners, including the lipid substrate, PtdIns4P, and/or downstream effectors, which might be regulated in their functionality by PtdIns(4,5)P2. The increased activity of recombinant PI4P 5-kinases lacking N-terminal domains is consistent with a previous report attributing autoinhibitory effects to the MORN repeat domain (Im et al., 2007).
Protein domains are mostly annotated based on their similarity to domains of known function or based on the occurrence of recognizable sequence motifs in other proteins. With regard to PI4P 5-kinases, a number of conserved protein domains have received positive annotations that suggest their involvement in certain processes, such as the MORN domain, which has been proposed to be involved in membrane association as well as the regulation of activity and enzyme–lipid-interactions, or the catalytic domain, which forms the catalytic center of the enzyme (Mueller-Roeber and Pical, 2002). In contrast, the Lin domain so far has not received positive functional assignment, being merely termed a linker between other domains of implied greater importance. Lin domains are characterized by substantial differences in primary or secondary structure, such as those between the Lin domains of AtPIP5K5 or NtPIP5K6-1 on one hand and that of AtPIP5K2 on the other (Figure 6), and it is our hypothesis that the lack of sequence conservation is the key to understanding the role of the Lin domain. In particular, dissimilar Lin-domains may provide a means for specific protein–protein interactions. According to this concept, alternative recruitment by dissimilar Lin domains of different PI4P 5-kinases might underlie the different effects PI4P 5-kinases, such as NtPIP5K6-1 or AtPIP5K2 exert in pollen tubes (Figures 1B–F). This notion is supported by the domain swapping experiments (Figure 7), in which the Lin domain of AtPIP5K2 was introduced into NtPIP5K6-1 together with the capability to induce tip swelling, whereas, reciprocally, the Lin domain of NtPIP5K6-1 introduced into AtPIP5K2 conferred the capability to induce secretion phenotypes. Protein–protein interactions of PI4P 5-kinases may be transient and may reflect dynamic changes in signaling metabolism. For instance, it has been reported that AtPIP5K2 interacts with small GTPases of the RabE family via its MORN domain and this interaction has been shown to affect localization in leaf epidermal cells (Camacho et al., 2009). In contrast, here we show full functionality of AtPIP5K2ΔNT–MORN in the overexpression setting, suggesting that the situation for cell types other than leaf epidermal cells may be different and the same enzyme may interact with other partners in pollen tubes. It is our current understanding that AtPIP5K2 is a multifunctional enzyme, such as the PI4P 5-kinases of Saccharomyces cerevisiae or N. crassa, that are recruited into different contexts by interacting with different partner proteins. Similar to plant PI4P 5-kinases, the yeast and N. crassa enzymes have evolved to contain additional regulatory domains at the N-terminus, or both at the N- and C-termini, respectively (Ischebeck et al., 2010a).
An interesting consequence of the proposed multifunctionality of AtPIP5K2 is the observation of altered overexpression effects upon deletion of N-terminal domains. N-terminal truncation of AtPIP5K2 resulted not only in an altered pattern of membrane association of AtPIP5K2ΔNT–MORN–Lin (Figure 2J), but upon overexpression also caused the appearance of secretion phenotypes only rarely (<1%) observed upon overexpression of full-length AtPIP5K2 (Figure 4C). A possible explanation is an inherent bifunctionality of AtPIP5K2 and the capability to alternatively act in the signaling context related to secretion phenotypes as well as in that related to tip swelling. Considering the hypothesis that specific protein–protein interactions are mediating the recruitment of PI4P 5-kinases into their correct signaling context, the Lin domain of AtPIP5K2 might be responsible for the association with factors controlling the cytoskeleton, whereas another, so far undefined domain would be responsible for the association of AtPIP5K2 with factors mediating the secretion phenotypes. Under the conditions used, the affinity of the Lin domain to its partners might be greater than that of possible other relevant domains to their partners, resulting in recruitment of full-length AtPIP5K2 being predominantly directed by the Lin domain. It follows that upon deletion of the Lin domain alternative, weaker interactions of other protein domains of AtPIP5K2 with other partners are enabled (Figure 4C). This concept is supported by the observation that pollen-expressed AtPIP5K2ΔNT–MORN–Lin was altered in the pattern of membrane association, but remained at the plasma membrane, suggesting that other domains than the Lin domain also contributed to the membrane association of AtPIP5K2. Furthermore, when Lin domains were swapped between AtPIP5K2 and NtPIP5K6-1, the resulting chimeric enzymes retained a substantial potential to exert the phenotypes caused by their respective parental enzymes (Figure 7), suggesting that the Lin domain is one, but not the only one, factor determining enzyme functionality in vivo.
From an evolutionary point of view, the accumulation of mutations in the Lin-domain as a protein–protein interaction module might enable interactions with an altered and expanded range of target proteins. Both the presence of several PI4P 5-kinase isoforms and the fact that the Lin-domain is not required for catalytic function of AtPIP5K2 makes the genomic region encoding this domain likely to accumulate mutations. The sliding windows analysis performed for orthologs of A. thaliana and A. lyrata (Figure 9) indicates that the genomic region encoding the Lin-domain of AtPIP5K2 displays an enhanced probability to accumulate mutations causing amino acid changes, much more so than the corresponding sequences encoding Lin-domains of other isoforms (Figure 9). This observation correlates well with the finding of an expanded functionality of AtPIP5K2 among the type B PI4P 5-kinases of Arabidopsis (Figure 1) and suggests that AtPIP5K2 might be in the process of acquiring altered functionality through the modulation of the Lin-domain that directs the enzyme into a new signaling context.
Overall, the data presented in this study demonstrate that the Lin domains are necessary for correct localization of PI4P 5-kinases and, therefore, are an important determinant of alternative in vivo functionality of the enzymes with regard to the control of pectin secretion or the actin cytoskeleton. Future experiments will be directed toward identifying other protein domains with roles in directing membrane recruitment of PI4P 5-kinases. The identification of downstream interaction partners of such domains will aid our understanding whether and how PtdIns(4,5)P2 might be channeled toward specific effectors that act in discrete signaling contexts.
Materials and Methods
cDNA constructs
The cDNA sequences of Arabidopsis PI4P 5-kinase genes AtPIP5K2 and AtPIP5K5 were amplified from cDNA prepared from young Arabidopsis inflorescences using the following primer combinations: AtPIP5K2: 5′-GATACATGTTGCGTGAACCGCTTGTTAGCGAAGAA-3′/5′-GATACATGTCGCCGTCTTCGATGAAGATTCT-3′; AtPIP5K5: 5′-GATTCATGAGCAAGGACCAAAGCTATGTTCTAAAAG-3′/5′-GATTCATGACATTGTCATCTGTGAAGACCTTGA-3′. PCR fragments were subcloned into the vector pGEM-Teasy (Promega) and sequenced. The resulting plasmids were designated as AtPIP5K2-pGEM-Teasy and AtPIP5K5-pGEM-Teasy, respectively. The cDNA sequence of NtPIP5K6 was isolated from a cDNA library of tobacco pollen tubes grown for 3 h cloned in the vector pGADGH, kindly provided by Prof. Dr. Benedikt Kost (Friedrich-Alexander-University Erlangen–Nürnberg, Germany). The library was screened by RT-PCR with degenerated primers corresponding to conserved regions of the catalytic domain of Arabidopsis PI4P 5-kinase sequences in combination with vector-specific primers. The full-length sequence of NtPIP5K6 was amplified with the primer combination 5′-GGTGGTTTTGTGGAGGGGAGAAAAGGAAGGG-3′/5′-CCTACTAGCTGGCAGTGAAAATCCTGTGTCATG-3′, cloned into the vector pJet1.2/blunt (Fermentas) and sequenced. This plasmid was designated NtPIP5K6-pJet.
The cDNA fragment for +2-EYFP (encoding EYFP plus two additional nucleotides) was amplified from a plasmid carrying the authentic clone provided by Dr. Martin Fulda (Georg-August-University Göttingen, Germany) using the following primer combination 5′-GATCGCGGCCGCATGGTGAGCAAGGGCGAG-3′/5′-GATCGATATCTTACTTGTACAGCTCGTCCATG-3′ and cloned into the vector pENTR2b (Invitrogen) as a NotI/EcoRV fragment, creating the plasmid pENTR-+2-EYFP. Prior to ligation, the control of cell death b (ccdb) gene was eliminated from pENTR2b by EcoRI digestion and religation. The cDNA fragment encoding full-length AtPIP5K5 was moved as a NotI/NotI fragment from the plasmid AtPIP5K5-pGEM-Teasy into pENTR-+2-EYFP. cDNA fragments encoding full-length AtPIP5K2 or full-length NtPIP5K6 were amplified from the plasmid AtPIP5K2-pGEM-Teasy or NtPIP5K6-pJet using the primer combinations: AtPIP5K2, 5′-GATCGTCGACATGATGCGTGAACCGCTTGTTAG-3′ (primer AtPIPK2-Sal_for)/5′-GATCGCGGCCGCCGCCGTCTTCGATGAAGATTCTGCTGATG-3′ (primer AtPIPK2-Not_rev); NtPIP5K6, 5′-GATCGTCGACATGAGCAAAGAATTTAGTGGCATTG-3′ (primer NtPIPK6-Sal_for)/5′-GATCGCGGCCGCCAGTGTCTTCTGCAAAAACTTTTAATAC-3′ (primer NtPIPK6-Not_rev) and cloned as SalI/NotI fragments into the vector pENTR-+2-EYFP. cDNA fragments encoding AtPIP5K2ΔNT–MORN (AtPIP5K2260–754) or AtPIP5K2ΔNT–MORN–Lin (AtPIP5K2344–754) were amplified from the plasmid AtPIP5K2-pGEM-Teasy using the following primer combinations: AtPIP5K2ΔMORN, 5-GATCGTCGACATGTTGGGGATTGAGAAGAATGAGTTGATTG-3′ (primer AtPIPK2ΔMORN_for)/AtPIPK2-Not_rev; AtPIP5K2ΔLin, 5′-GATCGTCGACATGTTGAAGAAACCTGGAGAGACGATATCT-3′(primer AtPIPK2ΔLin_for)/AtPIPK2-Not_rev. cDNA fragments encoding AtPIP5K5ΔNT–MORN (AtPIP5K5253–772) and AtPIP5K5ΔNT–MORN–Lin (AtPIP5K5370–772) were amplified from the plasmid AtPIP5K5-pGEM-Teasy using the following primer combinations: AtPIP5K5ΔMORN, 5′-GATCGTCGACATGCCGTCAGGAAATGAAGGGAATCTTG-3′/5′-GATCGCGGCCGCTATTGTCATCTGTGAAGACCTTGAA-3′ (primer AtPIPK5-Not_rev); AtPIP5K5ΔLin, 5′-GATCGTCGACATGAGGAAGCAAGGGGAGACGATATC-3′/AtPIPK5-Not_rev. cDNA fragments encoding NtPIP5K6ΔNT–MORN (NtPIP5K6241–775) or NtPIP5K6ΔNT–MORN–Lin (NtPIP5K6366–775) were amplified from the plasmid NtPIP5K6-pJet using the following primer combinations: NtPIP5K6ΔMORN, 5′-GATCGTCGACTCATGAAGCCATCAAGTTCAAGAACAGGTACTTTC-3′ (primer NtPIPK6ΔMORN_for)/NtPIPK6-Not_rev; NtPIP5K6ΔLin, 5′GATCGTCGACTCATGAAGAAGCCAGGACAAACAATATCCAAAG-3′ (primer NtPIPK6ΔLinker_for)/NtPIPK6-Not_rev. All fragments were cloned as SalI/NotI fragments into the vector pENTR-+2-EYFP and sequenced. The constructs were transferred by Gateway technology (Invitrogen) from pENTR2b to the pLatGW plasmid, an expression vector containing the tomato Lat52 promotor (Twell et al., 1990), an attR gateway cassette, and a cauliflower mosaic virus 35S terminator. The pLatGW vector was a gift from Prof. Dr. Wolfgang Dröge-Laser (Julius–Maximilians-University Würzburg, Germany).
Domain swap constructs between NtPIP5K6 and AtPIP5K2 were generated by overlap extension PCR (Horton et al., 1989). For the construct encoding NtPIP5K6_LinPIP5K2, cDNA fragments encoding NtPIP5K61–240 or NtPIP5K6366–775 were amplified with overlapping ends from the plasmid NtPIP5K6-pJet using the primer combinations: NtPIPK6-Sal_for/5′-CGCAACAATCAACTCATTCTTCTCAATCCCATAATAAGTTCCACTTTGCTCTCTAGAATC-3′and 5′-AAGAATCCTTGTTGTTTCAGCGGTGAGGCTAAGAAGCCAGGACAAACAATATCCAAAGGG-3′/NtPIPK6-Not_rev, respectively. The cDNA encoding AtPIP5K2260–343 was amplified using the primer combination 5′-ATTCTAGAGAGCAAAGTGGAACTTATTATGGGATTGAGAAGAATGAGTTGATTGTTGCG-3′/5′-CCCTTTGGATATTGTTTGTCCTGGCTTCTTAGCCTCACCGCTGAAACAACAAGGATTCTT-3′. The three amplicons were fused in a subsequent PCR reaction, using the primer combination NtPIPK6-Sal_for/NtPIPK6-Not_rev and the resulting PCR-fragment was cloned as a SalI/NotI fragment into the vector pENTR-+2-EYFP and sequenced. For the construct encoding AtPIP5K2_LinNtPIP5K6, cDNA fragments encoding AtPIP5K21–259 or AtPIP5K2344–754 were amplified from the plasmid AtPIP5K2-pGEM-Teasy using the primer combinations AtPIPK2-Sal_for/5′-ATCGAAAGTACCTGTTCTTGAACTTGATGGATCAAAGAAATTCCTCATGAGATTACTAC-3′and 5′-CAACCACATTGCATCAAAATTCAACCAACCAAGAAACCTGGAGAGACGATATCTAAAGG-3′/AtPIPK2-Not_rev, respectively. The cDNA encoding NtPIP5K6241–365 was amplified from the plasmid NtPIP5K6-pJet using the primer combination 5′-GTAGTAATCTCATGAGGAATTTCTTTGATCCATCAAGTTCAAGAACAGGTACTTTCGAT-3′/5′-CCTTTAGATATCGTCTCTCCAGGTTTCTTGGTTGGTTGAATTTTGATGCAATGTGGTTG-3′. The fusion-PCR of the three fragments was done with the primer combination AtPIPK2-Sal_for/AtPIPK2-Not_rev and the resulting product was cloned as a SalI/NotI fragment into the vector pENTR-+2-EYFP. After sequencing the constructs were transferred into the pLatGW plasmid by Gateway technology.
Constructs for heterologous expression in E. coli were created by moving the cDNA sequence encoding AtPIP5K2 as a PciI–PciI fragment from the plasmid AtPIP5K2-pGEM-Teasy into the NcoI-digested bacterial expression vector pETM-41 (EMBL protein expression and purification facility). The cDNA sequence of AtPIP5K5 was released from the plasmid AtPIP5K5-pGEM-Teasy and moved into pETM-41 as a BspHI/BspHI fragment. The cDNA sequence of NtPIP5K6 was amplified from the plasmid NtPIP5K6-pJet using the primer combination 5′-GATTCATGAGCAAAGAATTTAGTGGCATTGTGAAGGCCTGGGAG-3′/5′-GATTCATGACAGTGTCTTCTGCAAAAACTTTTAATACGAAGTCC-3′(primer NtPIPK6-BspH_rev) and cloned as BspHI/BspHI fragments into the NcoI site of pETM-41. cDNA fragments encoding AtPIP5K2260–754 or AtPIP5K2344–754 were amplified from the plasmid AtPIP5K2-pGEM-Teasy using the following primer combinations AtPIPK2ΔMORN_for/5′-GATCACATGTCGCCGTCTTCGATGAAGATTCTGCTGATG-3′(primer AtPIPK2-Pci_rev) and AtPIPK2ΔLin_for/AtPIPK2-Pci_rev, respectively, and cloned as PciI/PciI fragments into the NcoI site of pETM-41. cDNA fragments encoding AtPIP5K5253–772 and AtPIP5K5370–772 were amplified from the plasmid AtPIP5K5-pGEM-Teasy using the primer combinations 5′-GATTCATGAACCCGTCAGGAAATGAAGGGAATCTTG-3′/5′-GATTCATGACATTGTCATCTGTGAAGACCTTGA-3′ (primer AtPIPK5-BspH_rev) or 5′-GATTCATGAGGAAGCAAGGGGAGACGATATC-3′/AtPIPK5-BspH_rev, respectively, and cloned as BspHI/BspHI fragments into the NcoI site of pETM-41. cDNA fragments encoding NtPIP5K6241–775 or NtPIP5K6366–775 were amplified from the plasmid NtPIP5K6-pJet using the primer combinations NtPIPK6ΔMORN_for/NtPIPK6-BspH_rev or NtPIPK6ΔLinker_for/NtPIPK6-BspH_rev, respectively, and cloned as BspHI/BspHI fragments into the NcoI site of pETM-41. All clones were introduced in frame with the cDNA encoding the NT MBP and polyhistidine (His) tags of pETM-41.
Transient expression in tobacco pollen tubes
Mature pollen was collected from four to six tobacco (Nicotiana subsuper1cum) flowers of 8-week-old plants. Pollen was resuspended in liquid growth medium (Read et al., 1993), filtered onto cellulose acetate filters, and transferred to Whatman paper moistened with growth medium. Within 5–10 min of harvesting, pollen was transformed by bombardment with plasmid-coated 1-μm gold particles with a helium-driven particle accelerator (PDS-1000/He; Bio-Rad) using 1350 p.s.i. rupture disks and a vacuum of 28′′ of mercury. Gold particles (1.25 mg) were coated with 3–7 μg of plasmid DNA. After bombardment, pollen was resuspended in growth medium and grown for 5–14 h in small droplets of media directly on microscope slides. Digital images were taken at room temperature as described below.
Microscopy and imaging
Images were recorded using either an Olympus BX51 epifluorescence microscope or a Zeiss LSM 510 confocal microscope. For BX51, images were obtained at ×200 magnification using an F41-028 HQFilterset for Yellow GFP (Olympus), an Olympus ColorView II camera, and analySIS Docu 3.2 software (Soft Imaging Systems). For LSM 510, EYFP was excited at 514 nm and imaged using an HFT 405/514/633-nm major beam splitter (MBS) and a 530- to 600-nm band-pass filter. If not specified otherwise, images of central optical plains were obtained by confocal microscopy at ×630 magnification using the Zeiss LSM510 image acquisition system and software (v 4.0; Zeiss). Fluorescence images were contrast-enhanced by adjusting brightness and γ-settings using image processing software (Photoshop; Adobe Systems). Pollen tube fluorescence intensities were analyzed from digital images using analySIS Docu 3.2 software (Soft Imaging Systems). For the analysis of fluorescence intensities pollen tubes were imaged at identical laser intensities and exposure times.
Heterologous expression in E. coli
Recombinant enzymes were expressed in E. coli strain BL21-AI (Invitrogen) at 25°C for 18 h after induction with 1 mM isopropylthio-b-galactoside and 0.2% (w/v) l-arabinose. Cell lysates were obtained by sonification in a lysis buffer containing 50 mM Tris–HCl, 300 mM NaCl, 1 mM EDTA, and 10% (v/v) glycerol, pH 8.0.
Lipid kinase assays
Lipid kinase activity was assayed by monitoring the incorporation of radiolabel from γ[32P]ATP into defined lipid substrates (Avanti Polar Lipids) as described (Cho and Boss, 1995) using total extracts of BL-21-AI expression cultures. Recombinant expression levels were adjusted between individual cultures according to immunodetection of expressed MBP-tagged proteins (data not shown), allowing for the comparison of enzyme activities obtained with different cultures. Assays were performed with bacterial crude extracts; for one assay, 45 μg protein were used. Radiolabeled lipid reaction products were separated by thin-layer chromatography using silica S60 plates (Merck) and CHCl3:CH3OH:NH4OH:H2O (57:50:4:11, v/v/v/v) as a developing solvent (Perera et al., 2005) and visualized by autoradiography using Kodak X-Omat autoradiography film (Eastman Kodak). Reaction products were identified according to comigration with authentic standards (Avanti Polar Lipids) that were visualized as described (König et al., 2007, 2008).
Phosphatidylinositol-bisphosphate bands were scraped from thin-layer chromatography plates according to autoradiography, and the radiolabel incorporated in lipids present in the scraped silica powder was quantified using liquid scintillation counting (Analyzer Tricarb 1900 TR; Canberra Packard). Enzyme activities presented in Figure 3 represent the mean of at least three independent experiments, assayed in duplicates.
Phylogenetic analyses
To construct the phylogenetic tree, AtPIP5K amino acid sequences were aligned using the MAFFT multiple sequence alignment program with the L-INS-i option (Katoh et al., 2005). After manual editing of the alignment, ProtTest 3.0 was used for selection of the best-fit model of protein evolution (Abascal et al., 2005). Phylogenetic relationships based on amino acid sequences were inferred from maximum likelihood methods implemented in MEGA 5 (JTT + G + F, 1000 bootstrap replicates) as previously described (Tamura et al., 2011). The phylogenetic tree was mid-point rooted and edited with FigTree v1.3.13.
For analysis of KA/KS ratios and sliding window plots, orthologous A. thaliana and A. lyrata protein sequences were determined with the PLAZA platform4 (Proost et al., 2009) based on the Best hit family and Tree-based orthologs criteria. Homologous protein sequences were aligned using MAFFT with the L-INS-i option (Katoh et al., 2005). Codon alignments generated with PAL2-NAL (Suyama et al., 2006) were used to compute divergence levels (KA/KS ratios) with DnaSP v5 (Librado and Rozas, 2009) and the sliding window option (window size, 50 bp; step size, 10 bp). The following pairs of orthologous genes were compared: AtPIP5K1, At1g1980; AlPIP5K1, Al1g23460; AtPIP5K2, At1g77740; AlPIP5K2, Al2g26700; AtPIP5K3, At2g26420; AlPIP5K3, Al4g08780; AtPIP5K4, At3g56960; AlPIP5K4, Al5g27310; AtPIP5K5, At2g41210; AlPIP5K5, Al4g28160; AtPIP5K6, At3g07960; AlPIP5K6, Al3g08440; AtPIP5K7, At1g10900; AlPIP5K7, Al1g11060; AtPIP5K8, At1g60890; AlPIP5K8, Al2g05080; AtPIP5K9, At3g09920; AlPIP5K9, Al3g00840.
Accession numbers
AtPIP5K2, At1g77740; AtPIP5K5, At2g41210; NtPIP5K6-1, JQ219669.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Dr. Benedikt Kost (Friedrich-Alexander-University Erlangen–Nürnberg, Germany) for providing the tobacco pollen tube-specific cDNA library and Dr. Mareike Heilmann (Martin-Luther-University Halle–Wittenberg, Germany) for helpful discussion. We also thank Prof. Dr. Wolfgang Dröge-Laser (Julius–Maximilians-University Würzburg, Germany) and Dr. Martin Fulda (Georg-August-University Göttingen. Germany) for providing plasmids. Lastly, we thank Kristian Ullrich for his support with phylogenetic analyses. This work was supported by Emmy-Noether grant He3424/1-5 from the German Research Foundation (DFG, to Ingo Heilmann) and the DFG through the Special Research Project SFB 648 (TP B8, to Marcel Quint).
Footnotes
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 10.1093/bioinformatics/bti263 [DOI] [PubMed] [Google Scholar]
- Anderson R. A., Boronenkov I. V., Doughman S. D., Kunz J., Loijens J. C. (1999). Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274, 9907–9910 10.1074/jbc.274.38.27139 [DOI] [PubMed] [Google Scholar]
- Camacho L., Smertenko A. P., Perez-Gomez J., Hussey P. J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell. Sci. 122, 4383–4392 [DOI] [PubMed] [Google Scholar]
- Cho M. H., Boss W. F. (1995). Transmembrane signaling and phosphoinositides. Methods Cell Biol. 49, 543–554 10.1016/S0091-679X(08)61479-X [DOI] [PubMed] [Google Scholar]
- Gubbels M. J., Vaishnava S., Boot N., Dubremetz J. F., Striepen B. (2006). A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J. Cell. Sci. 119, 2236–2245 10.1242/jcs.02949 [DOI] [PubMed] [Google Scholar]
- Heilmann I. (2009). Using genetic tools to understand plant phosphoinositide signalling. Trends Plant Sci. 14, 171–179 10.1016/j.tplants.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 10.1016/0378-1119(89)90359-4 [DOI] [PubMed] [Google Scholar]
- Im Y. J., Davis A. J., Perera I. Y., Johannes E., Allen N. S., Boss W. F. (2007). The N-terminal membrane occupation and recognition nexus domain of Arabidopsis phosphatidylinositol phosphate kinase 1 regulates enzyme activity. J. Biol. Chem. 282, 5443–5452 10.1074/jbc.M611342200 [DOI] [PubMed] [Google Scholar]
- Ischebeck T., Seiler S., Heilmann I. (2010a). At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma 240, 13–31 10.1007/s00709-009-0093-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Vu L. H., Jin X., Stenzel I., Löfke C., Heilmann I. (2010b). Functional cooperativity of enzymes of phosphoinositide conversion according to synergistic effects on pectin secretion in tobacco pollen tubes. Mol. Plant 3, 870–881 10.1093/mp/ssq031 [DOI] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Heilmann I. (2008). Type B phosphatidylinositol-4-phosphate 5-kinases mediate pollen tube growth in Nicotiana tabacum and Arabidopsis by regulating apical pectin secretion. Plant Cell 20, 3312–3330 10.1105/tpc.108.059568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T., Stenzel I., Hempel F., Jin X., Mosblech A., Heilmann I. (2011). Phosphatidylinositol-4,5-bisphosphate influences Nt-Rac5-mediated cell expansion in pollen tubes of Nicotiana tabacum. Plant J. 65, 453–468 10.1111/j.1365-313X.2010.04435.x [DOI] [PubMed] [Google Scholar]
- Ishihara H., Shibasaki Y., Kizuki N., Wada T., Yazaki Y., Asano T., Oka Y. (1998). Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem. 273, 8741–8748 10.1074/jbc.273.15.8741 [DOI] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 10.1093/nar/gki198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S., Hoffmann M., Mosblech A., Heilmann I. (2008). Determination of content and fatty acid composition of unlabeled phosphoinositide species by thin layer chromatography and gas chromatography. Anal. Biochem. 197–201 10.1016/j.ab.2008.03.052 [DOI] [PubMed] [Google Scholar]
- König S., Mosblech A., Heilmann I. (2007). Stress-inducible and constitutive phosphoinositide pools have distinct fatty acid patterns in Arabidopsis thaliana. FASEB J. 21, 1958–1967 10.1096/fj.06-7887com [DOI] [PubMed] [Google Scholar]
- Kost B. (2008). Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 18, 119–127 10.1016/j.tcb.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N. H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145, 317–330 10.1083/jcb.145.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J., Fuelling A., Kolbe L., Anderson R. A. (2002). Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J. Biol. Chem. 277, 5611–5619 10.1074/jbc.M110775200 [DOI] [PubMed] [Google Scholar]
- Kunz J., Wilson M. P., Kisseleva M., Hurley J. H., Majerus P. W., Anderson R. A. (2000). The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol. Cell 5, 1–11 10.1016/S1097-2765(00)80398-6 [DOI] [PubMed] [Google Scholar]
- Kusano H., Testerink C., Vermeer J. E. M., Tsuge T., Shimada H., Oka A., Munnik T., Aoyama T. (2008). The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20, 367–380 10.1105/tpc.107.056119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lou Y., Gou J. Y., Xue H. W. (2007). PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19, 163–181 10.1105/tpc.106.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Moller S. G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8, 293–299 10.1038/sj.embor.7400902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer H. J., Munnik T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306 10.1146/annurev.arplant.54.031902.134748 [DOI] [PubMed] [Google Scholar]
- Mueller-Roeber B., Pical C. (2002). Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol. 130, 22–46 10.1104/pp.004770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera I. Y., Davis A. J., Galanopoulou D., Im Y. J., Boss W. F. (2005). Characterization and comparative analysis of Arabidopsis phosphatidylinositol phosphate 5-kinase 10 reveals differences in Arabidopsis and human phosphatidylinositol phosphate kinases. FEBS Lett. 579, 3427–3432 10.1016/j.febslet.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2009). PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21, 3718–3731 10.1105/tpc.109.071506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. D., Misra S., Boronenkov I. V., Anderson R. A., Hurley J. H. (1998). Structure of type IIbeta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell 94, 829–839 10.1016/S0092-8674(00)81741-9 [DOI] [PubMed] [Google Scholar]
- Read S. M., Clarke A. E., Bacic A. (1993). Stimulation of growth of cultured Nicotiana tabacum W 38 pollen tubes by poly(ethylene glycol) and Cu(II) salts. Protoplasma 177, 1–14 10.1007/BF01403393 [DOI] [Google Scholar]
- Seiler S., Plamann M. (2003). The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14, 4352–4364 10.1091/mbc.E02-07-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45, 960–967 10.1093/pcp/pch122 [DOI] [PubMed] [Google Scholar]
- Sousa E., Kost B., Malho R. (2008). Arabidopsis phosphatidylinositol-4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell 20, 3050–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I., Ischebeck T., König S., Holubowska A., Sporysz M., Hause B., Heilmann I. (2008). The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20, 124–141 10.1105/tpc.107.052852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. (2000). Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11–22 10.1016/S1097-2765(00)00003-4 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole J. M., Nielsen E. (2008). Phosphoinositides in plants: novel functions in membrane trafficking. Curr. Opin. Plant Biol. 11, 620–631 10.1016/j.pbi.2008.10.010 [DOI] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–713 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W., Vermeer J. E., Gadella T. W., Jr., Munnik T. (2007). Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. Plant J. 52, 1014–1026 10.1111/j.1365-313X.2007.03292.x [DOI] [PubMed] [Google Scholar]
- Vincent P., Chua M., Nogue F., Fairbrother A., Mekeel H., Xu Y., Allen N., Bibikova T. N., Gilroy S., Bankaitis V. A. (2005). A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J. Cell Biol. 168, 801–812 10.1083/jcb.200412074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yan A., Feijo J. A., Furutani M., Takenawa T., Hwang I., Fu Y., Yang Z. (2010). Phosphoinositides regulate clathrin-dependent endocytosis at the tip of pollen tubes in Arabidopsis and tobacco. Plant Cell 22, 4031–4044 10.1105/tpc.110.076760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]